Abstract
Selective androgen receptor modulators (SARMs) are androgen receptor (AR) ligands that induce anabolism while having reduced effects in reproductive tissues. In various experimental contexts SARMs fully activate, partially activate, or even antagonize the AR, but how these complex activities translate into tissue selectivity is not known. Here, we probed receptor function using >1000 synthetic AR ligands. These compounds produced a spectrum of activities in each assay ranging from 0 to 100% of maximal response. By testing different classes of compounds in ovariectomized rats, we established that ligands that transactivated a model promoter 40–80% of an agonist, recruited the coactivator GRIP-1 <15%, and stabilized the N-/C-terminal interdomain interaction <7% induced bone formation with reduced effects in the uterus and in sebaceous glands. Using these criteria, multiple SARMs were synthesized including MK-0773, a 4-aza-steroid that exhibited tissue selectivity in humans. Thus, AR activated to moderate levels due to reduced cofactor recruitment, and N-/C-terminal interactions produce a fully anabolic response, whereas more complete receptor activation is required for reproductive effects. This bimodal activation provides a molecular basis for the development of SARMs.
Keywords: Drug Action, Hormones, Nuclear Receptors, Steroid Hormone, Steroid Hormone Receptor, Transcription Regulation, Bone Formation, Sarcopenia, Virilization, Androgen
Introduction
Androgen production declines with age in both genders (1). Although secondary sexual traits such as facial hair are maintained, lean mass declines, and low testosterone levels are associated with a greater risk for several age-related diseases (2–4). An inevitable consequence of aging is the loss of bone and muscle mass, which results in part from a steady decline in anabolic and anti-catabolic hormone signaling (1). These lean mass and bone deficits confer higher risk for osteoporotic fractures, age-related sarcopenia, and loss of independence.
Genetic and pharmacological data have clearly established that androgen restoration therapies promote bone and muscle mass in post-menopausal women and aged men (5). However, androgens, as male sex hormones, can produce adverse effects, such as hirsutism, acne, deepening of the voice, and reproductive tissue changes (6). In addition, providing women with testosterone may increase the risk for breast cancer and other health problems due to its metabolic conversion to estradiol. Similarly treatment of men may also present a significant risk factor for benign prostate hyperplasia and prostate cancer (7).
Given these concerns, tissue-selective androgen receptor modulators (SARMs)13 that restore healthy bone and muscle but do not cause uterine, prostate, or sebaceous gland hypertrophy would be attractive therapies (8, 9). The fundamental idea of selective modulation of nuclear receptors is validated by the development of selective estrogen receptor modulators as ligands for the two estrogen receptors (ERα and ERβ). Raloxifene, for example, is a clinically used selective estrogen receptor modulator that slows bone loss and antagonizes estrogen actions in uterus and breast and reduces the risk of the development of estrogen receptor-dependent breast cancers (10, 11). Preclinical proof of concept for an anabolic SARM with reduced androgenization was established in animal models by several SARMs such as TZP-4238, S-40503, S-1, S-4, LGD2226, LGD2941, JNJ-28330835, and JNJ-37654032 (12–19). We developed a proof-of-concept SARM, TFM-4AS-1, that increases periosteal bone formation, cortical bone mass, and lean body mass as effectively as DHT but with markedly reduced effects on uterus, prostate, and the pilosebaceous unit (PSU) (20). However, as each SARM has a unique in vivo profile that is rarely linked to a specific in vitro activity profile, the relationship between transcriptional activity and physiological effect has not been established.
Androgens act by binding to the androgen receptor (AR), a member of the nuclear receptor superfamily. Upon ligand binding, AR interacts with DNA sequences or other proteins in the regulatory regions of target genes to recruit transcriptional cofactors and regulate gene expression. Although DHT maximally activates the full repertoire of androgen-responsive genes, synthetic AR ligands range from full agonists to complete antagonists depending on the gene and cellular context (21–23). These ligands exert complex activities presumably by differentially modulating the structure of AR co-factor recruitment sites such as the N-terminal-located activation function-1 (AF-1) and the C-terminal-located AF-2, which interact with transcriptional coregulators such as TIF2/SRC-2/GRIP-1/NCO-2 (24). AR is maximally active when the ligand promotes an interaction between the N- and C-terminal AR domains (the N/C interaction). Interestingly, mutations in the N- or C-terminal domain of AR that impair this interaction were found in incompletely virilized patients with partial androgen insensitivity; some of these mutations also inhibit the recruitment of transcriptional cofactors to AF-2 (25–27). These genetic data suggest that synthetic ligands that promote the transactivation activity of AR while exhibiting reduced ability to support the N/C interaction or cofactor recruitment could provide tissue selectivity in vivo.
Drug development requires the iterative sorting of thousands of compounds through in vitro assays followed by selection of a very small subset for in vivo validation. Thus, medicinal chemistry requires assays that efficiently identify compounds with the highest possible target selectivity and affinity that correspond to desired outcomes in animal models. The process of identifying clinically useful SARMs faces significant challenges as the search is not for the most potent receptor agonist or antagonist but rather for compounds that precisely manipulate transcriptional activities of the receptor producing tissue-selective actions in animals and humans. Currently, there has been no systematic information that defines relationships between the in vitro characteristics of androgens and their effects on tissues.
To establish a rational drug design paradigm for the discovery of novel SARMs, we employed biochemical assays that measure binding to endogenously expressed human AR (AR radioligand competition binding assays (ARBIND)), transactivation of a model promoter via endogenous human AR (TAMAR), AR AF-2 domain recruitment of the coactivator GRIP-1 (TRAF2), and the AR N/C interaction (VIRCON). In addition, we developed a set of gene expression surrogate biomarkers that are predictive for the long term effects of anabolic androgens on bone and in PSUs. This capability enabled testing of a large number of compounds with various in vitro profiles in rats for their effects on cortical bone formation, lean body mass, uterus growth, PSU hypertrophy, and seminal vesicle growth. We established a correlation between the in vitro actions of androgens and these physiological endpoints and, thus, provided a systematic and predictive approach to the development of SARMs. This led to the identification of the SARMs MK-0773 and 2-FPA. MK-0773 was suitable for therapeutic development, and Phase 1b clinical data have provided clinical support for this paradigm for SARM development.
MATERIALS AND METHODS
Reagents and Animals
All reagents were from Sigma unless noted. Rats were Sprague-Dawley from Taconic Farms (Tarrytown, NY) and were individually housed with ad libitum access to food and water. All procedures were in accordance with Institutional Care and Use Committee guidance. The compounds were synthesized in-house. Cell lines MDA-MB-453, COS-1, and CV-1 were from the American Type Culture Collection (ATCC), Manassas, VA.
Binding and Transcription Assays
Binding and transactivation assays used the human breast carcinoma cell line MDA-MB-453, which expresses endogenous AR (45, 46). ARBIND assays with 0.5 nm [3H]methyltrienolone (R1881, a potent AR agonist) were as previously described (28). TAMAR assays (transactivation modulation of AR) in 96-well plates used transient transfection of a modified mouse mammary tumor virus (MMTV) long terminal repeat promoter upstream of luciferase (MMTV-LUC). This MMTV has two direct repeat copies of a consensus glucocorticoid receptor response element between positions −88 and −190; these sequences are also recognized by AR (46, 47). The VIRCON (virilization counterscreen) assay, which measures the AR N-terminal domain/ligand binding domain (LBD) interaction (N/C) of rhesus AR (rhAR), was evaluated by a mammalian two-hybrid assay in CV1 cells as detailed in Schmidt et al. (20). The N/C interaction was evaluated as a ligand-mediated increase of luciferase reporter activity.
The TRAF2 (transcriptional recruitment to AF2) assay, which measures the activation mediated by the AF-2 domain and potentiated by the full-length rat GRIP-1 coactivator, was measured by a mammalian two-hybrid assay in similar manner to the VIRCON assay. The Gal4-DNA binding domain was fused with the LBD of rhAR (pm-rhAR-LBD, amino acids 637–895), and the VP16AD was fused to the rat GRIP-1 (pcDNA3-GRIP-1 plasmid). The luciferase reporter plasmid with 5 copies of GAL-4 DBD binding sites (Gal4 × 5X90-luciferase) was transfected into COS-1 cells with the pm-rhAR-LBD and pcDNA3-GRIP-1 plasmids, and the AF-2 function was evaluated as a ligand-dependent dose response.
Rodent Studies
Animal procedures were performed as described (48). Female rats were ovariectomized (OVX) at 12 weeks, and test compound administration began at least 8 weeks post-surgery for 24 days (20). Animals were randomized into groups (n = 10–16), and compounds dissolved in propylene glycol were administered by subcutaneous injection. At necropsy, uteri were dissected at the boundary of the cervix and weighed wet. Dorsal skin samples were taken for RNA analysis and processed for histomorphometry. Measurements of sebaceous gland size were assessed by using a Nikon Eclipse E1000M microscope/digital camera system and BioQuant software. Tibiae were collected for RNA processing and histological studies (20). Data were analyzed by one way analysis of variance, Fisher's protected least significant difference (Statview, Version 5.0). The effects of compounds on cortical bone formation, muscle, lean body mass (LBM), uterus, and PSUs are shown as % activity relative to DHT (3 mg/kg), which is set to 100%. Criteria for compound entry into animal studies included 1) AR binding affinities <50 nm, 2) a clear dose-response activity in TAMAR assay with an inflection point (IP) value <250 nm, and 3) a maximal transcriptional activity that is stable over 3 treatment doses. Typically, compounds were studied at three doses, and data were expressed as a percent of positive control (either DHT or the SARM TFM-4AS-1 depending on the endpoint) to allow direct comparison across experiments. For correlations between in vitro and in vivo measurements, the greatest measured response as a percentage of positive control was plotted against the compound's maximal in vitro responses.
Bone Histology
Anabolic androgens act on the periosteum, a site at which newly deposited bone tissue confers maximal increase in mechanical strength per unit of new bone tissue. Double calcein labeling was used to identify regions of periosteum undergoing mineralization in aged, long term post-OVX rats that were also treated with alendronate (5.6 μg/week) to reduce bone turnover. Bone formation rate (BFR), the most reliable osteoanabolic endpoint, is calculated by multiplying the extent of periosteal bone surface with double label by the mineral apposition rate (distance between labels/time between labels) (20).
Myoanabolism
Pretreatment base-line and 24-day post-treatment assessment of body composition was assessed by quantitative NMR (EchoMRI-500 NMR). From these measurements, the change in lean body mass (ΔLBM) was then calculated, corrected for vehicle changes, and expressed as %DHT value. Data were analyzed by one way analysis of variance, Fisher's PLSD (Statview, Version 5.0).
Male Reproductive Tract
Prostate and seminal vesicles were studied in 3–4-month-old 250–300-g rats after orchidectomy (ORX). Nine days after surgery, animals were injected (subcutaneously) daily with test compounds for 17 days, and the weight of the seminal vesicles (SVs) was compared with ORX rats treated with vehicle or DHT as a positive control.
RNA Measurements
To accelerate the bone and PSU evaluation, we developed gene-based biomarkers assays for measuring BFR and sebaceous gland hypertrophy. A genomic microarray analysis identified genes that were expressed proportionately by an increase in bone formation rate in the section of rat tibial diaphysis (bone qPCR); type II collagen (COL2A1) was induced by the increase in BFR, whereas stearoyl-CoA desaturase-1 (SCD1) was repressed in a manner inversely correlated with BFR. The rat DNA sequences used for the assays are: SCD-1 (forward, 5′-CCGTGGCTTTTTCTTCTCTCA-3′; reverse, 5′-GCTTTCCGCCCTTCTCTTTG-3′; probe, 6-FAM-5′-TGGGCTGCTTGTGCGCAAACACC-3′); collagen type II A1 (forward, 5′-AGTCGCTGGTGCTGCTGAC-3′; reverse, 5′-CCCTAATTTTCGGGCATCCT-3′; probe, 6-FAM-5′-TGCTCATCGCCACGGTCCTACAATG-3′). The endogenous control is 18 S RNA, Applied Biosystem #4308329.
Using a candidate gene approach complemented by microarrays, two genes, Scd3 (stearoyl-CoA desaturase 3), a marker of the sebaceous gland, and squalene epoxidase, which encodes an enzyme essential for producing cholesterol esters and squalene waxes present in sebum, were expressed commensurate with sebaceous gland hypertrophy (skin qPCR). After comparing the results of skin qPCR with histology over several experiments, it was determined that the gene-based readout provided sufficiently reliable information to replace histological analysis. The rat DNA sequences used for the assays are: squalene epoxidase (forward, 5′-CCCACTTCGTTGGCTTCATT-3′; reverse, 3′-ATGAGAACTGGACTGGGATCGA-3′; probe, 6FAM-5′-ACCACAGTTTAAAGCCAATTTCGCGGA-3′); SCD-3 (forward, 5′-TCTTTGACAGCCGGGTGTTT-3′; reverse, 5′-CGCCGTGGCTTTTTCTTCT-3′; probe, 6FAM-5′-CGCACAAGCAGCCAACCCACG-3′) normalization gene cyclophilin (forward, 5′-CAAATGCTGGACCAAACACAA-3′; reverse, 5′-GCCATCCAGCCACTCAGTCT-3′; probe VIC-5′-TGGTTCCCAGTTTTTTATCTGCACTGCCT-3′). After completion of dosing, tissues were collected, and RNA expression levels were measured by quantitative RT-PCR of total RNA. The RNA was collected and analyzed as previously described (49). Activity was calculated as % relative to DHT (3 mg/kg), which was set to 100%, except in bone, where the effects of 10 mg/kg 4AS-TFM-1 were set to 100%.
RESULTS
AR Ligands Exhibit Diverse Levels of Agonism in Three Transcription Assays
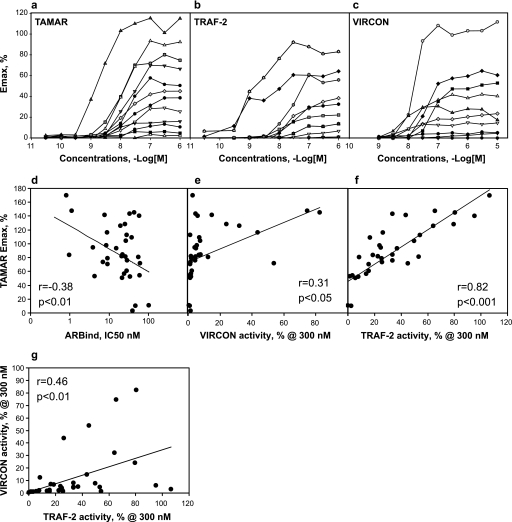
To identify novel SARMs for clinical development, we used the 4-aza-steroidal structure of 4AS-TFM-1 as a starting point for lead optimization. In addition, we also identified new leads in a single dose, high throughput screen of ∼1,000,000 compounds (data not shown). After lead identification, we executed several rounds of iterative medicinal chemistry to generate diverse libraries around each lead. Chemistry design targeted the generation of compounds that had high affinity binding to AR natively expressed in human MDA-MB-453 breast cancer cells (20, 28). Compounds that exhibited apparent binding affinity <300 nm with little or no displacement of radioligands from related steroid hormone receptors (estrogen receptor α, progesterone receptor, or glucocorticoid receptor at 2 μm) were tested in the TAMAR assay for their ability to stimulate transcription from the transiently transfected modified MMTV reporter gene (20, 28). This assay was chosen because in pilot experiments, TAMAR activities accurately represented the levels of agonism or antagonism of known AR ligands that matched their activities in animal studies or the clinic. In other systems this correlation was not always apparent (20). Interestingly, structurally diverse AR ligands exhibited maximal transactivation levels (Emax) ranging in a continuum from antagonist (0%) to full agonist (100%) relative to fully active doses of the AR agonists R1881 or DHT (representative data are shown in Fig. 1a). Partial agonism was not a function of poor potency, as compounds sustained their maximal activity over a wide range of concentrations, indicating a truly maximal response. To exclude the possibility that partial activity was due to poor cell permeability, we mixed partial agonists with R1881 and observed that these compounds dose-dependently inhibited R1881-mediated transactivation (20). Likewise, the compounds were subjected to the TRAF2 assay for measuring the ability of ligands to stimulate the recruitment of the transcriptional cofactor GRIP-1 to the AF-2 domain of AR using a mammalian two-hybrid assay in CV1 cells. Again, AR ligands exhibited a spectrum of activities ranging between agonism and antagonism (representative data are shown in Fig. 1b). In this case there was no synthetic bias toward low activity. For both VIRCON and TRAF2 assays, the Emax values were typically stable over ≥2 log units before exhibiting very high activity at a single concentration generally >3 μm. Thus, we excluded these apparently artifactual values and limited analysis of these data to values generated at 300 nm.
FIGURE 1.
AR-mediated transcription assays identify ligands with diverse maximal agonistic activities. Examples of dose response treatments of AR ligands with diverse properties on three types of AR-controlled transcription assays (a–c) are shown. Each dose data point is an average of four treatments. a, shown is transactivation of MMTV-luciferase reporter via endogenous human AR in MDA-MB-453 cells (TAMAR). b, shown is transactivation mediated by the AF-2 domain of AR and potentiated by the coactivator GRIP-1 in COS-1 cells (TRAF-2). c, shown is the ligand-induced interaction between the AR C terminus with the N terminus using mammalian two-hybrid assay in CV-1 cells (VIRCON). d–g, correlations between the actions of various AR-ligands in vitro assays are shown. d, shown is the correlation between the Emax values of compounds in TAMAR assay (MMTV transactivation) and the AR binding affinities of various AR ligands (ARBIND). Ligand displacement assays with native AR is endogenously expressed in MDA-MB-453 cells as described (28). e, shown is the correlation between Emax values for compounds in TAMAR assay and the activities in VIRCON assay at 300 nm. VIRCON measures the AR N-C interactions. f, shown is the correlation between Emax values for compounds in TAMAR assay and the activities in TRAF-2 assay at 300 mm. TRAF-2 measures the transactivation mediated by AF-2 domain of AR potentiated by the coactivator (GRIP-1). g, correlation between Emax values for compounds in VIRCON and TRAF2 assays is shown. The regression and p value for 38 compounds are indicated in each panel. The regression and p value for the compounds are indicated in each panel.
The ligands were next tested in a VIRCON assay for their ability to promote the N/C interaction using a mammalian two-hybrid assay in CV1 cells. Similar to the transactivation assay, AR ligands exhibited continuous levels of activities that ranged between that of an antagonist (0%) to that of an agonist (100%), (representative data are shown in Fig. 1c). Because we hypothesized that the N/C-terminal interaction is required for virilization, chemical modifications that led to reduced VIRCON activity were selected for further iteration, leading to a bias in the library toward low activity in this assay. In summary we generated and characterized a collection of selective AR ligands with varying activities across three functional measures of receptor activation.
Requirements for Inducing the N/C Interaction Differ from Transactivation and Coactivator Recruitment
We then asked if AR ligands acted similarly in all three measures of receptor activation or, rather, if each assay reflected distinct biochemical or structural effects. First, we asked if compounds with higher binding potencies were more likely to be full agonists by using the 38 compounds that were characterized fully in animal models (see below). Plotting binding potency (the IC50 in ARBIND) versus transactivation activity (Emax in TAMAR) revealed a low but significant negative correlation (r = −0.38, p < 0.01, Pearson), indicating that the most potent compounds were more likely to activate the MMTV promoter as effectively as DHT or R1881 (Fig. 1d). However, this analysis also demonstrated that binding potency alone does not explain transactivation activity. Next we compared the maximal activity in N/C interaction (VIRCON activities at 300 nm) to transactivation (Emax in TAMAR). Although a positive correlation was observed (r = 0.31, p < 0.05, Pearson), this relationship was due largely to two full agonists with high VIRCON and TAMAR values; removing them from the analysis eliminated a significant relationship (Fig. 1e). In fact, ligands with <20% maximal activity in VIRCON could be either full agonists, antagonists, or partially active in TAMAR (Fig. 1e). Thus, the N/C interaction assay provided data that was not redundant with MMTV transactivation, suggesting that these two transcriptional events require distinct aspects of receptor function. In contrast, a comparison of the cofactor recruitment assay (TRAF2 activities at 300 nm) to MMTV transactivation revealed a highly significant positive correlation (r = 0.82, p < 0.001) (Fig. 1f). Thus, these two assays provided largely redundant information and implicate coactivator recruitment as a major determinant of AR ability to activate the MMTV promoter. Finally we compared TRAF2 to VIRCON activities at 300 nm and again found these values to be significantly correlated (r = 0.46, p < 0.01) (Fig. 1g). This correlation appears to be due to several compounds not being highly active in either experiment; perusal of ligands with high TRAF2 values showed that VIRCON values could range from <5% to >50%. Thus, transactivation and cofactor recruitment experiments provided similar but not identical insights into the nature of the ligand-receptor interaction, whereas the N/C interaction assay appeared to measure different aspects of receptor function.
Gene Regulation Endpoints as Biomarkers for the Actions of Androgen on Bone Formation and Sebaceous Gland Hypertrophy
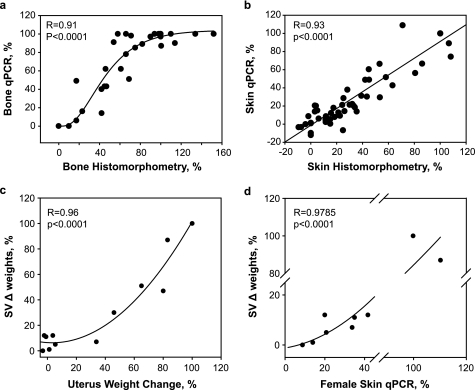
The above data allowed novel AR ligands to be binned into distinct categories depending on activity in these three assays. We then sought to determine the physiological relevance of displaying distinct in vitro properties. However, to study the full effects on androgens on sebaceous glands and cortical bone in rats required 24 days of dosing followed by 4–6 weeks of fixation, section preparation, and histomorphometric analyses. To expedite this evaluation we conducted genome-wide microarray analyses of gene expression in rats treated for 24 days with DHT, cyproterone acetate, bicalutamide, and selected SARMs such as TFM-4AS-1 and Cl-4AS-1 (20) in bone and skin. Gene expression changes were detected by competitive hybridization experiments between vehicle and treated RNA specimens to 25,000-feature microarrays. Transcript changes across several AR ligand treatments were tested for significant correlation with histomorphometric data from contralateral samples. Transcript changes that were highly positively or negatively correlated with the histological endpoints were selected for replication using qPCR. Using this strategy we found that both 3 mg/kg DHT and 10 mg/kg fully anabolic SARM TFM-4AS-1 induced type II collagen (Col2a1) cortical bone RNA and repressed stearoyl-CoA desaturase-1 (Scd1) RNA. Normalizing these two changes in gene expression relative to DHT activity as 100% and averaging them provided a rapid gene-based surrogate marker (bone qPCR) for bone histomorphometry. Plotting Bone qPCR values against BFR revealed a significant sigmoid correlation (r = 0.91, p < 0.0001 n = 36), in which bone qPCR values ≥100% of 3 or 10 mg/kg TFM-4AS-1 consistently indicated an anabolic response that is equivalent to that in intact rats.
We then developed gene-based biomarkers that correlated with the actions of androgens on sebaceous gland hypertrophy. Two genes, Scd3 (stearoyl-CoA desaturase 3), a marker of the sebaceous gland that is expressed higher in males than in females (29), and squalene epoxidase, which encodes an enzyme essential for producing cholesterol esters and squalene waxes present in sebum (30), were expressed commensurate with sebaceous gland hypertrophy (31). Measuring Scd3 and squalene epoxidase RNA relative to the effect of 3 mg/kg DHT (skin qPCR assay) were linearly correlated with the histological analyses of sebaceous gland in adjacent skin specimens (r = 0.93, p < 0.0001, n = 54, Fig. 2b). Therefore, the gene-based readouts provide sufficiently reliable information to replace histological analysis for the purposes of building structure-activity relationships.
FIGURE 2.
Correlations between androgen actions on tissues measured via the use of biomarkers and histological analysis in eugonadal female and male rats. The histological studies and the RNA levels were measured in the skin and bones of the same OVX rats (a and b). a, the % increase in bone formation rate in the femur of OVX rats determined by histomorphometric analysis is plotted against the average regulation of the RNA for SCD1 and Col2A1 (Bone qPCR). b, the % increased in sebaceous gland area in skin of OVX rats is plotted against the average induction of SCD3 and SLQE in skin RNA (Skin qPCR). The actions of androgens in OVX rats predict their actions on male reproductive tissues in ORX rats (c and d). c, effects of SARMs on changes in uterus weights in OVX rats are plotted against the changes in SV weights of ORX rats. d, effects of SARMS on changes in skin of OVX rats are plotted against the changes in SVs in ORX rats. Skin changes were measured by the skin qPCR biomarker method. The values changed induced by 3 mg/kg/day DHT treatment compared with the values obtained in vehicle treated rats was set to 100%. The regression and p values for the compounds are indicated in each panel.
Correlation between the Action of SARMs in OVX Rats and ORX Rats
We next considered whether the SARMs identified in OVX females would display reduced effects in the male sexual organs, a hallmark of SARMs. Thus, we measured the effects of selected ligands on seminal vesicle weight in orchidectomized rats that had been dosed for 17 days with varying amounts of compound. Plotting % change in uterine weight against % change in SV weight (each relative to 3 mg/kg DHT) revealed a significant nonlinear regression line (r = 0.963, p < 0.0001, Chapman, n = 14) (Fig. 2c). Similarly, plotting percent of changes in Skin qPCR to the changes in SV weights also revealed a significant nonlinear regression line (r = 0.979, p < 0.0001 n = 9) (Fig. 2d). Similar relationships with prostate weight were observed (data not shown). Thus, the OVX female rat data were predictive of reduced stimulatory activity in castrated male rats.
Developing a Rational Drug Development Paradigm
Having developed multiple rapid measures of androgenic activity in aged OVX rats, we next determined which transcriptional properties predicted tissue selectivity. Using a single animal model enabled simultaneous direct comparisons of the actions of SARMs in different tissues in animals experiencing uniform compound exposures. Furthermore, females are naïve to high concentrations of androgens and, thus, eliminate the concern that time post-castration would alter the sensitivity of the animals and complicate the data.
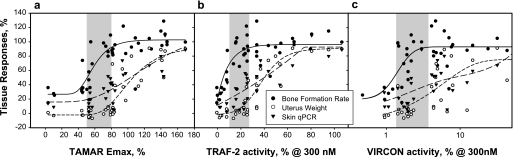
Selected AR ligands were tested in OVX rats at multiple doses for 24 days. At the end of dosing, plasma compound concentrations were measured over 24 h to ensure that sufficient ligand concentrations were present to produce a maximal response (i.e. the predicted free fraction of the drug exceeded the concentration theoretically required to saturate the AR). Thirty-eight compounds were selected for analyzing the relationship between the in vitro and in vivo activities. Plotting the maximal activities of these compounds in MMTV transactivation (TAMAR Emax) against bone anabolic activity resulted in a significant sigmoidal correlation between the two measurements (r = 0.88, p < 0.0001, Chapman) (Fig. 3a, solid line). Fully anabolic ligands possessed average TAMAR Emax values of 103.8 versus 45.8% for less than fully anabolic compounds, a highly significant difference (p < 0.001, Student's t test, defining full anabolism as bone formation values >70% of 3 mg/kg DHT). Similar comparisons against uterus weight produced a highly correlated sigmoid curve fit (r = 0.88, p < 0.0001, Chapman) (Fig. 3a, short-dash line). However, uterine stimulation required higher transactivation activity as only compounds with a TAMAR Emax value >75–80% produced significant increases in uterus weight. Comparing sebaceous gland enlargement by these compounds versus their TAMAR Emax produced a significant sigmoidal curve (r = 0.82, p < 0.0001, Chapman) (Fig. 3a, long-dash line). Similar to uterus weight, significant increases in sebaceous gland area were produced by compounds with Emax values >80%. Intriguingly, 72% of the compounds with Emax values of ∼50–80% produced full osteoanabolism, and among these, 57% produced lower increases in uterus weights or sebaceous gland area than the levels produced by DHT. Therefore, we concluded that compounds with TAMAR Emax values in the range of ∼50–80% (shaded area in Fig. 3a) were more likely to be tissue-selective. These data support the concept that partial agonism is a reliable, if imperfect, predictor of tissue selectivity, consistent with previous SARM discovery strategies (12–19, 32).
FIGURE 3.
Correlations between the actions of androgens on tissues and their activities in transcription assays. The Emax values of compounds in each of the three transcription assays, TAMAR (a), TRAF2 (b), and VIRCON (c), are plotted against each their maximal effects on the increases in BFR measured by histomorphometry (solid circles and solid regression line), uterus weight (open circles and long-dash regression line), and sebaceous gland area measured by skin qPCR (solid triangles and short-dash regression line). The effects on BFR, uterus, and skin in OVX rats are shown as % activity relative to DHT (3 mg/kg), which is set to 100%. The regression values are: TAMAR versus BFR r = 0.82, uterus weight r = 0.89, skin qPCR r = 0.88; TRAF-2 versus BFR r = 0.83, uterus weight r = 0.83, skin qPCR r = 0.85; VIRCON versus BFR r = 0.68, uterus weight r = 0.75, skin qPCR r = 0.70. The p values for all are <0.0005.
Next we sought to determine whether GRIP-1 recruitment to AF-2 predicted tissue selectivity. Plotting TRAF2 values at 300 nm against their bone anabolic activity produced an exponential correlation between the two measurements (r = 0.83, p < 0.0001) (Fig. 3b, solid line). Bone formation was stimulated maximally by compounds with TRAF2 activities >10%. Similar plotting of the TRAF2 data against uterus weights resulted in a highly correlated sigmoidal curve fit (r = 0.91, p < 0.0001, Chapman) (Fig. 3b, short-dash line). Compounds with TRAF2 activities of 25% or higher tended to increase uterus weight, whereas compounds with activities higher than ∼60% were fully uterotrophic. Plotting TRAF2 activities at concentrations of 300 nm against their actions on sebaceous gland areas revealed a linear curve fit (r = 0.83, p < 0.001; Fig. 3b, long-dash line) in which significant virilization occurred at TRAF2 values higher than ∼30%. Thus, compounds with TRAF2 values in the range of 10–30% (shaded areas in Fig.) were likely to be fully anabolic SARMs.
Finally we assessed the predictive value of monitoring the N/C interaction. In this assay the transition from inactive to fully active compound occurs within the range of 0–20% activity. Therefore, to increase resolution, we plotted the tissue activities of compounds against the log of the VIRCON activities at 300 nm. Comparing log10 VIRCON values against bone formation revealed a sigmoid relationship (r = 0.68, p < 0.0001, Chapman). Compounds with activities above ∼2% were fully anabolic (Fig. 3c, solid line). In contrast, the effects on sebaceous glands or uterus revealed flatter but highly correlated curves against uterus weight (r = 0.70, p < 0.0005; Fig. 3c, short-dashed line) and sebaceous gland area (r = 0.75, p < 0.0001; Fig. 3c, long-dashed line). Compounds with VIRCON Emax values below ∼7% produced limited effects on both sebaceous glands and uterus (Fig. 3c, shaded area). Thus, androgens with TAMAR Emax of 50–80%, TRAF2 Emax of 10–30%, and VIRCON Emax values <7% tended to display tissue selectivity.
The SARM Profile of 2-FPA
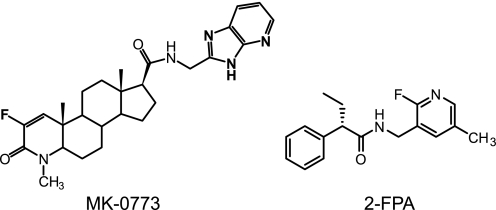
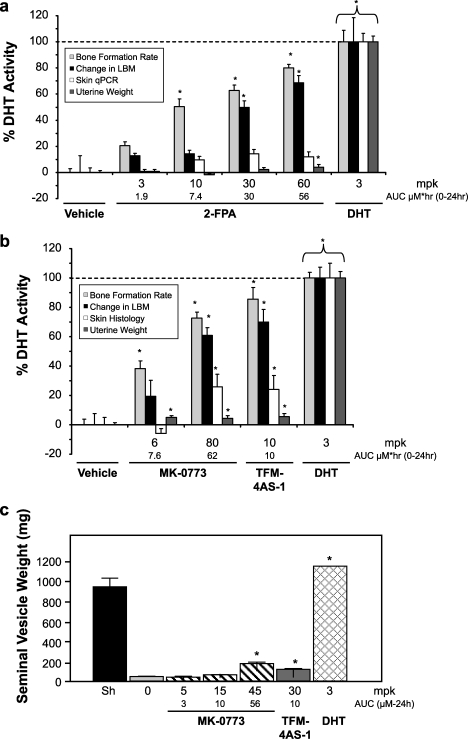
Having established predictors of tissue selectivity in both male and female rats, synthesis was directed to generate compounds with the optimal profile. One such compound is 2-FPA (Fig. 4), a non-steroidal ligand that selectively binds to AR (ARBIND IC50 = 31 nm with IC50 > 5 μm against other steroid hormone receptors and 5α-reductase) and is a partial agonist on the MMTV promoter (TAMAR IP = 293 nm; Emax = 72%) with low activity both in coactivator recruitment (TRAF2 Emax < 5%) and N/C interaction (VIRCON Emax 4%) (Table 1). 2-FPA was dosed daily to OVX rats (3–60 mg/kg) for 24 days and achieved dose-proportional increases in plasma exposures over 24 h of 1.9, 7.4, 30, and 56 μm·h (integrated area under a 24-h pharmacokinetic curve) that produced dose-related increases in cortical BFR (Fig. 5a). A parallel increase in LBM was observed by quantitative NMR. The anabolic effects of 2-FPA reached ∼80% of that produced by 3 mg/kg DHT, but unlike DHT, it did not increase uterus weight and had reduced effects on sebaceous glands measured by Scd3 and squalene epoxidase RNA induction and by histomorphometry (Fig. 5a). Thus, 2-FPA is a representative novel non-steroidal SARM identified by this paradigm.
FIGURE 4.
Chemical structures of MK-0773 and 2-FPA. Shown are chemical structures of MK-0773 ((4aS,4bS,6aS,7S,9aS,9bS,11aR)-3-fluoro-N-(3H-imidazo[4,5-b]pyridin-2-ylmethyl)-1,4a,6a-trimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide) and 2-FPA ((2S)-N-[(2-fluoro-5-methylpyridin-3-yl)methyl]-2-phenylbutanamide).
TABLE 1.
In vitro profiles of 2-FPA and MK-0773
| Compound | ARBIND IC50 | TAMAR |
TRAF2 |
VIRCON |
|||||
|---|---|---|---|---|---|---|---|---|---|
| IP | Emax | IP nm | 0.3 m | Emax | IP | 0.3 m | Emax | ||
| nm | nm | %a | nm | %a | %a | nm | %a | %a | |
| MK-0773 | 6.6 | 25 | 78 | 93 | 29 | 37 | 1,185 | 1.9 | 7.5 |
| 2-FPA | 31 | 293 | 72 | 3,408 | <5 | 25 | 41,940 | 4 | 36 |
| TFM-4AS-1 | 31 | 18 | 54 | 931 | 16 | 33 | 13,050 | 1.9 | 6.6 |
| R1881 | 0.79 | 0.33 | 127 | 1.1 | 88 | 99 | 22 | 120 | 120 |
| DHT | 1.1 | 4.7 | 148 | 1.8 | 66 | 99 | 200 | 73 | 116 |
a Values represents % agonism relative to the activity of 100 nm R1881.
FIGURE 5.
Tissue-selective effects of MK-0773 and 2-FPA in eugonadal rats. The effects of 2-FPA (a) and MK-0773 (b) and were measured in OVX rats that were treated at the given doses once daily for 24 days. Compound levels in plasma were measured in 3 separate animals at 0.25, 1, 2, 4, 8, and 24 h, and the area under the curve (AUC) was determined to allow comparison of 24 h exposures. Shown are the mean values for the various readouts of 10–12 animals + S.E. *, significantly different from ovariectomy + vehicle alone (p < 0.05, one way analysis of variance, Fisher's PLSD). All the data are expressed as percent of the activity obtained by treatment by 3 mg/kg/day DHT. Periosteal bone formation rate was measured by quantitative histomorphometry (percent double-labeled surface/total bone surface multiplied by mineral apposition rate). Uterine wet weight was used as a measure of androgen effects on reproductive organs. The change in LBM was measured by scanning the rats with quantitative NMR before and at the end of the treatments. The values for LBM are expressed as mean change from baseline compared with DHT. The effects on sebaceous gland area by MK-0773 were determined by quantitative histomorphometry of dorsal back skin. The effects on skin for 2-FPA were determined by skin qPCR assay. c, MK-0773 had a limited effect on seminal vesicles growth in ORX rats. ORX rats were treated daily with either vehicle, MK-0773, TFM-4AS-1, or DHT as indicated. For a comparison for normal androgen tone, a group of Sham (Sh) operated rats was treated with vehicle. The wet weights of the seminal vesicles were measured after treatments for 14 days. Compound levels in plasma were measured in 3 separate animals at 0.25, 1, 2, 4, 8, and 24 h, and the area under the curve was determined to allow comparison of 24 h exposures. Shown are the mean values of 10–16 animals ±S.E., *, significantly different from plus vehicle alone (p < 0.05, Kruskal-Wallis). mpk = mg/kg.
The SARM Profile of MK-0773
MK-0773 is a 4-aza-steroid that binds to AR with an IC50 = 6.6 nm (Fig. 4 and Table 1) and is a partial agonist in TAMAR (IP = 25 nm Emax = 78%) with TRAF2 Emax = 29% and VIRCON Emax = 2%. MK-0773 has favorable absorption, distribution, metabolism, and excretion properties across species, synthetic and formulation properties amenable to large scale clinical trials, minimal activity toward other receptors and enzymes, and low risk for drug-drug interactions and, thus, was selected for clinical development (Table 1) (31). The IC50 of MK-0773 binding to AR was increased 3.5-fold in the presence of 25% rat serum and 13-fold in the presence of 25% human serum, indicating that it binds to serum proteins. The affinity of MK-0773 for AR across species was evaluated using COS cells transfected with AR, and IC50 values were very similar in four species (rat, 0.50 nm; dog, 0.55 nm; rhesus, 0.45 nm; human, 0.65 nm). MK-0773 is selective for AR; no binding was detected to other nuclear receptors including glucocorticoid receptor and progesterone receptor (IC50 > 2 μm) nor was there any 5α-reductase inhibitor activity (IC50 > 10,000 nm). Finally, it possessed very low affinity to the MK-499 binding site on the human Ether-à-go-go-related gene cardiac potassium channel responsible for QT prolongation effects of several drugs (Ki = 49500 nm).
MK-0773 was dosed subcutaneously for 24 days in the OVX rat model (6 and 80 mg/kg) and produced plasma exposures over 24 h of 6.6 and 62 μm·h (Fig. 5b). This treatment produced exposure-related stimulatory effects on cortical BFR and LBM. The maximal anabolic effects of MK-0773 were equivalent to the SARM TFM-4AS-1 and ∼80% of 3 mg/kg DHT. Both MK-0773 and TFM-4AS-1 had <5% of the effect on uterus weight, and about 30–50% of the increase of sebaceous gland area produced by 3 mg/kg DHT. Next, ORX male rats were treated with daily subcutaneous dosing with MK-0773 (5, 15, and 80 mg/kg) for 17 days. The treatments produced plasma exposures over 24 h of 3, 10, and 56 μm·h. At the highest exposure tested, MK-0773 increased seminal vesicle weights by 12% that achieved by 3 mg/kg DHT (Fig. 5c and Table 2) and had similarly reduced effects on the prostate (data not shown). Taken together, MK-0773 exhibits the profile of an anabolic SARM with limited effects on the sebaceous glands and reproductive tracts of OVX and ORX rats.
TABLE 2.
Effects of MK-0773 on seminal vesicle weight in ORX rats
AUC, area under the curve.
| Compound | Dose | AUC | Seminal vesicle weighta |
|---|---|---|---|
| mg/kg | μm·h | % | |
| MK-0773 | 5 | 2.4 | 0 |
| MK-0773 | 15 | 10 | 2 |
| MK-0773 | 45 | 56 | 12 |
| TFM-4AS-1 | 10 | ∼10 | 7 |
a The effects of compounds on seminal vesicle are shown as % increase over vehicle with 3 mg/kg DHT set to 100%.
Effect of MK-0773 and 2-FPA on Cholesterol
In humans anabolic androgens reduce high density lipoprotein cholesterol by mechanisms that are not understood but have been proposed to reflect changes in lipid metabolism in adipose tissue (33). We, thus, asked if SARMs developed by our method would also reduce cholesterol levels in preclinical models. Treatments of OVX rats with MK-0773, 2-FPA, TFM-4AS-1, and DHT at anabolic doses for 24 days resulted in a similar decline in both total cholesterol and high density lipoprotein by 25–37% (Table 3). Thus, the partial agonism and tissue selectivity of MK-0773 does not translate into differential effects on lipid metabolism in OVX rats.
TABLE 3.
Effects of MK-0773 and 2-FPA treatments on serum cholesterol in OVX rats
Sprague-Dawley rats OVX at 12 weeks and age 20 weeks at the study initiation were treated with the indicated doses for 24 days. Rats were terminally bled, and serum was analyzed for lipids. HDL, high density lipoprotein.
| Total cholesterol | HDL | |
|---|---|---|
| mg/dl | mg/dl | |
| Experiment 1 | ||
| Vehicle | 126 ± 3.1 | 37 ± 1.6 |
| MK-0773, 5 mg/kg | 124 ± 6.9 | 32 ± 3.2 |
| MK-0773, 25 mg/kg | 109 ± 3.0a | 25 ± 1.3a |
| TFM-4AS-1, 10 mg/kg | 92 ± 2.5a | 23 ± 0.8a |
| DHT, 3 mg/kg | 81 ± 2.6a | 19 ± 0.9a |
| Experiment 2 | ||
| Vehicle | 131 ± 4.1 | 67 ± 2.1 |
| 2-FPA, 3 mg/kg | 126 ± 4.0 | 65 ± 2.4 |
| 2-FPA, 10 mg/kg | 122 ± 7.1 | 61 ± 3.5 |
| 2-FPA, 30 mg/kg | 98 ± 9.8a | 47 ± 4.2a |
| 2-FPA, 60 mg/kg | 81 ± 4.9a | 38 ± 2.3a |
| TFM-4AS-1, 10 mg/kg | 101 ± 7.3a | 48 ± 3.1a |
| DHT, 3 mg/kg | 86 ± 7.5a | 41 ± 3.1a |
a p < 0.01, versus vehicle, one way analysis of variance, Fisher's PLSD.
DISCUSSION
SARMs offer promise for treating osteoporosis, age-related sarcopenia, and other conditions where low androgen tone might contribute to disease susceptibility or progression. Thus, there has been intense interest in discovering SARMs, and several AR ligands with SARM-like properties have been reported (12–15, 17–19, 34), including TFM-4AS-1, a 4-aza-steroidal compound that formed the basis for our investigations (20). We previously demonstrated that SARMs are gene-selective agonists; that is, within a single cell type they induce some androgen-responsive genes as robustly as DHT and others partially or not at all (20). However, it is not known how a ligand interacting with a single receptor (the AR) within a uniform cellular context could regulate one AR-responsive gene but not another. Nor is there an understanding of how this gene selectivity manifests as tissue selectivity in vivo.
Given this limitation in our knowledge, most SARMs were identified as ligands that behaved neither like a full agonist nor a complete antagonist of an androgen-responsive promoter that was co-transfected with AR into a heterologous cell system. Potential SARMs were often assessed by comparing their effects on the levator ani, a sexually dimorphic androgen-responsive reproductive muscle that is used as a convenient surrogate for the skeletal muscle anabolism, to their ability to stimulate prostate or seminal vesicles growth in castrated male rats (12). Tissue selectivity in this type of model was sometimes confirmed in long term studies where musculoskeletal growth was measured directly. In an attempt to develop a rational design strategy, Bohl et al. (35) used three-dimensional quantitative structure activity relationships of androgens with the AR to identify structural features that are required for SARM activities. However, until now there have been no detailed studies where large numbers of structurally distinct SARMs were analyzed for differences in in vitro assays and compared with physiological effects in several androgen responsive tissues. These data, as reported here, illustrate rational discovery of tissue-selective nuclear receptor ligands and provide insight into the mechanism of SARM tissue selectivity.
To undertake such a study we developed a diverse chemical library of uncharacterized AR ligands and examined them with transcriptional assays designed to capture multiple aspects of receptor activation. Like all steroid hormone receptors, the AR is a modular protein with distinct functions attributable to specific domains (36, 37). The LBD serves a dual purpose as a high affinity site for androgens and as a cofactor recruitment and scaffolding domain. We measured the strength of the interaction between each compound and the receptor in competitive binding studies, whereas the ability of the ligand to confer the appropriate conformation for cofactor recruitment was measured in the TRAF2 GRIP-1 assay. The N-terminal domain of AR contains a ligand-independent activator function (AF-1) but also functions largely as ligand-dependent cofactor for the LBD, a function that we postulated, based on human genetic data, functions in reproductive physiology (25–27, 38). This receptor function was monitored in the VIRCON mammalian two-hybrid assay. The ability of ligands to activate endogenous AR in the TAMAR assay was used to capture all critical steps in receptor function, such as dissociation of AR from cytosolic heat shock proteins, translocation to the nucleus, DNA binding, and eventually nuclear export and receptor degradation. Presumably ligands that cannot induce these basic responses would not be anabolic.
We determined that partial agonism in transactivation and AF-2-mediated GRIP-1 recruitment when combined with greatly reduced ability to induce the N/C interaction is the hallmark of a SARM. Compounds that induced the N/C interaction effectively were fully physiologically active in both musculoskeletal and reproductive organs. This result suggests to us that the N/C interaction, a relatively unique feature of the AR, evolved to produce the full spectrum of male secondary sexual traits in response to high levels of potent androgens. However, as shown here and elsewhere (27, 39), even though the N/C interaction may modulate transcription, it is not absolutely required and does not appear to be required for aspects of androgen signaling that might be important in both genders, such as anabolism and lipoprotein metabolism. As such, measuring this interaction provides an efficient, mechanism-related means of discovering SARMs.
We also observed that GRIP-1 recruitment to the AF-2 domain did not provide significantly different information than transactivation alone. The strong linear relationship between these two measurements suggests that cofactor recruitment, potentially of GRIP-1 and related protein(s), is an obligate step in MMTV promoter activation in MDA-MB-453 cells. However peptide-based structural probe studies indicate that SARMs induce distinct conformations in the LBD (40) and interact differently than full agonists with critical amino acid residues with the LBD (35), although these differences were not apparent by x-ray crystallography (41). A recent set of experiments in prostate cancer cells indicated that unlike DHT, neither CBP/p300 (cAMP-response element-binding protein (CREB)-binding protein) nor CARM1 (coactivator-associated arginine methyltransferase 1) was recruited to AR, and although both DHT and SARMs recruited p160 (steroid receptor coactivator-1) cofactors, different protein-protein interaction sites were involved (42). Thus, it seems likely that SARMs induce changes in the LBD that not only affect the N/C interaction but also a myriad of other protein interactions, only one of which was directly measured in our GRIP-1 assay. It will be interesting to probe SARM libraries with more sensitive LBD assays to identify new classes of AR ligands and assess their physiological effects. Alternatively, mechanistically distinct SARMs could be discovered by employing genome-wide gene expression measurements from MDA-MB-453 or other cells to identify ligands with distinct gene selectivities.
Although SARMs can provide important insight into the function of AR and transcriptional regulation by nuclear receptors, the medical potential of these compounds is also of significant interest. By correlating transcriptional changes to histological measurements, we show that gene expression biomarkers can provide rapid quantitative indications of androgen action in various tissues. This discovery raises the possibility that AR ligands could be validated as SARMs in humans before the start of long clinical trials that expose large numbers of subjects to an unproven therapy, a process that was required for the clinical evaluation of selective estrogen receptor modulators. Such a methodology would allow for select compounds such as MK-0773 and 2-FPU to be rapidly assessed in subjects of different genders, ages, disease states, and endogenous androgen levels to identify the ligand properties ideal for these indications.
The detection of type II collagen mRNA in rat tibial diaphysis was unexpected, as its expression is selective for cartilage. It is possible that a transient induction of type II collagen mRNA is the result of an increase in tissue remodeling caused by the anabolic actions of androgens. Although quantitative measurements of type II collagen mRNA levels were very useful as a biomarker for the assessment of BFR, further study with specific collagen antibodies will be required to determine whether and where type II collagen protein is present during periosteal bone formation.
In summary, we describe in vitro properties of SARMs that predict tissue-selective effects in rodents. Because human data for MK-0773 are now available, the translatability of the animal model used in this work can be assessed. MK-0773 produced anabolism in both men and women in a 12-week Phase Ib trial while inducing no or very low effects on sebaceous glands, endometrium, or prostate (43, 44). Furthermore, the reduction in total cholesterol and high density lipoprotein we report here in rats was also observed in the clinic. Thus, AR ligands that can lead to promoter activation but do not robustly induce the N/C interaction will potentially have a therapeutic value. It will be interesting to test new classes of SARMs based on additional biochemical measurements to determine whether the AR can accommodate multiple modes of interaction, each with distinct medicinal potential.
Acknowledgments
With deep gratitude we recognize Gideon A. Rodan, may his soul rest in peace, for enthusiastic intellectual input and support to this project and our work. We give special thanks to Dwight A. Towler, who made this project a reality, for contributions to our work. We also thank Jeffrey H. Toney and M. Katherine Holloway for assistance in providing reagents and advice that helped start the SARM project.
This work was supported by the research and development budget of Merck & Co., Inc.
- SARM
- selective androgen receptor modulators
- AR
- androgen receptor
- rhAR
- rhesus AR
- N/C
- N- and C-terminal
- PSU
- pilosebaceous unit
- AF-1
- activation domain-1
- AF-2
- activation domain-2
- ARBIND
- AR binding assay
- TAMAR
- transactivation modulation of AR
- VIRCON
- virization counterscreen assay
- TRAF-2
- transcriptional recruitment to AF-2
- MMTV
- mouse mammary tumor virus
- qPCR
- quantitative PCR
- ORX
- orchidectomy
- OVX
- ovariectomy
- LBD
- ligand binding domain
- LBM
- lean body mass
- IP
- inflection point
- GRIP-1
- glutamate receptor-interacting protein 1
- R1881
- methyltrienolone
- 6-FAM
- 6-carboxyfluorescein
- BFR
- bone formation rate
- SV
- seminal vesicle
- SCD1
- stearoyl-CoA desaturase-1.
REFERENCES
- 1.Harman S. M., Metter E. J., Tobin J. D., Pearson J., Blackman M. R. (2001) J. Clin. Endocrinol. Metab. 86, 724–731 [DOI] [PubMed] [Google Scholar]
- 2.Bhasin S. (2003) J. Endocrinol. Invest. 26, 814–822 [DOI] [PubMed] [Google Scholar]
- 3.Isidori A. M., Giannetta E., Pozza C., Bonifacio V., Isidori A. (2005) J. Endocrinol. Invest. 28, 73–79 [PubMed] [Google Scholar]
- 4.Stanworth R. D., Jones T. H. (2009) Front Horm. Res. 37, 74–90 [DOI] [PubMed] [Google Scholar]
- 5.Bhasin S. (2003) J. Gerontol. A Biol. Sci. Med. Sci. 58, 1002–1008 [DOI] [PubMed] [Google Scholar]
- 6.Braunstein G. D. (2007) Fertil. Steril. 88, 1–17 [DOI] [PubMed] [Google Scholar]
- 7.Kaufman J. M. (2003) Aging Male 6, 166–174 [PubMed] [Google Scholar]
- 8.Segal S., Narayanan R., Dalton J. T. (2006) Expert Opin. Investig. Drugs 15, 377–387 [DOI] [PubMed] [Google Scholar]
- 9.Bhasin S., Jasuja R. (2009) Curr. Opin. Clin. Nutr. Metab. Care 12, 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper M. W., Flowers D. E., Huster W. J., Neild J. A., Harper K. D., Arnaud C. (1996) J. Bone Miner. Res. 11, 835–842 [DOI] [PubMed] [Google Scholar]
- 11.Vogel V. G., Costantino J. P., Wickerham D. L., Cronin W. M., Cecchini R. S., Atkins J. N., Bevers T. B., Fehrenbacher L., Pajon E. R., Jr., Wade J. L., 3rd, Robidoux A., Margolese R. G., James J., Lippman S. M., Runowicz C. D., Ganz P. A., Reis S. E., McCaskill-Stevens W., Ford L. G., Jordan V. C., Wolmark N. (2006) JAMA 295, 2727–2741 [DOI] [PubMed] [Google Scholar]
- 12.Allan G., Sbriscia T., Linton O., Lai M. T., Haynes-Johnson D., Bhattacharjee S., Ng R., Sui Z., Lundeen S. (2008) J. Steroid Biochem. Mol. Biol. 110, 207–213 [DOI] [PubMed] [Google Scholar]
- 13.Allan G. F., Tannenbaum P., Sbriscia T., Linton O., Lai M. T., Haynes-Johnson D., Bhattacharjee S., Zhang X., Sui Z., Lundeen S. G. (2007) Endocrine 32, 41–51 [DOI] [PubMed] [Google Scholar]
- 14.Fuse H., Fukumoto S., Sone H., Miyata Y., Saito T., Nakayama K., Takahashi H., Matsumoto T., Ogata E. (1997) J. Bone Miner. Res. 12, 590–597 [DOI] [PubMed] [Google Scholar]
- 15.Gao W., Kearbey J. D., Nair V. A., Chung K., Parlow A. F., Miller D. D., Dalton J. T. (2004) Endocrinology 145, 5420–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao W., Reiser P. J., Coss C. C., Phelps M. A., Kearbey J. D., Miller D. D., Dalton J. T. (2005) Endocrinology 146, 4887–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada K., Furuya K., Yamamoto N., Nejishima H., Ichikawa K., Nakamura T., Miyakawa M., Amano S., Sumita Y., Oguro N. (2003) Biol. Pharm. Bull. 26, 1563–1569 [DOI] [PubMed] [Google Scholar]
- 18.Miner J. N., Chang W., Chapman M. S., Finn P. D., Hong M. H., López F. J., Marschke K. B., Rosen J., Schrader W., Turner R., van Oeveren A., Viveros H., Zhi L., Negro-Vilar A. (2007) Endocrinology 148, 363–373 [DOI] [PubMed] [Google Scholar]
- 19.Riedmaier I., Tichopad A., Reiter M., Pfaffl M. W., Meyer H. H. (2009) J. Steroid Biochem. Mol. Biol. 114, 167–173 [DOI] [PubMed] [Google Scholar]
- 20.Schmidt A., Harada S., Kimmel D. B., Bai C., Chen F., Rutledge S. J., Vogel R. L., Scafonas A., Gentile M. A., Nantermet P. V., McElwee-Witmer S., Pennypacker B., Masarachia P., Sahoo S. P., Kim Y., Meissner R. S., Hartman G. D., Duggan M. E., Rodan G. A., Towler D. A., Ray W. J. (2009) J. Biol. Chem. 284, 36367–36376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong C., Kelce W. R., Sar M., Wilson E. M. (1995) J. Biol. Chem. 270, 19998–20003 [DOI] [PubMed] [Google Scholar]
- 22.Kallio P. J., Poukka H., Moilanen A., Jänne O. A., Palvimo J. J. (1995) Mol. Endocrinol. 9, 1017–1028 [DOI] [PubMed] [Google Scholar]
- 23.Pfahl M. (1993) Endocr. Rev. 14, 651–658 [DOI] [PubMed] [Google Scholar]
- 24.Ye X., Han S. J., Tsai S. Y., DeMayo F. J., Xu J., Tsai M. J., O'Malley B. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9487–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langley E., Kemppainen J. A., Wilson E. M. (1998) J. Biol. Chem. 273, 92–101 [DOI] [PubMed] [Google Scholar]
- 26.Thompson J., Saatcioglu F., Jänne O. A., Palvimo J. J. (2001) Mol. Endocrinol. 15, 923–935 [DOI] [PubMed] [Google Scholar]
- 27.Quigley C. A., Tan J. A., He B., Zhou Z. X., Mebarki F., Morel Y., Forest M. G., Chatelain P., Ritzén E. M., French F. S., Wilson E. M. (2004) Mech. Ageing Dev. 125, 683–695 [DOI] [PubMed] [Google Scholar]
- 28.Chen F., Knecht K., Leu C., Rutledge S. J., Scafonas A., Gambone C., Vogel R., Zhang H., Kasparcova V., Bai C., Harada S., Schmidt A., Reszka A., Freedman L. (2004) J. Steroid Biochem. Mol. Biol. 91, 247–257 [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y., Prouty S. M., Harmon A., Sundberg J. P., Stenn K. S., Parimoo S. (2001) Genomics 71, 182–191 [DOI] [PubMed] [Google Scholar]
- 30.Smith K. R., Thiboutot D. M. (2008) J. Lipid Res. 49, 271–281 [DOI] [PubMed] [Google Scholar]
- 31.Meissner R. S., et al. (2009) 237th ACS National Meeting, Salt Lake City, UT, Abst. Medi 32 [Google Scholar]
- 32.Manfredi M. C., Bi Y., Nirschl A. A., Sutton J. C., Seethala R., Golla R., Beehler B. C., Sleph P. G., Grover G. J., Ostrowski J., Hamann L. G. (2007) Bioorg. Med. Chem. Lett. 17, 4487–4490 [DOI] [PubMed] [Google Scholar]
- 33.Nantermet P., Harada S., Liu Y., Cheng S., Johnson C., Yu Y., Kimme D., Holder D., Hodor P., Phillips R., Ray W. J. (2008) Endocrinology 149, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 34.Mohler M. L., Bohl C. E., Jones A., Coss C. C., Narayanan R., He Y., Hwang D. J., Dalton J. T., Miller D. D. (2009) J. Med. Chem. 52, 3597–3617 [DOI] [PubMed] [Google Scholar]
- 35.Bohl C. E., Chang C., Mohler M. L., Chen J., Miller D. D., Swaan P. W., Dalton J. T. (2004) J. Med. Chem. 47, 3765–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centenera M. M., Harris J. M., Tilley W. D., Butler L. M. (2008) Mol. Endocrinol. 22, 2373–2382 [DOI] [PubMed] [Google Scholar]
- 37.Li J., Al-Azzawi F. (2009) Maturitas 63, 142–148 [DOI] [PubMed] [Google Scholar]
- 38.Holterhus P. M., Werner R., Struve D., Hauffa B. P., Schroeder C., Hiort O. (2005) Exp. Clin. Endocrinol. Diabetes 113, 457–463 [DOI] [PubMed] [Google Scholar]
- 39.Askew E. B., Gampe R. T., Jr., Stanley T. B., Faggart J. L., Wilson E. M. (2007) J. Biol. Chem. 282, 25801–25816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C. Y., McDonnell D. P. (2002) Mol. Endocrinol. 16, 647–660 [DOI] [PubMed] [Google Scholar]
- 41.Narayanan R., Mohler M. L., Bohl C. E., Miller D. D., Dalton J. T. (2008) Nucl. Recept. Signal. 6, e010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek S. H., Ohgi K. A., Nelson C. A., Welsbie D., Chen C., Sawyers C. L., Rose D. W., Rosenfeld M. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoch S. A., et al. (2008) 90th Annual Meeting of the Endocrine Society, San Francisco, CA, Abst. OR35–33 [Google Scholar]
- 44.Stoch S. A., et al. (2009) 91st Annual Meeting of the Endocrine Society, Washington, D.C., Abst. S21–24 [Google Scholar]
- 45.Hall R. E., Birrell S. N., Tilley W. D., Sutherland R. L. (1994) Eur. J. Cancer 30A, 484–490 [DOI] [PubMed] [Google Scholar]
- 46.Bettoun D. J., Scafonas A., Rutledge S. J., Hodor P., Chen O., Gambone C., Vogel R., McElwee-Witmer S., Bai C., Freedman L., Schmidt A. (2005) J. Biol. Chem. 280, 38898–38901 [DOI] [PubMed] [Google Scholar]
- 47.Schmidt A., Endo N., Rutledge S. J., Vogel R., Shinar D., Rodan G. A. (1992) Mol. Endocrinol. 6, 1634–1641 [DOI] [PubMed] [Google Scholar]
- 48.Nantermet P. V., Xu J., Yu Y., Hodor P., Holder D., Adamski S., Gentile M. A., Kimmel D. B., Harada S., Gerhold D., Freedman L. P., Ray W. J. (2004) J. Biol. Chem. 279, 1310–1322 [DOI] [PubMed] [Google Scholar]
- 49.Nantermet P. V., Masarachia P., Gentile M. A., Pennypacker B., Xu J., Holder D., Gerhold D., Towler D., Schmidt A., Kimmel D. B., Freedman L. P., Harada S., Ray W. J. (2005) Endocrinology 146, 564–578 [DOI] [PubMed] [Google Scholar]