Abstract
Katanin p60 (kp60), a microtubule-severing enzyme, plays a key role in cytoskeletal reorganization during various cellular events in an ATP-dependent manner. We show that a single domain isolated from the N terminus of mouse katanin p60 (kp60-NTD) binds to tubulin. The solution structure of kp60-NTD was determined by NMR. Although their sequence similarities were as low as 20%, the structure of kp60-NTD revealed a striking similarity to those of the microtubule interacting and trafficking (MIT) domains, which adopt anti-parallel three-stranded helix bundle. In particular, the arrangement of helices 2 and 3 is well conserved between kp60-NTD and the MIT domain from Vps4, which is a homologous protein that promotes disassembly of the endosomal sorting complexes required for transport III membrane skeleton complex. Mutation studies revealed that the positively charged surface formed by helices 2 and 3 binds tubulin. This binding mode resembles the interaction between the MIT domain of Vps4 and Vps2/CHMP1a, a component of endosomal sorting complexes required for transport III. Our results show that both the molecular architecture and the binding modes are conserved between two AAA-ATPases, kp60 and Vps4. A common mechanism is evolutionarily conserved between two distinct cellular events, one that drives microtubule severing and the other involving membrane skeletal reorganization.
Keywords: ATPases, Cytoskeleton, Endosomes, Membrane Trafficking, Microtubules
Introduction
Microtubules (MTs)2 are polymers of α- and β-tubulin heterodimers. MTs exist as networks that dynamically and rapidly reorganize during different phases of the cell cycle. Spontaneous growth as well as shortening at the ends is indispensable for functional rearrangement. For example, they form the mitotic spindle during M phase, which mediates chromosome segregation during cell division based on the nature of dynamic rearrangement of MTs (reviewed in Refs. 1, 2). Many cellular events involving MTs are driven not only by autonomous polymerization and dissociation of tubulin but also by MT-severing enzymes. These enzymes disassemble the MTs to promote large changes in the cytoskeleton in an ATP-dependent manner (3).
There are three known MT-severing enzymes, katanin, spastin, and fidgetin, all of which belong to type I AAA-ATPases (4–7). Katanin was first identified from sea urchin cytosol (8) and consists of two subunits as follows: a 60-kDa catalytic subunit (kp60) containing a single AAA domain, and an 80-kDa regulatory subunit (kp80) (9, 10). Both the subunits are genetically conserved among many higher eukaryotes. Katanin localizes at the centrosomes in an MT-dependent manner (11), which is probably required for recycling and for the poleward flux of tubulin in the spindle by disassembling MTs at their minus ends (12, 13). kp60 homologs are also found in plants, insects, and nematodes but not in yeasts.
kp60 has a common domain organization typical of a type I AAA-ATPase, which consists of an N-terminal substrate binding region followed by a single AAA domain at the C terminus. In general, AAA-ATPases are believed to act as protein unfoldases that promote various cellular events, including dissociation of protein complexes, MT severing, protein degradation, protein translocation across organelle membranes, vesicle fusions, and multivesicular body formation (reviewed in Refs. 14, 15).
Hartman and Vale (13) demonstrated that the N-terminal half of kp60 contains an MT binding region, although the presence of a structural MT binding domain was not proved. The importance of the N-terminal MT binding region of a plant kp60 ortholog has been recently reported (16). In our previous study, we successfully isolated a folded structural domain from the kp60 N-terminal region (termed kp60-NTD) (17). Although standard bioinformatics tools (e.g. PSI-BLAST (18), Pfam (19, 20), and SMART (21)) failed to detect any similarity between kp60-NTD and other known domains, more sensitive bioinformatics techniques (e.g. FORTE (22) and FUGUE (23)) can detect substantial similarities between kp60-NTD and MIT domains. MIT domains are small helical domains involved in protein-protein interactions that are conserved among Vps4, spartin, spastin, and some other proteins (24).
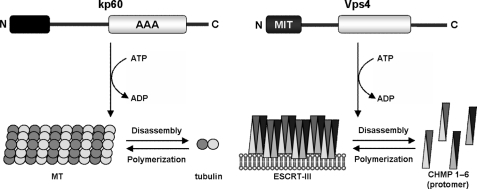
In this study, we present the solution structure of kp60-NTD. We show that this structure is closely related to that of the MIT domain. In this context, the overall molecular architecture of kp60 resembles other MIT domain-containing type I AAA-ATPases, such as the MT-severing enzyme spastin and the ESCRT-III disassembling enzyme Vps4 (Fig. 1A). Because the isolated kp60-NTD solely binds tubulin in vitro, the domain is a novel tubulin binding domain. Finally, the key residues of kp60-NTD for binding tubulin were determined. A model for MT binding is further discussed, which allows us to propose a model for the mechanism of MT severing by katanin.
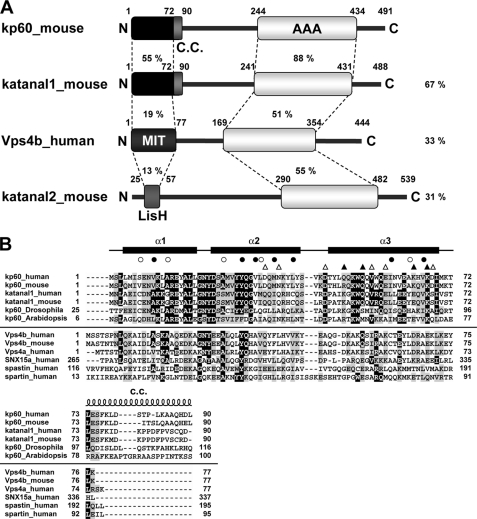
FIGURE 1.
Domain architectures and multiple sequence alignment of kp60s and proteins containing MIT domains. A, domain architectures of mouse kp60, katanal1 and -2, and human Vps4b. The amino acid identities of each domain and full-length proteins between kp60 and other proteins are indicated. C.C., coiled-coil; MIT, MIT domain; LisH, LIS1 homology domain; AAA, AAA domain. B, multiple sequence alignment of kp60-NTDs and related proteins with secondary structure elements of kp60-NTD. The secondary structure elements are shown at the top of the figure. The α-helices (α1–3) are represented as thick lines and the C.C. region as a coil. Filled and open circles above the alignments indicate well conserved and less conserved core residues, respectively (see Fig. 2A). Triangles indicate residues substituted with Ala for examining tubulin binding. (Filled triangle, involved in tubulin binding; open triangle, not involved.) Protein names and UniProtKB accession numbers are as follows: kp60 human (O75449); kp60 mouse (Q9WV86); kp60 Drosophila (Q9VN89); kp60 Arabidopsis (Q9SEX2); katanal1 human (Q9BW62); katanal1 mouse (Q8K0T4); Vps4b human (O75351); Vps4b mouse (P46467); Vps4a human (Q9UN37); SNX15a human (Q9NRS6); spartin human (Q8N0X7); and spastin human (Q9UBP0). The sequence alignment was generated by ClustalX (62).
EXPERIMENTAL PROCEDURES
Protein Techniques
Expression vectors for the recombinant GST-tagged form of kp60-NTDs of human and mouse were constructed using PRESAT vector methodology (17, 25). The fusion proteins were expressed in Escherichia coli BL21 (DE3), followed by affinity purification on glutathione-Sepharose (GE Healthcare), and were dialyzed. These fusion proteins were used for tubulin binding assays. For NMR spectroscopy, 2 liters of culture was incubated with [15N]ammonium chloride and [13C]glucose as the sole nitrogen and carbon sources, respectively, following a standard fermentation protocol at 25 °C. Divalent cation was present as a trace mineral during fermentation. Purification of 15N- and 13C-/15N-labeled kp60-NTDs was achieved by glutathione-Sepharose affinity chromatography followed by thrombin digestion, benzamidine-Sepharose chromatography, cation exchange chromatography using a SP-Sepharose column, and gel filtration using Superdex 75 column (GE Healthcare).
NMR Spectroscopy
Samples for NMR spectroscopy contained either 15N- or 13C-/15N-labeled kp60-NTD at concentrations of 0.5–0.9 mm in 5% D2O, 95% H2O, 20 mm sodium phosphate, and 1 mm EDTA with 50 mm NaCl/without NaCl (pH 6.5). Backbone and side chain assignments were obtained from 15N-heteronuclear single quantum coherence spectroscopy, 13C-heteronuclear single quantum coherence spectroscopy, HNCA, HNCO, HNCACB, CBCACONH, HCC(CO)NH, CC(CO)NH, and HCCH-total correlation spectroscopy spectra recorded at 25 °C, using Bruker Avance spectrometers (500 and 800 MHz, Avance; Bruker Biospin, Germany) equipped with cryomagnetic probes (26, 27). Data were processed using NMRPipe (28) and SPARKY (29) software. Interproton distances were obtained from three-dimensional 13C- and 15N-edited nuclear Overhauser effect spectroscopy spectra recorded with a 100-ms mixing time. Structures were calculated using a standard seven iteration cycle protocol of the program CYANA version 2.0.17 (30, 31). All nuclear Overhauser effect cross-peaks were selected manually using SPARKY. In total, 1723 meaningful nuclear Overhauser effect upper distance restraints were obtained, including 304 long range distances. Dihedral angle restraints were calculated on the basis of backbone atom chemical shifts (32) using the TALOS program. The 20 structures with the lowest restraint energies were selected and analyzed using MOLMOL (33) and PROCHECK-NMR software (Table 1) (34). No distance restraint was violated by more than 0.3 Å and no torsional restraint by more than 5.0°. All the figures were prepared using MOLMOL and PyMOL. The atomic coordinates of the 20 best kp60-NTD NMR structures have been deposited in the Protein Data Bank under accession code 2rpa. Chemical shift assignments have been deposited in the BioMagResBank under accession code 11075.
TABLE 1.
Experimental restraints and statistics for 20 structures of kp60-NTD
| Distance restraints | |
| Total no. of restraints | 1723 |
| Intraresidue | Unused |
| Sequential restraints (|i − j| = 1) | 831 |
| Medium range restraints (1 〈|i − j| ≤4) | 462 |
| Long range restraints (|i − j| >4) | 304 |
| Dihedral angle restraints | 126 |
| φ/ψ/χ | 63/63/0 |
| Hydrogen bond restraints | 0 |
| Statistics used for and obtained from the structure calculations | |
| Final Statistics (20/100) | |
| Cutoffs, distance (0.3 Å) and angle (3.0°) | |
| Maximum target function | 0.06 |
| Maximum violations | |
| Distance violation | 0.21 Å |
| Angle violation | 9.15° |
| Coordinate precision (residues 4–68) | |
| Backbone r.m.s.d. | 0.33 Å |
| Heavy atom r.m.s.d. | 0.83 Å |
| Ramachandran plot statistics (%) (all residues) | |
| Residues in most favored regions | 92.2 |
| Residues in additionally allowed regions | 7.1 |
| Residues in generously allowed regions | 0.1 |
| Residues in disallowed regions | 0.0 |
Mutation Studies and Tubulin Binding Assays
Ala-substituted mutants were prepared by PCR amplification of the entire expression plasmid for kp60-NTD (residues 1–72) according to a standard PCR mutagenesis method using QuikChange site-directed mutagenesis kit (Stratagene). Two complementary oligonucleotides with mutated sequences for each mutant were used as primers (supplemental Table 1). The resulting kp60-NTD genes were sequenced to confirm the mutations. All proteins were purified with glutathione-Sepharose (GE Healthcare) and dialyzed against a buffer containing 50 mm Tris-HCl and 150 mm NaCl (pH 7.5). For pulldown assay, 80 pmol of GST (negative control) or GST fusion proteins were mixed with 10 μl of glutathione-Sepharose 4B (GE Healthcare) in 100 μl of binding buffer containing 80 mm PIPES-KOH (pH 6.8), 0.5 mm EGTA, and 2 mm MgCl2 for 1 h at 4 °C. After washing the beads, 182 pmol (10 μg) of porcine tubulin (Cytoskeleton) was mixed in 200 μl of binding buffer for 2 h at 4 °C. The beads were washed three times, and the associated proteins were eluted with 50 mm Tris-HCl and 10 mm reduced glutathione (pH 7.5). The eluted proteins were resolved by SDS-PAGE and stained with silver.
Model Building
A molecular model of the complex of kp60-NTD with a tubulin tetramer was constructed manually using MOLMOL (33) on the basis of the complex between Vps4a-MIT and CHMP1a (PDB code 2jq9). First, the kp60-NTD structure determined in this study was superimposed onto the corresponding position of Vps4a-MIT. Then the tubulin tetramer, taken from PDB code 3du7, was superimposed onto the C-terminal helix of CHMP1a with the best one position selected out of the eight candidate positions of tubulin.
RESULTS
Structural Prediction and Sequence Analysis of kp60-NTD
Prior to structural determination, we extensively analyzed residues 1–90 of the N-terminal sequences of mouse and human kp60, which represent the sequences preceding the AAA domains, by both bioinformatics and biophysical methods (17). In brief, we found that these regions are genetically conserved only within a single subfamily of type I AAA-ATPase, corresponding to kp60 orthologs (Fig. 1B). Members of this family are found in mammals, other vertebrates, plants, insects, urchins, and nematodes, but not in yeasts or bacteria. It should be noted that some archaeal kp60s (e.g. gi: 13814089 and 223478990) that lack this N-terminal region are less well related to other kp60s, although a strong relationship is found for Vps4 orthologs. Thus, these archaeal kp60s may be better annotated as Vps4 homologs (35).
Focusing upon the AAA-ATPase domain and analyzing the domain level phylogenetic tree, the type I AAA-ATPases, including kp60, spastin, and Vps4, form a single cluster (7, 36). The kp60 orthologs with a conserved N-terminal region form a small subfamily, which is different from the Vps4 subfamily (supplemental Fig. 1). In some mammalian genomes (e.g. mouse, rat, and human), kp60-like A1s (katanal1s) are also conserved (Fig. 1). Katanal1s are very similar kp60 paralogs (∼67% sequence identity over the entire chain). Moreover, this region (residues 1–90) can be further divided into two parts as follows: a well conserved core region (residues 1–72) and the following less-conserved region (∼18 residues). The latter was a putative coiled-coil region, and the N-terminal region (residues 1–90) may form a dimer (17), whereas the first 72 residues behaved as an ideal “NMR ready” monomer. We call this region (residues 1–72) the core N-terminal domain (denoted kp60-NTD) and used it for further analysis.
Structure of kp60-NTD
kp60-NTD was analyzed by standard solution NMR techniques. All of the backbone and 96% of the nonexchangeable protons of the side chain signals were assigned. An ensemble of 20 structures with low CYANA target functions (Fig. 2A) was generated from 1723 experimental NMR constraints. These 20 structures satisfy the experimental constraints very well (Table 1). The stereochemical quality of the ensemble members is good, with all backbone ϕ/ψ angles occupying the most favored or additionally allowed regions of the Ramachandran plot (Table 1; supplemental Fig. 2). Excluding the disordered regions, i.e. the N-terminal region (residues 1–3 plus the preceding extra six residues of the tag) and the C-terminal region (residues 69–72), the r.m.s.d. values were 0.33 Å for backbone heavy atoms and 0.83 Å for all heavy atoms.
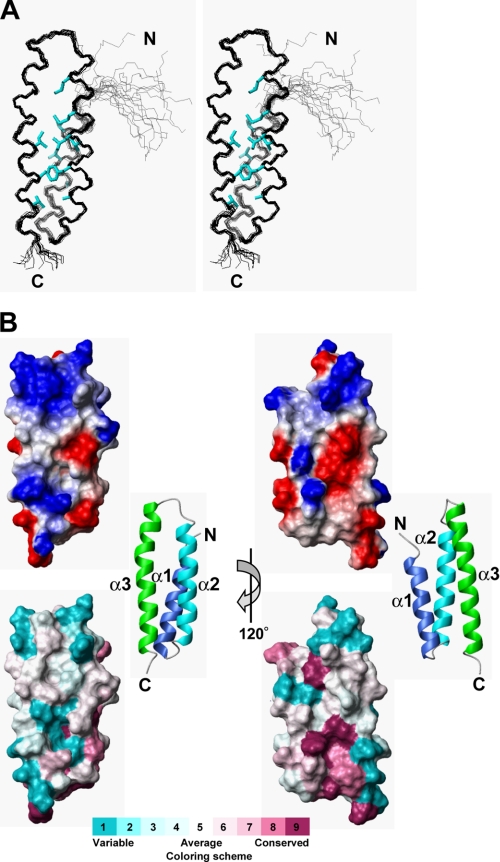
FIGURE 2.
Solution structure of kp60-NTD. A, stereo view of the best fit superposition of the 20 structures with lowest target functions. Side chains of buried residues with solvent accessibility less than 10% are shown (cyan). B, top, electrostatic surface potential mapped onto a van der Waals surface diagram. The color scale ranges between −20 kBT (red) to +20 kBT (blue), where kB is Boltzmann's constant and T is temperature. Bottom, sequence conservation among the kp60-NTDs is mapped on the surface. Conservative and variable residues are colored purple and cyan, respectively. The color codes were produced by ConSurf (63). Ribbon diagrams of the kp60-NTDs are shown in the middle. The surface composed of helices 2 and 3 is shown as the front view (left) and the rear view (right).
As shown in Fig. 2B, kp60-NTD is organized into antiparallel three-helix bundle that consists of helix 1 (4–19), helix 2 (23–41), and helix 3 (46–69). The secondary structure is shown in Fig. 1B along with its amino acid sequence. Helices 1 and 2 are connected by a very tight three-residue turn, whereas helices 2 and 3 are connected by a more flexible four-residue loop. Helices 2 and 3 are longer than helix 1, thereby exposing a large protrusion formed by helix 2 C terminus and helix 3 N terminus. These three helices are packed against one another nearly in parallel. The packing angles between the helices are similar as follows: 19.3° between 1 and 2, 21.1° between 2 and 3, and 26.1° between 1 and 3. Interhelical contacts mainly include hydrophobic side chain-side chain interactions. Core residues employed in these contacts are shown in Fig. 2A as well as in Fig. 1B. A total of 12 nonpolar contacts between helices 1 and 2, 17 between helices 2 and 3, and 5 between helices 1 and 3 were observed. The spatial arrangement of these three helices is nearly symmetric. The interhelical distances between helices 1 and 2 (5.0 Å) and helices 2 and 3 (5.5 Å) were shorter than that between helices 1 and 3 (6.5 Å). We found no obvious crevices or pockets on the surface of kp60-NTD. The kp60-NTD surface is highly charged (Fig. 2B).
Structural Similarities of kp60-NTD with MIT Domains and Other Tetratricopeptide Repeat Proteins
When the structure of kp60-NTD was subjected to DALI search (37), several MIT domains were first retrieved with Z-scores higher than 7.0, including NRBF-2 (PDB code 2crb, Z-score of 9.6, and r.m.s.d. of 1.6 Å), Vps4b (PDB code 1wr0, Z-score of 8.9, and r.m.s.d. of 2.7 Å), Vta1 (PDB code 2rkk, Z-score of 7.4, and r.m.s.d. of 2.4 Å), spastin (PDB code 3eab, Z-score of 7.2, and r.m.s.d. of 2.2 Å), and spartin (PDB code 2dl1, Z-score of 7.1, and r.m.s.d. of 2.4 Å). Thus, we first compared the structure of kp60-NTD with those of the MIT domains. Fig. 3 shows the structural comparisons between kp60-NTD and each of the MIT domains along with their sequence identity and structural fitness. Despite a low sequence similarity (10–19%), the kp60-NTD fold resembles those of the MIT domains, as shown by backbone r.m.s.d. of 2.2–2.7 Å for more than 67 residues from the secondary structural regions. Among these, spastin is the product of SPG4, which is mutated in the most common form of hereditary spastic paraplegia (4), and is involved with MT maintenance in axons (38, 39). Thus, the MIT domain of spastin is one of the closest homolog of kp60-NTD with regard to its physiological relevance to MT severing.
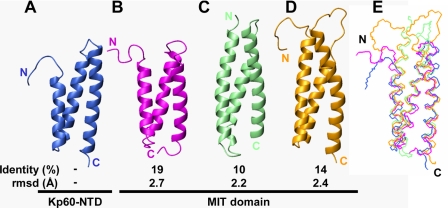
FIGURE 3.
Structural comparisons of kp60-NTD with MIT domains. Ribbon diagrams of the proteins are as follows: A, kp60 (PDB code 2rpa); B, Vps4b (PDB code 1wr0); C, spastin (PDB code 3eab); D, spartin (PDB code 2dl1). Identity (top, %) and r.m.s.d. (bottom, Å) between kp60-NTD and the MIT domains are also presented. E, superposition of kp60-NTD (blue), Vps4b-MIT (magenta), spastin-MIT (pale green), and spartin-MIT (orange).
After comparing the structures in detail, all the helices were well superimposed, although the loop between helices 2 and 3 was not (Fig. 3E). The structures of the kp60 tubulin-binding site, kp60-NTD, and the MIT domain were strikingly similar, although their sequence similarity was very low (∼19%). Thus, kp60-NTD is classified as a variant MIT domain. Because some of the MIT domains (e.g. spastin and spartin) are considered to bind microtubule (and/or tubulin), this structural similarity is not surprising.
One of the most characteristic features of the MIT domain is its unique hydrophobic core formed by conserved Ala residues, referred to as the “Ala zipper” (40, 41). These are thus identified as the key residues for the MIT domain signature (Ala-Xaa6-Ala-Xaa11-Ala-Xaa6-Ala). These conserved Ala residues are present along the buried surfaces of helices 1–3 facing each other, thereby forming a hydrophobic core. In kp60 and its closely related homologs, these key Ala residues are only partly conserved. For example, Ile-6 and Val-32 in mouse kp60-NTD correspond to the zipper-forming Ala residues (Ala-9 and Ala-35) in the human Vps4a-MIT domain. Although the MIT domain signature is not conserved in kp60-NTD, this domain is obviously a close variant of the MIT domain. This imperfect conservation of the MIT-domain signature may explain why methods such as PSI-BLAST (18) and HMMER (19) could not predict the structural similarity between kp60-NTD and the MIT domain. In the DALI search, we also found other proteins containing either twisted α-helical hairpins or tetratricopeptide repeat motifs with Z-scores higher than 5.0. For example, partial structures of glycine-tRNA synthetase α-chain (PDB code 1j5w, Z-score of 8.7, and r.m.s.d. of 2.2 Å), 14-3-3 protein Tau (PDB code 2btp, Z-score of 8.2, and r.m.s.d. of 2.4 Å), cyclophilin 40 (PDB code 1ihg, Z-score of 8.1, and r.m.s.d. of 2.7 Å), α-E-catenin (PDB code 1l7c, Z-score of 7.9, and r.m.s.d. of 2.0 Å), fkbp52 (PDB code 1p5q, Z-score of 7.5, and r.m.s.d. of 3.0 Å), invertase inhibitor Nt-CIF (PDB code 1rj1, Z-score of 6.3, and r.m.s.d. of 2.1 Å), and Hop (PDB code 1elr, Z-score of 5.2, and r.m.s.d. of 3.7 Å) were shown to resemble kp60-NTD (data not shown).
Tubulin Binding by kp60-NTDs
To examine the molecular function of kp60-NTD as an MT binding domain, we performed in vitro MT binding assays using polymerized MTs. Contrary to our expectation, we found that the amount of kp60-NTD co-sedimented with MTs was very low, at the limit of detectability (supplemental Fig. 3) (data not shown). However, kp60-NTD co-sedimented with medium size MTs (supplemental Fig. 3D). These results suggested that kp60-NTD might bind to oligomeric tubulin and/or MT fragments rather than enormous polymerized MTs. Thus, we did a pulldown assay using GST-tagged kp60-NTD with unpolymerized tubulin. In vitro tubulin binding activity of kp60-NTD was observed (Fig. 4A, lane 5).
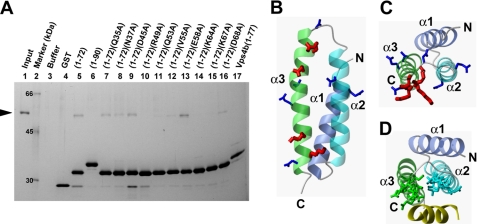
FIGURE 4.
Interactions of kp60-NTD with tubulin. A, pulldown assays of tubulin with GST-tagged kp60-NTDs of wild type and Ala mutants and Vps4b-MIT in vitro. Tubulin was used as the input. Molecular size is shown in lane 2. Only the buffer and the GST tag used as negative controls are shown in lanes 3 and 4. Recombinant proteins used for pulldown are indicated at the top of the gel. SDS-PAGE was silver-stained. B and C, side and top views of the ribbon diagram of kp60-NTD, respectively. Side chains of residues that were substituted with Ala are shown. In the pulldown assay, residues that were affected and unaffected by Ala mutations for tubulin binding are colored red and blue, respectively. D, top view of the ribbon diagram of the complex between Vps4-MIT and CHMP1a (yellow) (PDB code 2jq9). Side chains of the residues interacting between Vps4 and CHMP1a are indicated.
This tubulin binding activity varied with the length of the N-terminal domain. kp60-NTD (residues 1–72) binds tubulin, whereas kp60-NTD (residues 1–90) does not (Fig. 4A, lane 6). In our previous report, we showed that kp60-NTD (residues 1–90) formed a dimer using the coiled-coil region (residues 73–90) (17). Thus, dimer formation may hide the interface of kp60-NTD from tubulin. In addition, we found that the Vps4b-MIT domain (residues 1–77) did not bind tubulin (Fig. 4A, lane 17). Thus, the observed tubulin binding activity is specific for kp60-NTD.
Tubulin-binding Site of kp60-NTD
To determine the interfacial residues on kp60-NTD involved with tubulin recognition, we carried out mutagenesis experiments. Prior to these experiments, we attempted to identify the tubulin-interacting residues on kp60-NTD by NMR titration experiments and failed. We observed unexpected severe signal broadening even at very low tubulin concentration, which made further NMR analysis difficult (data not shown). Then, 10 residues from kp60-NTD (Gln-35, Asn-37, Asp-45, Arg-49, Gln-53, Val-55, Glu-58, Lys-64, Lys-67, and Asp-68) were selected, and each was substituted with Ala. These residues were carefully selected from the surface residues located on helices 2 and 3. The binding activities of mutants were examined by pulldown experiments (Fig. 4A, lanes 7–16).
The most significant effects were observed in mutations of residues on helix 3 as follows: Arg-49, Gln-53, Lys-64, and Lys-67. All of these side chains are hydrophilic and are exposed to the surface composed of helices 2 and 3 (Fig. 4, B and C). In addition, three of the four key residues are positively charged, suggesting an electrostatic interaction between kp60-NTD and tubulin. These residues were not conserved in the Vps4b-MIT domain as well as in the other MIT domains, such as spastin and spartin (Fig. 1B). This result is partially consistent with the inability of Vps4b-MIT to bind tubulin (Fig. 4A, lane 17). Because spastin and spartin can bind or regulate MTs (42, 43), this might indicate that these proteins bind MTs using regions other than the MIT domains. In contrast, mutants V55A, E58A, and D68A retained substantial tubulin binding activities (Fig. 4A, lanes 12, 13, and 16). These residues are also on helix 3, but are exposed to the surface composed of helices 1 and 3 or outside of helix 3 (Fig. 4, B and C). Similarly, residues in helix 2 (Gln-35 and Asn-37) and loop 2 (Asp-45) were not involved in tubulin binding (Fig. 4A, lanes 7–9).
We further examined whether full-length kp60s with or without mutation in the N-terminal domain bind tubulin. We generated GST-tagged full-length kp60 in E. coli. Prior to the binding assay, we confirmed that the recombinant full-length kp60s had ATPase activity, according to the protocol in the recent paper (supplemental Fig. 4A) (16). We then performed pulldown experiments using mutants R49A and K67A of full-length kp60. The full-length kp60 (wild type) bound tubulin, whereas the mutants lacked tubulin binding activities, as expected by the results of kp60-NTDs (supplemental Fig. 4B; Fig. 4A).
DISCUSSION
Structural and Functional Comparisons with Other Tubulin Binding Domains
In this study, we have determined the structure of a novel tubulin binding domain derived from the conserved region of kp60, which was classified as a variant MIT domain. To our knowledge, this is the first experimental evidence for the direct interaction between an isolated MIT domain and tubulin.
To date, structures of many MT and/or tubulin binding domains have been determined (supplemental Fig. 5) (44–49). Interestingly, all-α-helical protein domains are dominant in these with solved structures, which might be advantageous for interactions with MT and/or tubulin. In the MT structure, the only accessible surface of tubulin includes helices 11 and 12 and the C-terminal tail (49–51). Thus, for one of tubulin recognition, helix-helix interactions of tubulin binding domains are suggested, although there are many structures of the known MT-interacting proteins left unsolved.
Structural and Functional Similarities to Vps4
We identified the tubulin-binding interface of kp60-NTD, which is on the surface comprising helices 2 and 3 (Fig. 4B). This result is consistent with the studies by Stoppin-Mellet, in which a truncation mutant of Arabidopsis kp60 (AtKSS) that lacked the N-terminal 15 residues corresponding to helix 1 still retained MT severing activity (16). Surprisingly, the tubulin binding interface is very similar to the substrate (Vps2 and CHMP1a)-binding interfaces of the MIT domains of Vps4 (Fig. 4, C and D) (35, 52, 53). In other words, the common substrate-binding interfaces appear to be preserved between MT severing and membrane skeletal reorganization.
In studies of Vps4-MIT complexed with C-terminal regions of Vps2 or CHMP1a, the MIT domains use helices 2 and 3 as the interface for the α-helical peptides (35, 52). The residues involved with kp60-NTD-tubulin interaction are relatively conserved among kp60 orthologs (Fig. 1B), but they are not conserved between kp60-NTD and the other MIT domains, thus explaining why Vps4-MIT did not bind tubulin (Fig. 4A, lane 17). In contrast, another interface of Vps4-MIT for CHMPs has been reported, in which Vps4-MIT uses a shallow cleft between helices 1 and 3 for binding a proline-rich, CHMP6-derived peptide named MIT-interacting motif 2 (MIM2) (53). In the structure of kp60-NTD, there was no correspondence between helices 1 and 3, as the interhelical distance was substantially narrower (6.5 Å) than that of Vps4-MIT. We do not rule out the possibility that the interface composed of helices 1–3 serves as the binding site of other factors, such as katanin p80 and NDEL1 (54), both of which regulate the subcellular localization of kp60.
Conserved Macromolecular Disassembling Mechanisms between Vps4 and Kp60
Our findings indicate that the molecular architectures of kp60 and Vps4 are very similar in the following points: domain organization, structures of the N-terminal domains, and the relative locations of the interfaces for target proteins. Here, we propose some common features of the molecular mechanisms in different biological processes, MT severing and late endosomal luminal membrane budding, driven by kp60 and Vps4, respectively.
First, both the enzymes disassemble polymeric macromolecular complexes known as cytoskeleton and membrane skeleton. Second, these enzymes release protomers from macromolecular complexes (MT and ESCRT-III) in the cytoplasm depending on ATP hydrolysis. Finally, their N-terminal domains serve as adaptors to the protomers. The similarities of these mechanisms are illustrated in Fig. 5.
FIGURE 5.
Schematic diagram of architecture and molecular function similarities between kp60 and Vps4. kp60 catalyzes the disassembly of MT via N-terminal domain binding, which results in MT severing. Vps4 catalyzes the release of the ESCRT-III protomer via the MIT domain binding, which results in endosomal membrane invagination. For both biological events, the N-terminal domains serve as adaptors for the polymeric macromolecules, thereby disassembling either the cytoskeleton or the membrane skeleton in an ATP-dependent manner.
The ESCRT-III complex is composed of self-associating coiled-coil proteins (CHMP1–6), which form filamentous circular structures on the membrane surface (55). When Vps4 interacts with ESCRT-III filaments, it pulls out CHMP protomers from the filamentous circular structure. The residual filaments of ESCRT-III then reorganize into a smaller circular structure by self-association. Vps4 continues to pull protomers away, and the circular structure shrinks into a smaller wheel. Finally, this downsizing of the ESCRT-III circle results in membrane budding with concomitant alterations of the membrane structure. This model is known as the “concentric circle model” (56).
We propose that the early stage of MT severing might have a similar mechanism. kp60 pulls a tubulin α/β-dimer away from MT in an ATP-dependent manner. However, contrary to the ESCRT-III polymer, polymerization of tubulin dimers is restricted to the plus end of MT as well as to the GTP form of tubulin, whereas polymerization at the minus end is extremely slow (reviewed in Refs. 1, 57). It is expected that once tubulin is pulled away by kp60, it can no longer fill the gap on MT. If two or more tubulin dimers are pulled away from this gap, then MT may start severing, resulting in a catastrophe. The structural similarities between kp60 and Vps4 revealed in this study encourage us to propose a model for the molecular mechanism of MT severing.
Model for kp60-NTD Binding to Tubulin Oligomer
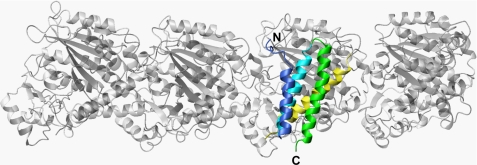
To assess the detailed mechanism of MT severing, we constructed a model for the complex between kp60-NTD and a tubulin tetramer (Fig. 6). Our study is confined to the interface of the kp60 N-terminal adaptor domain to its tubulin substrate, as we did not identify the kp60-binding site on tubulin. Nevertheless, numerous literature resources provide a basis for model construction as follows. As discussed previously, the major candidates of the structural elements of tubulin that are accessible from outside MT are helices 11 and 12 (49–51). Next, the similarity between the interfaces of kp60-NTD with tubulin and that of Vps4-MIT with CHMP1a suggests that a helix on tubulin, which is similar to the CHMP1a helix (residues 115–127) bound to Vps4-MIT (52), may serve as the binding site of kp60. Taking all the information into account, we propose a model for the tubulin + kp60-NTD complex (Fig. 6).
FIGURE 6.
Model of α-tubulin binding with kp60-NTD. Ribbon diagram of a model complex between kp60-NTD and a tubulin tetramer (gray) is shown. α-Tubulin helix 12, a putative interface of kp60-NTD, is colored yellow.
While constructing the model, the following points were hypothesized: (i) one of the last helices (helix 11 or 12) makes contact with kp60-NTD at its helix 2/3 interface; (ii) the relative position and orientation between kp60-NTD and one of the tubulin helices mimic those between Vps4-MIT and the CHMP1a helix; (iii) steric crash between kp60-NTD and tubulin should be avoided; and (iv) charge-charge interactions between kp60-NTD and the tubulin helix should be maximized.
As a result, we found the following four candidate positions on tubulin C-terminal helices for kp60-NTD binding: (i) helix 11 (residues 386–396); (ii) helix 11 (residues 390–400); (iii) helix 12 (residues 420–430); and (iv) helix 12 (residues 423–433). All these positions are present on both tubulin-α and tubulin-β. Finally, by assessing complementarity of charge interactions in the model, the final model was selected out of the eight candidate models. Its helix 12 (residues 420–430) of tubulin α3 binds with kp60-NTD by occupying the corresponding position of CHMP1a (residues 115–127) that binds Vps4-MIT (PDB code 2jq9) (Fig. 6; supplemental Fig. 6) (52).
Alternatively, we generated the model of kp60-NTD + tubulin tetramer complex based on the complex between spastin-MIT with CHMP1b (PDB code 3eab) in which spastin-MIT used helices 1 and 3 as the interface to CHMP1b (supplemental Fig. 7) (58). This model is not consistent with our mutation studies (Fig. 4). Consequently, we justified the modeling of helices 2 and 3 as the tubulin-binding site. Because CHMP1b serves as an adaptor of spastin but not a substrate, this alternative model suggests that the helix 1/3 surface of kp60-NTD is a putative binding site for kp80, an adaptor of kp60 to bound MT and/or tubulin.
In the model of Fig. 6, the direction of the pore of the hexameric AAA-ATPase domain, which follows C-terminal to kp60-NTD, may approach the C-terminal tail of tubulin. We further confirmed this idea by using the model structure of full-length hexameric kp60 complexes with tubulin (supplemental Fig. 8). The location is consistent with the hypothesis that the pore of the AAA domain “sucks in” the tubulin C-terminal tail upon ATP hydrolysis (threading model) (59, 60). In fact, the literature suggests that kp60 function requires its direct interaction with the C-terminal tail of tubulin. This is based on the observation that MT severing activity was abolished when MTs were pretreated with subtilisin (8). Additional evidence regarding the MT-severing mechanism of spastin, another related AAA-ATPase, may support this idea. Spastin also recognizes and pulls the tubulin C-terminal tail as an initial binding site that is indispensable for its MT severing activity (42, 61). The hypotheses derived from our complex model require confirmation by additional experimentation.
In conclusion, the structure and key residues of kp60-NTD provide new insights into the molecular mechanisms of how the enzyme severs MT. The similarities of the molecular mechanisms as well as of the domain organizations suggest that these are evolutionally conserved among type I AAA-ATPases, kp60 and Vps4, whose cellular functions are distinct.
Supplementary Material
Acknowledgment
We thank Kaori Satomura for technical assistance.
A part of this work was performed under the Cooperative Research Program of Institute for Protein Research, Osaka University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. 1–8, Table 1, and additional references.
- MT
- microtubule
- AAA
- ATPase associated with various cellular activities
- ESCRT
- endosomal sorting complexes required for transport
- GST
- glutathione S-transferase
- kp60
- katanin p60
- MIT domain
- microtubule interacting and trafficking domain
- r.m.s.d.
- root mean square deviation
- NTD
- N-terminal domain
- PIPES
- 1,4-piperazinediethanesulfonic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1.Desai A., Mitchison T. J. (1997) Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 2.Wittmann T., Hyman A., Desai A. (2001) Nat. Cell Biol. 3, E28–E34 [DOI] [PubMed] [Google Scholar]
- 3.Zhang D., Rogers G. C., Buster D. W., Sharp D. J. (2007) J. Cell Biol. 177, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C. S., Cruaud C., Dürr A., Wincker P., Brottier P., Cattolico L., Barbe V., Burgunder J. M., Prud'homme J. F., Brice A., Fontaine B., Heilig B., Weissenbach J. (1999) Nat. Genet. 23, 296–303 [DOI] [PubMed] [Google Scholar]
- 5.Errico A., Ballabio A., Rugarli E. I. (2002) Hum. Mol. Genet. 11, 153–163 [DOI] [PubMed] [Google Scholar]
- 6.Cox G. A., Mahaffey C. L., Nystuen A., Letts V. A., Frankel W. N. (2000) Nat. Genet. 26, 198–202 [DOI] [PubMed] [Google Scholar]
- 7.Frickey T., Lupas A. N. (2004) J. Struct. Biol. 146, 2–10 [DOI] [PubMed] [Google Scholar]
- 8.McNally F. J., Vale R. D. (1993) Cell 75, 419–429 [DOI] [PubMed] [Google Scholar]
- 9.Hartman J. J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R. D., McNally F. J. (1998) Cell 93, 277–287 [DOI] [PubMed] [Google Scholar]
- 10.McNally K. P., Bazirgan O. A., McNally F. J. (2000) J. Cell Sci. 113, 1623–1633 [DOI] [PubMed] [Google Scholar]
- 11.McNally F. J., Okawa K., Iwamatsu A., Vale R. D. (1996) J. Cell Sci. 109, 561–567 [DOI] [PubMed] [Google Scholar]
- 12.McNally F. J., Thomas S. (1998) Mol. Biol. Cell 9, 1847–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman J. J., Vale R. D. (1999) Science 286, 782–785 [DOI] [PubMed] [Google Scholar]
- 14.Lupas A. N., Martin J. (2002) Curr. Opin. Struct. Biol. 12, 746–753 [DOI] [PubMed] [Google Scholar]
- 15.Ogura T., Wilkinson A. J. (2001) Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 16.Stoppin-Mellet V., Gaillard J., Timmers T., Neumann E., Conway J., Vantard M. (2007) Plant Physiol. Biochem. 45, 867–877 [DOI] [PubMed] [Google Scholar]
- 17.Iwaya N., Goda N., Unzai S., Fujiwara K., Tanaka T., Tomii K., Tochio H., Shirakawa M., Hiroaki H. (2007) J. Biomol. NMR 37, 53–63 [DOI] [PubMed] [Google Scholar]
- 18.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., Griffiths-Jones S., Howe K. L., Marshall M., Sonnhammer E. L. (2002) Nucleic Acids Res. 30, 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., Studholme D. J., Yeats C., Eddy S. R. (2004) Nucleic Acids Res. 32, D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. (2004) Nucleic Acids Res. 32, D142–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomii K., Akiyama Y. (2004) Bioinformatics 20, 594–595 [DOI] [PubMed] [Google Scholar]
- 23.Shi J., Blundell T. L., Mizuguchi K. (2001) J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 24.Ciccarelli F. D., Proukakis C., Patel H., Cross H., Azam S., Patton M. A., Bork P., Crosby A. H. (2003) Genomics 81, 437–441 [DOI] [PubMed] [Google Scholar]
- 25.Goda N., Tenno T., Takasu H., Hiroaki H., Shirakawa M. (2004) Protein Sci. 13, 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki T., Lee W., Arrowsmith C. H., Muhandiram D. R., Kay L. E. (1994) J. Am. Chem. Soc. 116, 11655–11666 [Google Scholar]
- 27.Cavanagh J., Fairbrother W. J., Palmer A. G., 3rd, Rance M., Skelton N. J. (2007) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., pp. 535–673, Academic Press, San Diego [Google Scholar]
- 28.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29.Goddard T. D., Kneller D. G. (2004) SPARKY, Version 3, University of California, San Francisco [Google Scholar]
- 30.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 31.Guntert P. (2003) Prog. Nucleic Magn. Reson. Spect. 43, 105–125 [Google Scholar]
- 32.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 33.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 29–32 [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 35.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007) Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 36.Fröhlich K. U. (2001) J. Cell Sci. 114, 1601–1602 [DOI] [PubMed] [Google Scholar]
- 37.Holm L., Sander C. (1997) Nucleic Acids Res. 25, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherwood N. T., Sun Q., Xue M., Zhang B., Zinn K. (2004) PLoS. Biol. 2, e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans K. J., Gomes E. R., Reisenweber S. M., Gundersen G. G., Lauring B. P. (2005) J. Cell Biol. 168, 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takasu H., Jee J. G., Ohno A., Goda N., Fujiwara K., Tochio H., Shirakawa M., Hiroaki H. (2005) Biochem. Biophys. Res. Commun. 334, 460–465 [DOI] [PubMed] [Google Scholar]
- 41.Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White S. R., Evans K. J., Lary J., Cole J. L., Lauring B. (2007) J. Cell Biol. 176, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel H., Cross H., Proukakis C., Hershberger R., Bork P., Ciccarelli F. D., Patton M. A., McKusick V. A., Crosby A. H. (2002) Nat. Genet. 31, 347–348 [DOI] [PubMed] [Google Scholar]
- 44.Ravelli R. B., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M. (2004) Nature 428, 198–202 [DOI] [PubMed] [Google Scholar]
- 45.Slep K. C., Vale R. D. (2007) Mol. Cell 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishima M., Maesaki R., Kasa M., Watanabe T., Fukata M., Kaibuchi K., Hakoshima T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10346–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guasch A., Aloria K., Pérez R., Avila J., Zabala J. C., Coll M. (2002) J. Mol. Biol. 318, 1139–1149 [DOI] [PubMed] [Google Scholar]
- 48.Carter A. P., Garbarino J. E., Wilson-Kubalek E. M., Shipley W. E., Cho C., Milligan R. A., Vale R. D., Gibbons I. R. (2008) Science 322, 1691–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikkawa M., Hirokawa N. (2006) EMBO J. 25, 4187–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nogales E., Wolf S. G., Downing K. H. (1998) Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 51.Bodey A. J., Kikkawa M., Moores C. A. (2009) J. Mol. Biol. 388, 218–224 [DOI] [PubMed] [Google Scholar]
- 52.Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007) Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 53.Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. (2008) Dev. Cell 15, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 54.Toyo-Oka K., Sasaki S., Yano Y., Mori D., Kobayashi T., Toyoshima Y. Y., Tokuoka S. M., Ishii S., Shimizu T., Muramatsu M., Hiraiwa N., Yoshiki A., Wynshaw-Boris A., Hirotsune S. (2005) Hum. Mol. Genet. 14, 3113–3128 [DOI] [PubMed] [Google Scholar]
- 55.Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nickerson D. P., Russell M. R., Odorizzi G. (2007) EMBO Rep. 8, 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howard J., Hyman A. A. (2003) Nature 422, 753–758 [DOI] [PubMed] [Google Scholar]
- 58.Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., Hurley J. H. (2008) Nat. Struct. Mol. Biol. 15, 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., Neher S. B., Oakes E. S., Siddiqui S. M., Wah D. A., Baker T. A. (2004) Cell 119, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lum R., Tkach J. M., Vierling E., Glover J. R. (2004) J. Biol. Chem. 279, 29139–29146 [DOI] [PubMed] [Google Scholar]
- 61.Roll-Mecak A., Vale R. D. (2008) Nature 451, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armon A., Graur D., Ben-Tal N. (2001) J. Mol. Biol. 307, 447–463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.