Abstract
Stem cell biology offers advantages to investigators seeking to identify new therapeutic molecules. Specifically, stem cells are genetically stable, scalable for molecular screening, and function in cellular assays for drug efficacy and safety. A key hurdle for drug discoverers of central nervous system disease is a lack of high quality neuronal cells. In the central nervous system, α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate (AMPA) subtype glutamate receptors mediate the vast majority of excitatory neurotransmissions. Embryonic stem (ES) cell protocols were developed to differentiate into neuronal subtypes that express AMPA receptors and were pharmacologically responsive to standard compounds for AMPA potentiation. Therefore, we hypothesized that stem cell-derived neurons should be predictive in high-throughput screens (HTSs). Here, we describe a murine ES cell-based HTS of a 2.4 × 106 compound library, the identification of novel chemical “hits” for AMPA potentiation, structure function relationship of compounds and receptors, and validation of chemical leads in secondary assays using human ES cell-derived neurons. This reporting of murine ES cell derivatives being formatted to deliver HTS of greater than 106 compounds for a specific drug target conclusively demonstrates a new application for stem cells in drug discovery. In the future new molecular entities may be screened directly in human ES or induced pluripotent stem cell derivatives.
Keywords: Embryonic Stem Cell; Glutamate Receptors Ionotropic (AMPA, NMDA); Induced Pluripotent Stem (iPS) Cell; Neuron; Receptor Structure-Function; Stem Cell; HTS; Drug Discovery
Introduction
Cognitive impairment is a fundamental trademark of many CNS2 diseases such as Alzheimer disease, schizophrenia, and attention deficit hyperactivity disorder. Unfortunately few pharmaceutical agents have proven to be efficacious in treating cognitive impairment. As such, the medical need for new therapeutic approaches for improving cognitive deficits has emerged as an important frontier in treating central nervous system diseases. Glutamate mediates the vast majority of excitatory neurotransmission in the CNS with post-synaptic signaling regulated by either ligand-gated cation channels (ionotropic) or G-protein-coupled receptors (metabotropic). Ionotropic α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptors are tetrameric aggregates composed of combinations of one or more of four subunits (GluR1–4) that can each bind a glutamate neurotransmitter molecule. Upon binding of glutamate, the associated ion channel pore opens, allowing cations to cross the membrane and depolarize the postsynaptic cell. Termination of the postsynaptic signal is driven primarily by the removal of glutamate from the synaptic cleft, reducing the occupancy of the glutamate binding site and subsequent channel closure (deactivation). Additionally, the conduction pathway will close in the continued presence of bound agonist as a result of conformational changes secondary to glutamate binding (desensitization).
AMPA receptor pharmacology has identified agonists, competitive and non-competitive antagonists, and positive allosteric modulators (3). This latter class of pharmacologic agent, the positive allosteric modulator or AMPA potentiator, enhances agonist-evoked currents by slowing deactivation and/or desensitization but does not evoke a response when applied in the absence of agonist. These AMPA-potentiators demonstrate modulation of the excitatory post-synaptic currents in vitro and in vivo and are efficacious in animal cognition models such as novel object recognition (4) and radial arm maze (5, 6). AMPA potentiators are believed to increase cognition by enhancing synaptic transmission and plasticity and by stimulating release of neurotrophins (7).
The nootropic agents such as aniracetam and piracetam have been used clinically to treat cognitive impairment and have also been shown to enhance AMPA receptor function. However, the marginal efficacy of these first generation compounds has led to interest in developing agents that are more potent and selective enhancers of AMPA receptor function. Although several novel AMPA receptor potentiators meeting these criteria have advanced to clinical testing, none is yet approved, and there remains opportunity for new molecules of this general class (8–11).
Typically primary, transformed, or genetically engineered cell lines are used for developing cell-based assays in drug discovery. Stem cells offer distinct advantages because of their ability to differentiate into genetically and functionally normal cells (12). For AMPA receptors, this is particularly relevant due to the multitude of distinct AMPA receptor subtypes formed as mixtures of four subunits, each having flip and flop splice isoforms. Adding to this complexity are striking GluR subunit editing by double-stranded RNA, developmentally regulated alternative splicing, and transmembrane AMPA receptor regulatory proteins (TARPs) that modulate desensitization and pharmacology (1, 2).
From the perspective of developing a novel therapeutic agent, it is not clear that any one or more combinations of subunits and associated proteins is more or less desirable than the others. Therefore, mouse embryonic stem (mES) cells were differentiated into neuronal precursor cells that consistently expressed functional AMPA receptors. It was speculated that this approach would offer the potential to interrogate an array of AMPA receptors comprising different and possibly naturally occurring subunit combinations with the goal of casting a broad net in the HTS.
Herein we report the development and application of an mES cell-derived neuronal HTS that was used to assay more than 2.4 million small molecules. From this effort, small drug-like molecules were discovered and subjected to further evaluation to assess their functional and binding properties. In addition, selected molecules were tested in human ES cell-derived neurons to assess translation from mouse to human receptors. To our knowledge this is the first report of any stem cell-based HTS being employed to identify biologically active lead molecules for a pharmacologically relevant target. Importantly, this approach can potentially be used for other central nervous system drug targets that are not easily reconstituted using traditional expression systems.
EXPERIMENTAL PROCEDURES
Growth and Maintenance of ES Cells
The murine ES cell line used was E14-Sx1-16C, which has a targeted mutation in the Sox1 gene, a neuroectodermal marker that offers G418 resistance when the Sox1 gene is expressed (13). ES cells were maintained undifferentiated as previously described (14). The murine ES cells were grown in SCML medium that had a base medium of Knock-outTM DMEM (Invitrogen) supplemented with 15% ES-qualified fetal bovine serum (Invitrogen), 0.2 mm l-glutamine (Invitrogen), 0.1 mm minimum essential medium nonessential amino acids (Invitrogen), 30 μg/ml gentamicin (Invitrogen), 1000 μg/ml ESGRO (Chemicon), and 0.1 mm 2-mercaptoethanol (Sigma). ES cells were plated on gelatin-coated dishes (BD Biosciences) in SCML media that was changed daily, and the cells were dissociated with 0.05% trypsin-EDTA (Invitrogen) every other day.
Embryoid Body Formation
Before embryoid body (EB) formation, the ES cells were cultured in serum-free SCML media, replacing fetal bovine serum with knock-out serum replacement (KSR; Invitrogen). To form EBs, ES cells were dissociated into a single cell suspension, then 3 × 106 cells were plated in bacterial dishes (Nunc 4014) and grown as a suspension culture in 30 ml of NeuroEB-I medium that consisted of Knock-outTM DMEM (Invitrogen) supplemented with 10% KSR (Invitrogen), 0.2 mm l-glutamine (Invitrogen), 0.1 mm minimum essential medium nonessential amino acids (Invitrogen), 25 μg/ml gentamicin (Invitrogen), 1000 μg/ml ESGRO (Chemicon), 0.1 mm 2-mercaptoethanol (Sigma), and 150 ng/ml transferrin (Invitrogen). The plates were put on a Stovall Belly Button wave rocker in an atmospheric oxygen incubator with 6% CO2. The media was changed on day 2 of EB formation with NeuroEB-I and on day 4 with NeuroEB-II (NeuroEB-I plus 1 μg/ml mouse Noggin (R&D Systems)).
Neural Progenitor Selection and Expansion
On day 5 of EB formation, the EBs were dissociated with 0.05% trypsin-EDTA and replated at 4 × 106 cells/100 mm of laminin-coated tissue culture dishes in NeuroII-G418 medium that consisted of a base medium of a 1:1 mixture of Knock-outTM DMEM/F-12 supplemented with N2 supplements, 0.1 mm l-glutamine, and 25 μg/ml gentamicin, and NeuroBasal medium supplemented with B27 supplement, 0.1 mm l-glutamine, and 25 μg/ml gentamicin (all from Invitrogen). The base medium was supplemented with 10 ng/ml bovine FGF (Invitrogen), 1 μg/ml mouse Noggin, 500 ng/ml Sonic Hedgehog N terminus, 100 ng/ml FGF-8b (R&D Systems), 1 μg/ml laminin, and 200 μg/ml G418 (Invitrogen) for selection of neural progenitor cells expressing Sox-1. The plates were put in a humidified incubator with a gas mixture of 6% CO2 and 2% oxygen and were maintained under these conditions for 6 days. During this 6-day selection period, the NeuroII media were changed daily. On day 6, the surviving neural progenitor foci were dissociated with 0.05% trypsin EDTA, and the cells were plated at a density of 1.5 × 106 cells/100 mm of laminin-coated dish in NeuroII-G418 medium. The cells were dissociated every other day for expansion and prepared for cryopreservation at passage 3 or 4. The cryopreservation medium contained 50% KSR, 10% dimethyl sulfoxide (DMSO) (Sigma), and 40% NeuroII-G418 medium. Neural progenitors were cryopreserved at 4 × 106 cells/ml in 1 ml/cryovial, placed in a controlled rate freezer overnight, and transferred to an ultra-low freezer or liquid nitrogen for long term storage.
mES Cell-derived Neural Progenitor in Vitro Differentiation
Cryopreserved murine ES cell-derived neural progenitors were thawed by the rapid thaw method in a 37 °C water bath. The cells were transferred from the cryovial to three 100-mm laminin-coated tissue culture dishes that already contained NeuroII-G418 and equilibrated in a 2% oxygen incubator. The media was changed with fresh NeuroII-G418 the next day. The cells were dissociated every other day as described above for expansion. To differentiate the neural progenitor cells, the cells were dissociated as described above and resuspended in differentiation medium NeuroIII containing a 4:1 ratio of the NeuroBasal medium/B27:DMEM/F-12/N2 supplemented with 1 μm cAMP (Sigma), 200 μm ascorbic acid (Sigma), 1 μg/ml laminin (Invitrogen), and 10 ng/ml brain-derived neurotrophic factor (R&D Systems). The cells were plated at 2,000, 4,000, 6,000, 8,000, and cells/well in 384-well plates coated with poly d-lysine (BD Biosciences) to determine the optimal density for the screen. Cells were plated at a density of 5.0 × 104 cells in 24-well plates for immunocytochemistry and on glass coverslips for electrophysiology studies. The plates were incubated in 6% CO2 and 2% oxygen to complete the differentiation process for 0–12 days before characterization by immunocytochemistry.
Immunocytochemistry
Indirect immunocytochemistry was applied to neural precursors that were plated in 24-well laminin/poly d-lysine-coated plates as described. Cells were fixed in 4% paraformaldehyde and 3% sucrose in PBS for 15 min at room temperature on days 0, 3, 5, 7, 9, and 12 of differentiation. Fixative was removed, and cells were washed three times in PBS. Fixed cells from each time point were stored at 4 °C. When all time points were fixed, PBS was removed, and blocking agent was applied. Blocking was carried out overnight at room temperature in 5% bovine serum albumin (Roche Applied Science Fraction V Protease free catalogue #100 350), 5% normal donkey serum (Jackson ImmunoResearch Laboratories catalogue #017-000-121) in PBS that contained 0.1% Triton X (Sigma). Primary antibodies were incubated for 2 h in blocking solution. Antibodies used were rabbit anti-MAP2ab (1:500), mouse anti-TuJ1 (1:100), mouse anti-TH (1:500), rabbit anti-GluR1 (2 μg/ml), rabbit anti-GluR2/3 (2 μg/ml), rabbit anti-GluR4 (2 μg/ml), rabbit anti-ChAT (1:500), goat anti-ChAT (1:500), mouse anti-GFAP (1:700), mouse anti-synaptophysin (1:500), mouse anti-Nestin (1:300), rabbit anti-musashi (1:300), rabbit anti-VMAT2 (1:200), rabbit anti-DBH (1:500), mouse anti-RT97 (1:200), rabbit anti Trk A (1:300), rabbit anti Substance P (1:500) (all from Chemicon), rabbit anti-γ-aminobutyric acid (1:500), mouse anti MAP2ab (1:500), and rabbit anti-5HT (1:500) (all from Sigma), rabbit anti CGRP (1:200) (Bachem), rabbit anti-Iba1 (1:200) (Wako), biotin-IB4 (1:500) (Molecular Probes, Eugene, OR), and primary isotype controls mouse anti-IgG1 (1:300) and rabbit IgG (1:1000) (Southern Biotech) and mouse IgG2a (1:300) (Chemicon). After incubation of primary antibodies, cells were washed three times with blocking solution, then secondary antibodies were applied. Secondary antibodies and their dilution factors in blocking solution were Alexa 488 donkey anti-mouse, Alexa 488 donkey anti-rabbit, Alexa 594 donkey anti-rabbit, Alexa 594 donkey anti-goat, Alexa 594 donkey anti-mouse, and Avidi-Alexa 594. Nuclear counter stain was Hoechst 33342. The secondary antibodies and nuclear counter stain were incubated for 1 h, and cells were washed 3 times with blocking solution followed by filling the wells with PBS. Fixed and stained cells were analyzed on a Nikon Eclipse 300 with epifluorescence with quantitation using a Cellomics microarray scan.
Patch Clamp Electrophysiology of ES Cell-derived Neurons
Agonist-evoked whole-cell currents were recorded from mES cell-derived neurons or rat cortical neurons that were isolated from E15 rats and maintained in culture for up to 2 weeks. Cells were continuously superfused at room temperature with a solution containing 140 nm NaCl, 5 nm KCl, 2 nm CaCl2, 1 nm MgCl2, 10 nm HEPES, 10 nm glucose (pH 7.4 with NaOH). Patch pipettes were filled with an internal pipette solution containing 126 nm CsSO3CH3, 10 nm CsCl, 4 nm NaCl, 1 nm MgCl2, 0.5 nm CaCl2, 5 nm EDTA, 10 nm HEPES, 3 nm ATP-Mg, 0.3 nm GTP-Na, 4 nm phosphocreatine (pH 7.2 with CsOH), and resistances ranged from 3 to 6 megaohms. Analog signals were acquired with an Axopatch 200B amplifier, filtered at ⅕ the sampling frequency, digitized, stored, and measured using pCLAMP software (Molecular Devices), and data were expressed as mean ± S.E.
Fluorimetric Imaging Plate Reader (FLiPR) Assay Using Mouse ES Cell-derived Neurons
The Molecular Devices automated FLiPR assay for measuring intracellular Ca+ fluxes is a commonly used platform technology for screening agonists and antagonists of G-protein coupled receptors (15, 16). For the HTS, the adherent mES cell-derived neural cultures were plated into 384-well poly-d-lysine-coated tissue culture dishes (BD Biosciences) by the automated SelecT (The Automation Partnerships, Wilmington, DE) at a cell density of 6 × 103 cells/well in differentiation medium (see above). Before assay, the cells were washed and incubated in 4 μm Fluo 3 AM (Molecular Probes) dye-loading solution for 1 h at 37 °C. After loading with fluorescent dye, cells were washed in assay buffer to remove residual dye. Subsequently, the cells were placed in assay buffer with 1, 3, 10, 30, or 100 μm AMPA for determining the concentration of AMPA required to initiate the reaction for 30 min at room temperature. The plates were then placed on the FLiPR, and 10 μm solutions of the chemical library in DMSO were dispensed into the 384 wells through the FLiPR pipette head. For validation of the AMPA response before running the HTS assays, the cells were assayed with established low and high impact potentiators IDRA-21, (17), cyclothiazide (CTZ), (18) LY-451646, (19–21), CX-546, (22) CX-516, (23), and S-18986 (24, 25) after exposure to 30 μm AMPA. Fluorescence activity data were measured as relative fluorescence unit change over background per well after the addition of chemical ligands. All data are reported as % response and standardized to the response at 10 μm CTZ set at 100%.
Human ES Cell Culture and Neuronal Differentiation
The methods used to derive the human neural progenitor cells, hNPTM (ArunA Biomedical Inc., Athens GA), were described elsewhere (26). Briefly, WA09 hESCs were expanded on mouse embryonic fibroblast cells in media containing DMEM/F-12 medium (Invitrogen) was supplemented with 20% KSR (Invitrogen), 1× minimum Eagle's medium, and 4 ng/ml FGF2. MEF medium (DMEM (Invitrogen) supplemented with 20% fetal bovine serum (Hyclone), 1× minimum Eagle's medium, 10 ng/ml human leukemia inhibitory factor, 0.1 mm β-mercaptoethanol (Invitrogen), and 4 ng/ml FGF2), then harvested with trypsin and plated on polyornithine/laminin-coated (5 μg/ml) tissue culture dishes. The cells are cultured in defined hESC medium without FGF2 for 1 week followed by 1 week in derivation medium which is hESC growth medium lacking fetal bovine serum and KSR but supplemented with N2 (Invitrogen). Finally, the cells were cultured and expanded with proliferation medium for 1 week. NeuroBasal medium (Invitrogen) and B27 (1×) was supplemented with 20 ng/ml FGF2 (Sigma) and 10 ng/ml LIF (Chemicon). At about 10 days of derivation, neural rosettes (in vitro representation of neural tubes) started to appear and eventually covered the entire plate. These cells were expanded as NPTM cells and were then allowed to differentiate before performing FLiPR assays in differentiation media (NeuroBasal medium (Invitrogen) and B27 (1×) supplemented with 10 ng/ml leukemia inhibitory factor (Chemicon).
Human hN2TM Cells for FLiPR Assays
The hN2TM cells were plated into 96-well plates providing triplicate determinations at each dose of each test compound using Molecular Devices calcium 4 assay kit (R8142). After the loading incubation, after 1 h at 37 °C, 5% CO2, the plates were brought to room temperature for 0.5 h. Half-log dilutions were made of each test compound before the assay such that all dilutions had equal concentrations of DMSO (0.3%) then transferred to hN2TM cell dye-loaded plates. The control wells included CTZ 32 μm + AMPA 100 μm (agonist), CTZ + buffer, and AMPA + buffer. Test compounds and agonist challenge were added to the cell plates using the Flexstation 3 plate reader (Molecular Devices). Fluorescence activity data were measured as relative fluorescence units. Dose response curves were fit to a three parameter model with a Hill coefficient of 1 using prism software. Total area under the curve for responses was normalized such that the average response to 100 μm AMPA in the presence of 32 μm cyclothiazide was defined as 100%, and the average response to buffer + 3% DMSO was defined as 0 (this average was calculated on a per plate basis).
RESULTS
mES Neuronal Differentiation and Validation
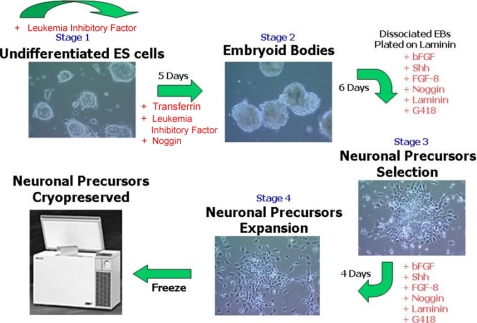
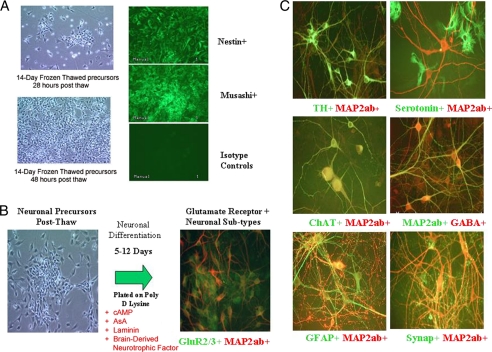
A robust and scalable method for neuronal differentiation of murine ES cells was developed that utilized a mES cell line with the targeted Sox 1 selection and inclusion of specific growth factors (Shh, bovine FGF, Noggin) in the differentiation media (Fig. 1). Of note, in the four-stage differentiation protocol is a 6-day selection in the growth factor mixture with G418 of Sox1-positive cells from dissociated embryoid bodies followed by further expansion of the neural progenitor cells for 4 days in selection and growth factor supplemented media. A single batch, 400–500 vials containing 4 × 106 cells/vial, of mES cell-derived neural progenitor cells was expanded and cryopreserved to ensure sufficient cellular substrate to deliver a HTS of >106 compounds. The neural progenitor cells when thawed maintained expansion ability and expression for neuronal precursor cell specific markers, Musashi and Nestin (Fig. 2A). Differentiation into more terminally differentiated neural subtypes was directed in NeuroIII media supplemented with cAMP, ascorbic acid, laminin, and brain-derived neurotrophic factor as described. Immunocytochemistry and functional studies were performed on days 0, 3, 5, 7, 9, and 12 post-plating. Importantly, upon differentiation neuronal subtypes expressing the glutamate receptor targets were identified (Fig. 2B). The differentiation protocol demonstrated various morphologic and immunocytochemistry positive neural subtype-specific markers, including TH, serotonin, GFAP, ChAT, γ-aminobutyric acid, synaptophysin, and glutamate receptors (Fig. 2C).
FIGURE 1.
Illustration of the derivation of mouse ES cell-derived neuron differentiation and expansion protocol for Sox1-selected neuronal precursors that allowed scaled growth of glutamate receptor positive neuronal cells for high throughput screening. The concentrations of the factors used at the various steps of the differentiation protocol are found under “Experimental Procedures.” bFGF, bovine fibroblast growth factor.
FIGURE 2.
Validation of mES-derived neuronal precursor and neuronal phenotypes. A, demonstration of neuronal precursor cells phenotype and expansion post-thawing is shown. Cells demonstrated expansion as evidenced by increased cell concentration 18 and 48 h after thaw. The expanded cells maintained expression of neuronal precursor markers Nestin and Musashi when compared with isotype controls. B, the mouse ES cell-derived neurons express glutamate receptors, the target for the AMPA potentiation HTS after neuronal differentiation of banked neuronal precursors cells. C, the differentiation protocol results in the differentiation of pan-neuronal subtypes as indicated by immunocytochemistry for dopaminergic, serotonergic, GABAergic, and astrocyte markers.
mES-derived Neuronal Cell-based Assay for AMPA Potentiation
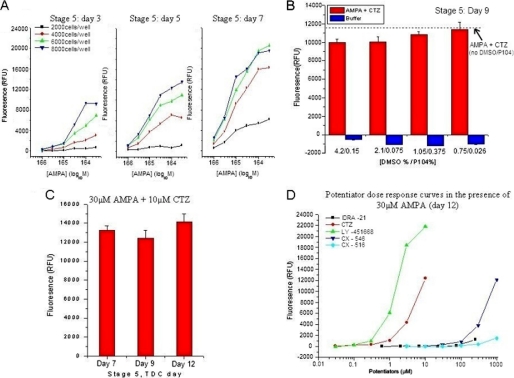
The mES cell neuronal in vitro differentiation and reproducible expression of neuronal cellular markers led us to assess for functional AMPA receptors using a Ca2+ flux assay that could be automated for HTS. It was found that the addition of 100 μm AMPA when added alone did not evoke a measurable increase in fluorescence using our assay conditions, presumably due to insufficient Ca2+ flux. However, robust Ca2+ flux was routinely evoked by the same concentration of AMPA after a short (∼2 min) pretreatment with the AMPA potentiator, CTZ (EC50 = 7.6 ± 0.4 μm). The response to AMPA in the presence of CTZ was completely abolished by the AMPA receptor antagonist 6-cyano-7-nitroquinoxaline-2,3-dione, suggesting the presence of functional AMPA receptors in the cell population. The assay conditions and responses AMPA in the presence of 10 μm CTZ was optimized in 384-well plates with respect to cell density and duration after plating. A maximal assay response was obtained with 6000 cells per well, and the signal response was reproducibly observed from day 7 to day 12 post-plating, providing a 5-day assay window (Fig. 3). Accordingly, the mES-derived neurons were assayed with established small molecule standards for signal potentiation by 30 μm AMPA. Dose responses curves were obtained for known AMPA receptor potentiators with a range of potency and intrinsic efficacy (CTZ, CX-546, and IDRA-21) in the stem cell-derived neurons (Fig. 3D). These results indicate that the cells are capable of identifying active compounds with similar rank order potency as compared with literature reports using either primary neurons or expressed receptor systems (27, 28). As a result, all novel chemical leads identified in this assay were subjected to additional biological characterization.
FIGURE 3.
Optimization and validation of mouse ES cell-derived neuronal for AMPA potentiation HTS. A, cell density optimization using pan-neuro II media is shown. Comparison of AMPA responses in with 2,000, 4,000, 6,000, and 8,000 cells/well in the presence of 10 μm cyclothiazide resulted in optimal responsiveness at 6,000 cells/well across screening time window. B, mouse ES cell-derived neurons demonstrate good responsiveness and tolerance to chemical library preservatives P104/DMSO at varying concentrations. Analysis was done using stage 5 cells at 9 days post-thawing. C, mouse ES cell-derived neurons demonstrate no significant difference at concentrations of 30 μm AMPA + 10 μm CTZ in signal size from day 7–12 post plating. D, potentiator dose-response curves in the presence of 30 μm AMPA in post-thaw day 12 neurons are shown. Mouse ES cell-derived neurons demonstrated a dose-response potency in the same rank order as was observed in rat primary neuronal cells with established standards for AMPA potentiation. Data are from McNeish (47) with permission. RFU, relative fluorescence units; TDC, time differentiation culture.
mES Differentiation for HTS
To format the AMPA screen in a 384-well plate format, neural progenitor cells were dissociated and pooled into one cell suspension that was loaded into a flask for the SelecT automated robotic plating (described under “Experimental Procedures”). The neural progenitor cells were plated into 384-well poly d-lysine-coated dishes and transferred into the low oxygen incubator for the differentiation process. Using established standards for AMPA potentiation, the best neural population for HTS was determined. Pan-neural population of cells provided a distribution of multiple neural subtypes (GABAergic, dopaminergic, glutamatergic, serotonergic, cholinergic, etc.) as well as the necessary support cells such as astrocytes and glial cells (Fig. 2C). It should be noted that in every differentiated population of neural cells, some neural progenitor cells remain as evidenced by Nestin and Musashi-positive cells (not shown). Although slight performance differences between batches of mES cell-derived neural progenitors were observed, the differentiation of the progenitor cells and the functional output of the differentiated neural population within a single batch were consistent and reproducible.
HTS Results and Medicinal Chemistry/Library Generation
A HTS of the Pfizer collection of ∼2.4 million molecules was conducted using the mES cell-derived neuronal cells. Compounds were identified as screening hits by meeting the following two criteria; 1) they did not evoke a calcium signal in the absence of AMPA, and 2) they enhanced activity of a subsequent challenge of AMPA to at least 10% that achieved by 10 μm CTZ. The median Z′ (a statistical measure of the suitability of the assay for the HTS setting) for the overall HTS was 0.57, indicating a highly predictive assay. However, even more compelling were the percent effect statistics for the 2.4 million compounds, where percent effect is calculated based upon no compound and 100% effect compound-positive control wells using 10 μm CTZ that was included on each plate. The overall standard deviation was 3.64% with a mean of 0.46. Thus, three times the standard deviation plus the mean yielded ∼11%, which guided our hit threshold criteria to be set at ≥10% effect with a significant p value for the primary HTS.
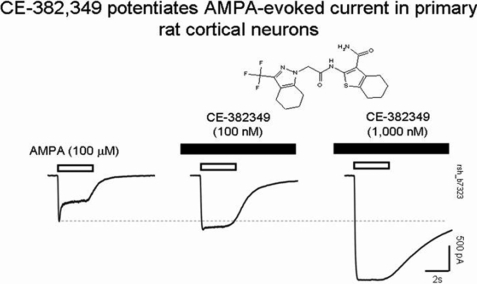
Of the 5911 screening hits, 37 were confirmed as positive hits upon retesting in triplicate at 32 μm using a cutoff of ≥30% of the CTZ response upon a challenge with AMPA but no signal before the addition of AMPA. Finally, 7 compounds were found to have EC50 values <10 μm in follow-up concentration-response testing. These 7 compounds represented 2 novel molecular series as well as known AMPA potentiator chemical space. One of these novel active molecules, compound CE-382349, contains both trifluoromethylpyrazole and thiophenecarboxamide motifs, which are linked via an amide spacer group. In the confirmation screen run in triplicate, CE-382349 showed 108% effect at 32.0 μm standardized to the response at 10 μm CTZ (set at 100%), with an EC50 between 9.9 and 13.3 μm. Recently, compounds containing this general structure were disclosed as having similar activity by Organon Laboratories Ltd (29). Functional activity of CE-382349 as a potentiator of AMPA receptors was verified in an electrophysiology assay using primary cultures of rat hippocampal neurons in a patch clamp assay (Fig. 4). The addition of CE-382349 in the absence of AMPA produced no detectable change in current. However, peak AMPA-evoked currents were increased by ∼44% (n = 3) and 163% (n = 2) in the presence of 100 and 1000 nm CE-382349, respectively. Notably, the addition of CE-382349 inhibited the fraction of current that desensitized during AMPA challenge, an activity similar to that report for other AMPA potentiators such as cyclothiazide and biarylpropylsulfonamides.
FIGURE 4.
Potentiation of AMPA-evoked responses by CE-382,349. Whole-cell currents were recorded from primary cultures of rat cortical neurons. Responses were evoked by 100 μm AMPA applied for 2 s once every 90 s either in the absence (left) or presence of either 100 nm (middle) or 1000 nm (right) CE-382349. Note that CE-382349 increased the peak inward current and slowed the current decay during the agonist challenge.
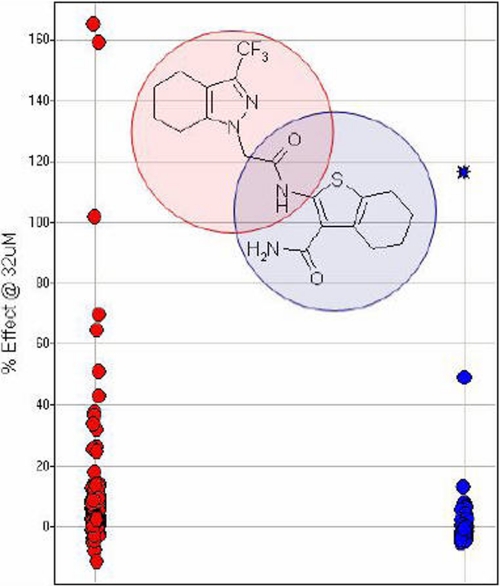
To assess the ability of this assay to detect structurally similar and novel compounds, structural features of CE-382349 that are important to its AMPA activity were assessed by individually varying the two heterocyclic motifs of the molecule. These analogs were screened at a single high concentration of 32 μm for their ability to potentiate AMPA-evoked responses in the Ca2+ flux assay. The structure activity relationship of these analogs suggests that varying the thiophenecarboxamide unit may represent the more promising avenue for identifying AMPA potentiators with enhanced efficacy (Fig. 5), and these results verify the robustness of this assay to detect and differentiate novel compounds. This conclusion was further supported by disclosures from GlaxoSmithKline claiming trifluoromethylpyrazoles as having AMPA potentiator activity (30–35).
FIGURE 5.
Hit follow-up library modification of CE-382,349. The red circles represent compounds from a library holding the pyrazole fragment constant; the blue circles are from a library holding the thiophene fragment constant. CE-382,349 is represented as a blue star. All the data are represented as % effect at 32 μm CE-382,349 versus the effect observed with the AMPA potentiator standard, 10 μm cyclothiazide.
Validation of New AMPA Potentiators in Human ES Cell-derived Neurons
One concern with using mouse cells in primary screening is that the output from the HTS would not adequately translate to native human receptors. Although it could not be expected that AMPA receptors of cells derived from mouse and human would have identical pharmacology, it was important to understand the extent of the difference. Therefore, the WA09 human ES cell line was differentiated into neuroepithelial cells that expressed positive cellular markers and demonstrated positive AMPA and CTZ responses in cellular assays (Fig. 6). A selection of 21 compounds including literature standards and proprietary compounds were evaluated in a Ca2+ fluorescence-based assay using neuronal differentiated cells derived from human ES cells (see “Experimental Procedures”). Using identical assay conditions, 17 of 21 compounds yielded similar results either having EC50 values less than 4-fold different or EC50 values greater than the highest tested concentration of 32 μm in both assays based on a single determination from the human-derived neuronal cells (Table 1). Although the exact reason for the difference in potency in the remaining 4 compounds is not known, only two of those, or about 10%, were greater than 20-fold different between the two cell-based systems. The overall concordance between the mouse and human ES cell-based assays was deemed sufficiently high to support follow-up medicinal chemistry optimization approaches using cultures derived from mouse ES cells.
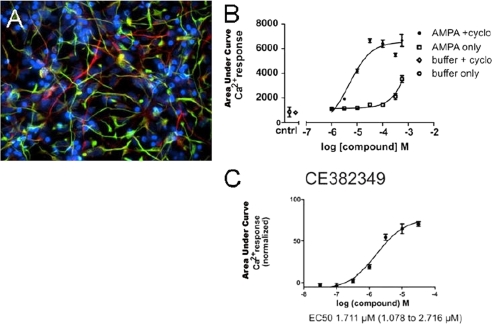
FIGURE 6.
Human ES cell-derived neuronal (hN2TM) cells used in the FLiPR assay demonstrate AMPA potentiation with CE-382,349. Results are shown two weeks after placing NP cells in differentiation media that lacks FGF2. A, ≈70% of hN2 TM cells are MAP2-positive (red), and nestin was also still present (green). B, hN2 TM cells were responsive to AMPA, and CTZ potentiated the AMPA response in hN2 TM cells. cntrl, control. C, CE-382,349 demonstrates AMPA potentiation in hN2 TM cells with an EC50 of ∼1.7 μm. cyclo, cyclothiazide.
TABLE 1.
Functional potency of 21 compounds determined using neuronal cells derived from human and mouse ES cells
Compounds are a mixture of publicly disclosed and proprietary chemical matter. Potencies from human ES cell-derived neurons were determined from a single assay where each concentration was run in triplicate. NSD, no signal detected up to 32 μm.
| Compound | hES cells (EC50) | mES cells |
-Fold difference (EC50/EC50) | ||
|---|---|---|---|---|---|
| Mean (EC50) | S.E. | n | |||
| μm | μm | μm | |||
| PF-4366288 | 0.005 | 0.54 | 0.19 | 9 | 110 |
| PF-4540725 | 0.02 | 2.7 | 0.64 | 4 | 164 |
| PF-4531686 | 0.14 | 2.6 | 0.80 | 5 | 19 |
| PF-4489045 | 0.14 | 0.22 | 0.24 | 12 | 2 |
| PF-4440438 | 0.32 | 3.8 | 0.81 | 3 | 12 |
| PNU-173092 | 0.54 | 0.29 | 1 | 2 | |
| PF-4353782 | 0.77 | 3.1 | 0.72 | 5 | 4 |
| PF-4713786 | 1.1 | 1.2 | 0.24 | 2 | 1 |
| CE-382349 | 1.7 | 6.8 | 3.6 | 3 | 4 |
| PNU-172723 | 1.7 | 1.5 | 1 | 1 | |
| LY-451646 | 1.9 | 4.1 | 0.69 | 14 | 2 |
| PF-4401962 | 2.2 | 0.57 | 1 | 4 | |
| PF-4595320 | 2.3 | 3.1 | 1 | 1 | |
| PF-4599919 | 6.9 | 20 | 5.6 | 3 | 3 |
| PF-4603900 | 6.9 | 3.6 | 1 | 2 | |
| Cyclothiazide | 16 | 7.6 | 0.40 | 94 | 2 |
| PF-4713783 | NSD | NSD | 2 | ||
| PF-2326666 | NSD | NSD | 2 | ||
| S18986 | NSD | NSD | 3 | ||
| CX516 | NSD | NSD | 2 | ||
| PF-4572523 | NSD | NSD | 3 | ||
DISCUSSION
The overriding goal of drug discovery is to rapidly identify novel compounds with the potential for superior clinical efficacy and safety. Typically, in vitro and in vivo models are identified and optimized to ensure that targets are validated, lead molecules are rapidly identified, and potential candidates are selected for characterization in human clinical trials. Stem cells have a more innate cellular background than typical exogenous expression systems that may provide a more predictive cell-based screening strategy for small molecule lead identification. Herein, we have described a mouse ES cell-based approach to primary screening that resulted in the identification of novel small molecule AMPA-type glutamate receptor potentiators. Although AMPA potentiators are of interest in enhancing cognition for several CNS disorders, this stem cell-based screening strategy could be used in the identification of substrates for other CNS targets. Currently, artificially engineered cells or primary cells from fetal rat brains are used for CNS assays, and although many successful cell-based HTS have been developed, many have not been optimal. In the case of G-protein-coupled receptors, for instance, a number of important variables may exist with these systems including stoichiometry of the target receptors versus the endogenous cell populations and proper signaling within the engineered cells. The mES cell-derived neurons provide a novel alternative for identifying AMPA potentiators that exploits endogenously expressed glutamate receptors that potentially represent the range and stoichiometry of subunit combinations and splice isoforms. The HTS identified two novel chemical structures with positive AMPA activity that were confirmed in secondary assays and in human ES cell-derived neuron FLiPR assay. Further evaluation of these newly identified structures and related analogs as leads in the development of new therapies for cognition enhancement continues and will be reported in due course.
One concern of stem cell based screen was that the cells would express multiple receptors and signaling pathways that could elicit a Ca2+ signal in the FLiPR assay. This could produce an unmanageable number of false positives in a single addition assay. However, the criteria used for selecting compounds that were inactive when added alone, but demonstrated enhanced Ca2+ flux when evoked by AMPA, appeared to eliminate the vast majority of compounds that acted as either direct agonist of AMPA receptors or on receptors that also couple to Ca2+. Therefore, stem cell-based screening assays are likely to be useful for target mechanisms that can be isolated from other systems in the stem cell derivatives, such as the case of allosteric modulators. Advances in automation and miniaturization have resulted in HTS of chemical libraries, often exceeding a million individual molecules, and is the major lead-generating technology employed in the pharmaceutical industry. In this work we coupled the advantages of stem cell biology and HTS to identify new lead molecules in an important area of unmet medical need. Specifically, a murine embryonic stem cell-derived assay has been developed to screen a compound file containing more than two million compounds. Particularly noteworthy is the fact that heretofore no stem cell-based assay has been employed in a HTS on the order of 106 compounds or more. The work described here was accomplished using mouse ES cells; however, recent advances suggest that hES cell-derived neurons can be reproducibly scaled and quality-controlled during differentiation to consider moving directly to primary HTS in human cells (26). If possible, this may eliminate translational differences between murine and human cellular generated data, as was observed in this study where 4/21 hits showed a 10× affinity difference between mouse and human cells. Recently, investigators have reported moving directly to human ES cells to enable phenotypic screening of thousands of compounds (36–38).
In addition, researchers have demonstrated the ability to reprogram somatic cells that have properties similar to embryonic cells. These induced pluripotent stem (iPS) cells were first described in murine cells (39) and were followed by the derivation of human iPS cell lines (40, 41). Human iPS cells have been achieved for known genetic diseases (42). Like ES cells, iPS cells have been differentiated into clinically relevant cells types including neurons (43), cardiomyocytes (44), hepatocytes (45), and insulin-producing cells (46). Accordingly, iPS cells may soon deliver a range of disease specific stem cells for primary HTS and secondary screening of validated targets.
In summary, this report demonstrates a breakthrough in drug discovery technology that applies stem cell biology with high throughput screening of very large chemical collections or libraries. It is expected that stem cells will have an expanded role in future HTS in the biopharmaceutical industry. Also, we predict that in the near future the extension of the work described here will be applied to human ES/iPS stem cells leading to further efficiencies in drug discovery and the delivery of safer and effective new medicines.
Acknowledgments
We acknowledge the excellent technical support of Pfizer biologists and chemists including Bill Blake, Keith Haskell, Linyan He, Mary Bauchmann, Steve Hawrylik, Jim Duerr, Justin Oborski, Chet Siok, Nandini Patel, and Deane Nason and from Kate Hodges of Aruna Biomedical.
Footnotes
- CNS
- central nervous system
- AMPA
- α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate
- HTS
- high-throughput screen(ing)
- ES
- embryonic stem
- mES
- mouse ES
- iPS
- induced pluripotent stem
- FLiPR
- fluorimetric imaging plate reader
- CTZ
- cyclothiazide
- DMEM
- Dulbecco's modified Eagle's medium
- EB
- embryoid body
- KSR
- knock-out serum replacement
- GABAergic
- aminobutyric acid-ergic
- FGF
- fibroblast growth factor
- PBS
- phosphate-buffered saline
- TH
- tyrosine hydroxylase
- GFAP
- glial fibrillary acidic protein
- ChAT
- cholinesterol acelyltransferase
- GABA
- γ-aminobutyric acid.
REFERENCES
- 1.Schoft V. K., Schopoff S., Jantsch M. F. (2007) Nucleic Acids Res. 35, 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato A. S., Siuda E. R., Nisenbaum E. S., Bredt D. S. (2008) Neuron 59, 986–996 [DOI] [PubMed] [Google Scholar]
- 3.Kew J. N., Kemp J. A. (2005) Psychopharmacology 179, 4–29 [DOI] [PubMed] [Google Scholar]
- 4.Silvers J. M., Harrod S. B., Mactutus C. F., Booze R. M. (2007) J. Neurosci. Methods 166, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olton D. S. (1987) Physiol. Behav. 40, 793–797 [DOI] [PubMed] [Google Scholar]
- 6.Levin E. D. (1988) Neurosci. Biobehav. Rev. 12, 169–175 [DOI] [PubMed] [Google Scholar]
- 7.Lynch G. (2006) Curr. Opin. Pharmacol. 6, 82–88 [DOI] [PubMed] [Google Scholar]
- 8.Lynch G. (2004) Curr. Opin. Pharmacol. 4, 4–11 [DOI] [PubMed] [Google Scholar]
- 9.Black M. D. (2005) Psychopharmacology 179, 154–163 [DOI] [PubMed] [Google Scholar]
- 10.Francotte P., de Tullio P., Fraikin P., Counerotte S., Goffin E., Pirotte B. (2006) Recent Pat. CNS Drug Discov. 1, 239–246 [DOI] [PubMed] [Google Scholar]
- 11.O'Neill M. J., Dix S. (2007) IDrugs 10, 185–192 [PubMed] [Google Scholar]
- 12.McNeish J. (2004) Nat. Rev. Drug Discov. 3, 70–80 [DOI] [PubMed] [Google Scholar]
- 13.Lang R. J., Haynes J. M., Kelly J., Johnson J., Greenhalgh J., O'brien C., Mulholland E. M., Baker L., Munsie M., Pouton C. W. (2004) Eur. J. Neurosci. 20, 3209–3221 [DOI] [PubMed] [Google Scholar]
- 14.Roach M. L., McNeish J. D. (2002) Methods Mol. Biol. 185, 1–16 [DOI] [PubMed] [Google Scholar]
- 15.Robas N. M., Fidock M. D. (2005) Methods Mol. Biol. 306, 17–26 [DOI] [PubMed] [Google Scholar]
- 16.Miret J. J., Zhang J., Min H., Lewis K., Roth M., Charlton M., Bauer P. H. (2005) J. Biomol. Screen 10, 780–787 [DOI] [PubMed] [Google Scholar]
- 17.Zivkovic I., Thompson D. M., Bertolino M., Uzunov D., DiBella M., Costa E., Guidotti A. (1995) J. Pharmacol. Exp. Ther. 272, 300–309 [PubMed] [Google Scholar]
- 18.Donevan S. D., Rogawski M. A. (1998) Neuroscience 87, 615–629 [DOI] [PubMed] [Google Scholar]
- 19.Mackowiak M., O'Neill M. J., Hicks C. A., Bleakman D., Skolnick P. (2002) Neuropharmacology 43, 1–10 [DOI] [PubMed] [Google Scholar]
- 20.Rorick-Kehn L. M., Hart J. C., McKinzie D. L. (2005) Psychopharmacology 183, 226–240 [DOI] [PubMed] [Google Scholar]
- 21.Bai F., Bergeron M., Nelson D. L. (2003) Neuropharmacology 44, 1013–1021 [DOI] [PubMed] [Google Scholar]
- 22.Lynch G., Granger R., Ambros-Ingerson J., Davis C. M., Kessler M., Schehr R. (1997) Exp. Neurol. 145, 89–92 [DOI] [PubMed] [Google Scholar]
- 23.Lauterborn J. C., Lynch G., Vanderklish P., Arai A., Gall C. M. (2000) J. Neurosci. 20, 8–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desos P., Lepagnol J. M., Moran P., Lestage P., Cordi A. A. (1996) J. Med. Chem. 39, 197–206 [DOI] [PubMed] [Google Scholar]
- 25.Lebrun C., Pillière E., Lestage P. (2000) Eur. J. Pharmacol. 401, 205–212 [DOI] [PubMed] [Google Scholar]
- 26.Shin S., Mitalipova M., Noggle S., Tibbitts D., Venable A., Rao R., Stice S. L. (2006) Stem Cells 24, 125–138 [DOI] [PubMed] [Google Scholar]
- 27.Miu P., Jarvie K. R., Radhakrishnan V., Gates M. R., Ogden A., Ornstein P. L., Zarrinmayeh H., Ho K., Peters D., Grabell J., Gupta A., Zimmerman D. M., Bleakman D. (2001) Neuropharmacology 40, 976–983 [DOI] [PubMed] [Google Scholar]
- 28.Baumbarger P. J., Muhlhauser M., Zhai J., Yang C. R., Nisenbaum E. S. (2001) J. Pharmacol. Exp. Ther. 298, 86–102 [PubMed] [Google Scholar]
- 29.Gillen K. J., Jamieson C., Maclean J. K. F., Moir E. M., Rankovic Z. (2008) PCT International Appl. WO2008/003452 A1 [Google Scholar]
- 30.Bradley D. M., Chan W. N., Ward S. E. (2008) PCT International Appl. WO2008/148836 A1 [Google Scholar]
- 31.Bradley D. M., Thewlis K. M., Ward S. E. (2008) PCT International Appl. WO2008/148832 A1 [Google Scholar]
- 32.Bertheleme N., Bradley D. M., Cardullo F., Merlo G., Pozzan A., Scott J. S., Thewlis K. M., Ward S. E. (2008) PCT International Appl. WO 2008/113795 A1 [Google Scholar]
- 33.Bradley D. M., Chan W. N., Harrison S. A., Thewlis K. M., Ward S. E. (2008) PCT International Appl. WO 2008/110566 [Google Scholar]
- 34.Bradley D. M., Chan W. N., Thewlis K. M., Ward S. E. (2008) PCT International Appl. WO 2008/053031 A1 [Google Scholar]
- 35.Bradley D. M., Chan W. N., Harrison S., Thatcher R., Thewlis K. M., Ward S. E. (2007) PCT International Appl. WO 2007/107539 A1 [Google Scholar]
- 36.Desbordes S. C., Placantonakis D. G., Ciro A., Socci N. D., Lee G., Djaballah H., Studer L. (2008) Cell Stem Cell 2, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borowiak M., Maehr R., Chen S., Chen A. E., Tang W., Fox J. L., Schreiber S. L., Melton D. A. (2009) Cell Stem Cell 4, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S., Borowiak M., Fox J. L., Maehr R., Osafune K., Davidow L., Lam K., Peng L. F., Schreiber S. L., Rubin L. L., Melton D. (2009) Nat. Chem. Biol. 5, 258–265 [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 40.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 41.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 42.Park I. H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M. W., Cowan C., Hochedlinger K., Daley G. Q. (2008) Cell 134, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L. (2009) Nat. Biotechnol. 27, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Wilson G. F., Soerens A. G., Koonce C. H., Yu J., Palecek S. P., Thomson J. A., Kamp T. J. (2009) Circ. Res. 104, e30–e41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Z., Cai J., Liu Y., Zhao D., Yong J., Duo S., Song X., Guo Y., Zhao Y., Qin H., Yin X., Wu C., Che J., Lu S., Ding M., Deng H. (2009) Cell Res. 19, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 46.Zhang D., Jiang W., Liu M., Sui X., Yin X., Chen S., Shi Y., Deng H. (2009) Cell Res. 19, 429–438 [DOI] [PubMed] [Google Scholar]
- 47.McNeish J. D. (2007) Curr. Opin. Pharmacol. 7, 515–520 [DOI] [PubMed] [Google Scholar]