Abstract
STEAP4 is a plasma membrane metalloreductase involved in the transport of iron and copper. Recently, STEAP4 was implicated in promoting insulin sensitivity by acting in white adipose tissue to control the production of inflammatory cytokines such as interleukin 6. Indeed, the loss of STEAP4 expression in mice leads to increased production of inflammatory cytokines in visceral white adipose tissue and systemic insulin resistance. In this study, we demonstrate that in mouse liver STEAP4 is produced at significant levels and that steap4 transcription is induced by interleukin 6. We further demonstrate that the steap4 gene is a direct target of phosphorylated STAT3 in mouse liver. In addition, hepatic STEAP4 expression is regulated by feeding and fasting, and obesity leads to the induction of STEAP4 expression in the liver. Interestingly, the regulation of STEAP4 in both feeding and fasting and the obese state appears to require the transcription factor CCAAT/enhancer-binding protein α that may act in concert with STAT3 as they both bind to the proximal steap4 promoter in vivo. Taken together, these data suggest the transcriptional regulation of hepatic STEAP4 may play a critical role in the response to nutritional and inflammatory stress and contributes to the protective effect of STEAP4 in vivo.
Keywords: Chromatin/Immunoprecipitation/ChIP, Cytokines/Interleukins, Transcription/C/EBP, Transcription/Regulation, Transcription/STAT, STEAP4
Introduction
Insulin resistance in multiple tissues is a hallmark of type 2 diabetes and occurs in part through impairment of insulin signaling in target tissues. Obesity in mice and humans results in increased production of cytokines in the adipose tissue and can contribute to insulin resistance (1–4). In addition, increased oxidative stress is also known to decrease insulin sensitivity (5, 6). Thus, an understanding of regulatory pathways that lead to insulin resistance is critical to the management of diabetes.
The transcription factor STAT3 plays a role in glucose homeostasis by negatively regulating hepatic gluconeogenic gene expression (7–9). STAT3 is activated by phosphorylation in response to cytokines such as IL-6,2 translocates to the nucleus as a dimer, and regulates transcription by interacting with promoter regions of target genes. STAT3 exists as two isoforms STAT3α and STAT3β, which are splice variants of the same gene (10–12). STAT3β is considered a dominant negative form of STAT3, in that it lacks a transactivation domain and competes for DNA with STAT3α, thereby inhibiting transcription (10). Because STAT3 regulates gluconeogenic gene expression in the liver, the goal of this study was to identify novel STAT3 targets in the liver that may play a role in the development of the metabolic syndrome. In this study, we discuss the identification and regulation of STEAP4 (six transmembrane epithelial antigen of prostate 4) as a new target of STAT3 in the liver.
STEAP4 (also known as TIARP or STAMP2) is a member of a family of metalloreductases that are involved in the reduction and transport of iron and copper (13). In humans, STEAP4 is expressed in the prostate, placenta, lung, heart, bone marrow, adipose tissue, and fetal liver (14–16), and it is elevated in prostate cancer (15). It is also expressed to a lower extent in adult human liver (15). In rodents, STEAP4 is expressed predominantly in white adipose tissue (WAT) (17, 18). It is also expressed in brown adipose tissue, liver, heart, kidney, and skeletal muscle (17). STEAP4 is expressed in differentiating 3T3-L1 adipocytes, and expression increases with progression of differentiation (17). STEAP4 is predominantly located in the plasma membrane, but it has also been detected in cytoplasmic vesicles and found to be associated with the Golgi apparatus (15, 17). The association with vesicles suggested a secretory or endocytotic role for STEAP4.
Recently, Wellen et al. (18) demonstrated that STEAP4 plays a role in preventing insulin resistance in mice, as mice lacking steap4 become insulin-resistant and exhibit impaired insulin signaling in visceral WAT and liver. Genetically obese ob/ob mice were found to have increased levels of STEAP4 mRNA in the visceral WAT, and ob/ob mice lacking steap4 have increased blood glucose as compared with ob/ob controls (18). Furthermore, a lack of STEAP4 in 3T3-L1 adipocytes and in visceral WAT in mice led to increased production of IL-6, which in turn could contribute to insulin resistance. Two studies conducted in humans demonstrated that STEAP4 is expressed in WAT (14, 16), but these studies showed opposing results with human STEAP4 expression being either increased (14) or decreased (16) in obese patients. Taken together, these results demonstrate that STEAP4 could be an important player in insulin signaling, and the regulation of STEAP4 expression is important for maintaining glucose homeostasis.
In this study, we identify steap4 as a novel target of STAT3 in the liver. Given the protective effect of hepatic STAT3 in whole-body metabolism (7, 8), it is likely that the induction of steap4 by this pathway plays an important role in glucose homeostasis. In addition, we show that C/EBPα also regulates hepatic expression of STEAP4 during feeding, whereas both C/EBPα and STAT3 regulate expression of steap4 in the presence of high levels of IL-6, and this coordinated regulation may occur in obese mice to elevate hepatic STEAP4 levels to confer a protective effect. Our data suggest that increased hepatic expression of steap4 plays a protective role in maintaining hepatic insulin signaling in the presence of inflammation and obesity.
EXPERIMENTAL PROCEDURES
Mouse Experiments
All experiments involving the use of mouse models were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center, and all mice were purchased from The Jackson Laboratory. Mice were fed a rodent chow diet (Harlan Teklad F6 Rodent Diet) unless otherwise stated. For IL-6 experiments, 10-week-old male C57BL6 mice were housed in single cages for 1 week at the animal facility at Beth Israel Deaconess Medical Center. Mice were handled daily to acclimatize them to minimize stress during the experiment. Mice were fasted for 16 h overnight and then given either saline or IL-6 (100 ng/mouse) injections intraperitoneally for 1 h. Mice were euthanized using CO2, and the livers were collected, snap-frozen in liquid nitrogen, and stored at −80 °C until further use. For adenovirus experiments where STAT3β was overexpressed, liver samples that had been collected in a previous study (9) were used. For adenovirus experiments where C/EBPα was overexpressed, 9-week-old male C57BL/6 mice were injected with either Ad-GFP (obtained from the Harvard Gene Therapy Initiative, 1 × 109 plaque-forming units/mouse) or Ad-C/EBPα (obtained from Vector Laboratories, 5 × 109 plaque-forming units/mouse) through the tail vein. These doses were determined to be the minimum amount of each virus required for substantial expression of GFP or C/EBPα, as detected by Western blot. On day 4 following injections, half of the mice in each group were fasted overnight. On day 5, livers were collected and used for gene expression analysis. For fed-fasted-refed experiments, 16-week-old C57BL6 wild type mice were maintained in single cages for a week at Beth Israel Deaconess Medical Center. Mice were either fed ad libitum, fasted for 16 h, or fasted for 16 h followed by a 2-h re-feed prior to euthanasia. Tissues were collected as described above. IL-6 knock-out mice were obtained from The Jackson Laboratory at 10 weeks of age and maintained at Beth Israel Deaconess Medical Center for 6 weeks in single cages, and the experiment was done at 16 weeks of age. 10-Week-old ob/ob and wild type C57BL6 control mice were housed in single cages for a week prior to the experiment. Mice were either fed ad libitum or fasted for 16 h prior to euthanasia. For the high fat diet (HFD) experiment, 17-week-old C57BL6 mice that were fed a 60% fat diet (D12492, Research Diets) from week 6 of age and corresponding chow-fed controls were maintained in single cages. Mice were either fed ad libitum or fasted for 36 h prior to euthanasia.
Gene Expression Analysis
Tissues were homogenized in STAT60 reagent (TelTest Inc) using a TissueLyser (Qiagen) at 30 Hz for 2 min. RNA was extracted according to the manufacturer's protocol. For gene expression profiling by microarray analysis, isolated total RNA was further purified with the RNeasy mini kit per the manufacturer's instructions (Qiagen). Total RNA (200 ng) was then used for GeneChip analysis. Preparation of terminally labeled cDNA, hybridization to genome-wide murine Gene Level 1.0 ST GeneChips (Affymetrix), and scanning of the arrays were carried out according to the manufacturer's protocols. RMA signal extraction, normalization, and filtering were performed as described previously (19) using a custom chip description file (version 11.0.1 (20). A variation filter was applied for selecting informative (i.e. significantly varying) genes. The filtering criteria for the exemplary data sets required an interquantile range of >0.5 and at least one sample with expression intensity of >100.
Statistical Group Comparisons
To calculate differential gene expression between individual sample groups, we performed a statistical comparison using the limma package as described previously (19). Briefly, limma estimates the fold change between predefined sample groups by fitting a linear model and using an empirical Bayes method to moderate the standard errors of the estimated log-fold changes for each probe set (21). A multiple testing correction based on the false discovery rate was performed to produce adjusted p values. All calculations were performed using the statistical software “R.” For further analysis of gene expression, quantitative PCR was performed. cDNA was synthesized using a reverse transcriptase kit (BD Biosciences) from 1 μg of RNA except for epididymal white adipose tissue in Fig. 4, where 0.625 μg of RNA was used because of insufficient RNA amounts. cDNA was diluted as needed, and mRNA levels of steap4, socs3, pepck, or c/ebpα were measured using gene-specific TaqMan assays (Applied Biosystems) and the Stratagene MX3000 thermocycler. Target gene mRNA levels were normalized to 18 S rRNA, TATA-binding protein, or cyclophilin mRNA levels in each sample. Different genes were chosen for normalizing expression based on the fact that expression of some of these genes changed in some tissues under the conditions being studied. For every tissue, the normalizing gene that showed no significant variation between groups was selected.
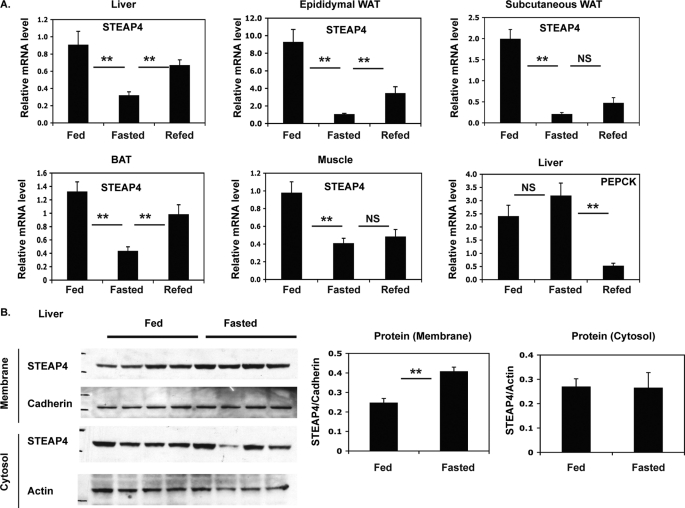
FIGURE 4.
Changes in steap4 expression with feeding and fasting. C57BL6 mice were fed ad libitum, fasted for 16 h, or fasted for 16 h and refed for 2 h and then sacrificed (n = 8 per group). A, relative steap4 mRNA expression levels in the liver, epididymal WAT, subcutaneous WAT, brown adipose tissue (BAT), and muscle as measured by qPCR are shown. Liver phosphoenolpyruvate carboxykinase (pepck) mRNA levels were measured by qPCR. B, STEAP4 protein levels in the liver were detected by Western blot in crude membrane and cytosol fractions and quantified. **, p ≤ 0.001; NS, not significant.
Western Blotting and Immunoprecipitations
For detecting proteins other than STEAP4, 50 mg of liver was homogenized in 1 ml of lysis buffer (Cell Signaling Technology) containing protease and phosphatase inhibitors using a TissueLyser for 1 min at 30 Hz. Homogenates were centrifuged at 16,000 × g for 20 min at 4 °C, and the supernatant was transferred to a fresh tube. Proteins were resolved either using 10% NuPAGE BisTris gels (Invitrogen) or 8% Tris-Tricine gels and transferred to nitrocellulose membrane. Blots were probed for phospho-STAT3 (anti phospho-STAT3 Tyr-705, Cell Signaling Technology) or C/EBPα (Santa Cruz Biotechnology) and visualized using the ECL-Plus kit (ThermoScientific). For total STAT3 and actin, blots used to detect P-STAT3 and C/EBPα were stripped in 0.1 m glycine and re-probed for total STAT3 (K-15, Santa Cruz Biotechnology) or actin (Sigma).
To detect STEAP4 expression, a crude membrane fraction was enriched as follows. 100 mg of liver was Dounce-homogenized in homogenization buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 250 mm sucrose, protease and phosphatase inhibitors) and then passed through a 23.5-gauge needle three times. The resulting homogenate was centrifuged at 1000 × g for 10 min at 4 °C to remove nuclei. The supernatant was centrifuged at ∼150,000 × g in an ultracentrifuge for 15 min at 4 °C. The resulting supernatant was saved as the cytosolic fraction, and the pellet was resuspended in 1× RIPA buffer and sonicated to ensure that membrane proteins were in solution. STEAP4 protein from both fractions was detected by Western blot as described above using an anti-STEAP4 antibody (Novus Biologicals). The blots were stripped and re-probed for pan-cadherin (membrane; antibody from Abcam) and actin (cytosol; antibody from Sigma). Band intensities were measured using ImageJ software (Public Domain, developed at the National Institute of Mental Health, Bethesda) and STEAP4 levels were normalized.
For immunoprecipitations, liver was homogenized as described above, and 100 μl of lysate was incubated overnight with anti-STAT3 (C20, Santa Cruz Biotechnology) in 900 μl of lysis buffer. Immunoprecipitated complexes were captured using protein A-agarose (ThermoScientific), washed three times in lysis buffer, and resolved on 10% NuPAGE gels. Western blotting was done as described above.
Chromatin Immunoprecipitation
ChIP assays were performed essentially as described previously (9). Briefly, 80 mg of liver was minced in phosphate-buffered saline and cross-linked using DSG (ThermoScientific) for 45 min at room temperature followed by cross-linking with 1% formaldehyde for 10 min. Cross-linked liver was then sonicated to yield chromatin and centrifuged to remove debris. Chromatin was pooled from four livers for each group to obtain a representative sample, and immunoprecipitations were performed in duplicate. The chromatin was pre-cleared using protein A-agarose and then incubated overnight with 4 μg of the appropriate antibody. Complexes were captured using protein A-agarose and washed; cross-links were reversed at 65 °C for 4 h, and DNA was extracted using phenol/chloroform/isoamyl alcohol followed by ethanol precipitation. DNA was air-dried and resuspended in water, and specific target promoters were amplified by PCR. PCR amplification of the β-globin locus was used as a control. Primer sequences are available upon request. PCR products were resolved on 2% agarose gels by electrophoresis, and images were captured using a Gel Doc (Bio-Rad). Band intensities were measured using the ImageJ software. Immunoprecipitation signals were averaged and calculated as a percent of the PCR signal from the input for each chromatin sample. For STAT3α versus STAT3β competition experiments, two STAT3 antibodies were used. The C-20 antibody detects only STAT3α, whereas the K-15 antibody detects both STAT3α and STAT3β.
Plasmids
To construct pGL3-steap4, −757 to +26 bp of the putative mouse steap4 promoter was PCR-amplified from a BAC (RP24-107D17) and cloned into pCR2.1 (Invitrogen). The steap4 promoter was then subcloned into pGL3 using KpnI-XhoI sites present in both vectors and sequenced. The three STAT3 sites in the pGL3-steap4 construct were mutated from TT(N4–5)AA to AA(N4–5)AA using the QuikChange multisite-directed mutagenesis kit (Stratagene) using a single primer per mutation based on the manufacturer's protocol. Single mutants were obtained by using one primer in the PCR, whereas double and triple mutations were obtained by using a combination of primers. All mutants were sequenced to confirm that the correct sequence was mutated and that no additional nonspecific mutations were generated. pCDNA3.1-C/EBPα was obtained from Dr. Ormond MacDougald (University of Michigan).
Cell Culture and Transient Transfections
HepG2 cells were routinely maintained in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose supplemented with 10% fetal bovine serum, antibiotics, and glutamine at 37 °C, 5% CO2. HepG2 cells were seeded in 12-well culture plates and allowed to become confluent. Cells were then placed in Opti-MEM and transfected with 100 ng of the appropriate luciferase construct per well using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For C/EBPα co-expression experiments, 100 ng of pCDNA3.1-C/EBPα was transfected with 50 ng of the luciferase construct and 350 ng of empty pCDNA3.1. Media containing the transfection mix were removed after 6 h and replaced with fresh Opti-MEM. Approximately 16 h post-transfection, cells were treated with IL-6 (20 ng/ml) for 6 h and then lysed in 200 μl of Passive Lysis Buffer (Promega). Luciferase activity was measured using a luciferase assay kit and luminometer (Promega). Luciferase values were normalized to total protein levels in each sample using a BCA assay kit (ThermoScientific). Luciferase reporter experiments were performed at least three times, and data from a representative experiment are shown.
Statistics
To determine whether differences between groups were statistically significant (other than the microarray experiment), we used the Student's t test, and p ≤ 0.05 was considered significant, as indicated by *. For experiments where p ≤ 0.001, significance is indicated by **.
RESULTS
steap4 Is a Target of STAT3 in the Liver
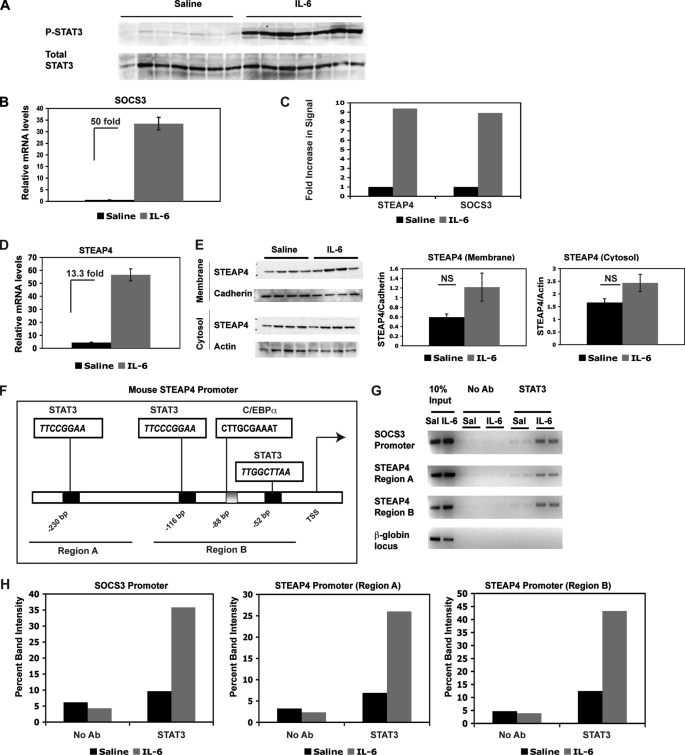
To identify novel targets of STAT3 in the liver, a microarray experiment was performed using mRNA from saline versus IL-6-treated C57BL6 mouse livers. As demonstrated previously (9), and now repeated, IL-6 induced STAT3 phosphorylation and a 50-fold increase of socs3 mRNA in the liver after 1 h of treatment (Fig. 1, A and B). P-STAT1 was not induced in IL-6-treated livers (data not shown). Three samples from each treatment group were used in a microarray experiment to identify differentially expressed genes. Among the transcripts that showed the largest changes in gene expression between saline- and IL-6-treated samples, we identified steap4, a plasma membrane-bound metalloreductase that has previously been implicated to play a role in insulin sensitivity. steap4 mRNA was induced 9.4-fold in IL-6-treated samples, whereas socs3 was induced 8.9-fold in the microarray experiment (as shown in Fig. 1C). steap4 mRNA levels were also measured by qPCR analysis, and a 13.3-fold increase was detected by this method (Fig. 1D). STEAP4 protein levels were then measured by Western blot analysis in the crude membrane fraction as well as cytosol. A trend toward increased STEAP4 proteins levels was detected in both membrane and cytosol fractions (Fig. 1E). Importantly, protein levels were measured from the same livers that had been used for qPCR analysis at the 1-h time point, so it is likely that the STEAP4 protein level increases further at a later time point after IL-6 treatment. IL-6 has been shown previously to increase both steap4 mRNA and protein levels in 3T3-L1 adipocytes (22), and protein expression was delayed compared with mRNA induction.
FIGURE 1.
steap4 expression is induced in mouse liver by IL-6. A, Western blot of P-STAT3 and total STAT3 in livers of mice treated with saline or IL-6 (n = 7 per group). B, qPCR analysis of socs3 mRNA in the same livers as in A. C, graphical representation of fold increase in signal in steap4 and socs3 as detected on microarrays (n = 3, p < 0.0001). D, qPCR analysis of steap4 mRNA levels in the same livers as in A (n = 7). E, Western blot analysis of STEAP4 protein levels in crude membrane fraction and cytosol (n = 4). Band intensities were measured using ImageJ, and STEAP4 levels were normalized to cadherin and actin levels in the two fractions, respectively. F, schematic representation of the mouse steap4 promoter depicting potential STAT3- and C/EBPα-binding sites. Regions that were amplified in ChIP assays are underlined and marked as region A and region B. G, ChIP assays demonstrating binding of STAT3 to the socs3 and steap4 promoters in vivo. Band intensities were measured and graphed (H). NS, not significant; Ab, antibody.
We then examined the steap4 promoter for potential STAT3-binding sites because STAT3 functions predominantly as a transcription factor that binds to promoter regions to activate transcription. We identified three potential STAT3-binding sites in the proximal steap4 promoter that conform to the known consensus sequences of TT(N4)AA and TT(N5)AA (Fig. 1F). We also identified a potential C/EBPα-binding site, as the steap4 promoter was previously shown to be regulated in 3T3L1 adipocytes by C/EBPα (18). ChIP assays were used to determine whether STAT3 bound to the steap4 promoter using a double cross-linking method that we have described previously (9). Two regions of the steap4 promoter were amplified by PCR, region A, containing the most distal STAT3 site at −230 bp, and region B, containing the two proximal STAT3 sites at −116 bp and −52 bp, and the C/EBPα-binding site at −88 bp. As expected, STAT3 was detected at the socs3 promoter and STAT3 recruitment increases with IL-6 treatment (Fig. 1G). In a similar manner, STAT3 was also detected at both regions of the steap4 promoter, and recruitment increased with IL-6 treatment. These results demonstrate that steap4 is a target of STAT3 in the liver and that IL-6-activated STAT3 increases transcription of steap4 by binding to the promoter region.
Identification of the STAT3-binding Site That Is Critical for Induction of steap4 by IL-6
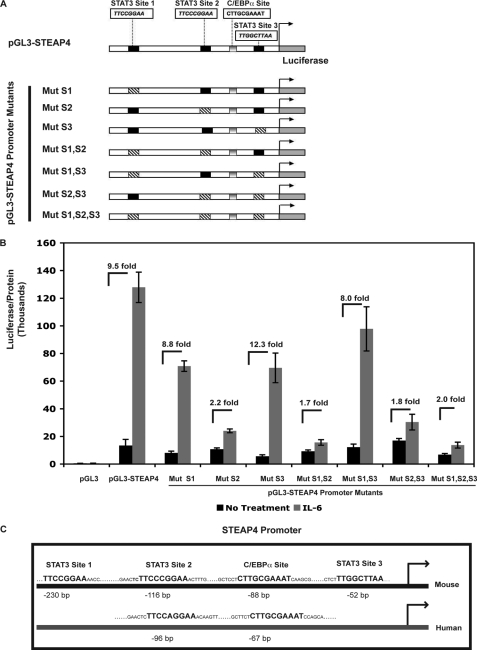
ChIP assays demonstrated STAT3 binding to the steap4 promoter at both regions amplified by PCR. However, the PCR signal of region A was weaker than that of region B, suggesting that perhaps not all sites were bound by STAT3. To determine which STAT3 sites were important for IL-6-mediated induction of steap4, a reporter construct containing −757 to +26 of the mouse steap4 promoter upstream of the firefly luciferase gene was generated. Mutations were introduced into each of the three potential STAT3 sites to obtain single mutants, all combinations of double mutants, and a triple mutant, as depicted in Fig. 2A. These luciferase constructs were transfected into HepG2 cells, which have previously been shown to have functional STAT3 signaling (23). IL-6 treatment resulted in 9.5-fold activation of the wild type steap4 promoter. Although mutations in site 1 and site 3 had a minimal effect, a mutation in site 2 resulted in a substantial decrease in induction of the steap4 promoter by IL-6 (Fig. 2B). Double mutants containing a mutation in site 2 and the triple mutant also showed decreased activity, suggesting that site 2 is critical for IL-6 induction of steap4. A comparison of the mouse and human STEAP4 promoters revealed that site 2 is conserved between the two promoters, as is the C/EBPα-binding site (Fig. 2C). Sites 1 and 3 in the mouse steap4 promoter are not detected in the human STEAP4 promoter. These results suggest that site 2 is bound by STAT3 and is required for induction by IL-6 and that the signal detected in region A in ChIP assays (Fig. 1) may only reflect binding at site 2, because regions A and B are not very far apart. Therefore, for all subsequent ChIP analysis in this study, we focused on region B of the steap4 promoter.
FIGURE 2.
Characterization of STAT3-binding sites in the mouse steap4 promoter. A, schematic representation of pGL3-steap4 promoter construct and constructs with mutated STAT3 sites that were generated. B, HepG2 cells were transfected with the various luciferase reporter constructs and treated with IL-6. Luciferase activity was measured and normalized to the total protein concentration in the sample (n = 3). C, schematic representation of the mouse and human steap4 promoters demonstrating conservation of STAT3 site 2 and the C/EBPα-binding site. Mut, mutant.
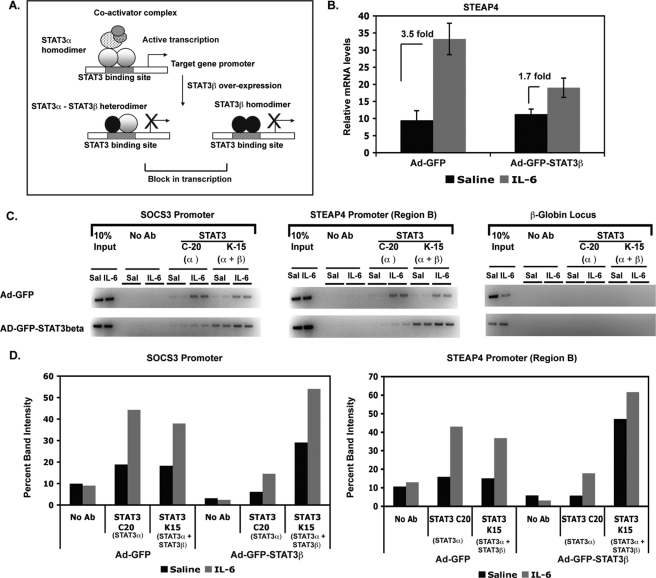
STAT3 Is Required in Vivo for IL-6 Induction of steap4
We have previously demonstrated that overexpressing STAT3β, the dominant negative form of STAT3, results in decreased transcriptional activity of STAT3 in mouse liver (9). STAT3β lacks the transactivation domain present in STAT3α and is thought to block transcription by STAT3α by competing for DNA-binding sites (as outlined in Fig. 3A). We utilized liver samples generated in our previous study where STAT3β was overexpressed in the livers of mice to determine whether STAT3 transcriptional activity was required for steap4 induction. In control virus-infected livers (Ad-GFP), steap4 was induced 3.5-fold, whereas livers infected with STAT3β-virus (Ad-GFP-STAT3β) showed only 1.7-fold induction of steap4 (Fig. 3B), demonstrating that transcriptional activity of STAT3 is required for steap4 induction by IL-6. ChIP assays were performed using virus-infected livers to determine whether STAT3α binding to the steap4 promoter in vivo is compromised in STAT3β-overexpressing livers resulting in decreased transcription. To achieve this, we used two antibodies against STAT3, the C-20 antibody that recognizes only STAT3α and the K-15 antibody that recognizes both STAT3α and STAT3β. As shown in Fig. 3C, in Ad-GFP livers, STAT3α is bound to both the socs3 and steap4 promoters (C-20 ChIPs), and the pattern of STAT3 binding is similar in K-15 ChIPs. However, in Ad-GFP-STAT3β livers, very little STAT3α is detected at both promoters (C-20 ChIPs), whereas an increase in both basal and IL-6-induced total STAT3 binding is detected in K-15 ChIPs. This suggests that in the presence of excess STAT3β, STAT3α recruitment decreases at these promoters, and STAT3β recruitment increases. These results demonstrate that STAT3α and STAT3β compete for DNA at the socs3 and steap4 promoters in vivo and that preventing STAT3α from binding to DNA results in decreased transactivation of the steap4 promoter.
FIGURE 3.
STAT3 transcriptional activity is required in vivo for steap4 mRNA induction by IL-6. A, schematic representation of competition between STAT3α and STAT3β at a STAT3 target promoter. STAT3β lacks the transactivation domain, functions as a dominant negative form of STAT3, and blocks transactivation by STAT3α. B, qPCR analysis of steap4 mRNA expression in mouse livers infected with either Ad-GFP or Ad-GFP-STAT3β and treated with IL-6 (n = 8–11). C, ChIP assays demonstrating binding of STAT3α and STAT3β to the socs3 and steap4 promoters in GFP- or STAT3β-overexpressing livers. The C-20 STAT3 antibody (Ab) recognizes only STAT3α, whereas the K-15 antibody recognizes both STAT3α and STAT3β. A control PCR for the β-globin locus is also shown. Band intensities were measured and graphed (D).
steap4 Expression Changes with Nutritional Status in Multiple Tissues
steap4 mRNA levels have been shown previously to decrease with fasting and increase with feeding in visceral adipose tissue of mice (18). We assessed changes in steap4 mRNA levels in several tissues and found that steap4 mRNA decreases after a 16-h fast in the liver, epididymal, subcutaneous, and brown adipose tissue depots and muscle of C57BL6 mice (Fig. 4A). A 2-h re-feeding period after a 16-h fast results in an increase in steap4 mRNA, although it does not reach statistical significance in subcutaneous WAT and muscle. STEAP4 protein levels in the liver were determined by Western blot using fed versus fasted liver samples, and surprisingly, STEAP4 proteins levels increased in the crude membrane fraction with fasting (Fig. 4B). This increase in STEAP4 protein was unexpected because mRNA levels decrease during fasting.
IL-6 and Phosphorylation of STAT3 Do Not Play a Role in Nutritional Regulation of steap4 Expression
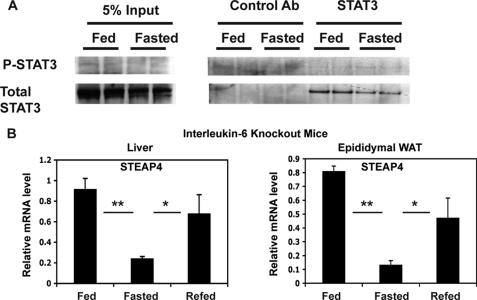
It has been demonstrated that the phosphorylation of STAT3 changes with feeding and fasting in the liver because of changes in acetylation of STAT3 (24). We therefore hypothesized that STAT3 phosphorylation during feeding may be involved in nutritional regulation of steap4. However, we could not consistently detect significant levels of P-STAT3 by immunoprecipitation and Western blot analysis of liver protein lysates (Fig. 5A). Because it was possible that P-STAT3 levels may have been down-regulated in the samples we examined, we did not rule out the possibility that IL-6 may mediate the increase in STEAP4 through STAT3. To further evaluate a role for STAT3 in this process, we hypothesized that IL-6 levels may increase systemically or locally in response to feeding and thereby increase expression of STEAP4 in peripheral tissues as it has been shown previously that insulin signaling in the brain results in a local hepatic increase of IL-6 and activation of STAT3 in the liver (7). However, in il-6 knock-out mice, there is a similar pattern of regulation of steap4 expression in liver and epididymal WAT as in wild type mice with feeding and fasting (Fig. 5B). Moreover, steap4 mRNA levels in il-6 knock-out mice are similar to the levels seen in wild type mice (data not shown). Therefore, IL-6 does not play a role in fed-fast regulation of steap4 expression in wild type lean mice.
FIGURE 5.
Lack of a role for STAT3 phosphorylation and IL-6 in fed-fast regulation of steap4 expression. A, P-STAT3 levels were detected by Western blot after immunoprecipitation of STAT3 from liver lysates (n = 2). B, qPCR analysis of steap4 mRNA levels in liver and epididymal WAT in interleukin-6 knock-out mice (n = 4). *, p ≤ 0.05; **, p < 0.001.
C/EBPα Regulates Nutritional Regulation of steap4 Expression
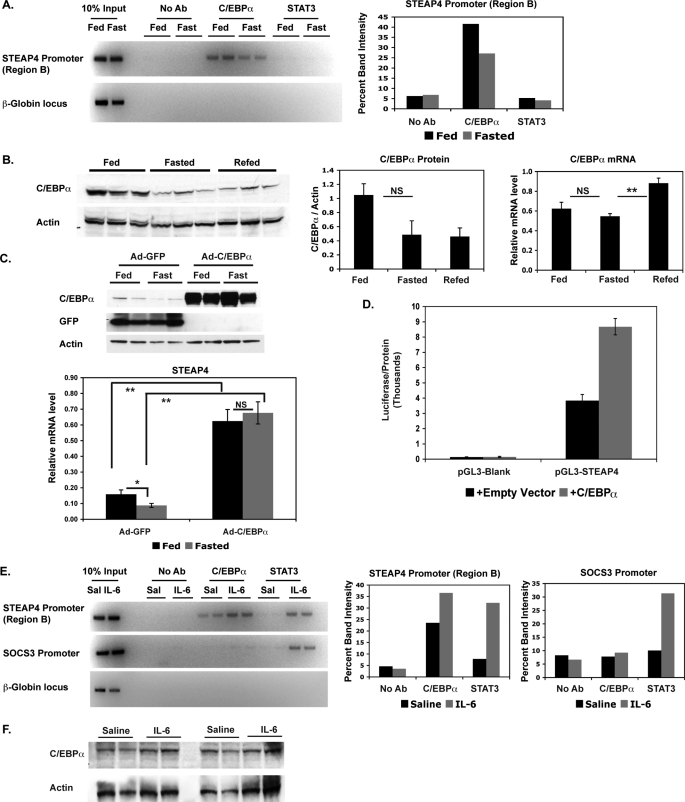
Because we had determined that STAT3 phosphorylation was unlikely to play a role in nutritional regulation of steap4 expression in the liver, we then considered other transcription factors that might be involved in this regulation. C/EBPα and liver X receptor have been suggested to regulate steap4 (18), and tumor necrosis factor-α treatment also regulates steap4 expression (17, 18), probably through NFκB activation. Using ChIP assays, we detected C/EBPα at the steap4 promoter, and a clear decrease in binding is seen in fasted samples compared with fed samples (Fig. 6A). As expected, based on the lack of STAT3 phosphorylation, we did not detect changes in STAT3 binding to the steap4 promoter under these conditions. Preliminary ChIP experiments did not detect any liver X receptor or NFκB at the steap4 promoter under either fed or fasted conditions (data not shown). These results suggest that C/EBPα recruitment to the steap4 promoter plays a major role in steap4 regulation.
FIGURE 6.
C/EBPα binds to the steap4 promoter during feeding and binding decreases during fasting. A, ChIP assays were performed using livers from mice that were either fed or fasted to detect the presence of C/EBPα and STAT3 at the steap4 promoter or the control β-globin locus. Band intensities were measured and graphed. B, C/EBPα protein levels in the liver were detected by Western blot (n = 3); c/ebpα mRNA levels were measured by qPCR analysis (n = 8). C, Western blot (C/EBPα, GFP, and actin) and qPCR (STEAP4) results from livers of mice overexpressing either GFP (control) or C/EBPα (n = 6–10) (D). HepG2 cells were transfected with pGL3-steap4 with or without co-transfected C/EBPα, and luciferase activity was measured and normalized to total protein levels (n = 4). E, ChIP assays were performed to detect the presence of C/EBPα and STAT3 at the steap4 and socs3 promoters in livers from mice treated with saline or IL-6; band intensities were measured and graphed. F, C/EBPα and actin protein levels in saline- and IL-6-treated livers were detected by Western blot (n = 4). *, p < 0.05; **, p < 0.001; NS, not significant; Ab, antibody.
We then determined whether c/ebpα protein and mRNA levels changed during fasting and re-feeding. C/EBPα protein levels decreased during fasting and remained low at 2 h after refeeding (Fig. 6B). However, c/ebpα mRNA levels showed only a slight decrease during fasting and a significant increase during re-feeding. This suggests that c/ebpα is down-regulated at the protein level during fasting and that there is a compensatory increase in c/ebpα mRNA levels during re-feeding. C/EBPα protein appears low in 2-h re-fed samples, but it is possible that these levels increase at a later time point, given the increase in mRNA levels. These results suggest that decreased expression of c/ebpα during fasting contributes to the reduction of C/EBPα at the steap4 promoter, resulting in down-regulation of steap4 expression.
To confirm the role of C/EBPα in nutritional regulation of hepatic STEAP4 in vivo, we overexpressed C/EBPα in the liver using adenovirus. C/EBPα-overexpressing livers show elevated expression of steap4 as compared with control GFP virus-infected livers (Fig. 6C, 3.6-fold increase in fed and 6.2-fold increase in fasted livers). In addition, there is no significant difference in steap4 mRNA levels between fed and fasted livers when C/EBPα is overexpressed, whereas in GFP-overexpressing livers, steap4 mRNA decreases with fasting. These results confirm that the level of C/EBPα protein in the liver is important for the nutritional regulation of steap4 expression. The role of C/EBPα in the regulation of STEAP4 is further supported by the fact that co-expression of the pGL3-STEAP4 reporter construct with C/EBPα in HepG2 cells resulted in a 2.2-fold increase in reporter activity (Fig. 6D).
Because IL-6-activated STAT3 binds to the steap4 promoter and increases transcription, we wanted to determine whether there were any changes in C/EBPα at the steap4 promoter with IL-6 treatment. ChIP analysis of the steap4 promoter showed that C/EBPα increased at the steap4 promoter but not at the socs3 promoter with IL-6 treatment (Fig. 6E). However, C/EBPα protein levels were not increased significantly with IL-6 treatment (Fig. 6F). Therefore, both feeding and IL-6 treatment resulted in increased C/EBPα at the steap4 promoter, but the mechanism by which this occurs differs in the two cases.
steap4 Expression Levels Are Increased in ob/ob Mice
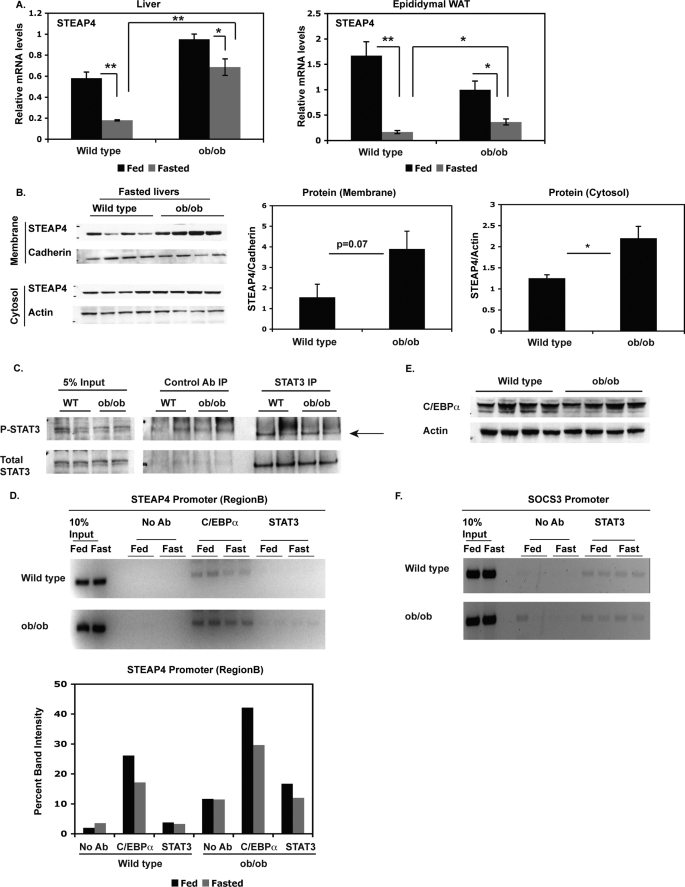
As steap4 mRNA levels are known to be elevated in visceral adipose tissue of ob/ob mice compared with lean control mice (18), we next determined steap4 expression levels in the livers of ob/ob mice compared with lean controls at the mRNA and protein levels. steap4 mRNA levels are significantly elevated in the livers of ob/ob mice in the fasted state (3.8-fold) and to a smaller degree in the fed state (1.6-fold, Fig. 7A). steap4 mRNA is also elevated in epididymal WAT in fasted ob/ob mice, although the increase is not as large as that seen in the liver (2.2-fold). STEAP4 protein levels in fasted ob/ob livers are also elevated compared with lean control livers; therefore, STEAP4 protein levels in ob/ob mice mirror mRNA levels (Fig. 7B). Because ob/ob mice are known to have elevated circulating IL-6 levels (25, 26), and these mice have previously been shown to have P-STAT3 in the liver and WAT (26), we hypothesized that the increased IL-6 and P-STAT3 contribute to elevated STEAP4 levels in ob/ob livers. Immunoprecipitation and Western blot analysis did not detect elevated P-STAT3 in ob/ob livers (Fig. 7C). ChIP analysis of the steap4 promoter revealed very weak binding of STAT3 to the steap4 promoter, and we were not able to detect an increase in STAT3 occupancy in ob/ob livers (Fig. 7D). STAT3 is detected at the socs3 promoter, demonstrating that the ChIP assay is functional (Fig. 7F), but there is no increased recruitment of STAT3 in ob/ob livers. As seen previously, C/EBPα was present at the steap4 promoter, and C/EBPα occupancy was slightly elevated in ob/ob livers in both fed and fasted conditions (Fig. 7D). This increase in C/EBPα at the steap4 promoter was not due to an increase in C/EBPα protein levels (Fig. 7E). These results mirror the effect of IL-6 treatment in lean mice, in that C/EBPα increases at the steap4 promoter without an apparent increase in C/EBPα protein levels.
FIGURE 7.
steap4 expression is increased in ob/ob mice. A, wild type (WT) and ob/ob mice were either fed ad libitum or fasted for 16 h and sacrificed. steap4 mRNA levels were measured in liver and epididymal WAT by qPCR analysis (n = 4). B, STEAP4 protein levels were detected by Western blot in fasted wild type and ob/ob livers. Band intensities were measured and shown as a graph (n = 4). C, P-STAT3 levels in fasted wild type or ob/ob livers were detected by Western blot after immunoprecipitating STAT3 from liver lysates (n = 2). D, ChIP assays were performed using livers from wild type and ob/ob mice to detect the presence of C/EBPα and STAT3 at the steap4 promoter. E, C/EBPα and actin protein levels in fasted wild type and ob/ob livers were detected by Western blot (n = 4). F, STAT3 was detected at the socs3 promoter in wild type and ob/ob mice by ChIP assays. *, p < 0.05; **, p < 0.001; Ab, antibody.
steap4 Expression Levels Are Increased in HFD-fed Mice
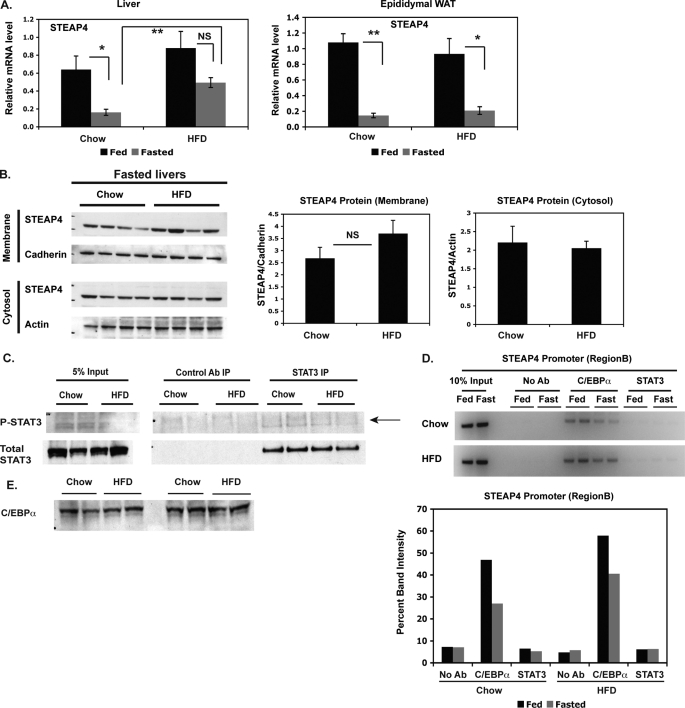
Because STEAP4 levels were elevated in ob/ob mice, we hypothesized that a similar increase would be seen in a different mouse model of obesity, such as diet-induced obesity. steap4 mRNA levels were significantly elevated in fasted but not fed livers of HFD-fed mice compared with lean chow-fed controls. No significant differences were seen in epididymal WAT in this model (Fig. 8A). STEAP4 protein levels also appeared to be higher in the crude membrane fraction in HFD-fed mice (Fig. 8B), again mirroring the increase in mRNA. Similar to ob/ob mice, we were unable to detect elevated P-STAT3 levels in fasted HFD-fed livers (Fig. 8C) or an increase in STAT3 at the steap4 promoter (Fig. 8D). An increase in C/EBPα was seen at the steap4 promoter (Fig. 8D) without an increase in C/EBPα protein levels (Fig. 8E), similar to the results seen in ob/ob mice and lean mice treated with IL-6.
FIGURE 8.
steap4 expression is increased in DIO mice. A, chow-fed and HFD-fed mice were either fed ad libitum or fasted for 36 h and then sacrificed. steap4 mRNA levels were measured in liver and epididymal WAT by qPCR analysis (n = 4). B, STEAP4 protein levels were detected by Western blot in fasted chow-fed and HFD-fed mouse livers. Band intensities were measured and graphed (n = 4). C, P-STAT3 levels in fasted chow-fed or HFD-fed mouse livers were detected by Western blot after immunoprecipitating STAT3 from liver lysates (n = 2). D, ChIP assays were performed using livers from chow-fed and HFD-fed mice to detect the presence of C/EBPα and STAT3 at the steap4 promoter. E, C/EBPα protein levels in fasted chow-fed and HFD-fed livers were detected by Western blot (n = 4). *, p < 0.05; **, p < 0.001; NS, not significant; Ab, antibody.
DISCUSSION
An aberrant change in the expression of genes involved in glucose homeostasis is one of the causes of insulin resistance, which eventually leads to diabetes. Therefore, tight regulation of the expression of genes involved in insulin signaling is critical to maintaining normal glucose homeostasis. The transcription factor STAT3 plays an important role in glucose homeostasis because it negatively regulates gluconeogenic gene expression in the liver (7–9). This has been demonstrated both genetically and more recently biochemically, as STAT3 interacts specifically with the regulatory elements controlling the expression of the gluconeogenic genes phosphoenolpyruvate carboxykinase and Glc-6-Pase. Because mice that lack hepatic STAT3 have a number of metabolic defects, including obesity and insulin resistance (8), we hypothesized that STAT3 also regulates other key metabolic targets in the liver. In this study, we determined that STAT3 regulates steap4 in mouse liver (Fig. 1), a protein that has previously been shown to promote insulin sensitivity in adipose tissue (18). This suggests that STAT3 may promote insulin sensitivity in the liver through the regulation of several target genes that are beneficial to metabolism.
To establish that steap4 is a bona fide target of STAT3, we used in vivo ChIP to show direct recruitment of STAT3 to a proximal region in the steap4 promoter. Furthermore, using a steap4 promoter reporter construct, we demonstrate that the STAT3-binding site at −116 bp (that is conserved in humans) clearly mediates this function. Additional support for the relevance of STAT3 in the in vivo regulation of hepatic steap4 expression is shown in experiments where STAT3β is overexpressed in the liver. Here, steap4 induction by IL6 is blocked because STAT3β is preferentially recruited to the STAT3 site in the steap4 promoter (Fig. 3). Taken together these data clearly establish steap4 as novel target of STAT3 in the liver.
To examine the potential physiological role of the STAT3 induction of steap4, we first looked at feeding and fasting mouse models. Indeed, steap4 mRNA expression is dramatically reduced in the liver in a similar fashion to WAT. In contrast, there is no decrease in STEAP4 protein levels in fasted livers. STEAP4 protein levels appear to increase in the membrane fraction, whereas the cytosolic fraction remains the same. It remains possible that there is an increase in STEAP4 translocation to the membrane with fasting, and there may be an increase in translation of STEAP4 protein to compensate for the fall in mRNA levels. STEAP4 protein levels may decrease with prolonged fasting, but this remains to be tested.
As STAT3 phosphorylation has been described to occur during feeding (24), we hypothesized that its dephosphorylation during fasting could explain the fall in steap4 mRNA levels. However, in both immunoprecipitation and ChIP assays, we could not detect evidence of P-STAT3 in the fed state, suggesting that P-STAT3 is not the mediator of nutritional change in steap4 gene expression. Because systemic IL-6 levels are high in obesity and increased hepatic P-STAT3 has been found in obese mice, we next examined steap4 regulation in obese mouse models (ob/ob and high fat diet induced obesity). Indeed, both steap4 mRNA and protein levels are increased in these models consistent with a potential protective role of STEAP4. However, again we could not detect evidence of increased STAT3 phosphorylation or recruitment to the steap4 promoter. Thus, although it is unlikely that STAT3 plays a role in nutritional regulation of steap4, we cannot rule out this possibility because P-STAT3 has been detected previously in livers from both fed and obese mice (24, 26). Alternatively, STAT3 signaling may be inhibited by SOCS3 in a negative feedback loop following induction of socs3 (27, 28), and it is therefore possible that although STAT3 was phosphorylated at an earlier stage in obesity, it was eventually down-regulated.
Because C/EBPα had been previously described as a regulator of the steap4 promoter in fat cells (18), we determined whether it could mediate the nutritional changes in hepatic STEAP4. Indeed, in the liver, changes in C/EBPα recruitment to the steap4 promoter are likely to mediate decreased expression of steap4 during fasting. Conversely, C/EBPα recruitment to the steap4 promoter is enhanced in obese livers and likely also plays a role in the elevation of hepatic STEAP4 in obesity. Interestingly, C/EBPα recruitment is also enhanced by IL-6 treatment and coincides with increased STAT3 recruitment. Additionally, IL-6 knock-out mice do not have altered levels of steap4 mRNA as compared with wild type mice. Given these observations, we suggest that C/EBPα regulates steap4 expression via both IL-6-dependent and IL-6-independent mechanisms. When C/EBPα protein levels increase, such as during feeding or when overexpressed, steap4 expression increases. When IL-6 levels increase, C/EBPα recruitment to the steap4 promoter increases and enhances transcription in a manner that is independent of total C/EBPα protein levels. Interestingly, C/EBPα can be phosphorylated by p38 MAPK on serine 21, resulting in increased transcriptional activity of C/EBPα (29), and IL-6 activates (phosphorylates) p38 MAPK in hepatocytes (30). Phospho-p38 levels are elevated in livers of ob/ob and db/db mice and in the livers of rats fed a high fat diet (29, 31, 32). Thus, it is possible that the increased recruitment of C/EBPα to the steap4 promoter could be explained by its phosphorylation. It is possible that when IL-6 levels reach a certain threshold in the serum, both STAT3 and C/EBPα drive transcription of steap4 and may synergize at the steap4 promoter.
Although the mechanism by which STEAP4 exerts its protective effects in the liver is not clear, in WAT it appears to function by controlling IL-6 production (18). The STEAP proteins are required for uptake of iron and copper because both metals have to be reduced prior to release from the endosome (as reviewed in Ref. 33). High levels of iron and copper in the serum are associated with diabetes, and both metals participate in the Fenton reaction to generate reactive oxygen species that can then lead to insulin resistance (5, 6). In addition, it has been shown that iron depletion in HepG2 cells and in vivo results in increased insulin signaling (34). Serum copper levels and reactive oxygen species have been shown to be elevated in db/db mice compared with nondiabetic controls (35). Chelating serum copper in db/db mice reduces reactive oxygen species and copper levels, while increasing insulin sensitivity and glucose tolerance (35). It is possible that in inflammatory states such as obesity, steap4 expression is increased by both STAT3 and C/EBPα to properly utilize excessive iron and copper to prevent the formation of reactive oxygen species and subsequent insulin resistance.
In summary, this study demonstrates that steap4 is regulated by both nutritional and inflammatory signals in the liver. Although C/EBPα regulates expression of steap4 in response to nutritional changes, both C/EBPα and STAT3 play a role in steap4 regulation under inflammatory conditions. Given that STAT3 regulates hepatic gluconeogenic gene expression (7–9) and that we have now established that steap4 is also a STAT3 target, it is very likely that STAT3 is a key metabolic integrator that regulates transcription of several genes involved in glucose homeostasis.
Acknowledgments
We thank Dr. Inna Astapova and Kaila Holtz for their help with experiments. We thank Dr. Ormond MacDougald for the pCDNA3.1-C/EBPα construct, and Dr. Jim Cormier for suggestions regarding the preparation of crude membranes from tissue.
This work was supported by the Smith Family Pinnacle Award from the American Diabetes Association and a grant from Takeda Pharmaceuticals.
- IL-6
- interleukin 6
- WAT
- white adipose tissue
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GFP
- green fluorescent protein
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative PCR
- C/EBPα
- CCAAT/enhancer-binding protein α
- HFD
- high fat diet.
REFERENCES
- 1.Hotamisligil G. S., Shargill N. S., Spiegelman B. M. (1993) Science 259, 87–91 [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil G. S., Arner P., Caro J. F., Atkinson R. L., Spiegelman B. M. (1995) J. Clin. Invest. 95, 2409–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr. (2003) J. Clin. Invest. 112, 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., Nakayama O., Makishima M., Matsuda M., Shimomura I. (2004) J. Clin. Invest. 114, 1752–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houstis N., Rosen E. D., Lander E. S. (2006) Nature 440, 944–948 [DOI] [PubMed] [Google Scholar]
- 7.Inoue H., Ogawa W., Asakawa A., Okamoto Y., Nishizawa A., Matsumoto M., Teshigawara K., Matsuki Y., Watanabe E., Hiramatsu R., Notohara K., Katayose K., Okamura H., Kahn C. R., Noda T., Takeda K., Akira S., Inui A., Kasuga M. (2006) Cell Metab. 3, 267–275 [DOI] [PubMed] [Google Scholar]
- 8.Inoue H., Ogawa W., Ozaki M., Haga S., Matsumoto M., Furukawa K., Hashimoto N., Kido Y., Mori T., Sakaue H., Teshigawara K., Jin S., Iguchi H., Hiramatsu R., LeRoith D., Takeda K., Akira S., Kasuga M. (2004) Nat. Med. 10, 168–174 [DOI] [PubMed] [Google Scholar]
- 9.Ramadoss P., Unger-Smith N. E., Lam F. S., Hollenberg A. N. (2009) Mol. Endocrinol. 23, 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldenhoven E., van Dijk T. B., Solari R., Armstrong J., Raaijmakers J. A., Lammers J. W., Koenderman L., de Groot R. P. (1996) J. Biol. Chem. 271, 13221–13227 [DOI] [PubMed] [Google Scholar]
- 11.Maritano D., Sugrue M. L., Tininini S., Dewilde S., Strobl B., Fu X., Murray-Tait V., Chiarle R., Poli V. (2004) Nat. Immunol. 5, 401–409 [DOI] [PubMed] [Google Scholar]
- 12.Yoo J. Y., Huso D. L., Nathans D., Desiderio S. (2002) Cell 108, 331–344 [DOI] [PubMed] [Google Scholar]
- 13.Ohgami R. S., Campagna D. R., McDonald A., Fleming M. D. (2006) Blood 108, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arner P., Stenson B. M., Dungner E., Näslund E., Hoffstedt J., Ryden M., Dahlman I. (2008) J. Clin. Endocrinol. Metab. 93, 2249–2254 [DOI] [PubMed] [Google Scholar]
- 15.Korkmaz C. G., Korkmaz K. S., Kurys P., Elbi C., Wang L., Klokk T. I., Hammarstrom C., Troen G., Svindland A., Hager G. L., Saatcioglu F. (2005) Oncogene 24, 4934–4945 [DOI] [PubMed] [Google Scholar]
- 16.Zhang C. M., Chi X., Wang B., Zhang M., Ni Y. H., Chen R. H., Li X. N., Guo X. R. (2008) Acta Pharmacol. Sin. 29, 587–592 [DOI] [PubMed] [Google Scholar]
- 17.Moldes M., Lasnier F., Gauthereau X., Klein C., Pairault J., Fève B., Chambaut-Guérin A. M. (2001) J. Biol. Chem. 276, 33938–33946 [DOI] [PubMed] [Google Scholar]
- 18.Wellen K. E., Fucho R., Gregor M. F., Furuhashi M., Morgan C., Lindstad T., Vaillancourt E., Gorgun C. Z., Saatcioglu F., Hotamisligil G. S. (2007) Cell 129, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Astapova I., Lee L. J., Morales C., Tauber S., Bilban M., Hollenberg A. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19544–19549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., Watson S. J., Meng F. (2005) Nucleic Acids Res. 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article 3 [DOI] [PubMed] [Google Scholar]
- 22.Fasshauer M., Kralisch S., Klier M., Lossner U., Bluher M., Chambaut-Guérin A. M., Klein J., Paschke R. (2004) FEBS Lett. 560, 153–157 [DOI] [PubMed] [Google Scholar]
- 23.Wegenka U. M., Buschmann J., Lütticken C., Heinrich P. C., Horn F. (1993) Mol. Cell. Biol. 13, 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie Y., Erion D. M., Yuan Z., Dietrich M., Shulman G. I., Horvath T. L., Gao Q. (2009) Nat. Cell Biol. 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harkins J. M., Moustaid-Moussa N., Chung Y. J., Penner K. M., Pestka J. J., North C. M., Claycombe K. J. (2004) J. Nutr. 134, 2673–2677 [DOI] [PubMed] [Google Scholar]
- 26.Klover P. J., Clementi A. H., Mooney R. A. (2005) Endocrinology 146, 3417–3427 [DOI] [PubMed] [Google Scholar]
- 27.Nicholson S. E., Willson T. A., Farley A., Starr R., Zhang J. G., Baca M., Alexander W. S., Metcalf D., Hilton D. J., Nicola N. A. (1999) EMBO J. 18, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr R., Willson T. A., Viney E. M., Murray L. J., Rayner J. R., Jenkins B. J., Gonda T. J., Alexander W. S., Metcalf D., Nicola N. A., Hilton D. J. (1997) Nature 387, 917–921 [DOI] [PubMed] [Google Scholar]
- 29.Qiao L., MacDougald O. A., Shao J. (2006) J. Biol. Chem. 281, 24390–24397 [DOI] [PubMed] [Google Scholar]
- 30.Zauberman A., Zipori D., Krupsky M., Ben-Levy R. (1999) Oncogene 18, 3886–3893 [DOI] [PubMed] [Google Scholar]
- 31.Gum R. J., Gaede L. L., Heindel M. A., Waring J. F., Trevillyan J. M., Zinker B. A., Stark M. E., Wilcox D., Jirousek M. R., Rondinone C. M., Ulrich R. G. (2003) Mol. Endocrinol. 17, 1131–1143 [DOI] [PubMed] [Google Scholar]
- 32.Li S. Y., Liu Y., Sigmon V. K., McCort A., Ren J. (2005) Diabetes Obes. Metab. 7, 448–454 [DOI] [PubMed] [Google Scholar]
- 33.Knutson M. D. (2007) Nutr. Rev. 65, 335–340 [DOI] [PubMed] [Google Scholar]
- 34.Dongiovanni P., Valenti L., Ludovica Fracanzani A., Gatti S., Cairo G., Fargion S. (2008) Am. J. Pathol. 172, 738–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka A., Kaneto H., Miyatsuka T., Yamamoto K., Yoshiuchi K., Yamasaki Y., Shimomura I., Matsuoka T. A., Matsuhisa M. (2009) Endocr. J. 56, 699–706 [DOI] [PubMed] [Google Scholar]