Abstract
Phosphoinositides play key roles in regulating membrane dynamics and intracellular signaling in eukaryotic cells. However, comparable lipid-based signaling pathways have not been identified in bacteria. Here we show that Mycobacterium smegmatis and other Actinomycetes bacteria can synthesize the phosphoinositide, phosphatidylinositol 3-phosphate (PI3P). This lipid was transiently labeled with [3H]inositol. Sensitivity of the purified lipid to alkaline phosphatase, headgroup analysis by high-pressure liquid chromatography, and mass spectrometry demonstrated that it had the structure 1,2-[tuberculostearoyl, octadecenoyl]-sn-glycero 3-phosphoinositol 3-phosphate. Synthesis of PI3P was elevated by salt stress but not by exposure to high concentrations of non-ionic solutes. Synthesis of PI3P in a cell-free system was stimulated by the synthesis of CDP-diacylglycerol, a lipid substrate for phosphatidylinositol (PI) biosynthesis, suggesting that efficient cell-free PI3P synthesis is dependent on de novo PI synthesis. In vitro experiments further indicated that the rapid turnover of this lipid was mediated, at least in part, by a vanadate-sensitive phosphatase. This is the first example of de novo synthesis of PI3P in bacteria, and the transient synthesis in response to environmental stimuli suggests that some bacteria may have evolved similar lipid-mediated signaling pathways to those observed in eukaryotic cells.
Keywords: Bacteria, Bacterial Metabolism, Inositol Phospholipid, Lipid, Phosphatidylinositol, Phosphatidylinositol Signaling, Phosphoinositide, Stress Response
Introduction
Lipids are part of complex signaling networks in eukaryotic cells, controlling numerous biological events. Major membrane lipids such as phosphoinositides, sphingolipids, cholesterol, and glycerophospholipids are all involved in the generation of second messengers. One class of phosphoinositide signaling molecules, phosphatidylinositol 3-phosphate (PI3P),6 was first identified in mammalian fibroblast cells in 1988 (1) and is now recognized as a ubiquitous and functionally important phosphoinositide that acts as a second messenger mediating various cellular processes such as endocytic membrane trafficking, motility, and cytokine release (2–4). The synthesis of PI3P is mediated by a family of phosphatidylinositol 3-kinases (PI3K), with the class III PI3K Vps34 being present in all eukaryotes examined so far (5), suggesting an ancient origin of phosphoinositide signaling.
In striking contrast, inositol lipid signaling has never been described in bacteria. Bacterial membranes are mainly composed of glycerophospholipids. For example, the plasma membrane of Escherichia coli is composed of three major glycerophospholipids: phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin. Sphingolipids, sterols, and certain glycerophospholipids such as phosphatidylserine and phosphatidylinositol (PI) are rarely found in bacteria (6). Interestingly, some bacteria contain phosphoinositide kinases and phosphatases that are capable of producing and hydrolyzing PI3P or PI4-phosphate (PI4P). For example, a protein/lipid kinase purified from Legionella micdadei was shown to phosphorylate PI (7). However Legionella lacks the capacity to synthesize PI (8), and it is likely that this kinase phosphorylates host PI. A similar modulation of host PI metabolism has been suggested to occur during Salmonella enterica infection by a bacterial phosphoinositide phosphatase SopB (9). In this case, SopB was proposed to dephosphorylate PI 3,4,5-P3 and PI 3,5-bisphosphate to generate PI3P and modulate host vesicular trafficking. Finally, Rhizobium leguminosarum, a nitrogen-fixing plant symbiont, possesses a phosphotransferase activity that can generate PI4P by transferring phosphate from Lipid A to PI. However, neither the substrate PI nor the product PI4P was detected in the bacterial membrane, leading the authors to suggest that PI4P is synthesized only during nodulation or that the enzyme utilizes exogenous PI from the host (10). Thus, while some bacteria can express inositol-lipid kinases/phosphotransferases and phosphatases, phosphorylated PI species have not yet been detected in bacteria.
Actinomycetes are a diverse group of Gram-positive bacteria that include important human pathogens such as Mycobacterium tuberculosis and Mycobacterium leprae and the industrially important species, Corynebacterium glutamicum. Unlike most other bacteria, they possess a canonical pathway of de novo PI biosynthesis (11–16), and PI is a major component of the plasma membrane. Furthermore, PI also acts as a membrane anchor for abundant glycolipids, such as PI mannosides (PIMs) and lipoarabinomannan (17). The gene encoding PI synthetase is essential for the growth of Mycobacterium smegmatis, a model organism of mycobacteria, suggesting that PI or its downstream products are essential (18). The abundance of PI and the capacity of mycobacteria to utilize PI for further elaborations led us to ask if these bacteria produce phosphorylated PIs, in a way similar to eukaryotic cells. Here we show that M. smegmatis synthesizes PI3P in response to environmental stimuli. Our study represents the first example of de novo synthesis of PI3P in bacteria and indicates that eukaryote-like lipid-mediated signaling may occur in some bacteria.
EXPERIMENTAL PROCEDURES
Metabolic Labeling
M. smegmatis strain mc2155 (19) was grown at 37 °C in Middlebrook 7H9 broth (Difco), supplemented with 0.2% (w/v) glucose, 0.2% (v/v) glycerol, and 15 mm NaCl. All experiments were performed on cultures harvested at mid-log growth phase. For metabolic labeling, 30 ml of bacterial culture was centrifuged (2,500 × g, 10 min), and the cell pellet was resuspended at 0.25 g/ml in Sauton's minimal medium pre-warmed at 37 °C. After incubation at 37 °C for 15 min, labeling was initiated by addition of [3H]inositol (Amersham Biosciences, 16.0 Ci/mmol, final concentration of 50 μCi/ml) and continued for 5 min. The culture was then chased by dilution in 10 volumes of various solutions (containing 1 mm non-radioactive inositol) as indicated. Aliquots were collected at the indicated time points and snap-frozen in liquid nitrogen prior to analysis. After thawing, bacteria were rapidly centrifuged, and [3H]inositol-labeled lipids were extracted twice in chloroform/methanol (2:1, v/v) and once in chloroform/methanol/water (1:2:0.8, v/v). Dried lipids were suspended in 1-butanol/water (2:1, v/v), and lipids in the upper 1-butanol-rich phase were analyzed by high-performance thin layer chromatography (HPTLC) on aluminum-backed Silica Gel 60 sheets (Merck) developed in chloroform/methanol/1 m ammonium acetate/13 m ammonia/water (180:140:9:9:23, v/v, solvent 1).
Oxalic Acid-impregnated HPTLC
HPTLC plates were prepared as described (20). Lipid extract from [3H]inositol-labeled M. smegmatis was loaded side by side with 2 μg of phosphoinositides from bovine brain (Sigma-Aldrich). The HPTLC plate was developed by chloroform/methanol/14.6 m ammonia/water (90:70:4:16, v/v, solvent 2). Bovine brain phosphoinositides were visualized by iodine vapor and contours of PI, PI monophosphate (PIP), and PI bisphosphates were marked on the HPTLC plate.
Enzymatic Treatments
Phospholipid species were purified by HPTLC, and silica scrapings were extracted in chloroform/methanol/water (10:10:3, v/v). Lipids were further purified by 1-butanol/water phase partition. Lipids (1000 dpm) were resuspended in 50 mm Tris-HCl (pH 9.0), 1 mm MgCl2, 0.16% (w/v) sodium deoxycholate (25 μl) and treated with alkaline phosphatase (E. coli C75, Takara, 0.4 unit) at 37 °C for 1 h. Lipids were recovered by 1-butanol/water phase partition and re-analyzed by HPTLC (solvent 1).
Preparation of Yeast and Leishmania Lipid Standards
A strain of Saccharomyces cerevisiae defective in de novo synthesis of inositol (RSY1156) (21) was grown to mid-log growth phase in SC broth, starved for inositol for 2 h, and incubated with [3H]inositol for 6 h. Lipids were extracted as previously described (20). For inositol bisphosphate standards (Fig. 2E), a mixture of inositol 1,3-bisphosphate and inositol 1,4-bisphosphate was prepared from the yeast lipids and mixed with commercially available [3H]inositol 1,4-bisphosphate to give a similar height for both peaks. Leishmania mexicana was metabolically labeled with [3H]inositol, and lipids were extracted as described before (22).
FIGURE 2.
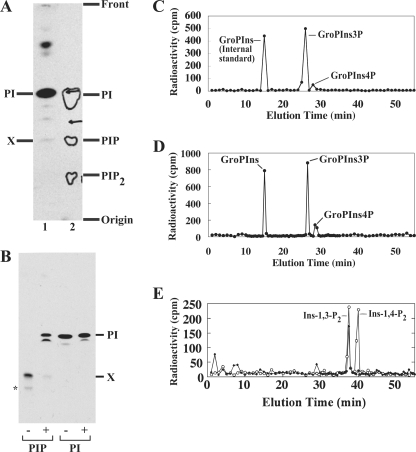
Structural characterization of Lipid X. A, migration of [3H]inositol-labeled Lipid X (lane 1) on an oxalic acid-impregnated HPTLC plate in comparison to bovine brain phosphoinositides (lane 2). Contours in lane 2 indicate PI, PIP, and PI bisphosphate (PIP2) from brain phosphoinositides, which were visualized by iodine vapor. B, PI and Lipid X were metabolically labeled with [3H]inositol, purified by preparative HPTLC, treated with alkaline phosphatase, and resolved by HPTLC. −, mock; +, alkaline phosphatase. *, a minor contaminant, which may be a PI3P variant. C, [3H]inositol-labeled Lipid X was deacylated, and released GroPIns phosphates were fractionated by HPLC. As an internal standard, GroPIns (generated by deacylating [3H]PI) was coinjected. D, co-elution of mycobacterial Lipid X with the yeast PI3P standard. [3H]Inositol-labeled Lipid X was deacylated as in C. [3H]Inositol-labeled yeast PI/PIP standards were prepared as described under “Experimental Procedures” and deacylated to produce a mixture of GroPIns, GroPIns3P, and GroPIns4P. The yeast standard mixture containing GroPIns (926 cpm), GroPIns3P (355 cpm), and GroPIns4P (206 cpm) was co-injected into the HPLC column with mycobacterial Lipid X (GroPIns3P/GroPIns4P candidates; 570 cpm/66 cpm). Recovery from the yeast GroPIns peak was 90% (830 cpm). E, inositol polyphosphates were generated by deglyceration of the GroPIns3P peak derived from Lipid X and fractionated by HPLC. Open circles, standards; closed diamonds, deglycerated M. smegmatis PI3P headgroup.
HPLC
HPTLC-purified phospholipids were resuspended in 200 μl of methanol/25% ammonia (1:1, v/v), and incubated at 37 °C overnight. The glycerophosphoinositols (GroPIns) were then recovered by phase partition in 1-butanol/50 mm HEPES (pH 7.4) (2:1, v/v). The lower aqueous phase was dried on a SpeedVac concentrator and analyzed by HPLC (Merck-Hitachi L-6210), using a Partisil 10-SAX-250A column (Alltech) fitted with a guard column. GroPIns was eluted in a stepwise linear gradient of ammonium phosphate (0–5 min, 0 mm; 5–45 min, 0–150 mm; 45–52 min, 150–250 mm; and 52–64 min, 250–1250 mm) at a flow rate of 1 ml/min. For some experiments, GroPIns was further treated for deglyceration using an established protocol (23). Briefly, the glycerol moiety was oxidized in 10 mm periodate (25 °C, 30 min). After quenching excess periodate by 250 mm ethylene glycol (25 °C, 30 min), 0.46% 1-aminopiperidine was added to eliminate aldehydes (25 °C, 3 h). The sample was purified by passage through a Dowex AG50W column, and the eluate was analyzed by HPLC.
Mass Spectrometry of PI3P
M. smegmatis cell pellets were extracted in chloroform/methanol (2:1, v/v) containing 0.25% HCl. Insoluble material was removed by centrifugation, and the supernatant was partitioned by addition of 1 n HCl. The lower organic phase was dried under N2 and dissolved in chloroform/methanol/water (10:10:3, v/v, 0.5 g wet pellet equivalent per ml). Samples were directly infused (10 μl/min) into the electrospray source of a 6410 Triple Quadrupole mass spectrometer (Agilent Technologies). The capillary voltage was set at 4000 V, and the gas temperature and flow rate were set to 250 °C and 8 liters/min, respectively. The nebulizer was set at 25 p.s.i. The fragmentator for precursor ion search was set to 80 V and collision energy to 42 eV. Negative ion precursor scanning was performed using m/z 241 and m/z 321 to detect the PI and PIP species, respectively (24). MS/MS experiments were conducted with fragmentator voltages of 80, 90, or 100 V and collision energies of 22, 32, 42, 47, or 50 eV.
Preparation of Cell Membranes
Wild-type M. smegmatis and an inositol auxotroph (Δino1) (25) were grown in 7H9 broth, supplemented with 0.2% (w/v) glucose, 0.2% (v/v) glycerol, 15 mm NaCl and, in the case of Δino1, 1 mm inositol and 20 μg/ml kanamycin. M. smegmatis Δino1 cells were suspended in the same growth medium without inositol 2 h prior to the preparation of the cell lysates to allow accumulation of CDP-diacylglycerol (CDP-DAG). Cells were harvested by centrifugation, washed in 50 mm HEPES/NaOH (pH 7.4) twice, and suspended in a lysis buffer containing 50 mm HEPES/NaOH (pH 7.4), 15% glycerol, 2 mm EGTA, and Complete Protease Inhibitor Mixture (EDTA-free, Roche Applied Science). Cells were lysed by probe sonication (15 cycles of 30 s with a 30-s interval on ice to keep the lysate cold) at a power setting of 5 (Astrason Ultrasonic Liquid Processor, Misonix, Farmingdale, NY). Unlysed cells and large cell debris were removed by centrifugation (3,000 × g, 10 min × 2), and a cell membrane/cell wall fraction was obtained by ultracentrifugation (257,000 × g, 30 min, 4 °C). The pellet was resuspended in lysis buffer and used for cell-free radiolabeling experiments.
Cell-free Biosynthesis of Phosphorylated PI
An aliquot of cell membrane suspension was supplemented with 5 mm MgCl2, 2 mm EGTA, and other components as indicated in each experiment. After 5 min of pre-warming at 37 °C, the reaction was started by the addition of 1.2 μCi of [3H]inositol. The incubation was stopped at 60 min by the addition of chloroform/methanol (1:1, v/v) to achieve a final ratio of chloroform/methanol/water (10:10:3, v/v). Extracted lipids were then subjected to 1-butanol/water phase partition, and purified lipids were fractionated by HPTLC (solvent 1).
RESULTS
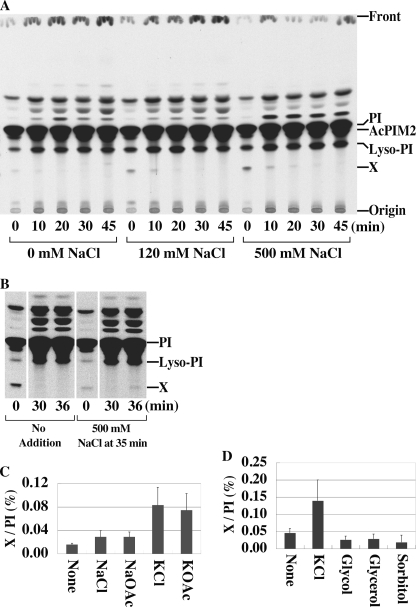
M. smegmatis was pulse-labeled with [3H]inositol, resulting in the rapid labeling of previously characterized inositol-lipids, including PI, lyso-PI, and the PIM species, AcPIM2 (25). A minor, unidentified species, termed Lipid X, was also labeled in these experiments (Fig. 1A). The synthesis of this species appeared to be transient, because label was rapidly lost during the chase (Fig. 1A). Synthesis of Lipid X was also stimulated by the presence of high salt (Fig. 1A). To confirm that the synthesis of Lipid X was stimulated by changes in extracellular NaCl, M. smegmatis was pulse-labeled with [3H]inositol in low salt medium, then chased in the same low salt medium containing excess non-radioactive inositol to deplete labeled Lipid X (Fig. 1B, 30 min). Addition of salt (500 mm NaCl) to these prelabeled cells resulted in a second pulse of Lipid X synthesis within 1 min indicating resynthesis from prelabeled inositol lipids (Fig. 1B). Potassium salts (KCl and KOAc) were even more effective at stimulating Lipid X synthesis, whereas non-ionic solutes such as glycol, glycerol, or sorbitol had no effect on Lipid X synthesis (Fig. 1, C and D). Taken together, these results suggest that Lipid X is rapidly synthesized from pre-existing inositol-lipids and turned over in response to changes in external salt concentrations.
FIGURE 1.
A new [3H]inositol-labeled lipid in M. smegmatis. A, cells were pulse-labeled with [3H]inositol and chased in 25 mm Tris buffer (pH 7.5)/1 mm inositol containing different concentrations of NaCl. AcPIM2, acyl-phosphatidylinositol dimannoside. The radiolabeled species directly above PI is likely to be Ac2PIM1 (35). Other less polar species have not been characterized. B, pulse-chase labeling was conducted in the absence of NaCl as described in A. Cells were then exposed to 500 mm NaCl at 35 min. C and D, cells were labeled with [3H]inositol as in B, and then exposed to increases in the concentration of different salts (300 mm) or non-ionic solutes (600 mm) at 35 min. Cells were harvested after 5 min and label in Lipid X normalized to that in PI. Averages of triplicates with standard deviations are shown.
The structure of Lipid X was investigated by HPTLC analysis of the native lipid, and HPLC analysis of the polar headgroup. [3H]Lipid X migrated close to authentic PIP on HPTLC using an oxalic acid-impregnated silica gel plate, that allows clear separation of PI, PIP, and PI bisphosphate species (Fig. 2A). Following treatment with alkaline phosphatase, Lipid X was converted to a faster migrating species with similar HPTLC mobility to PI (Fig. 2B). These analyses suggested that Lipid X is a PI monophosphate. HPLC analysis of the polar headgroup of phosphoinositides is a well established method to determine in which position the inositol ring is phosphorylated (20). Lipid X was treated with mild base to remove fatty acyl groups, and the released GroPIns headgroup was analyzed by anion-exchange HPLC. Two peaks were resolved, containing 89.6% and 10.4% of the radiolabel (Fig. 2C). The retention time of the major peak was similar to that of a PI3P headgroup released from yeast PIP standards. To confirm co-elution, we injected the polar headgroups of Lipid X together with the polar headgroups of the yeast PI/PIP standards (Fig. 2D). The Lipid X peaks co-eluted precisely with the yeast GroPIns headgroups as assessed by recovery of radioactivity in each peak. Specifically, the peak corresponding to glycerophosphoinositol 3-phosphate (GroPIns3P) contained 945 cpm, corresponding to the sum of yeast GroPIns3P (355 cpm) and the major peak of Lipid X (570 cpm) (102% recovery), whereas 252 cpm was recovered from the glycerophosphoinositol 4-phosphate (GroPIns4P) peak, representing 93% of the sum of yeast GroPIns4P (206 cpm) and the minor peak of M. smegmatis Lipid X (66 cpm). These data confirmed that the major and minor polar headgroups of Lipid X correspond to GroPIns3P and GroPIns4P, respectively. To provide further evidence that the major headgroup carries a phosphate at the 3-OH of inositol, we eliminated the glycerol moiety of the major GroPIno3P by periodate treatment, producing inositol polyphosphates. Inositol polyphosphates show different retention times from their GroPIns counterparts, and a new peak originating from the major GroPIns headgroup of Lipid X co-migrated precisely with authentic inositol 1,3-bisphosphate (Fig. 2E). Taken together, these data established Lipid X as PI3P.
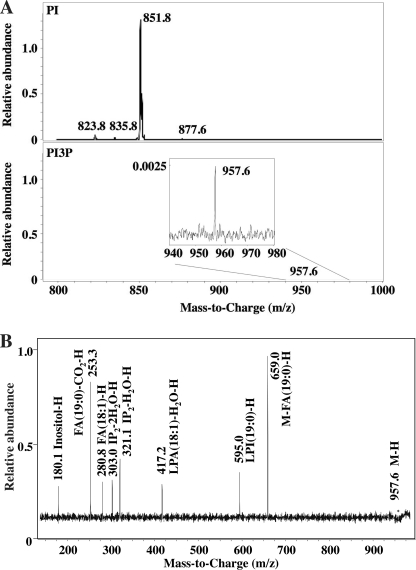
We next determined the precise mass and abundance of mycobacterial PI3P by liquid chromatography-MS. A characteristic ion at m/z 321 represents the dehydrated diphosphate inositol headgroup derived from PIP. We used this ion and conducted precursor ion scanning of mycobacterial lipid extracts. Strikingly, we detected only a single molecular species of PIP at m/z 957.6. The molecular weight of this species suggested that it contains acyl chains with a combined length of 37 carbons and one double bond (37:1) (Fig. 3A, lower panel). MS/MS analysis of the m/z 957.6 ion generated characteristic product ions at m/z 321.1 (IP2-H2O-H), 303.0 (IP2-2H2O-H), 280.8 (18:1 fatty acid-H), 253.3 (19:0 fatty acid-CO2-H), and 417.2 (lyso phosphatidic acid-H2O-H with 18:1 fatty acid) (Fig. 3B). These analyses suggest that the major molecular species of mycobacterial PI3P carries 19:0 (tuberculostearic) and 18:1 fatty acids.
FIGURE 3.
Mass spectrometric analysis of PI3P. A, precursor ion scanning MS for molecular species of PI (upper panel) and PI3P (lower panel). B, MS/MS of m/z = 957.6 from PI3P fraction. The mass spectra obtained with different fragmentator voltages and collision energies were merged to form the composite mass spectrum. FA, fatty acid; IP, inositol phosphate; and LPA, lysophosphatidic acid.
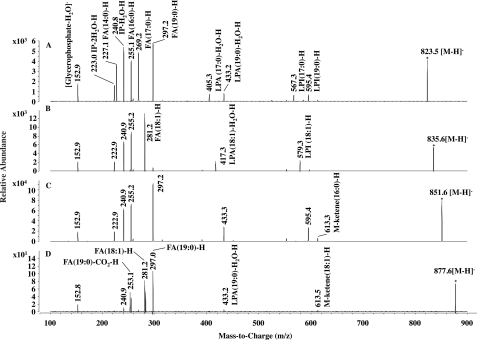
Addition of a phosphate to the inositol moiety increases the molecular weight of PI by 80 mass units, suggesting that the PI3P species at m/z 957.6 was derived from a PI species with m/z ∼878. A minor molecular species of PI representing <∼1% of the total PI pool was detected with this molecular weight (m/z 877.6 in Fig. 3A, upper panel). We examined the fatty acyl chain composition of each PI species (Fig. 4). A minor peak at m/z 823.5 represented a mixture of two PI species, containing either C17:0/C16:0 or C19:0/C14:0 (Fig. 4A). Another minor PI species at m/z 835.6 carried C18:1/C16:0 fatty acids (Fig. 4B), whereas the major molecular species of PI (m/z 851.8) contained C19:0 and C16:0 (Fig. 4C). Finally, the PI species at m/z 877.6 carried C19:0 and C18:1 acyl chains (Fig. 4D), being consistent with the fatty acid composition of PI3P. This minor PI species may represent a precursor and/or product of PI3P synthesis and turnover (see “Discussion”).
FIGURE 4.
MS/MS of M. smegmatis PI species. A–D, fragment ions from m/z = 823.5, m/z = 835.6, m/z = 851.6, m/z = 877.6, respectively. Note that PI from m/z = 823.5 is a mixture of C19:0/C14:0-PI and C17:0/C16:0-PI. The fragmentator was set at 100 V with a collision energy of 45.0 eV and 72 scans. FA, fatty acid; IP, inositol phosphate; LPA, lysophosphatidic acid; and LPI, lyso-PI.
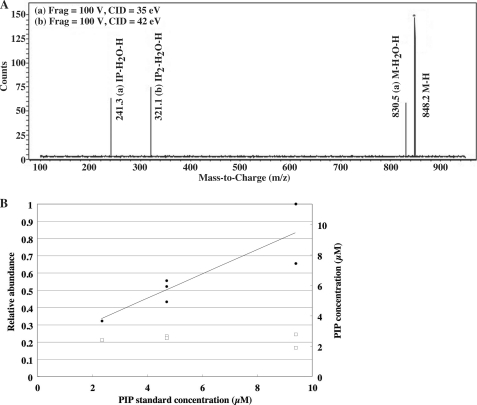
We next estimated the quantity of PI3P in mycobacterial lipid extracts by liquid chromatography-MS multiple reaction monitoring with an internal standard (Fig. 5). We used a synthetic amino-PI3P (m/z 848.2) as an internal standard and confirmed the production of the characteristic dehydrated diphosphate inositol ion at m/z 321 (Fig. 5A). We then analyzed a fixed concentration of mycobacterial PI3P spiked with increasing concentrations of amino-PI3P (Fig. 5B). Responses increased linearly with the increasing concentrations of amino-PI3P. Using this standard curve, mycobacterial PI3P (based on a fixed concentration) was estimated to be present at a concentration of 2.3 μm. This equates to ∼5 μg/g wet cell pellet under standard growth condition, corresponding to ∼8 × 104 PI3P molecules per cell or ∼1.6% of total PI in the plasma membrane.
FIGURE 5.
Endogenous PI3P concentration estimated from the relative abundance of synthetic PI3P used as an internal standard. A, the parent (Q1) → product ion (Q3) transition of m/z 848 → 321 was confirmed for the synthetic PI3P standard (C37H73O16P2N1). The fragmentator was set at 100 V with a collision energy of 35.0 or 42.0 eV. The mass spectra obtained with different collision energies were merged to form the composite mass spectrum. B, the PI3P fraction from M. smegmatis (open squares) was spiked with the synthetic PI3P standard (closed circles) at the indicated concentration. Electrospray ionization-MS/MS was used to measure transition pairs in selective multiple reaction monitoring mode. The fragmentator was set at 100 V with a collision energy of 25.0 eV and scanned 200–300 times each.
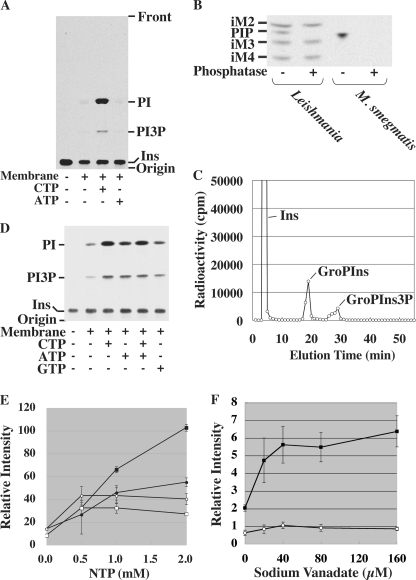
To examine the enzymatic characteristics of PI3P synthesis, we developed a cell-free system for mycobacterial PI3P biosynthesis. Incubation of the membrane fraction with [3H]PI and phosphate donors under various conditions did not produce PI3P (data not shown). Given the unusual lipid composition of mycobacterial PI3P, we examined whether endogenous PI might be a better substrate for the putative PI3P synthase. The mycobacterial membrane fraction was incubated with [3H]inositol and various nucleotides. Supplementation with CTP, but not ATP, resulted in robust synthesis of both PI and PI3P (Fig. 6A). PI3P synthesized in this cell-free system was sensitive to alkaline phosphatase (Fig. 6B). To determine if the phosphate is linked to 3-OH of inositol residue, the crude lipid extract was deacylated and subjected to HPLC analysis. As shown in Fig. 6C, we identified a peak corresponding to GroPIns3P in addition to GroPIns derived from PI. These data suggest that PI3P synthesis was reconstituted in a cell-free system. The requirement for CTP may reflect the need for this nucleotide in the synthesis of CDP-DAG, the lipid acceptor for PI synthetase, or as the donor substrate for PI3K. To distinguish between these two possibilities, we took advantage of an M. smegmatis Δino1 mutant that is auxotrophic for inositol (25) and prepared membranes after inositol starvation. Using this Δino1 cell-free system, synthesis of PI was reconstituted by the addition of [3H]inositol alone, without added NTPs (Fig. 6D), suggesting that CDP-DAG accumulated during starvation. Significantly, PI3P was also synthesized under these conditions, suggesting that the phosphate donor for PI3P synthesis was endogenously available and the requirement for exogenous CTP in the wild-type membrane system was primarily to allow synthesis of CDP-DAG. Addition of CTP, ATP, and to a lesser extent GTP, enhanced PI and PI3P synthesis by the Δino1 membranes. CTP was the most efficient substrate to produce PI, being consistent with its role in generating CDP-DAG. Addition of CTP and ATP together did not show synergistic effects on PI3P synthesis (Fig. 6D, compare lanes 3 and 5), suggesting that CTP was sufficient for PI3P synthesis as well. Adding increasing concentrations of CTP to the Δino1 membranes stimulated PI synthesis, with concomitant increase in PI3P synthesis, although the stimulatory effect on PI3P synthesis started to plateau at 1 mm (Fig. 6E). Addition of ATP also stimulated PI and PI3P synthesis, but to a lesser extent, and concentrations above 0.5 mm had no effect on the synthesis of either phospholipid (Fig. 6E). These results suggest that PI3P synthesis is more efficient when it is coupled to relatively lower rates of de novo PI synthesis. When PI synthesis rates become high, PI3P synthesis starts to plateau. Although both CTP and ATP stimulate PI and PI3P synthesis, the precise phosphate donor for the PI3P synthesis remains undefined.
FIGURE 6.
Synthesis of PI3P in a mycobacterial cell-free system. A, the membrane fraction from a wild-type strain was incubated with [3H]inositol and various nucleotides (2 mm). Ins, inositol. B, PI3P was synthesized in a cell-free system, and a crude lipid extract was treated with 11 units of alkaline phosphatase from bovine intestinal mucosa (Sigma-Aldrich) in a buffer containing 1 m CHES (pH 9.8), 1 mm MgCl2, and 0.008% Triton X-100 at 37 °C for 2 h. Lipid extracts from [3H]inositol-labeled L. mexicana was used as a control, showing that alkaline phosphatase digested PIP specifically without digesting glycoinositol phospholipids (iM2, iM3, and iM4) (22). C, PI3P was prepared by incubating wild-type membranes with [3H]inositol and CTP. Crude lipid extracts were deacylated and applied to an HPLC. Detection of GroPIns3P peak indicated the cell-free synthesis of PI3P in addition to the synthesis of PI (GroPIns peak). In some experiments, as shown here, the elution time of GroPIns was slightly delayed, but GroPIns3P consistently eluted 10–11 min after the elution of GroPIns. Ins, free unincorporated inositol carried over from the cell-free radiolabeling. D, the membrane fraction from the Δino1 mutant was incubated with [3H]inositol and various nucleotides (2 mm). E, reactions were performed as in D with various concentrations of nucleoside triphosphates (NTPs), and plotted as relative radioactivities. Filled symbols, CTP; open symbols, ATP; squares, PI; circles, PI3P. Averages of triplicates with standard deviations are shown. F, the effect of sodium orthovanadate was examined using the system as in D. No nucleotide was added. Averages of triplicates with standard deviations are shown. Open circle, PI; closed square, PI3P.
Because metabolic labeling experiments showed only transient accumulation of PI3P, it appeared reasonable to speculate that PI3P levels are controlled by the balance of a synthase/kinase and a phosphatase. We tested if kinase inhibitors can inhibit mycobacterial PI3P synthesis. Wortmannin and LY294002, two well known inhibitors of PI kinases, showed no effect on mycobacterial PI3P synthesis in the cell-free system (data not shown). Similarly, cell-free PI3P synthesis was unaffected by staurosporine, a more general kinase inhibitor, and ATPγS, a non-hydrolyzable ATP analog (data not shown). In contrast, addition of sodium orthovanadate, a potent phosphatase inhibitor, to the cell-free system resulted in increased PI3P labeling (Fig. 6F), suggesting that PI3P levels are controlled in part by a catabolic pathway.
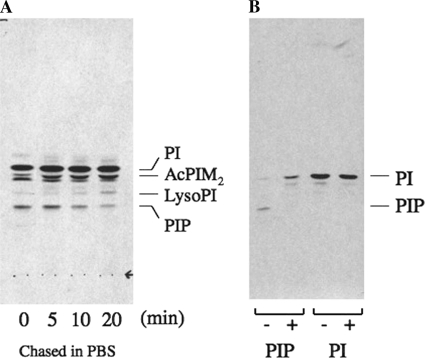
Finally, we wanted to examine if PIP synthesis is conserved in other bacteria. Corynebacteria are a group of bacteria that include medically and industrially important bacteria. They belong to the order Actinomycetales and are evolutionary related to mycobacteria. We radiolabeled C. glutamicum with [3H]inositol and analyzed extracted lipids by HPTLC. We identified a lipid species with the same HPTLC mobility as mycobacterial PI3P (Fig. 7A). It was sensitive to alkaline phosphatase treatment, and the dephosphorylated product migrated with the same Rf value as PI (Fig. 7B). These data indicate that corynebacteria produce a PIP species. When radiolabeled species were chased in phosphate-buffered saline, the PIP species decreased in quantity while the level of radiolabeled PI remained unchanged (Fig. 7A). These data together suggest that synthesis of PIP species is a conserved feature of Actinomycetales.
FIGURE 7.
Synthesis of PIP in C. glutamicum. A, transient synthesis of PIP after chase in phosphate-buffered saline. C. glutamicum was grown to a mid log phase and harvested. The cell pellet was rinsed and incubated in Sauton's minimal medium for 15 min. Radiolabeling with [3H]inositol was performed at 30 °C for 5 min and chased in phosphate-buffered saline for the time indicated. B, alkaline phosphatase treatment of C. glutamicum PIP. PIP or PI was purified from HPTLC plates and subjected to digestion with (+) or without (−) alkaline phosphatase. Note that PIP was converted to a less polar species consistent with PI.
DISCUSSION
In this study, we have demonstrated that mycobacteria transiently synthesize a single molecular species of PI3P in response to salt stress. To our knowledge, this study represents the first identification of polyphosphoinositides in prokaryotes.
Both Gram-negative and -positive bacteria are known to modulate their membrane phospholipid composition in response to salt or osmotic stress (26). The levels of anionic phospholipids such as phosphatidylglycerol or cardiolipin increase, whereas levels of zwitterionic phospholipids such as phosphatidylethanolamine decrease. These changes are thought to represent important adaptations to changes in the environment. Mycobacterial PI3P synthesis is clearly different from these gross changes of the plasma membrane lipid composition, because PI3P represents less than a few percent of the total membrane phospholipids, and the induction of PI3P synthesis by salt stress only results in a transient accumulation of PI3P. These observations seem more consistent with the possibility that mycobacterial PI3P plays a role in signaling. In fact, PI3P is known to mediate signaling in response to salt stress in mammalian and plant cells (27, 28), and such a function of PI3P may be conserved in mycobacteria. To further explore this possibility, we are currently searching for effector molecules, which may mediate the downstream signaling.
The M. smegmatis genome contains putative paralogues of eukaryotic PI3K and inositol monophosphatase located in tandem (MSMEG_4191 and MSMEG_4190, respectively). However, deleting these genes had no effect on PI3P biosynthesis (data not shown). Indeed, the M. tuberculosis homolog of MSMEG_4190 has recently been suggested to encode a 3′-phosphoadenosine-5′-phosphatase involved in sulfur metabolism (29). To gain further insights into mycobacterial PI3P metabolism, we reconstituted PI3P synthesis and catabolism in a cell-free system. Our analyses suggested that PI3P is synthesized by an unusual kinase or phosphotransferase system and is catabolized by a vanadate-sensitive phosphatase. Inhibitors of eukaryotic PI kinases, such as Wortmannin and LY294002, showed no effects on mycobacterial PI3P synthesis. Although Wortmannin-insensitive kinases involved in PIP synthesis have been described in various eukaryotic cells (30–32), these observations are also consistent with the existence of a novel phosphorylation mechanism for the synthesis of mycobacterial PI3P. Because we could not uncouple PI and PI3P synthesis in our cell-free system, it is currently difficult to pinpoint the phosphate donor for PI3P synthesis. Further studies are required to characterize the enzymatic mechanisms of PI3P synthesis, and more importantly identify the enzymes involved in PI3P metabolism.
Fatty acid composition of mycobacterial PI3P appeared to be homogeneous, carrying C19:0/C18:1 fatty acids. The 19:0 fatty acid is likely to be a 10-methylstearic acid, known as tuberculostearic acid, which is a characteristic fatty acyl moiety of mycobacterial phospholipids (33, 34). Interestingly, the fatty acid composition of PI3P was different from the major PI species, carrying C19:0/C16:0. These analyses suggest that the enzyme responsible for synthesis of PI3P only utilizes a minor pool of PI, or that PI3P synthesized from bulk PI undergoes fatty acid remodeling. It is noteworthy that PIM species, a class of glycosylated PIs, predominantly carry C19:0/C16:0 fatty acids (35, 36), apparently utilizing the bulk PI for their synthesis. Therefore, the unusual fatty acid composition of PI3P is a novel and unique aspect of mycobacterial PI metabolism. PI species with distinctive fatty acid compositions appear to be utilized for PIP synthesis in mammalian and yeast cells (37, 38). It remains unknown why and how phosphoinositides with distinct fatty acid compositions are produced, but it is interesting that some features are shared between the eukaryotic and mycobacterial PI3P metabolic pathways. Taken together, it is tempting to speculate that the functions of mycobacterial PI3P are evolutionary related to those of eukaryotic counterparts.
Acknowledgments
We thank Keiko Kinoshita (Osaka University) for assistance, Dr. Thusitha Rupasinghe (Metabolomics Australia, Bio21 Institute, Melbourne) for mass spectrometry analysis, Dr. Leon Helfenbaum (University of Melbourne) for provision of yeast inositol auxotroph strains, and Prof. Andrew Holmes (University of Melbourne, Bio21 Institute) for providing synthetic amino-PI3P.
Footnotes
- PI3P
- phosphatidylinositol 3-phosphate
- PI4P
- phosphatidylinositol 4-phosphate
- CDP-DAG
- CDP-diacylglycerol
- HPLC
- high-pressure liquid chromatography
- HPTLC
- high-performance TLC
- MS
- mass spectrometry
- MS/MS
- tandem MS
- PI
- phosphatidylinositol
- PI3K
- phosphatidylinositol 3-kinases
- PIM
- phosphatidylinositol mannoside
- GroPIns
- glycerophosphoinositol
- GroPIns3P
- glycerophosphoinositol 3-phosphate
- GroPIns4P
- glycerophosphoinositol 4-phosphate
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- PIP
- phosphatidylinositol monophosphate.
REFERENCES
- 1.Whitman M., Downes C. P., Keeler M., Keller T., Cantley L. (1988) Nature 332, 644–646 [DOI] [PubMed] [Google Scholar]
- 2.Kutateladze T. G. (2006) Biochim. Biophys. Acta 1761, 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falasca M., Maffucci T. (2006) Arch. Physiol. Biochem. 112, 274–284 [DOI] [PubMed] [Google Scholar]
- 4.Lindmo K., Stenmark H. (2006) J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 5.Engelman J. A., Luo J., Cantley L. C. (2006) Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 6.Geiger O., González-Silva N., Lüpez-Lara I. M., Sohlenkamp C. (2010) Prog Lipid Res. 49, 46–60 [DOI] [PubMed] [Google Scholar]
- 7.Saha A. K., Dowling J. N., Mukhopadhyay N. K., Glew R. H. (1989) J. Bacteriol. 171, 5103–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finnerty W. R., Makula R. A., Feeley J. C. (1979) Ann. Intern Med. 90, 631–634 [DOI] [PubMed] [Google Scholar]
- 9.Hernandez L. D., Hueffer K., Wenk M. R., Galán J. E. (2004) Science 304, 1805–1807 [DOI] [PubMed] [Google Scholar]
- 10.Basu S. S., York J. D., Raetz C. R. (1999) J. Biol. Chem. 274, 11139–11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G. F., Chater K. F., van Sinderen D. (2007) Microbiol. Mol. Biol. Rev. 71, 495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan P. J., Lehane D. P. (1971) Lipids 6, 401–409 [DOI] [PubMed] [Google Scholar]
- 13.Yano I., Furukawa Y., Kusunose M. (1969) J. Bacteriol. 98, 124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabaud H., Tisnovska H., Vilkas E. (1971) Biochimie 53, 55–61 [DOI] [PubMed] [Google Scholar]
- 15.Kataoka T., Nojima S. (1967) Biochim. Biophys. Acta 144, 681–683 [DOI] [PubMed] [Google Scholar]
- 16.Brennan P., Ballou C. E. (1968) Biochem. Biophys. Res. Commun. 30, 69–75 [DOI] [PubMed] [Google Scholar]
- 17.Brennan P. J. (2003) Tuberculosis 83, 91–97 [DOI] [PubMed] [Google Scholar]
- 18.Jackson M., Crick D. C., Brennan P. J. (2000) J. Biol. Chem. 275, 30092–30099 [DOI] [PubMed] [Google Scholar]
- 19.Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]
- 20.Dove S. K., Michell R. H. (1999) in Signal Transduction: A Practical Approach (Milligan G. ed) 2nd Ed., pp. 255–280, Oxford University Press, Oxford [Google Scholar]
- 21.Doering T. L., Schekman R. (1996) EMBO J. 15, 182–191 [PMC free article] [PubMed] [Google Scholar]
- 22.Ralton J. E., McConville M. J. (1998) J. Biol. Chem. 273, 4245–4257 [DOI] [PubMed] [Google Scholar]
- 23.Brearley C. A., Hanke D. E. (1997) in Signalling by Inositides: A Practical Approach (Shears S. ed) pp. 1–32, IRL Press at Oxford University Press, Oxford [Google Scholar]
- 24.Hsu F. F., Turk J. (2000) J. Am. Soc. Mass Spectrom 11, 986–999 [DOI] [PubMed] [Google Scholar]
- 25.Haites R. E., Morita Y. S., McConville M. J., Billman-Jacobe H. (2005) J. Biol. Chem. 280, 10981–10987 [DOI] [PubMed] [Google Scholar]
- 26.Romantsov T., Guan Z., Wood J. M. (2009) Biochim. Biophys. Acta 1788, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irarrazabal C. E., Burg M. B., Ward S. G., Ferraris J. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leshem Y., Seri L., Levine A. (2007) Plant J. 51, 185–197 [DOI] [PubMed] [Google Scholar]
- 29.Hatzios S. K., Iavarone A. T., Bertozzi C. R. (2008) Biochemistry 47, 5823–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameroni E., De Virgilio C., Deloche O. (2006) J. Biol. Chem. 281, 38139–38149 [DOI] [PubMed] [Google Scholar]
- 31.Lemke L. E., Paine-Murrieta G. D., Taylor C. W., Powis G. (1999) Cancer Chemother. Pharmacol. 44, 491–497 [DOI] [PubMed] [Google Scholar]
- 32.Lymn J. S., Gallagher K. L., Clunn G. F., Fexby S. E., Patel M. K., Hughes A. D. (2003) FEBS Lett. 555, 591–596 [DOI] [PubMed] [Google Scholar]
- 33.Lennarz W. J., Scheuerbrandt G., Bloch K. (1962) J. Biol. Chem. 237, 664–671 [PubMed] [Google Scholar]
- 34.Spielman M. A. (1934) J. Biol. Chem. 106, 87–96 [Google Scholar]
- 35.Morita Y. S., Patterson J. H., Billman-Jacobe H., McConville M. J. (2004) Biochem. J. 378, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., Jackson M. (2002) J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 37.Wenk M. R., Lucast L., Di Paolo G., Romanelli A. J., Suchy S. F., Nussbaum R. L., Cline G. W., Shulman G. I., McMurray W., De Camilli P. (2003) Nat. Biotechnol. 21, 813–817 [DOI] [PubMed] [Google Scholar]
- 38.Milne S. B., Ivanova P. T., DeCamp D., Hsueh R. C., Brown H. A. (2005) J. Lipid Res. 46, 1796–1802 [DOI] [PubMed] [Google Scholar]