Abstract
Purine transport is essential for malaria parasites to grow because they lack the enzymes necessary for de novo purine biosynthesis. The Plasmodium falciparum Equilibrative Nucleoside Transporter 1 (PfENT1) is a member of the equilibrative nucleoside transporter (ENT) gene family. PfENT1 is a primary purine transport pathway across the P. falciparum plasma membrane because PfENT1 knock-out parasites are not viable at physiologic extracellular purine concentrations. Topology predictions and experimental data indicate that ENT family members have eleven transmembrane (TM) segments although their tertiary structure is unknown. In the current work, we showed that a naturally occurring polymorphism, F394L, in TM11 affects transport substrate Km. We investigated the structure and function of the TM11 segment using the substituted cysteine accessibility method. We showed that mutation to Cys of two highly conserved glycine residues in a GXXXG motif significantly reduces PfENT1 protein expression levels. We speculate that the conserved TM11 GXXXG glycines may be critical for folding and/or assembly. Small, cysteine-specific methanethiosulfonate (MTS) reagents reacted with four TM11 Cys substitution mutants, L393C, I397C, T400C, and Y403C. Larger MTS reagents do not react with the more cytoplasmic positions. Hypoxanthine, a transported substrate, protected L393C, I397C, and T400C from covalent modification by the MTS reagents. Plotted on an α-helical wheel, Leu-393, Ile-397, and Thr-400 lie on one face of the helix in a 60° arc suggesting that TM11 is largely α helical. We infer that they line a water-accessible surface, possibly the purine permeation pathway. These results advance our understanding of the ENT structure.
Keywords: Membrane Proteins, Nucleoside, Nucleoside Nucleotide Transport, Parasite Metabolism, Protein Chemical Modification, Purine, Accessibility, Cysteine, Equilibrative, Malaria
Introduction
Malaria is a major public health problem in the developing world. Worldwide there are 300–500 million cases of malaria per year that result in over 1 million deaths per year, mostly of children (1, 2). The unicellular parasite Plasmodium falciparum causes the most severe form of malaria. It is transmitted via a mosquito vector. Artemisinin-based combination therapy is the treatment of choice but concerns are growing that resistance to artemisinin is developing in southeast Asia as it did for chloroquine in the past (3–5). Thus, there is an urgent need to identify potential targets for new antimalarial drugs.
During its 48 h intraerythrocytic life cycle, P. falciparum replicates its 23 Mb genomic DNA 16–32 times. Given the large requirement for purine nucleotides for DNA synthesis, considerable interest has focused on the purine salvage pathway as a possible drug target because P. falciparum is a purine auxotroph, incapable of de novo purine biosynthesis (6). Transmembrane purine transporters perform the essential first step by moving purines from the erythrocyte cytoplasm to the parasite cytoplasm. The P. falciparum genome contains four putative members of the equilibrative nucleoside transporter (ENT)3 gene family (7, 8). The P. falciparum equilibrative nucleoside transporter 1 (PfENT1) is the primary purine transporter (9–16). PfENT1 knock-out parasites are not viable during in vitro culture at physiologic purine concentrations but can be rescued by growth at supraphysiologic purine levels (13, 15). Thus, a PfENT1 inhibitor might be an effective antimalarial drug (17). The ENTs might also serve to transport antimalarial drugs that target purine salvage pathway enzymes into the parasite cytoplasm so that they can reach their target proteins (18).
The ENT transporters are generally capable of transporting multiple purine nucleosides and nucleobases and many also transport pyrimidines as well (19, 20). Elucidating the structure of the ENT family would help to explain the structural basis of transporter specificity and facilitate the design of specific inhibitors. The ENT family members are predicted to have eleven transmembrane (TM) segments with a cytoplasmic N terminus and extracellular C terminus (21, 22). Mutations in multiple transmembrane segments have been shown to affect substrate affinity and/or inhibitor efficacy including residues in TM1 (M33, hENT1), TM2 (M89 and L92, hENT1), TM4 (G154, hENT1 = C140, rENT2; S160, hENT1; K155, CfNT2), TM8 (F334, N338, hENT1), and TM11 (L442, hENT1 = I429, CeENT1; S469 & T478, LdNT1.1) (23–31). For many of these residues it is not known whether they line the permeation pathway or lie elsewhere in the protein where the mutations alter the protein structure leading to the observed functional effects of the mutations. There have been several attempts to generate structural models of ENT family proteins. One generated a homology model of the PfENT1 transporter based on the structure of the prokaryotic glycerol-3-phosphate transporter, a member of the major facilitator superfamily (32, 17, 33), and the other used the Rosetta protein structure prediction software to generate an ab initio model of LdNT1.1 (34). The organization of the transmembrane segments was similar in both models supporting the hypothesis that the ENTs may be members of the major facilitator superfamily (34). Interestingly, in the three ab initio models generated, the positions of ten of the eleven transmembrane segments were similar in all three models. Only the position of TM11 was significantly different among the three models presented (34). In one model it lined the permeation pathway, and in two others it was located on the protein periphery. A recent report identified a mENT1 alternate splice variant that lacked exon 11, which encodes TM9 to TM11. They reported that Δexon-11-mENT1 is capable of performing nucleoside transport (35). This raises the question of whether this portion of the protein is essential for protein assembly, targeting or purine transport. Thus, there is considerable uncertainty regarding the structure and functional role of TM11 in ENTs. We have utilized the substituted cysteine accessibility method (SCAM) to explore the structure and function of TM11.
SCAM provides a systematic approach to identify the residues in a transmembrane segment that line an aqueous transport pathway or ion channel (36–44). In this approach, the reactivity of engineered cysteine (Cys) residues with water-soluble, sulfhydryl-specific reagents is assayed. The reagents that we have used are derivatives of methanethiosulfonate (MTS) that react with ionized thiolates (-S−) 109 times faster than with the unionized thiols (-SH) (45, 37). Only Cys that are at least transiently on the water-accessible protein surface will ionize to any significant extent. Thus, this approach allows us to determine the water-surface accessibility of residues in a transmembrane segment. This approach was previously applied to the TM5 segment of the Leishmania donovani nucleoside transporter 1 (46) and TM4 of the Crithidia fasciculate nucleoside transporter 2 (31), revealing the water-accessible faces on these transmembrane segments.
The existence of a polymorphism in the PfENT1 TM11 segment F394L had been noted in the literature (9, 10), but the functional consequences have not been investigated. This polymorphism is located in the middle of a highly conserved GXXXG motif (Fig. 1). GXXXG motifs are frequently found at sites of helix-helix interactions and are often important for protein folding and assembly (47–50). We demonstrated that the polymorphism had functional effects of the adenosine and hypoxanthine Km. To understand the structural basis for the functional effect of a polymorphism in the PfENT1 TM11 segment we utilized SCAM to investigate the TM11 segment.
FIGURE 1.
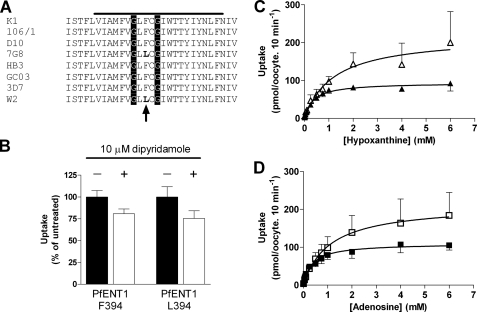
Amino acid sequence alignment of the residues in and flanking the TM11 segment of various ENT homologues. Solid line above the sequences indicates the predicted extent of TM11. Numbers to the right of each sequence indicate the amino acid number of the final residue. The absolutely conserved glycines in the GXXXG motif are highlighted by the gray stripes. Species origin of sequence indicated by initial letters, m, mouse; r, rat; h, human; Ce, C. elegans; Ld, L. donavani; Pf, P. falciparum. Residues in bold in species other than P. falciparum indicate mutants studied by other laboratories and discussed in the Introduction. The bold, italicized letters in the P. falciparum sequence indicate the positions where MTS reagents reacted with Cys substitution mutants. The vertical arrow indicates the location of the F/L polymorphism at position 394 in the P. falciparum sequence.
EXPERIMENTAL PROCEDURES
Identification of Naturally Occurring PfENT1 Polymorphisms
To search for polymorphisms within the PfENT1 gene, we PCR amplified PfENT1 from the genomic DNA of 11 separate parasite strains. PCR primers 5′-GTGCTGTTTACATATATATTAATAGG-3′ and 5′-GAGGAGATATATACGAAATTTAC-3′ (Sigma-Aldrich, St. Louis, MO) were designed to anneal to the genomic DNA regions flanking the 5′- and 3′-ends of the PfENT1 gene, respectively. Genomic DNA samples were derived from the following commonly studied parasite strains (geographic origin): HB3 (Honduras), 3D7 (The Netherlands), K1 (Thailand), W2 (Indochina), 106/1 (Sudan), D10 (Papua New Guinea), and 7G8 (Brazil). In addition to these strains, genomic DNA from clinical isolates derived from Madagascar, Malaysia, and Santa Lucia were also PCR amplified. Finally, genomic DNA from the GC03 parasite strain, a progeny of the genetic cross of the HB3 and Dd2 parasite strains (51) was also examined. The PCR-amplified PfENT1 region from each strain was sequenced at the Albert Einstein College of Medicine DNA Sequencing Facility.
PfENT1 Plasmid and Site-specific Mutations
For these experiments, we used the PfENT1-pXOON plasmid containing the P. falciparum 3D7 strain pfent1 gene in the pXOON plasmid (52) that we used previously (53, 16). The PfENT1-pXOON plasmid was linearized with NheI, and capped mRNA was prepared as described previously (16).
To study the functional effect of the naturally occurring polymorphisms that we identified, we introduced the mutations F36S, L133F, and F394L individually into the PfENT1-pXOON plasmid. Cysteine substitution mutations were generated, one at a time, from position 390 to position 405 within the PfENT1 TM11 protein sequence. Position 395 was not altered, as PfENT1 contains an endogenous cysteine at this position. In addition to these mutants, the double mutant C395S/I397C was created. For the HA-tagged constructs the epitope sequence (YPYDVPDYA) was added to the C terminus of PfENT1 by insertional mutagenesis. All mutations were generated using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) and the entire pfent1 cDNA was verified by DNA sequencing.
Expression of PfENT1 in Xenopus oocytes- Female Xenopus laevis were purchased from Nasco (Fort Atkinson, WI). Stage V-VI oocytes were defolliculated with a 60 or 75 min incubation in 2 mg/ml Type 1A collagenase (Sigma-Aldrich) in OR2 (82.5 mm NaCl, 2 mm KCl, 1 mm MgCl2, and 5 mm HEPES; pH adjusted to 7.5 with NaOH). Oocytes were washed thoroughly in OR2 and stored at 16 °C in SOS medium: 82.5 mm NaCl, 2.5 mm KCl, 1 mm MgCl2, 5 mm HEPES, pH 7.5 with 100 international units/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B (Invitrogen, Carlsbad, CA) and 5% horse serum (Sigma-Aldrich). 24 h post-isolation, oocytes were injected either with 23 nl of 1 ng/nl pfent1 capped mRNA or diethylpyrocarbonate (DEPC)- treated water. Nucleoside/nucleobase uptake assays were performed 3 or 4 days post-injection.
Western Blot of Total and Membrane Protein
Three days post-injection, ∼70 oocytes for each condition were transferred to a dish containing calcium-free buffer (115 mm NaCl, 2.5 mm KCl, 1.8 mm MgCl2, 10 mm HEPES, pH 8) and reacted with 0.5 mg/ml sulfo-NHS-LC biotin for 30 min at room temperature to biotinylate cell surface proteins. The oocytes were rinsed with biotin-free buffer and transferred to Tris-NaCl buffer (100 mm NaCl, 20 mm Tris, pH 7.4) to remove excess biotin. Oocytes were lysed by trituration in Tris-NaCl buffer containing 1% Triton X-100, 0.5% deoxycholate, 10 mm N-ethylmaleimide and HALT protease inhibitor mixture (Pierce), 20 μl/oocyte, solubilized by rotating at 4 °C for 1 h and centrifuged three times at 16,000 × g for 20 min at 4 °C to remove yolk. An aliquot from each sample was saved for the total protein fraction, while the remaining amount from each sample was incubated with streptavidin beads (Pierce) by rotating for 3 h at 4 °C. For the membrane protein fraction, the proteins were eluted from the streptavidin beads by incubation at 37 °C for 10 min in 4× SDS sample buffer. Sample buffer was also added to the total protein fraction. Proteins were separated by SDS-PAGE on a 4–15% Tris-gradient gel (Bio-Rad) and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The blot was blocked with 5% milk, and probed using a mouse monoclonal anti-hemagglutinin (HA) epitope primary antibody (1:1000 dilution) (Covance, Princeton, NJ), and a goat anti-mouse horseradish peroxidase-conjugated IgG secondary antibody (1:1000 dilution) (Pierce). The blot was imaged using ECL Enhanced Chemiluminescence Substrate (Thermo Scientific, Rockford, IL) with a Fluorochem 8000 gel imaging system (Alpha Innotech/Cell Biosciences, Santa Clara, CA).
PfENT1-mediated Purine Uptake Studies
To measure PfENT1-mediated purine uptake, three to five PfENT1-expressing oocytes were added to room temperature E1 buffer (140 mm NaCl, 2.8 mm KCl, 2 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, pH 7.4) containing either [8-3H]adenosine (23 Ci/mmol) or [2, 8-3H]hypoxanthine (30 Ci/mmol) and incubated at room temperature for 10 min. Assays were terminated by five washes with ice-cold E1 buffer, followed by solubilization of the oocytes individually with 200 μl of 5% SDS. Uptake of radiolabeled substrate was determined with a Wallac WinSpectral 1414 Liquid Scintillation counter. All data points represent the average of at least ten individual oocytes derived from three separate oocyte isolations unless indicated otherwise.
To account for non-PfENT1 mediated uptake or nonspecific binding, all experiments were performed in parallel with DEPC-water injected oocytes or uninjected oocytes. For all of the substrates tested, no significant difference was observed between DEPC-water injected or uninjected oocytes. For this reason, these data sets were pooled to represent background uptake values. Background values were subtracted before analysis.
Substrate Vmax and Km data were calculated from the concentration dependence of substrate uptake using the Michaelis-Menten equation (GraphPad Prism 5.0, La Jolla, CA). To determine whether Cys substitutions altered PfENT1 transport function, uptake of 100 nm [8-3H]adenosine (23 Ci/mmol) (10 min) was measured in oocytes expressing the individual Cys mutants and wild-type PfENT1, as described above, and normalized to the WT uptake in a given batch of oocytes. % uptake = (Cys-mutant adenosine uptake/WT adenosine uptake) × 100. Significant differences compared with wild-type between normalized uptake values were determined by a one-way ANOVA (p < 0.05) using Dunnett's multiple comparison post-hoc test (p < 0.05).
Substituted Cysteine Accessibility Experiments
To examine the water-surface accessibility of residues within PfENT1 TM11, we assayed the reactivity of Cys substitution mutants with MTS reagents. Oocytes expressing each of the TM11 Cys mutants were washed briefly in OR2 and treated for 2 min with either 2 mm MTS reagent in E1 buffer (MTS-treated) or E1 buffer alone (mock-treated). Oocytes were washed 3–5 times with room temperature E1 buffer to remove MTS reagent and transferred to a solution containing 100 nm [8-3H]adenosine (23 Ci/mmol) in E1 buffer, to measure PfENT1-mediated adenosine uptake as described above. The percent inhibition due to MTS reagent application was calculated as [1 − (adenosine uptake after MTS reagent/adenosine uptake in untreated oocytes)] × 100. MTS-reactive Cys mutants were defined as those where the extent of MTS reagent induced inhibition of adenosine uptake was significantly greater than the effect on WT by a one way ANOVA using Dunnett's multiple comparison post hoc test (p < 0.05).
The MTS reagents used included, in order of increasing size, methyl methanethiosulfonate (MMTS), MTS-ethylammonium (MTSEA+), MTS-ethyltrimethylammonium (MTSET+), MTS-ethylsulfonate (MTSES−), MTS-ethylammonium-biotin (MTSEA-biotin), and MTS-tetramethylrhodamine (MTS-TMR) (Biotium, Haywood, CA). MTSET+ and MTSES− are of comparable size. To ensure that the MTS reagents did not hydrolyze prior to their application, MTSEA-biotin and MTS-TMR were prepared as a 200 mm stock solution in DMSO while MMTS was purchased as a 10 m stock solution in DMSO. These reagents were diluted into E1 buffer at appropriate concentrations immediately before use. Solvent controls were used where appropriate. MTSET+, MTSES−, and MTSEA+ were all dissolved directly in E1 buffer immediately before use.
Hypoxanthine Protection Experiments
For the Cys mutants that reacted with an MTS reagent, we investigated whether co-application of hypoxanthine, a PfENT1 transported substrate, with the MTS reagent would protect the Cys mutant from covalent modification. Oocytes were pretreated with a saturating concentration of hypoxanthine (>4 mm) and a range of concentrations of an MTS reagent. After a 2-min pretreatment, the oocytes were washed five times with room-temperature E1 buffer before being placed in 100 nm [8-3H]adenosine for the 10 min uptake experiment as described above. Uptake values were then transformed to a percentage of uptake in oocytes in the absence of MTS reagent (100%), or uninjected oocytes in the absence of MTS reagent (0%). The inhibition data were fit by nonlinear regression analysis using a one site competition equation, %Maximal Uptake = (max uptake − min uptake)/(1 + 10[inhibitor]−logIC50), where the min uptake was constrained to be greater than zero and max uptake was determined to be the best fit by the program using GraphPad Prism 5.0 software (La Jolla, CA). Protection is inferred if the MTS reagent IC50 value was increased in the presence of hypoxanthine compared with the MTS reagent IC50 in the absence of hypoxanthine.
Data Analysis
All data were analyzed using Microsoft Excel and GraphPad Prism 5.0 software. Each data point represents the values from at least three experiments + S.E. unless otherwise indicated. All experiments were performed in oocytes from at least two separate frogs, unless otherwise indicated. Significant differences were determined using either a one-way ANOVA and Dunnett's post-hoc test or Student's t test where appropriate. A paired two tailed t test was used to compare the effects of dipyridamole.
RESULTS
Previous studies have noted the existence of polymorphisms within the pfent1 gene but the functional consequences of these polymorphisms have never been studied (9). To corroborate these reports and to search for other polymorphisms, we sequenced the pfent1 gene in eleven P. falciparum strains originating around the world (gift of Dr. David Fidock, Columbia University). We considered the 3D7 sequence as the wild type. We identified a C1182G polymorphism, however we note that this polymorphism alters the amino acid at position 394 in the W2 and 7G8 strains from phenylalanine to leucine (Fig. 2A). A previous study (9) described the presence of a C1154G polymorphism within PfENT1 and suggested that this mutation leads to a phenylalanine to leucine mutation at position 385 in the primary protein sequence. Upon comparing Carter's published W2 pfent1 sequence and our own W2 pfent1 sequence, we find they are identical. We surmise that the Carter study correctly identified the polymorphism within this region of the protein, but incorrectly annotated its position. In addition to the F394L mutation, we identified other polymorphisms that lead to the following mutations; F36S (HB3, GC03, Malayan), V129I (HB3, GC03, Malayan), and L133F (Malayan).
FIGURE 2.
Effect of polymorphism at position 394 on PfENT1 function. A, amino acid sequences of the region in and flanking PfENT1 TM11 from eight distinct P. falciparum strains. The specific strain is indicated to the left, the solid bar above indicates the predicted extent of TM11. The glycines in the absolutely conserved GXXXG motif are highlighted by the vertical stripes, and the 394 position of the polymorphism is indicated by the vertical arrow below the sequences. Note that 7G8 and W2 have a leucine at position 394, whereas all of the other strains have a phenylalanine. B, effect of 10 μm dipyridamole on 1 μm [3H]adenosine uptake by oocytes expressing either Phe-394 or Leu-394 PfENT1 expressed as a percent of the untreated. The differences in the presence and absence of 10 μm dipyridamole are not significant by paired two-tailed Student's t test. Of note, the dipyridamole Ki for the sensitive human ENT1 transporter is 48 nm (25). C, concentration dependence of [3H]hypoxanthine uptake by oocytes expressing either Phe-394 (open triangles) or Leu-394 (solid triangles). Lines represent Michaelis-Menten fits to the data. D, concentration dependence of [3H]adenosine uptake by oocytes expressing either Phe-394 (open squares) or L394 (solid squares). Lines represent Michaelis-Menten fits to the data.
To determine whether these polymorphisms exert effects upon PfENT1, we generated the F36S, L133F and F394L mutations in a Xenopus oocyte pfent1 expression vector. Because of the conservative nature of the V129I polymorphism, this position was not studied further. Wild-type and mutant PfENT1 were expressed in Xenopus oocytes, and the kinetics of [3H]adenosine and [3H]hypoxanthine uptake were measured to determine the Km and Vmax for the wild-type (3D7 strain) and mutant transporters. We found that the F394L mutation caused a significant (p < 0.05) change in the PfENT1 substrate Km for both adenosine and hypoxanthine (Table 1) causing a 3-fold reduction in the Km (Fig. 2, C and D). The Vmax of the F394L mutant was 2-fold lower than wild-type PfENT1. The F36S and L133F mutants did not alter the adenosine Km but resulted in increases in the adenosine Vmax. It should be noted that the changes in Vmax could arise either because of changes in the transport rate of individual transporters or changes in the level of expression in the plasma membrane. Our experiments did not distinguish between these possibilities.
TABLE 1.
Km and Vmax for adenosine and hypoxanthine transport by PfENT1 and several naturally occurring polymorphisms
| Adenosine |
Hypoxanthine |
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | pmol/oocyte | μm | pmol/oocyte | |
| PfENT1 3D7a | 1,080 ± 570b | 212 ± 40 | 1,204 ± 620 | 218 ± 42 |
| PfENT1 L394 | 364 ± 108 | 110 ± 8 | 355 ± 73 | 94 ± 5 |
| PfENT1 F133 | 975 ± 187 | 328 ± 22 | 1,370 ± 398 | 255 ± 29 |
| PfENT1 S36 | 2,254 ± 595 | 574 ± 67 | 2,128 ± 805 | 387 ± 75 |
a PfENT1 3D7 is Phe-394, Leu-133, and Phe-36.
b Values are mean ± S.E.
A previous study of human and Caenorhabditis elegans ENTs (29) demonstrated that mutagenesis at a position in those proteins analogous to PfENT1 L393 (Fig. 1) altered both transport Km as well as sensitivity to the nucleoside transport inhibitor dipyridamole. The proximity of the F394L polymorphism to the position identified by Visser et al. suggested that mutation at position 394 might sensitize PfENT1 to dipyridamole. Furthermore, initial characterizations of PfENT1 by Carter et al. (9) reported sensitivity to 10 μm dipyridamole, while Parker et al. (10) reported insensitivity to dipyridamole, and it was suggested that the amino acid difference at this position might be responsible for modulating dipyridamole sensitivity. However, we find that 10 μm dipyridamole did not significantly inhibit the [3H]adenosine uptake by oocytes expressing either wild-type (p = 0.06) or F394L (p = 0.21) PfENT1 (Fig. 2B). Of note, for the Ki for the dipyridamole-sensitive human ENT1 is 48 nm (25).
Based upon hydropathy plot analysis, position 394 is located within the TM11 segment of PfENT1. This residue lies in the middle of a highly conserved GXXXG motif found in many ENT family members (Fig. 1, position 394 indicated by arrow). The functional effect of the F394L mutation and the presence of a conserved structural motif within this region of TM11 suggested that this transmembrane domain could play an important role in the structure and function of PfENT1. To further study this region of PfENT1, we utilized the substituted cysteine accessibility method (SCAM) to probe the water surface accessibility of residues in TM11 surrounding position 394 (54, 37).
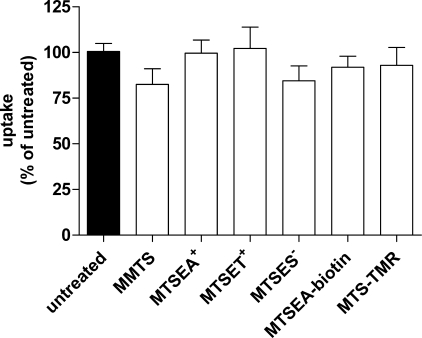
Wild-type PfENT1 contains 11 endogenous Cys residues. We first examined the effect on [3H]adenosine uptake of various sulfhydryl-reactive MTS reagents on the wild-type PfENT1 expressed in Xenopus oocytes. We used six MTS reagents that varied in terms of size and charge. A 2-min application of 2 mm MMTS, MTSEA+, MTSET+, MTSES−, MTSEA-biotin, or MTS-TMR (MTS reagents are listed in order of increasing size, except for MTSET+ and MTSES−, which are of similar size) had no significant effect on subsequent [3H]adenosine uptake (Fig. 3). We infer that the endogenous Cys are either inaccessible to reaction with the MTS reagents tested or that reaction had no functional effect on adenosine transport. Regardless of the interpretation, this finding suggests that functional effects observed following MTS treatment of PfENT1 TM11 Cys mutants are not likely to be due to modification of the endogenous PfENT1 Cys.
FIGURE 3.
Effect of a 2-min application of various MTS reagents on subsequent 10-min [3H]adenosine uptake by oocytes expressing WT PfENT1. Each uptake value is represented as a percent of uptake by untreated oocytes. Mean ± S.E. of three experiments is shown. One way ANOVA showed no significant differences between the means (p = 0.59).
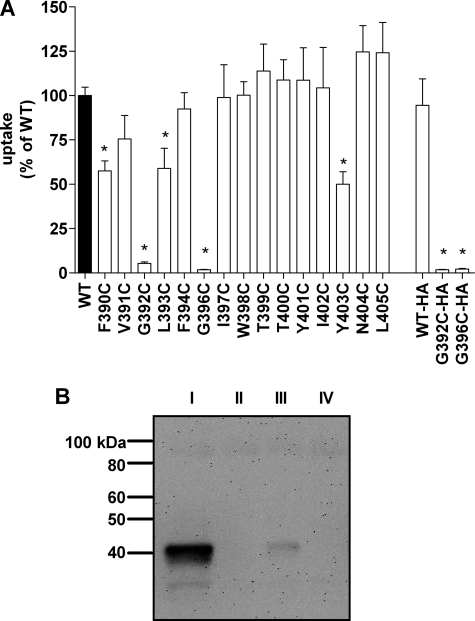
We introduced Cys substitutions, one at a time into the 15 residues from Phe-390 to Leu-405 with the exception of position 395, which is one of the endogenous Cys in the wild-type sequence. To use changes in substrate uptake as an assay for covalent modification of engineered Cys by the MTS reagents the Cys mutants must have a measurable rate of substrate transport. Therefore, we assessed the impact of the Cys mutations on nucleoside transport function. We expressed each of the Cys substitution mutants individually in Xenopus oocytes and measured the uptake of [3H]adenosine. Cys substitution mutations at positions F390C, L393C, and Y403C reduced the adenosine uptake by about 50% compared with wild type (Fig. 4A). Because we only measured substrate uptake at a single substrate concentration we cannot elucidate the mechanistic basis for the reduction in substrate uptake that we observed in these three Cys mutants. However, the important issue is that they transport [3H]adenosine at a rate sufficient to assay effects of MTS reagent application. At positions Gly-393 and Gly-396, the two highly conserved glycine residues in the GXXXG motif, mutation to Cys abolished adenosine uptake (Fig. 4A). For all other positions examined, Cys substitution mutagenesis had no statistically significant effect upon adenosine transport.
FIGURE 4.
Effect of TM11 Cys substitution mutants on PfENT1 transport function and protein expression. A, [3H]adenosine uptake by Cys substitution mutant expressing oocytes as a percent of uptake by wild-type PfENT1-expressing oocytes. * indicates uptake significantly different from WT by one way ANOVA using Dunnett's multiple comparison post hoc test (p < 0.05). HA indicates constructs with a C-terminal HA epitope tag. Mean ± S.E. of three experiments is plotted. B, Western blot of total cellular proteins from oocytes expressing HA epitope-tagged constructs of WT (I), G392C (II), and G396C (III) PfENT1, and uninjected oocytes (IV) is shown. Blot probed with mouse anti-HA primary antibody and a goat anti-mouse horseradish peroxidase-conjugated IgG secondary antibody was visualized by chemiluminescence.
The dramatic decrease in PfENT1 mediated adenosine transport in the G392C or G396C mutants could be explained by disruption of either 1) protein folding and expression or 2) transport function. To determine whether these mutations affected protein expression level, we generated an HA-epitope tagged version of the wild-type, G393C and G396C constructs. Addition of the HA epitope tag had little effect on adenosine uptake compared with the corresponding untagged constructs (Fig. 4A). We assayed the PfENT1 expression levels in whole oocyte protein preparations using Western blots probed with an anti-HA primary antibody. Our attempts to isolate membrane protein fractions by surface biotinylation and subsequent pull down with streptavidin-agarose beads gave no detectable chemiluminescent signal on a Western blot. However, Western blots of total oocyte protein showed that the G393C and G396C mutants caused a significant reduction in PfENT1 protein expression levels compared with wild type (Fig. 4B). This reduction in PfENT1 expression level likely explains the near absence of adenosine uptake for these two mutants.
For the 13 Cys mutants that displayed functional adenosine uptake we initially screened for surface accessibility with a 2-min application of 2 mm MTSEA-biotin. We chose to screen initially with MTSEA-biotin due to its comparable size to purine nucleosides. We hypothesized that if MTSEA-biotin reacted with an accessible Cys residue it would be more likely than the smaller MTS reagents to exhibit a functional effect upon subsequent PfENT1 transport properties.
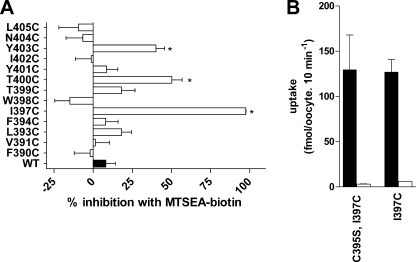
To determine whether MTSEA-biotin reacted with the introduced Cys mutants, we assayed the effect of applying this reagent upon subsequent PfENT1 mediated [3H]adenosine uptake. We found that MTSEA-biotin application caused a significant inhibition of subsequent [3H]adenosine uptake for the mutants I397C, T400C, and Y403C (Fig. 5A). As a control to ensure that the MTS reagents were not reacting with the endogenous TM11 Cys, Cys-395, we generated a double mutant C395S/I397C. The effect of MTSEA-biotin was similar on I397C and on the double mutant C395S/I397C (Fig. 5B). This confirms that the effects of MTSEA-biotin modification were due to reaction with the engineered Cys and not due to reaction with the nearby endogenous Cys-395. From these findings, we infer that positions 397, 400, and 403, are, at least transiently, on the water-accessible protein surface.
FIGURE 5.
Effect of a 2-min application of 2 mm MTSEA-biotin on subsequent [3H]adenosine uptake by oocytes expressing the TM11 Cys substitution mutants. A, percent inhibition and S.E. for each mutant is shown. * indicate mutants where MTSEA-biotin application caused significantly greater inhibition than the effect on WT by one way ANOVA using Dunnett's multiple comparison post hoc test (p < 0.05). B, control for the presence of the endogenous Cys at position 395. Black bars, [3H]adenosine uptake by untreated oocytes; white bars, [3H]adenosine uptake by MTSEA-biotin treated oocytes. Note that the double mutant does not affect adenosine uptake (black bars), and the effect of a 2-min application of 2 mm MTSEA-biotin is the same in the double mutant C395S/I397C and in the single mutant I397C (white bars). This experiment, panel B, was performed in oocytes from a single frog.
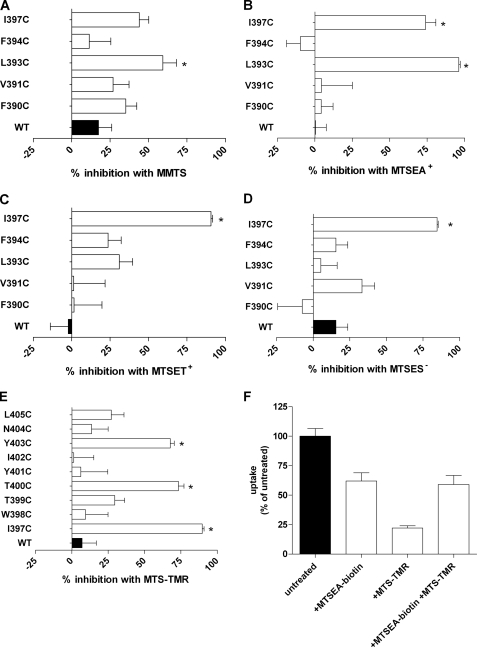
For the remainder of the positions tested, no change in [3H]adenosine uptake was noted following MTSEA-biotin treatment. There are several potential explanations for these apparently non-reactive residues. They could be inaccessible to reaction or they could undergo reaction but have no functional effect. We hypothesized that near the intracellular end of TM11 there might not be sufficient space for the fairly large MSTEA-biotin molecule to reach a water accessible Cys. For this reason, we examined the accessibility of the residues from position Phe-390 to Ile-397 with the smaller MTS reagents MMTS and MTSEA+. A 2-min application of 2 mm MMTS only caused significant inhibition of subsequent [3H]adenosine uptake at L393C (Fig. 6A). MTSEA+ caused significant inhibition of subsequent [3H]adenosine uptake in the mutants L393C and I397C but had no significant effects on the other Cys mutants (Fig. 6B).
FIGURE 6.
Effect of a 2-min application of various MTS reagents on subsequent [3H]adenosine uptake by oocytes expressing subsets of the TM11 Cys substitution mutants. The x-axis shows percent inhibition of subsequent [3H]adenosine uptake. Effect of 2 mm application of: A, MMTS; B, MTSEA+; C, MTSET+; D, MTSES−; E, MTS-TMR. F, effects of sequential application of MTSEA-biotin and MTS-TMR on subsequent [3H]adenosine uptake by oocytes expressing the T400C mutant PfENT1. The figure in panel F illustrates the results from one batch of oocytes. Similar results were obtained in a second batch of oocytes from a different frog.
To probe the electrostatic environment along the access pathway and in proximity to the more cytoplasmic engineered TM11 Cys mutants we tested the effects of the positively charged MTSET and the negatively charged MTSES on the Cys substituted for Phe-390 to Ile-397. These reagents are of comparable size. Both reacted with I397C but with none of the other more cytoplasmic Cys mutants (Fig. 6, C and D). This suggests that their size limited their access to L393C that reacted with the smaller, but positively charged MTSEA but not the larger MTSET.
At the extracellular end of TM11, we hypothesized that there might be sufficient room to accommodate an MTSEA-biotin modified Cys without having a functional effect. Therefore, we tested the effect of applying the larger MTS reagent, MTS-TMR, on the Cys substitution mutants from Ile-397 to Leu-405. A 2-min application of 2 mm MTS-TMR significantly inhibited subsequent [3H]adenosine uptake of the same mutants as MTSEA-biotin, I397C, T400C, and Y403C (Fig. 6E). For the T400C mutant we noted that the 2-min application of MTS-TMR caused significantly greater inhibition of subsequent [3H]adenosine uptake than the application of MTSEA-biotin. To determine whether the MTSEA-biotin reaction had gone to completion we first applied 2 mm MTSEA-biotin for 2 min, washed the oocytes and then applied 2 mm MTS-TMR for 2 min. The subsequent application of MTS-TMR caused no additional inhibition of adenosine uptake (Fig. 6F). We infer that the MTSEA-biotin reaction had gone to completion and that the greater inhibition caused by MTS-TMR was due to its larger size compared with MTSEA-biotin.
We infer that the MTS reactive Cys mutants are, at least transiently, on the water-accessible protein surface because the MTS reagents react about 109 times faster with the ionized thiolates than with an unionized thiol (45); only Cys on the water-accessible surface will ionize to any significant extent. Whether they line the purine permeation pathway or some other water-accessible surface in the transmembrane domain is difficult to establish. We sought to determine whether substrate transport via PfENT1 would affect the MTS reagent reaction rates with the reactive Cys mutants. We assayed the extent of inhibition as a function of MTS reagent concentration applied in the absence or presence of >4 mm hypoxanthine, a saturating concentration of a purine nucleobase transported by PfENT1. We used hypoxanthine for the protection experiments to avoid effects on cytoplasmic adenosine metabolism that would alter subsequent adenosine uptake (16). The presence of >4 mm hypoxanthine during the MTS reagent application significantly increased the MTS reagent IC50 for the L393C, I397C, and T400C mutants (Fig. 7 and Table 2). In contrast, the Y403C mutant was not protected from reaction with MTS-TMR by the presence of >4 mm hypoxanthine (Fig. 7D and Table 2). Thus, the presence of a saturating concentration of hypoxanthine protected a subset of the Cys mutants from reaction with MTS reagent.
FIGURE 7.
Hypoxanthine, a transported substrate, protects some of the MTS-reactive Cys mutants from covalent modification by the MTS reagents. Oocytes expressing the MTS-reactive PfENT1 mutants: A, L393C; B, I397C; C, T400C; and D, Y403C. Oocytes were incubated for 2 min in solutions containing increasing concentrations of MTS reagents either in the absence (open triangles) or presence of >4 mm hypoxanthine (filled squares). The oocytes were washed, and the subsequent [3H]adenosine uptake was measured. Uptake as a percent of no MTS pretreatment is plotted as a function of MTS reagent concentration. Each point is the mean ± S.E. of 5 oocytes from one experiment; in some cases the S.E. is smaller than the symbol. Note that in panels A–C the MTS inhibition curve in the presence of hypoxanthine is displaced to higher concentrations, which indicates that the hypoxanthine protected the Cys from reaction. No protection is observed for Y403C in panel D.
TABLE 2.
IC50 for MTS reagents reacting with PfENT1 TM11 Cys mutants in the absence and presence of >4 mm hypoxanthine
| Mutant | MTS-reagent | −Hypoxanthine |
+Hypoxanthine |
|---|---|---|---|
| IC50 | IC50 | ||
| μm | μm | ||
| L393C | MTSEA | 70 ± 21 | 685 ± 109 |
| I397Ca | MTSEA-biotin | 4 ± 2a | 82 ± 12a |
| T400C | MTS-TMR | 17 ± 4 | 357 ± 237 |
| Y403C | MTS-TMR | 88 ± 24 | 75 ± 32 |
a For I397C, values come from two batches of oocytes and are the mean ± range/2. All other values are mean ± S.E. from three independent experiments.
DISCUSSION
There has been little investigation of the role of the TM11 segment in the structure and function of equilibrative nucleoside transporters. Based on the presence of a naturally occurring functional polymorphism in the P. falciparum PfENT1 TM11 segment and its location in the middle of a highly conserved GXXXG motif we performed a substituted cysteine accessibility study of this transmembrane segment. Mutation of either glycine residue in the GXXXG motif to Cys caused a significant reduction in protein expression level. We did not investigate whether this was due to a reduced synthesis rate or an increased degradation rate. Increased degradation could arise due to protein misfolding or failure of correct assembly. The TM11 GXXXG motif is present in most ENT family members. GXXXG motifs are frequently found at helix-helix interaction sites where the presence of the glycines creates a patch on the surface of the helix where access to the backbone is unhindered by amino acid side chains (47–50). This creates a “hole” in the helix surface where a “knob” formed by large amino acid side chains from an adjacent helix can insert to form a stable helix-helix interaction site. The importance of GXXXG motifs was first identified as the basis of dimerization in glycophorin and has subsequently been shown to serve an important role in the folding and assembly of transmembrane segments in numerous integral membrane proteins (47–50). At present we do not know whether the helix interacting with the GXXXG motif is one of the other ten helices in the same PfENT1 molecule or whether it is a dimer interface site either to form a homodimer with another PfENT1 molecule or a heterodimer with some other protein that would have to be common to P. falciparum and Xenopus oocytes. Nevertheless, the interaction via the GXXXG motif appears to be critical for PfENT1 expression.
The SCAM study of TM11 identified four MTS reactive Cys substitution mutants, L393C, I397C, T400C, and Y403C. Based on the preferential reactivity of MTS reagents with ionized thiolates and the assumption that only water-accessible Cys will ionize to any significant extent, we infer that the corresponding amino acids lie on the water-accessible protein surface. When plotted on an α-helical wheel (Fig. 8), three of these residues, Leu-393, Ile-397, and Thr-400, lie on one face of the helix within an arc of 60°. This suggests that the secondary structure of much of TM11 is α helical. The most extracellular residue, Tyr-403, does not lie on this face of the TM11 helix. It may be accessible to reaction with the MTS reagents because it extends above the plane of the membrane. Modification of a Cys substituted for Tyr-403 may have a functional effect because it lies on the helix face between the two conserved glycines, although at a more extracellular level. If this helix face forms an important structural interaction with another transmembrane helix then modification of this Cys by the larger MTS reagents may disrupt this interaction causing effects on protein dynamics and indirectly on transport function.
FIGURE 8.
α-Helical wheel and net representations of the amino acids in the PfENT1 TM11 segment. Color code: MTS-reactive residues, red; mutation affects function, green; Cys mutation alters protein expression, blue; endogenous Cys, yellow. The dashed line indicates the MTS-reactive face of the TM11 helix.
Whereas MTSEA-biotin reacted with the more extracellular of the MTS-reactive Cys mutants, it did not react with the most cytoplasmic MTS-reactive mutant, L393C. The ability of the smaller MTS reagents, MMTS and MTSEA+ to react with L393C suggests that local steric factors, either in close proximity to this residue or along the access pathway, may limit the access of the larger reagents to this residue. None of the MTS reagents had an effect on the next more cytoplasmic residue on this helix face, F390C suggesting that it may be inaccessible to the MTS reagents, most likely due to tight protein packing in the region of this residue. It is important, however, to interpret apparently “non-reactive” Cys mutants with caution because we are using a functional assay to determine chemical reaction. The substituted Cys may be inaccessible for reaction or it may react but covalent modification of the residue may have no functional effect. We have seen examples of silent reaction in other proteins that we have studied (55). The lack of effect of the MTS reagents on the N404C mutant may be an example of a silent reaction. N404C lies at the extracellular end of TM11 on the same helix face as the MTS reactive residues (Fig. 8). We cannot rule out the possibility that N404C is close enough to the extracellular end that there is sufficient space to accommodate the MTS-TMR-modified Cys residue without causing steric interactions with other parts of the protein or with the purine as it enters the transport pathway. This could result in covalent modification of the Cys with no apparent functional consequences.
The transported substrate hypoxanthine protected three of the reactive Cys mutants, L393C, I397C, and T400C, from modification by MTS reagents but did not protect Y403C. The protected Cys mutants lie on one side of the TM11 α-helix. This suggests that the corresponding wild-type amino acids may line part of the nucleobase permeation pathway and that the presence of hypoxanthine sterically protected them from reaction with the MTS reagents. However, we cannot exclude the possibility that hypoxanthine transport induced a conformational change in the protein that reduced the access of the MTS reagent to these engineered Cys residues. Further work will be necessary to determine whether TM11 lines part of the substrate permeation pathway. In a recent publication ab initio models ofLdNT1.1 were generated using the Rosetta software (34). Three models were assembled computationally. The major difference among the three models was in the position of the TM11 segment. Model 2, which positions TM11 in the permeation pathway lining, was favored by Valdés et al. (34) because mutations of two residues in the LdNT1.1 TM11 segment S469F (aligned with PfENT1 A388, Fig. 1) and T478F (aligned with PfENT1 I397, Fig. 1) crippled transport. Our current results support the hypothesis that a subset of TM11 residues line the permeation pathway. Of note, the PfENT1 residues Ala-388 and Ile-397 are aligned with those mutated in the report of Valdés et al. (34); they lie on opposite sides of the TM11 helix (Fig. 8A). Our results indicate that Ile-397 is on the water-accessible protein surface in the permeation pathway, whereas Ala-388 lies on the opposite side of the TM11 helix away from the permeation pathway. The I397C mutant displayed similar adenosine transport to wild type but covalent modification of I397C by the larger MTS reagents, MTS-TMR, MTSEA-biotin, MTSET, and MTSES caused almost complete inhibition of adenosine transport, consistent with a phenylalanine at this position disrupting LdNT1.1 function. Modification of I397C by the smaller reagent, MMTS, only caused a 50% decrease in adenosine transport suggesting that the thiomethyl modification of the engineered Cys was better tolerated. MMTS-modified Cys would occupy a similar volume as the native isoleucine residue but lack the β-branched structure. The β-branched side chain limits the potential rotamer conformations when the residue is in an α-helix. Thus, the increased rotamer flexibility of the MMTS-modified Cys compared with the native isoleucine might account for the observed effects on adenosine transport.
Recent work (35) reported that expression in PK15-NTD cells of an alternative splice variant of the mENT1 transporter lacking exon 11, which encodes TM9–11, yielded functional nucleoside transport and NBMPR binding. It should be noted that the PK15-NTD cells retain endogenous nucleoside transport mechanisms whose existence complicates the interpretation of the Δexon-11-mENT1 heterologous expression experiments (56). Mouse ENT1 has the conserved GXXXG motif in TM11 (Fig. 1). Further work will be necessary to reconcile our findings that mutation of either of the glycines in the GXXXG motif significantly reduced protein expression and eliminated transport with the reported result that the exon 11 deletion variant was functional.
The naturally occurring polymorphism that provided the impetus for this study is at position 394. A Cys substituted at this position was not reactive with any of the MTS reagents tested. It lies in the middle of the GXXXG motif but on the opposite side of the TM11 helix from the two glycines (Fig. 8). Mutation of phenylalanine to leucine at this position had an effect on the Km and Vmax for adenosine transport. This position is probably buried in the protein interior interacting with another transmembrane segment. The effects of the mutation are probably indirect, because of an alteration in the protein structure that affects the position of residues that interact with the transported substrates. Thus, one cannot conclude that because mutation at a given position affects the apparent affinity for substrate or inhibitor that the amino acid at that position directly interacts with the substrate or inhibitor.
The current work has demonstrated that residues from TM11 likely form a portion of the permeation pathway lining. We have also shown that the highly conserved GXXXG motif has an important role in the folding and assembly of ENTs. Further work on the structure of the ENTs will help to elucidate the basis for substrate and inhibitor specificity and the conformational changes that the protein undergoes during the transport process. This will aid in the identification and design of specific PfENT1 inhibitors that might serve as a new generation of antimalarial drugs.
Acknowledgments
We thank Minghao Liu for technical assistance with some of the experiments and Moez Bali, Nicole McKinnon, Rishi Parikh, and M. Belen Cassera for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health NIGMS Medical Scientist Training Program Grant T32-GM007288 (to P. M. R.).
- ENT
- equilibrative nucleoside transporter
- CeENT
- C. elegans ENT
- hENT
- human ENT
- LdNT1.1
- L. donovani Nucleoside Transporter 1.1
- mENT
- mouse ENT
- rENT
- rat ENT
- MTS
- methanethiosulfonate
- MMTS
- methylmethanethiosulfonate
- MTSEA
- MTS-ethylammonium
- MTSES
- MTS-ethylsulfonate
- MTSET
- MTS-ethyltrimethylammonium
- MTS-TMR
- MTS-tetramethylrhodamine
- NBMPR
- S6-(4-nitrobenzyl)-mercaptopurine riboside
- PfENT1
- P. falciparum equilibrative nucleoside transporter 1
- SCAM
- substituted cysteine accessibility method
- TM
- transmembrane
- HA
- hemagglutinin
- TM
- transmembrane
- ANOVA
- analysis of variance
- WT
- wild type.
REFERENCES
- 1.Cox J., Hay S. I., Abeku T. A., Checchi F., Snow R. W. (2007) Trends Parasitol. 23, 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellems T. E., Hayton K., Fairhurst R. M. (2009) J. Clin. Invest. 119, 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eastman R. T., Fidock D. A. (2009) Nat. Rev. Microbiol. 7, 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mita T., Tanabe K., Kita K. (2009) Parasitol. Int. 58, 201–209 [DOI] [PubMed] [Google Scholar]
- 5.Plowe C. V. (2009) Trans. R. Soc. Trop. Med. Hyg 103, Suppl. 1, S11–S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downie M. J., Kirk K., Mamoun C. B. (2008) Eukaryot. Cell 7, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin R. E., Henry R. I., Abbey J. L., Clements J. D., Kirk K. (2005) Genome Biol. 6, R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin R. E., Ginsburg H., Kirk K. (2009) Mol. Microbiol. 74, 519–528 [DOI] [PubMed] [Google Scholar]
- 9.Carter N. S., Ben Mamoun C., Liu W., Silva E. O., Landfear S. M., Goldberg D. E., Ullman B. (2000) J. Biol. Chem. 275, 10683–10691 [DOI] [PubMed] [Google Scholar]
- 10.Parker M. D., Hyde R. J., Yao S. Y., McRobert L., Cass C. E., Young J. D., McConkey G. A., Baldwin S. A. (2000) Biochem. J. 349, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rager N., Mamoun C. B., Carter N. S., Goldberg D. E., Ullman B. (2001) J. Biol. Chem. 276, 41095–41099 [DOI] [PubMed] [Google Scholar]
- 12.Downie M. J., Saliba K. J., Howitt S. M., Bröer S., Kirk K. (2006) Mol. Microbiol. 60, 738–748 [DOI] [PubMed] [Google Scholar]
- 13.El Bissati K., Zufferey R., Witola W. H., Carter N. S., Ullman B., Ben Mamoun C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downie M. J., Saliba K. J., Bröer S., Howitt S. M., Kirk K. (2008) Int. J. Parasitol. 38, 203–209 [DOI] [PubMed] [Google Scholar]
- 15.El Bissati K., Downie M. J., Kim S. K., Horowitz M., Carter N., Ullman B., Ben Mamoun C. (2008) Mol. Biochem. Parasitol. 161, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riegelhaupt P. M., Cassera M. B., Fröhlich R. F., Hazleton K. Z., Hefter J. J., Schramm V. L., Akabas M. H. (2010) Mol. Biochem. Parasitol. 169, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin S. A., McConkey G. A., Cass C. E., Young J. D. (2007) Curr. Pharm. Des. 13, 569–580 [DOI] [PubMed] [Google Scholar]
- 18.Schramm V. L. (2004) Nucleosides Nucleotides Nucleic Acids 23, 1305–1311 [DOI] [PubMed] [Google Scholar]
- 19.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. (2004) Pflugers Arch. 447, 735–743 [DOI] [PubMed] [Google Scholar]
- 20.Landfear S. M., Ullman B., Carter N. S., Sanchez M. A. (2004) Eukaryot. Cell 3, 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths M., Beaumont N., Yao S. Y., Sundaram M., Boumah C. E., Davies A., Kwong F. Y., Coe I., Cass C. E., Young J. D., Baldwin S. A. (1997) Nat. Med. 3, 89–93 [DOI] [PubMed] [Google Scholar]
- 22.Sundaram M., Yao S. Y., Ingram J. C., Berry Z. A., Abidi F., Cass C. E., Baldwin S. A., Young J. D. (2001) J. Biol. Chem. 276, 45270–45275 [DOI] [PubMed] [Google Scholar]
- 23.Yao S. Y., Sundaram M., Chomey E. G., Cass C. E., Baldwin S. A., Young J. D. (2001) Biochem. J. 353, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SenGupta D. J., Lum P. Y., Lai Y., Shubochkina E., Bakken A. H., Schneider G., Unadkat J. D. (2002) Biochemistry 41, 1512–1519 [DOI] [PubMed] [Google Scholar]
- 25.Visser F., Vickers M. F., Ng A. M., Baldwin S. A., Young J. D., Cass C. E. (2002) J. Biol. Chem. 277, 395–401 [DOI] [PubMed] [Google Scholar]
- 26.Endres C. J., Sengupta D. J., Unadkat J. D. (2004) Biochem. J. 380, 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SenGupta D. J., Unadkat J. D. (2004) Biochem. Pharmacol. 67, 453–458 [DOI] [PubMed] [Google Scholar]
- 28.Endres C. J., Unadkat J. D. (2005) Mol. Pharmacol. 67, 837–844 [DOI] [PubMed] [Google Scholar]
- 29.Visser F., Baldwin S. A., Isaac R. E., Young J. D., Cass C. E. (2005) J. Biol. Chem. 280, 11025–11034 [DOI] [PubMed] [Google Scholar]
- 30.Visser F., Sun L., Damaraju V., Tackaberry T., Peng Y., Robins M. J., Baldwin S. A., Young J. D., Cass C. E. (2007) J. Biol. Chem. 282, 14148–14157 [DOI] [PubMed] [Google Scholar]
- 31.Arendt C. S., Ullman B. (2010) J. Biol. Chem. 285, 6024–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramson J., Kaback H. R., Iwata S. (2004) Curr. Opin. Struct. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 33.Law C. J., Maloney P. C., Wang D. N. (2008) Annu. Rev. Microbiol. 62, 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdés R., Arastu-Kapur S., Landfear S. M., Shinde U. (2009) J. Biol. Chem. 284, 19067–19076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robillard K. R., Bone D. B., Park J. S., Hammond J. R. (2008) Mol. Pharmacol. 74, 264–273 [DOI] [PubMed] [Google Scholar]
- 36.Akabas M. H., Kaufmann C., Archdeacon P., Karlin A. (1994) Neuron 13, 919–927 [DOI] [PubMed] [Google Scholar]
- 37.Karlin A., Akabas M. H. (1998) Methods Enzymol. 293, 123–145 [DOI] [PubMed] [Google Scholar]
- 38.Javitch J. A., Shi L., Liapakis G. (2002) Methods Enzymol. 343, 137–156 [DOI] [PubMed] [Google Scholar]
- 39.Hou Z., Ye J., Haska C. L., Matherly L. H. (2006) J. Biol. Chem. 281, 33588–33596 [DOI] [PubMed] [Google Scholar]
- 40.Kaback H. R., Dunten R., Frillingos S., Venkatesan P., Kwaw I., Zhang W., Ermolova N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueckler M., Makepeace C. (2008) J. Biol. Chem. 283, 11550–11555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Ye L., McKinney C. C., Feng M., Maloney P. C. (2008) Biochemistry 47, 5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slugoski M. D., Ng A. M., Yao S. Y., Lin C. C., Mulinta R., Cass C. E., Baldwin S. A., Young J. D. (2009) J. Biol. Chem. 284, 17281–17292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verselis V. K., Trelles M. P., Rubinos C., Bargiello T. A., Srinivas M. (2009) J. Biol. Chem. 284, 4484–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts D. D., Lewis S. D., Ballou D. P., Olson S. T., Shafer J. A. (1986) Biochemistry 25, 5595–5601 [DOI] [PubMed] [Google Scholar]
- 46.Valdés R., Vasudevan G., Conklin D., Landfear S. M. (2004) Biochemistry 43, 6793–6802 [DOI] [PubMed] [Google Scholar]
- 47.Russ W. P., Engelman D. M. (2000) J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 48.Doura A. K., Kobus F. J., Dubrovsky L., Hibbard E., Fleming K. G. (2004) J. Mol. Biol. 341, 991–998 [DOI] [PubMed] [Google Scholar]
- 49.Senes A., Engel D. E., DeGrado W. F. (2004) Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 50.Harrington S. E., Ben-Tal N. (2009) Structure 17, 1092–1103 [DOI] [PubMed] [Google Scholar]
- 51.Wellems T. E., Panton L. J., Gluzman I. Y., do Rosario V. E., Gwadz R. W., Walker-Jonah A., Krogstad D. J. (1990) Nature 345, 253–255 [DOI] [PubMed] [Google Scholar]
- 52.Jespersen T., Grunnet M., Angelo K., Klaerke D. A., Olesen S. P. (2002) BioTechniques 32, 536–538 [DOI] [PubMed] [Google Scholar]
- 53.Cassera M. B., Hazleton K. Z., Riegelhaupt P. M., Merino E. F., Luo M., Akabas M. H., Schramm V. L. (2008) J. Biol. Chem. 283, 32889–32899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akabas M. H., Stauffer D. A., Xu M., Karlin A. (1992) Science 258, 307–310 [DOI] [PubMed] [Google Scholar]
- 55.Bera A. K., Chatav M., Akabas M. H. (2002) J. Biol. Chem. 277, 43002–43010 [DOI] [PubMed] [Google Scholar]
- 56.Hoque K. M., Chen L., Leung G. P., Tse C. M. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1988–1995 [DOI] [PubMed] [Google Scholar]