Abstract
This minireview focuses on structural studies that have provided insights into our current understanding of thick filament regulation in muscle. We describe how different domains in the myosin molecule interact to produce an inactive “off” state; included are head-head and head-rod interactions, the role of the regulatory light chain, and the significance of the α-helical coiled-coil rod in regulation. Several of these interactions have now been visualized in a wide variety of native myosin filaments, testifying to the generality of these structural motifs across the phylogenetic tree.
Keywords: Myosin, Protein Phosphorylation, Protein Structure, Skeletal Muscle, Smooth Muscle, Coiled Coil, Microscopy, Regulation, Regulatory Light Chain
Introduction
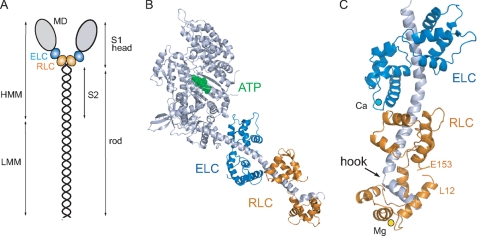
The publication of the crystal structure of the globular head region of skeletal muscle myosin (subfragment 1 (S1)3) in 1993 (1) made it possible for the first time to understand how biological motors function at the atomic level (Fig. 1, A and B). By fitting, the S1 head structure into macromolecular complexes of actin filaments decorated with S1, Rayment et al. (2) were able to propose the first detailed mechanism for the generation of force and movement in motile cells. This exciting new structural framework, together with impressive advances in determining the displacement and force produced by a single molecular motor, inspired a Biophysical Discussions meeting in 1994. We discussed the role of the light chains in the so-called “lever arm” of the myosin head (3). Here, we summarize some of the major developments in the regulation of class II myosins in the intervening 15 years.
FIGURE 1.
Structure of the myosin molecule. A, schematic diagram of the myosin molecule. The N-terminal region of the myosin heavy chain forms the globular motor domain (MD; gray), which contains the sites for ATP and actin binding. The ELC (blue) and the RLC (orange) stabilize a single α-helical polypeptide chain at the C terminus of the motor domain. The remainder of the heavy chain forms an ∼160-nm α-helical coiled-coil rod, which gives rise to the filamentous properties of class II myosins. Proteolysis of myosin produces a soluble subfragment (HMM), consisting of the S1 head and the adjacent S2 rod, and the insoluble light meromyosin (LMM) fragment responsible for myosin assembly. The molecular mass of the myosin heavy chain is ∼200 kDa; the RLC and ELC are each ∼20 kDa. B, ribbon representation of scallop myosin S1 (Protein Data Bank code 1QVI). The nucleotide (ATP; green) pocket is located at the lower end of a large cleft that serves as a communication pathway between actin and the nucleotide-binding site. C, ribbon diagram of the scallop regulatory domain (Protein Data Bank code 1WDC), with Ca2+ (cyan) bound to domain 1 of the ELC and Mg2+ (yellow) bound to the N terminus of the RLC. The α-helical myosin heavy chain (gray) has a sharp bend or hook near the C terminus (6, 7).
Background
The discovery that phosphorylation of a single serine residue (Ser19) in one of the two classes of light chains, the so-called regulatory light chain (RLC), is entirely sufficient to activate smooth and nonmuscle myosin II was known as early as 1975 (Ref. 4 and reviewed in Ref. 5). Phosphorylation also occurs in the homologous light chains (LCs) of skeletal and cardiac muscle myosins, but the effect on activity is relatively minor compared with smooth muscle myosin, and regulation takes place primarily through calcium activation of the thin filament. The second class of LCs is often referred to as the essential LC (ELC) because of its strong association with the myosin heavy chain. Although the crystal structure of S1 highlighted the importance of the LCs in stabilizing a long α-helical region at the C terminus of the globular catalytic domain, it did little to reveal the mysteries of how a single post-translational modification could switch “on” the active site at a distance of >10 nm!
The first structural insight into how LCs might contribute to regulation came from the crystal structure of the regulatory domain of scallop myosin, a class II molluscan myosin that is thick filament-regulated by calcium binding to the ELC (Fig. 1C) (6). An unusual feature of the calcium-binding site in domain I of the ELC is that it requires specific interactions with both the RLC and the myosin heavy chain for stabilization because the isolated ELC does not bind calcium. Thus, all three chains are involved in regulation; the absence of calcium destabilizes these linkages, leading to a more flexible lever arm and an inactive catalytic domain.
One region in the regulatory domain that merits more attention is the unusual sequence and sharp bend of ∼60° that interrupts the long α-helical heavy chain near the head-rod junction. This so-called “hook” is composed of highly hydrophobic residues (WQW) that are embedded in the hydrophobic core of the N-terminal lobe of the RLC (Fig. 1C). It is notable that even though chicken skeletal S1 is not considered a regulated myosin, the LC-binding domain shares considerable structural homology with the scallop regulatory domain: it too has a hook region characterized by the sequence WPW that is similarly enfolded by the hydrophobic core of the RLC (1, 7). All class II myosins appear to have a signature of hydrophobic residues clustered in this region of the heavy chain, and they all bind the RLC rather than the closely related calmodulin. If the RLC is removed by chemical reagents or by mutagenesis, the heavy chain self-associates into large aggregates that are unable to assemble into the highly ordered helical structure of a thick filament (8). This behavior probably accounts for the loss of myofibrillar assembly and embryonic lethality observed in the zebrafish upon disruption of its single cardiac-specific RLC gene (9). Unconventional myosins all use calmodulin (with an occasional additional ELC) to stabilize the α-helical C terminus of the motor domain; the exact number of calmodulins depends on the length of the lever arm. The RLC is restricted to the class II myosins, which have a 1:1 stoichiometry between the RLC and the heavy chain from yeast to humans. The necessity for this specialized structure at the head-rod junction may be related to the asymmetric head-head interaction involved in the inactive state.
Regulation of Contractility by Head-Head Interactions
Although the main structural features of a myosin head seemed to be preserved across species as diverse as Dictyostelium discoideum and chicken, the structural basis for the inactive “off” state was not well understood. Removal of calcium in molluscan myosins and dephosphorylation in thick filament-regulated vertebrate myosins clearly affected transmission of information from the lever arm to the active site, but how this coupling was achieved remained unknown. Electron microscopic images of metal-shadowed smooth muscle and nonmuscle myosins under conditions favoring an inactive state showed that the normally extended α-helical rod folded at ∼50 and 100 nm from the invariant proline that marks the head-rod junction (Fig. 2A) (10–13). The heads appeared to be pointed downward toward the rod, resulting in a compact folded conformation that sedimented at 10 S (Svedberg) rather than at the 6 S value characteristic of the highly elongated conformation of muscle myosin (Fig. 2A) (13). Importantly, the 10 S structure was shown to have virtually no enzymatic activity, and the nucleotide-binding site contained tightly bound ADP and phosphate (14, 15). This off state requires two heads and a minimum length of rod; S1 and single-headed myosin are essentially active (16, 17), but the resolution of the electron micrographs was too low to determine any details of the intramolecular interactions needed to stabilize the 10 S structure.
FIGURE 2.
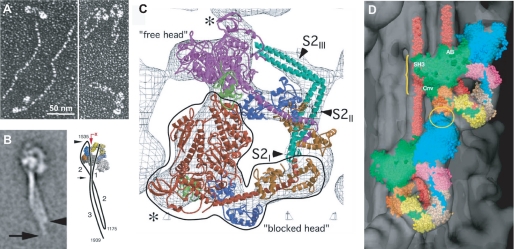
Inhibited configuration of the myosin molecule. A, electron micrographs of smooth muscle myosin rotary-shadowed with platinum at high (extended 6 S form) and low (folded 10 S form) ionic strength (13). B, electron microscopy of folded smooth muscle myosin molecules by negative staining and single-particle image processing (22). The accompanying diagram shows the two bends in the rod near residues 1175 and 1535 to form the 10 S structure. A photoactivated probe (the red arrow points to RLC Cys108) can cross-link the rod to the LC (43). C, electron cryomicroscopy of two-dimensional crystalline arrays of unphosphorylated smooth muscle HMM on a lipid monolayer surface (19). The outlined blocked head (red) interacts with the converter (green) and the ELC (blue) on the free head (magenta). The assignment for the S2 density is uncertain and more likely follows a path between the heads as indicated by the negatively stained molecules in B. D, atomic model of smooth muscle HMM (see C) fitted into the three-dimensional reconstruction of a tarantula muscle myosin filament obtained by electron cryomicroscopy and single-particle imaging techniques (25). The blocked and free heads are colored green and blue, respectively, in this representation. Regions of possible interactions are between the ELC and free heads (yellow ellipse) and between the blocked motor domain and S2 (yellow bracket). SH3, Src homology 3 domain; AB, actin binding; Cnv, converter.
The big breakthrough came in ∼2000 with the visualization of the myosin heads in two-dimensional crystalline arrays of expressed, dephosphorylated, and thiophosphorylated smooth muscle heavy meromyosin (HMM; the soluble two-headed subfragment of myosin) (Fig. 1A) on positively charged lipid monolayers (18). The two-dimensional projections at 2.3 nm resolution in negative stain revealed distinct structural differences between the two states of RLC phosphorylation: the active form of the HMM heads showed intermolecular interactions with neighboring molecules, whereas the inactive form showed clear intramolecular head-head interactions that immediately suggested a mechanism for inhibition (18).
A subsequent study of dephosphorylated HMM by electron cryomicroscopy of unstained frozen-hydrated specimens (∼2.0 nm resolution) resulted in a three-dimensional image reconstruction that revealed more details of this asymmetric head interaction (Fig. 2C) (19). By fitting the crystal structure of the motor domain plus the ELC (20) into the density of these crystalline arrays, it could be shown that the ATPase activity of one head is “blocked” by positioning its actin-binding interface on the converter and ELC region of the neighboring so-called “free head.” The binding to the converter domain prevents the domain movements needed for phosphate release and activation. This three-dimensional structure also showed a segment of the rod (subfragment 2 (S2)) in close proximity to the heads, but the exact location was uncertain. Although the asymmetric head structure of dephosphorylated HMM heads provided an attractive molecular mechanism for the inhibited state and suggested why phosphorylation would disrupt these multiple protein interactions, the structure was at first received with some skepticism because of the charged lipid surface. However, when the same structure was seen with two-dimensional crystals of intact folded 10 S myosin, which packs very differently on the lipid monolayer surface, it was difficult to escape the conclusion that this asymmetric head-head interaction was an inherent property of a regulated myosin molecule (21). Moreover, a recent electron microscopic study using single-particle averaging of negatively stained smooth muscle myosin molecules confirmed the head-head interaction in 10 S molecules and showed that the folded rod starts at the junction between the two heads and follows a path that interacts with the blocked head before rejoining the other tail segments (Fig. 2B) (22).
Relaxed State in Native Thick Filaments
At physiological ionic conditions, striated muscle myosin forms a highly ordered filamentous structure whose backbone does not change with the state of phosphorylation. Unlike striated muscles, smooth muscle cells need to maintain a high degree of mechanical plasticity to adapt to the wide range of muscle lengths and volumes in hollow organs such as the bladder and uterus (23). Consequently, a plausible but still unproven hypothesis is that smooth muscle myosin filaments are dynamic structures that can recruit monomers from a large pool of 10 S myosin when more or longer filaments are needed and, conversely, shorten their length by a similar mechanism. Under in vitro conditions of low ionic strength and ATP, dephosphorylated myosin filaments are not stable and immediately dissociate to the 10 S conformation. Phosphorylation is needed to convert the folded 10 S to the extended 6 S structure, which then spontaneously assembles into filaments (13). This labile property of smooth muscle myosin makes it extremely difficult to study dephosphorylated heads in a filamentous environment. A more propitious model system for studying thick filament structure has been the phosphorylation-regulated striated muscle filaments from tarantula muscles, which are longer and thicker than vertebrate filaments and contain a paramyosin core (24).
The most compelling demonstration for the existence of the head-head interaction in the relaxed dephosphorylated filamentous state has come from the three-dimensional reconstruction of tarantula thick filaments using electron cryomicroscopy to preserve the native unstained structure (25). Using a single-particle approach that avoids the need for perfect helical symmetry in traditional helical reconstructions (26), it was possible to improve the apparent resolution to ∼2.5 nm from the 5 nm resolution previously obtained for negatively stained tarantula thick filaments. This higher resolution made it possible to computationally fit the atomic model of smooth muscle HMM into the appropriate regions of density in the three-dimensional reconstruction. It was remarkable to see how well the model generated by Taylor and co-workers (19) fit into the reconstruction of this distantly related, invertebrate striated muscle filament (Fig. 2D). Evidence for the generality of this motif has come from the published structures of vertebrate cardiac myosin filaments from mouse (27) and rabbit (28) muscles, which also showed the asymmetric two-headed smooth muscle myosin structure although at lower resolution (∼4 nm). Unlike tarantula filaments, not all the heads in the striated muscle filaments could be fitted with the smooth muscle HMM model due to perturbations of helical symmetry and the increased mobility of heads in vertebrate thick filaments. Nevertheless, the head-head interactions seen in the cardiac muscle filaments received independent support from visualization of the folded 10 S conformation in isolated cardiac and skeletal muscle myosin molecules by electron microscopy (29). It is clear that the heads of unregulated myosins (vertebrate striated) can undergo interactions similar to those of regulated myosins (vertebrate smooth and nonmuscle) in the relaxed (bound ADP·Pi) state. However, the intramolecular interactions are far weaker in a thin filament-regulated myosin, and blebbistatin, a noncompetitive inhibitor of ATPase activity, was added to stabilize the heads in the relaxed conformation in both isolated molecules (29) and filaments (27).
The asymmetric head motif has now been found in a number of other species under relaxing conditions, including scallop muscle myosin filaments (30) and Limulus (horseshoe crab) muscle thick filaments (31), adding further support to the hypothesis that intramolecular head-head interactions exist in the off state across the phylogenetic tree (32). This striking new structure explains many earlier puzzling observations, foremost among them being that the biochemical properties of skeletal muscle myosin were relatively unchanged upon phosphorylation and failed to explain why the sequence of a phosphorylatable serine was conserved throughout evolution. Physiological experiments on skinned fast twitch muscle fibers had shown a small but significant temperature-dependent increase in force production at low levels of calcium upon RLC phosphorylation (33). Electron microscopy of rabbit skeletal muscle thick filaments indicated increased disorder in the surface array of phosphorylated cross-bridges compared with the ordered heads in relaxed filaments (34). These observations led Sweeney et al. (33) to propose that phosphorylation shifts the distribution of cross-bridges away from the thick filament backbone toward the thin filament, thereby optimizing the rate of force production and muscle performance. The central concept of this mechanism for the role of LC phosphorylation in vertebrate striated muscles was essentially correct, but it lacked the structural information implicating intramolecular head-head interactions in the relaxed state of the thick filament.
Role of the α-Helical Coiled-coil Rod in Myosin Regulation
There is general agreement that the rod stabilizes the 10 S conformation and enhances the ability of the heads to trap ADP and Pi at the active site (15). Possible intra- and intermolecular interactions between the heads and S2 (the soluble one-third of the ∼160-nm long rod nearest to the heads) (Fig. 1A) are suggested by the fitting of atomic models into three-dimensional reconstructions of tarantula and Limulus thick filaments (Fig. 2D) (25, 31).
Insights into the functional role of the S2 rod segment (other than a simple joining of two heads) have come from an investigation of the minimum length of S2 required to regulate activity in expressed smooth muscle HMM (35). The α-helical coiled-coil S2 sequence has a periodic repeat of hydrophobic apolar residues at positions a and d within 7 residues (the so-called “heptad” repeat of the form abcdefg) that accounts for the “knobs-into-holes” packing of two α-helices. The remaining residues in the heptad tend to be charged; some favor interchain ionic interactions stabilizing the coiled coil (e and g), whereas the outermost residues (b, c, and f) may lead to protein-protein interactions necessary for filament formation. Expressed HMM constructs consisting of 2, 7, or 15 heptads past the invariant proline marking the head-rod junction were analyzed for their ability to regulate ATPase activity by phosphorylation (35). To stabilize the dimeric form of these truncated constructs, the GCN4 leucine zipper (a transcriptional activator containing only 32 amino acids but a preponderance of leucine residues) was added to the C terminus of the HMM constructs. Although the 2- and 7-heptad HMM dimers had some degree of regulation (3–5-fold change in ATPase activity between the phosphorylated and dephosphorylated states), an HMM construct consisting of 15 heptads was able to achieve a fully regulated state (>20-fold) characteristic of HMM prepared by chymotryptic digestion.
One interpretation of these findings is that the heads in the inhibited state need to interact with the charged surface of the rod to maintain the off state. A segment of 15 heptads approximates the length of the myosin head. This interaction may be more dependent on the pattern of charged residues than on a specific sequence of polar residues based on the unexpected finding that a striated muscle rod sequence could replace the smooth muscle rod in a chimeric HMM construct without any loss of regulation (35). The ability to interchange isoform-specific rod sequences has become more comprehensible by the finding that the crystal structure of 15-heptad smooth muscle S24 is very similar to the coiled-coil atomic structure of 18-heptad human cardiac S2 (36); a common motif is a pronounced unwinding of the coiled coil at the N terminus of regulated smooth muscle S2. An unwinding of the two α-helices at the N-terminal region has also been observed in a shorter (∼7-heptad) fragment from the scallop myosin rod, suggesting that this instability near the head-rod junction may be an important feature of class II myosins (37). Another interesting characteristic of the S2 crystal structures is the departure from strict 2-fold symmetry due to axial staggering of core residues between the two helices; an asymmetric S2 structure may well be a prerequisite for an asymmetric head-head interaction (36).
An unwound but α-helical head-rod junction appears to be an essential element for optimal mechanical function as well as regulation of enzymatic activity. When a leucine zipper was engineered into the rod region immediately adjacent to the heads (0-heptad zip), the power stroke of wild-type smooth muscle HMM was reduced from a unitary step size of ∼10 nm to <1 nm as measured by an optical trap assay (38). Moving the leucine zipper to 15 heptads beyond the conserved invariant proline restored the step size, leading to the conclusion that flexibility at the head-rod function is inherent in the design of class II motors.
Consistent with the structural and biophysical studies, computational normal mode analysis has suggested that uncoiling of 2–3 heptads at the head-rod junction is necessary for coupling the torsional motions of the S1 heads with the rod to achieve an optimal inhibited state (39). Molecular modeling has also provided a key insight into how ATP binding can cause dephosphorylated smooth muscle myosin filaments to dissociate to the 10 S state in vitro. The strain introduced by twisting the separated myosin heads into a compact asymmetric structure may be propagated along the rod so that even the assembly-competent light meromyosin is subject to small distortions that destabilize the rod-rod interactions in the thick filament backbone (39).
Role of the Regulatory Light Chain
The folded core structure of the RLC has been seen in crystal structures of chicken skeletal myosin (1) and scallop myosin (6), but unfortunately, ∼20 residues of the N-terminal domain are too disordered to be visible. What information we have has come mainly from mutational analysis, cross-linking, spectroscopic measurements, and molecular dynamics. Mutational studies showed that the region encompassing Lys11–Arg16 was particularly important in stabilizing the 10 S conformation (40). Phosphorylation of Ser19 disrupts these multiple weak, predominantly ionic interactions between the heads in the off state. EPR studies on spin-labeled RLC bound to myosin (41), coupled with molecular dynamic simulations (42), suggested that the disruption by phosphorylation is caused by a transition from a disordered to a more ordered helical structure that is stabilized by a salt bridge between Ser19 and Arg16.
Extensive photo-cross-linking studies have attempted to characterize putative interactions between the RLC and rod in unphosphorylated (10 S) myosin. Cross-links have been found between a photoactivated probe on RLC Cys108 (43) and residues spanning region 1500–1600 of the rod, consistent with a folded structure (Fig. 2B). Cross-links have also been shown between the N-terminal domain of one RLC and the C-terminal domain of the partner RLC on the neighboring head in the 10 S conformation (44), but these cross-linked peptides are incompatible with the geometry of the asymmetric head-head structure (21). Given the dynamic nature of the myosin subunits involved in regulation, it is conceivable that the cross-linking reaction may have captured one of the less populated states in the equilibrium between the on and off states.
Perspectives
One of the most challenging goals for understanding the mechanism of regulation is to obtain a high resolution structure of a two-headed myosin molecule in the inhibited state. Such a crystallographic structure would provide the first opportunity to see in detail the interactions between the heads and with the rod. It might even be possible to visualize the elusive N-terminal domain of the RLC and to determine how it contributes to the stabilization of the off state.
An area of rapid progress has been in high resolution electron cryomicroscopy, with the aim of analyzing less than perfectly ordered helical structures to near-atomic resolution. This would clearly benefit the visualization of filamentous myosin interactions, especially in the more disordered vertebrate muscle thick filaments, where perturbations are thought to arise from the presence of accessory proteins such as titin and myosin-binding protein C (27, 28).
Although our understanding of muscle regulation has made impressive progress during the last decade, many aspects of this complex cellular mechanism remain poorly understood. In addition to more crystallographic and electron microscopic studies, techniques such as single-molecule biophysics and molecular dynamics may reveal transitory states that cannot be captured by current structural approaches.
This work was supported, in whole or in part, by National Institutes of Health Grants AR53975 (to S. L.) and HL38113 (to K. M. T.). This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
U. B. Nair, M. A. Rould, and K. M. Trybus, personal communication.
- S1
- subfragment 1
- RLC
- regulatory light chain
- LC
- light chain
- ELC
- essential LC
- HMM
- heavy meromyosin
- S2
- subfragment 2.
REFERENCES
- 1.Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. (1993) Science 261, 50–58 [DOI] [PubMed] [Google Scholar]
- 2.Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. (1993) Science 261, 58–65 [DOI] [PubMed] [Google Scholar]
- 3.Lowey S., Trybus K. M. (1995) Biophys. J. 68, 120S–126S; Discussion 126S–127S [PMC free article] [PubMed] [Google Scholar]
- 4.Adelstein R. S., Conti M. A. (1975) Nature 256, 597–598 [DOI] [PubMed] [Google Scholar]
- 5.Adelstein R. S., Eisenberg E. (1980) Annu. Rev. Biochem. 49, 921–956 [DOI] [PubMed] [Google Scholar]
- 6.Xie X., Harrison D. H., Schlichting I., Sweet R. M., Kalabokis V. N., Szent-Györgyi A. G., Cohen C. (1994) Nature 368, 306–312 [DOI] [PubMed] [Google Scholar]
- 7.Houdusse A., Cohen C. (1996) Structure 4, 21–32 [DOI] [PubMed] [Google Scholar]
- 8.Pastra-Landis S. C., Lowey S. (1986) J. Biol. Chem. 261, 14811–14816 [PubMed] [Google Scholar]
- 9.Rottbauer W., Wessels G., Dahme T., Just S., Trano N., Hassel D., Burns C. G., Katus H. A., Fishman M. C. (2006) Circ. Res. 99, 323–331 [DOI] [PubMed] [Google Scholar]
- 10.Trybus K. M., Huiatt T. W., Lowey S. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 6151–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onishi H., Wakabayashi T. (1982) J. Biochem. 92, 871–879 [DOI] [PubMed] [Google Scholar]
- 12.Craig R., Smith R., Kendrick-Jones J. (1983) Nature 302, 436–439 [DOI] [PubMed] [Google Scholar]
- 13.Trybus K. M., Lowey S. (1984) J. Biol. Chem. 259, 8564–8571 [PubMed] [Google Scholar]
- 14.Cross R. A., Cross K. E., Sobieszek A. (1986) EMBO J. 5, 2637–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross R. A., Jackson A. P., Citi S., Kendrick-Jones J., Bagshaw C. R. (1988) J. Mol. Biol. 203, 173–181 [DOI] [PubMed] [Google Scholar]
- 16.Trybus K. M. (1994) J. Biol. Chem. 269, 20819–20822 [PubMed] [Google Scholar]
- 17.Cremo C. R., Sellers J. R., Facemyer K. C. (1995) J. Biol. Chem. 270, 2171–2175 [DOI] [PubMed] [Google Scholar]
- 18.Wendt T., Taylor D., Messier T., Trybus K. M., Taylor K. A. (1999) J. Cell Biol. 147, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendt T., Taylor D., Trybus K. M., Taylor K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4361–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez R., Freyzon Y., Trybus K. M., Cohen C. (1998) Cell 94, 559–571 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Wendt T., Taylor D., Taylor K. (2003) J. Mol. Biol. 329, 963–972 [DOI] [PubMed] [Google Scholar]
- 22.Burgess S. A., Yu S., Walker M. L., Hawkins R. J., Chalovich J. M., Knight P. J. (2007) J. Mol. Biol. 372, 1165–1178 [DOI] [PubMed] [Google Scholar]
- 23.Seow C. Y. (2005) Am. J. Physiol. Cell Physiol. 289, C1363–C1368 [DOI] [PubMed] [Google Scholar]
- 24.Crowther R. A., Padrón R., Craig R. (1985) J. Mol. Biol. 184, 429–439 [DOI] [PubMed] [Google Scholar]
- 25.Woodhead J. L., Zhao F. Q., Craig R., Egelman E. H., Alamo L., Padrón R. (2005) Nature 436, 1195–1199 [DOI] [PubMed] [Google Scholar]
- 26.Egelman E. H. (2000) Ultramicroscopy 85, 225–234 [DOI] [PubMed] [Google Scholar]
- 27.Zoghbi M. E., Woodhead J. L., Moss R. L., Craig R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2386–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Khayat H. A., Morris E. P., Kensler R. W., Squire J. M. (2008) J. Struct. Biol. 163, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung H. S., Komatsu S., Ikebe M., Craig R. (2008) Mol. Biol. Cell 19, 3234–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Khayat H. A., Morris E. P., Squire J. M. (2009) J. Struct. Biol. 166, 183–194 [DOI] [PubMed] [Google Scholar]
- 31.Zhao F. Q., Craig R., Woodhead J. L. (2009) J. Mol. Biol. 385, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung H. S., Burgess S. A., Billington N., Colegrave M., Patel H., Chalovich J. M., Chantler P. D., Knight P. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6022–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweeney H. L., Bowman B. F., Stull J. T. (1993) Am. J. Physiol. Cell Physiol. 264, C1085–C1095 [DOI] [PubMed] [Google Scholar]
- 34.Levine R. J., Kensler R. W., Yang Z., Stull J. T., Sweeney H. L. (1996) Biophys. J. 71, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trybus K. M., Freyzon Y., Faust L. Z., Sweeney H. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blankenfeldt W., Thomä N. H., Wray J. S., Gautel M., Schlichting I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17713–17717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Brown J. H., Reshetnikova L., Blazsek A., Farkas L., Nyitray L., Cohen C. (2003) Nature 424, 341–345 [DOI] [PubMed] [Google Scholar]
- 38.Lauzon A. M., Fagnant P. M., Warshaw D. M., Trybus K. M. (2001) Biophys. J. 80, 1900–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tama F., Feig M., Liu J., Brooks C. L., 3rd, Taylor K. A. (2005) J. Mol. Biol. 345, 837–854 [DOI] [PubMed] [Google Scholar]
- 40.Ikebe M., Ikebe R., Kamisoyama H., Reardon S., Schwonek J. P., Sanders C. R., 2nd, Matsuura M. (1994) J. Biol. Chem. 269, 28173–28180 [PubMed] [Google Scholar]
- 41.Nelson W. D., Blakely S. E., Nesmelov Y. E., Thomas D. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4000–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espinoza-Fonseca L. M., Kast D., Thomas D. D. (2007) Biophys. J. 93, 2083–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olney J. J., Sellers J. R., Cremo C. R. (1996) J. Biol. Chem. 271, 20375–20384 [DOI] [PubMed] [Google Scholar]
- 44.Wahlstrom J. L., Randall M. A., Jr., Lawson J. D., Lyons D. E., Siems W. F., Crouch G. J., Barr R., Facemyer K. C., Cremo C. R. (2003) J. Biol. Chem. 278, 5123–5131 [DOI] [PubMed] [Google Scholar]