Abstract
The RNA-binding protein human antigen R (HuR) has been implicated in apoptosis in multiple ways. Several studies have shown that in response to a variety of stresses HuR promotes the expression of proapoptotic mRNAs, whereas others reported its regulatory effect on antiapoptotic messages. We recently showed that in response to severe stress, HuR is cleaved to generate two cleavage products (CPs), HuR-CP1 (24 kDa) and HuR-CP2 (8 kDa), by which it promotes apoptotic cell death. Here, we show that this cleavage event is dependent on protein kinase RNA (PKR). Surprisingly, although in response to the apoptotic inducer staurosporine PKR itself is not phosphorylated, PKR triggers the cleavage of HuR via its downstream effector FADD that in turn activates the caspase-8/caspase-3 pathway. This effect, however, does not require the phosphorylation of the eukaryotic translation initiation factor 2α. Additionally, we observed that these HuR-CPs are sufficient to trigger cell death in the absence of activation of the PKR pathway. Therefore, our results support a model whereby in response to lethal stress, PKR, without being phosphorylated, activates the FADD/caspase-8/caspase-3 pathway to trigger HuR cleavage, and the HuR-CPs are then capable of promoting apoptosis.
Keywords: Apoptosis, Caspase, Protein Kinase RNA (PKR), RNA-binding Protein, RNA Turnover, ELAV, Human Antigen R (HuR)
Introduction
One of the most important posttranscriptional regulators of gene expression is the RNA-binding protein human antigen R (HuR).3 HuR associates with the 3′ untranslated region of multiple messages involved in a diverse array of cellular process, ranging from cell development and tissue differentiation, to cell survival and apoptotic death (1). The specific elements with which HuR binds are termed AU-rich elements, which confer instability upon the mRNA transcripts in which they are contained (2). HuR has actually been shown to play a stabilizing role when bound to AU-rich element-containing mRNAs, although it has also been shown to regulate the localization and translation of its mRNA targets as well (1, 3, 4).
In the last five years, several groups have published evidence showing that HuR is involved in cell survival and death, by regulating pro- and antiapoptotic mRNAs (1, 5–8). Our group was also studying the proapoptotic role of HuR when we discovered that in response to severe stress, HuR is exported to the cytoplasm with its protein partner pp32/PHAPI and is then cleaved by caspases-3 and -7 to generate two cleavage products (HuR-CP1 and HuR-CP2) (9). Although only 10–15% of HuR is cleaved (9, 10), it has also been shown that only ∼10% of total HuR is typically found in the cytoplasm due to its nucleocytoplasmic shuttling (11, 12). This cleavage may be prevented by mutating the 226th amino acid of HuR (Asp226) to an alanine (HuRD226A), and we observed that by preventing the generation of the HuR-CPs, cell death is delayed (9). Additionally, we recently showed that the 10–15% cleavage of HuR is essential to promote muscle fiber formation (10), further demonstrating the functional relevance of HuR cleavage. Our results also suggested that the promotion of apoptosis seen via HuR cleavage may be due to enhancement of apoptosome activity, possibly by disrupting the PHAPI·HuR complex.
The apoptosome is a large complex of Apaf-1 protein, along with procaspase-9, dATP, and cytochrome c molecules. During intrinsically induced apoptosis, cytochrome c is released from the mitochondria and activates the apoptosome, which leads to the generation of active caspase-9 (13, 14). Caspase-9 is an initiator cysteine protease which, when cleaved from procaspase-9 to become active, can activate downstream caspases such as -3 and -7 (effector caspases), which will process multiple targets, leading to cell death (13, 14). Moreover, the ability for the apoptosome to generate active caspase-9 has been shown to be potentiated or perturbed by additional factors. Prothymosin α is an inhibitor of apoptosomal activity, whereas a complex of PHAPI, heat shock protein 70 (Hsp70), and cellular apoptosis susceptibility protein is capable of promoting the activity of the apoptosome (15, 16).
Apoptosis may also be extrinsically induced in response to stress. This typically involves the binding of extracellular death ligands (e.g. Fas ligand) to cognate cell surface receptors (e.g. Fas), which triggers the recruitment of intracellular adaptor proteins, such as FADD, to activate the initiator caspase-8 (17). External stresses such as oxidative stress and drug treatment induce apoptosis by also activating the stress kinase protein kinase RNA (PKR), which then promotes cell death in a FADD/caspase-8-dependent manner (18–20). Caspase-8 is particularly interesting because it can directly activate caspase-3 while also promoting cytochrome c release from the mitochondria by activating the Bcl-2 protein Bid, leading to apoptosome activation (21). This demonstrates that through the activation of the apoptosome, cross-talk between the two apoptotic pathways (intrinsic and extrinsic) may occur to amplify the initial death stimulus.
Although it has been established that the cleavage of HuR represents an important step in the proapoptotic role that this protein plays (9), the signal transduction pathway(s) responsible for this event are still unknown. In this study, we identified the PKR/FADD/caspase-8-mediated signal transduction pathway as one of the main molecular processes responsible for the cleavage of HuR and show that the generation of HuR-CPs itself can trigger apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa CCL-2 cells (American Type Culture Collection), as well as PKR (generous gift from N. Sonenberg, McGill University, Montreal, Canada), GCN2, PERK knock-out, and wt mouse embryonic fibroblasts (MEFs) (kind donation from D. Ron, Skirball Institute, New York, NY) were grown and maintained in Dulbecco's modified Eagle's medium containing l-glutamine (Sigma-Aldrich) with fetal bovine serum (10%) and penicillin/streptomycin (Sigma-Aldrich). S51A eukaryotic translation initiation factor 2α (eIF2α) knock-in MEFs (generously donated by R. Kaufman, Howard Hughes Medical Institute, University of Michigan Medical Center, Ann Arbor, MI) were grown and maintained in the same medium as described for HeLa, supplemented with nonessential amino acids and β-mercaptoethanol (Invitrogen). FADD knock-out MEFs (kindly donated by Dr. T. Mak, Princess Margaret Hospital, Toronto, ON, Canada) were grown in the same medium as HeLa, supplemented with β-mercaptoethanol. Small interfering RNA (siRNA) duplexes (control (22), siRNA·FADD (Santa Cruz Biotechnology), and siRNA·PKR (Qiagen)) were transfected as described previously when at ∼50% confluence, according to the manufacturer's instructions. Samples were harvested 48 h following the transfection.
Preparation of Cell Extracts and Immunoblotting
Cells were scraped into their medium, spun down for 5 min at 1500 rpm and at 4 °C in a Thermo IEC Centra CL3R centrifuge, after which the cell pellet was washed with phosphate-buffered saline and spun a second time. Cell pellets were then resuspended on ice in lysis buffer, containing 50 mm HEPES (pH 7), 150 mm NaCl, 10% glycerol, 1% Triton X-100, 10 mm sodium pyrophosphate, 100 mm NaF, 1 mm EGTA, 1.5 mm MgCl2, 0.1 mm sodium orthovanadate, and complete EDTA-free protease inhibitors (Roche Applied Science). Western blotting was done as described previously (11) using the following antibodies to target the following factors: HuR (23), Ras-GAP SH3 domain-binding protein (G3BP) (24), FADD (Millipore), Thr(P)446-PKR (Abcam), caspase-8, Ser(P)51-eIF2α, Ser(P)73-c-Jun, (Thr(P)202/Tyr(P)204)-ERK1/2, Thr(P)334-MAPKAPK-2, PKR, eIF2α, caspase-3 (Cell Signaling), GST, α-tubulin (Sigma-Aldrich), and poly(ADP-ribose)polymerase (PARP; R&D Systems).
Antennapedia (AP)-conjugated Protein Production
AP-HuR-CP1-GST and AP-HuR-CP2-GST were generated by cloning HuR-CP1 and HuR-CP2 fragments into the AP/GST vector as described previously (22). The amplification of HuR-CP1 and HuR-CP2 was done with the following primers: AP-HuR-CP1-GST forward, 5′-GGCGCGGCCGCATCTAATGGTTATGAAGACCAC-3′; AP-HuR-CP1 reverse, 5′-GGCCTCGAGGTGATCGACGCCCATGGG-3′; AP-HuR-CP2-GST forward, 5′-GGCGCGGCCGCACACATGAGCGGGCTCTCT-3′; AP-HuR-CP2 reverse, 5′-GGCCTCGAGTTTGTGGGACTTGTTGGTTTT-3′. NotI/XhoI restriction digest fragments of HuR-CP1 and -CP2 PCR products were inserted into the NotI/XhoI sites of the Gateway AP-GST vector to produce AP-HuR-CP1-GST and AP-HuR-CP2-GST. The AP-tagged proteins were expressed and produced as described previously (22) except for the following modifications: the proteins were eluted in a buffer containing 300 mm Tris (pH 8.8) and 120 mm NaCl, with 10 mm glutathione for the first elution and 20 mm glutathione for further elutions.
Cell Treatments and Assessing Cell Death
Polyinosinic-polycytidylic acid (poly(I-C)), SP600125, SB203580, okadaic acid (Calbiochem), and Z-IETD-fmk (BD Pharmingen) were used to treat cells as described in the figure legends. Cells were treated with staurosporine (STS; Sigma-Aldrich) at either a 1 μm or a 100 nm concentration. AP-conjugated proteins were also given to cells at either a 75 or 150 nm dose 1 h prior to a 3-h treatment of 100 nm STS, as described in figure legends. AP-GST, AP-HuR-GST, AP-HuR-CP1-GST, and AP-HuR-CP2-GST proteins were produced and purified as described above.
Cell death was assessed by Annexin V staining and by Western blotting. Alternatively, cells were washed with phosphate-buffered saline and stained with Annexin V-FITC (BioVision, Inc.) according to the manufacturer's protocol, and counted by flow cytometry. Western blot analysis, as described above, was performed with quantification of caspase-3 and PARP cleavage products using ImageQuant software, by normalizing band intensities against those seen in the loading controls. Statistical analysis to determine significance was calculated using two-tailed, unpaired t tests and was performed using both online GraphPad software and Microsoft Excel software.
RESULTS
Cleavage of HuR Is Dependent on PKR, FADD, and Caspase-8
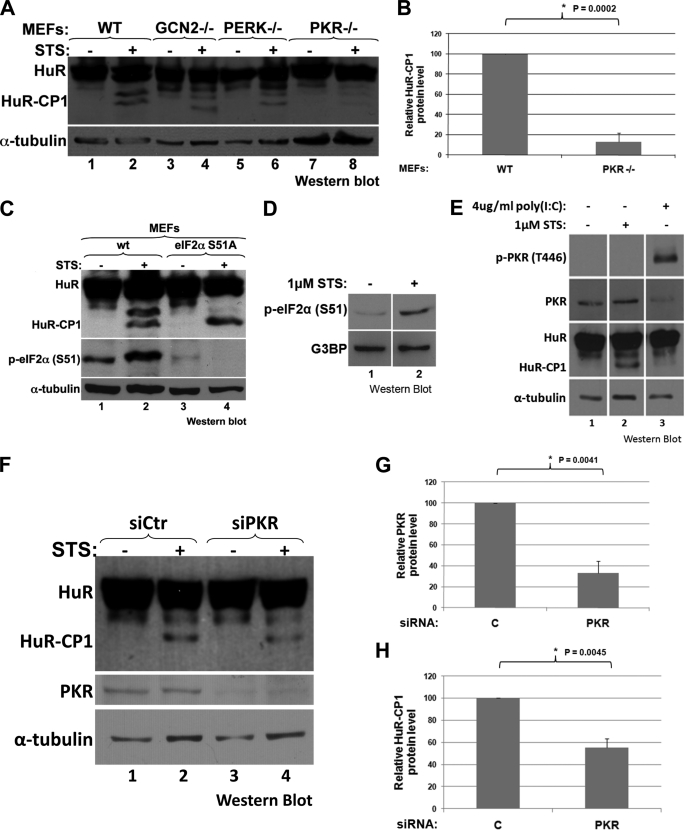
It is well established that in response to extrinsic and intrinsic signals the cell activates common pathways that collaborate to trigger cell death (13). In response to a variety of stresses, cells can activate four stress kinases, GCN2 (amino acid starvation), heme-regulated inhibitor (iron deficiency), PERK (ER stress), and PKR (viral activity and external stress), which are all capable of phosphorylating eIF2α, leading to a significant reduction in general translation (25). Under different conditions of stress, each of these four kinases may be activated and by regulating translation, allow the cell to choose between either addressing the stress or dying. Because the cleavage of HuR correlates with its proapoptotic activity (9), we investigated whether one of the ubiquitous stress response kinases, GCN2, PERK, or PKR (heme-regulated inhibitor is normally only highly expressed in blood cells), could be involved in the caspase-mediated cleavage of HuR. To achieve this, we treated MEFs expressing or not GCN2, PERK, or PKR with 1 μm STS (a known apoptotic inducer) (9) for 6 h and monitored the cleavage of HuR. Intriguingly, although the GCN2−/−, PERK−/−, and wtMEF cells all had comparable levels of HuR cleavage, the PKR−/− cells produced a much fainter band for HuR-CP1 after STS treatment (Fig. 1A). Upon quantification, we found that this corresponded to more than a 6-fold decrease in HuR cleavage (Fig. 1B). Of note, we observed two cleavage bands in MEF cells for HuR, which may be due to a post-translational modification of HuR-CP1.
FIGURE 1.
Cells lacking PKR show significantly less cleavage of HuR in response to stress. A and B, HuR cleavage is reduced by ∼85% in the absence of PKR. MEF cells with knock-out of GCN2, PERK, or PKR (or wtMEFs) were treated for 6 h with 1 μm STS, or not, and their total cell extracts were analyzed by Western blotting. Antibodies against HuR and α-tubulin (loading control) were used. Band intensities for HuR-CP1 and the loading control were quantified, and mean values were graphed (B) with error bars representing the S.E. of three independent experiments, a representative sample of which is shown in A. The asterisk denotes that the observed decrease is significant, with p = 0.0002. C, HuR cleavage does not depend on eIF2α phosphorylation. wtMEFS, or MEFs in which eIF2α was knocked out and then rescued with a knock-in of nonphosphorylatable S51A eIF2α, were treated, or not, as in A and analyzed by Western blotting, probing for HuR, phospho (p)-eIF2α (Ser51), and α-tubulin (loading control). D and E, eIF2α, but not PKR, is phosphorylated in HeLa cells in response to STS. HeLa cells were treated with STS or poly(I-C) (4 μg/ml with Lipofectamine 2000 for 4 h), and their extracts were analyzed by Western blotting, probing for p-eIF2α (Ser51) and G3BP (loading control) (D), or PKR, p-PKR (Thr446) and α-tubulin (loading control) (E). F–H, HuR cleavage is reduced in HeLa cells with PKR knockdown. HeLa cells were transfected with siRNA against PKR or control (C) siRNA and were treated, or not, for 3 h with 1 μm STS, 48 h following transfection. Cell lysates were analyzed by Western blotting, probing for HuR, PKR, and α-tubulin (loading control). Band intensities for PKR (G) and HuR-CP1 (H) were determined as described for B and graphed in G and H, with the asterisks denoting that the observed decreases are significant, with p = 0.0041 (G) and p = 0.0045 (H). All blots shown (C–F) are representative of at least three independent experiments.
This result is consistent with a previous report demonstrating the involvement of PKR in promoting apoptosis in an eIF2α-dependent manner (26). It has been shown that following PKR activation, a temporal progression occurs whereby the NF-κB pathway is originally activated, after which point eIF2α is phosphorylated, leading to cell death. We thus wished to determine how PKR may signal the cleavage of HuR. PKR employs a number of downstream pathways to effect many different actions on the cell once activated (18), so we proceeded to screen these to determine whether impairing the specific pathways could reduce the cleavage of HuR in response to STS. We saw that in wtMEF cells, STS treatment causes an increase in phosphorylation of eIF2α (Fig. 1C, lanes 1 and 2). Despite this, MEFs with a nonphosphorylatable eIF2α knock-in (S51A) still showed HuR cleavage in response to STS (Fig. 1C, lane 4), suggesting that eIF2α phosphorylation is not important for the cleavage of HuR.
Next, we assessed the implication of PKR in the cleavage of HuR in HeLa cells, where cleavage was first observed (9). We investigated whether PKR itself is phosphorylated under conditions in which HuR is cleaved. We observed that although eIF2α is phosphorylated in response to STS (Fig. 1D), PKR is not, as shown by an anti-Thr(P) 446 PKR antibody (Fig. 1E, lane 2). Additionally, treating HeLa cells with poly(I-C), a known activator of PKR phosphorylation (31), failed to trigger HuR cleavage (Fig. 1E, lane 3). These observations are consistent with the fact that the phosphorylation of eIF2α is not required for the cleavage of HuR (Fig. 1C), indicating that PKR promotes HuR cleavage in a phosphorylation-independent manner. Of note, the antibody against total PKR recognizes an epitope near Thr446, which may have prevented the recognition of total PKR upon poly(I-C) treatment (Fig. 1E, lane 3). To confirm the requirement of PKR in the cleavage of HuR during the apoptotic response, we depleted PKR expression in HeLa cells using specific siRNAs. As in PKR−/− MEFs, we observed that when PKR is knocked down (∼67%, Fig. 1G), HeLa cells exposed to 1 μm STS for 3 h showed a reduction (∼45%) in HuR cleavage compared with controls (Fig. 1F, compare lanes 2–4 and H).
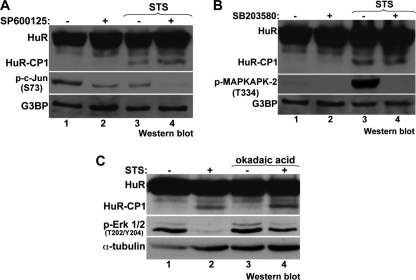
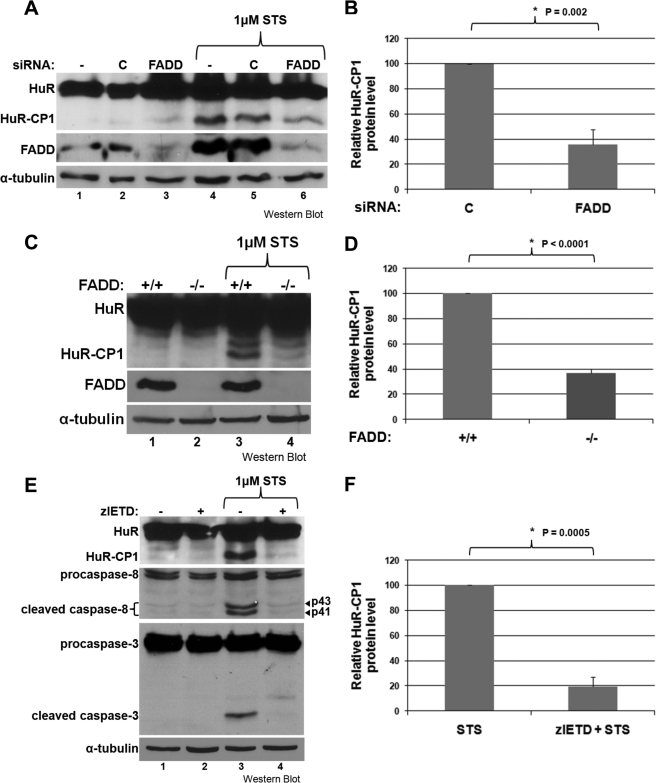
We then proceeded to look at other downstream pathways of PKR (JNK, p38, and protein phosphatase 2A (PP2A)) (18, 27) using chemical inhibitors. We specifically targeted the activities of JNK, p38, and PP2A, which have all been shown to be activated by PKR (18, 27). Inhibiting each of these factors in the presence of STS did not however cause a decrease in HuR cleavage (Fig. 2, A–C). As expected, inhibition of either JNK or p38 reduced the phosphorylation of c-Jun and MAPKAPK-2, respectively (28, 29). Inhibition of PP2A resulted in an increase in levels of phospho-ERK1/2, a target of PP2A (30). One remaining PKR-activated pathway that has been shown to trigger apoptosis directly is through the activation of FADD, which then recruits procaspase-8, leading to its activation (31, 32). We verified whether FADD is necessary for HuR cleavage using siRNA in HeLa cells (Fig. 3A) and FADD knock-out MEFs (Fig. 3C) and observed that in the absence of FADD, HuR cleavage is significantly reduced (Fig. 3, A–D). We then looked at the cleavage of HuR in response to stress when the activity of caspase-8, the downstream effector of FADD, is blocked. Treating HeLa cells with a caspase-8-specific inhibitor (Z-IETD-fmk) (33) in the presence of STS led to a significant reduction in HuR cleavage (Fig. 3, E and F). These data suggest that it is through FADD and caspase-8 that PKR promotes the cleavage, and consequentially, the apoptotic-role, of HuR. To support this, we also looked at the activation of caspase-3, which is downstream of caspase-8, and saw that it is not activated when caspase-8 is inhibited (Fig. 3E). This corresponds with our previous data showing that HuR cleavage depends on caspase-3 (9). Together, these observations argue that HuR cleavage is a downstream activity of PKR and is one that enables PKR to enhance apoptotic response. Of note, by comparing Western blots demonstrating HuR cleavage (Figs. 1–3), we observed that the pattern of cleavage differs between cell types, possibly due to cell-type or species-specific modifications of HuR.
FIGURE 2.
HuR cleavage does not depend on the activities of JNK, p38, or PP2A. HeLa cells were treated with inhibitors against JNK (25 μm SP600125 for 1 h), p38 (20 μm SB203580 for 1 h), or PP2A (100 nm okadaic acid for 1 h), then with 1 μm STS for 3 h. Extracts were analyzed by Western blotting, probing for G3BP or α-tubulin (as loading controls), HuR, and either p-c-Jun (Ser73) to validate efficacy of JNK inhibition (A), p-MAPKAPK-2 (Thr334) to validate efficacy of p38 inhibition (B), or p-ERK1/2 (Thr202/Tyr204) to validate efficacy of PP2A inhibition (C). All blots shown are representative of at least three independent experiments.
FIGURE 3.
HuR is cleaved in a FADD- and caspase-8-dependent manner. A and B, knockdown of FADD in HeLa cells reduces HuR cleavage. HeLa cells were transfected with siRNA against FADD or control (C) siRNA and 48 later were treated or not with 1 μm STS for 3 h. Cell extracts were analyzed by Western blotting looking for HuR, FADD, and α-tubulin (loading control) expression levels. C and D, MEFs with knocked-out expression of FADD exhibit reduced cleavage of HuR. MEF cells lacking FADD were treated with 1 μm STS for 6 h, or not, and extracts were analyzed by Western blotting, probing for HuR, FADD, and α-tubulin (loading control). E and F, HuR cleavage depends on the activity of caspase-8. HeLa cells were treated for 30 min with 25 μm caspase-8 specific inhibitor, Z-IETD-fmk, followed by 3 h of 1 μm STS. Extracts were analyzed by Western blotting, using antibodies against HuR, caspase-8 (which recognizes procaspase-8 as well as the two cleaved/active isoforms) and α-tubulin (loading control). For quantifications (B, D, and F), the band intensities of HuR-CP1 and loading controls of the Western blots were determined, and mean values were graphed with error bars representing the S.E. of at least three independent experiments, representative samples of which are shown (A, C, and E). Asterisks denote significant decreases in HuR-CP1 generation, with p = 0.002 (B), p < 0.0001 (D), and p = 0.0005 (F).
HuR Cleavage Products Induce Apoptosis Downstream of PKR/FADD/Caspase-8
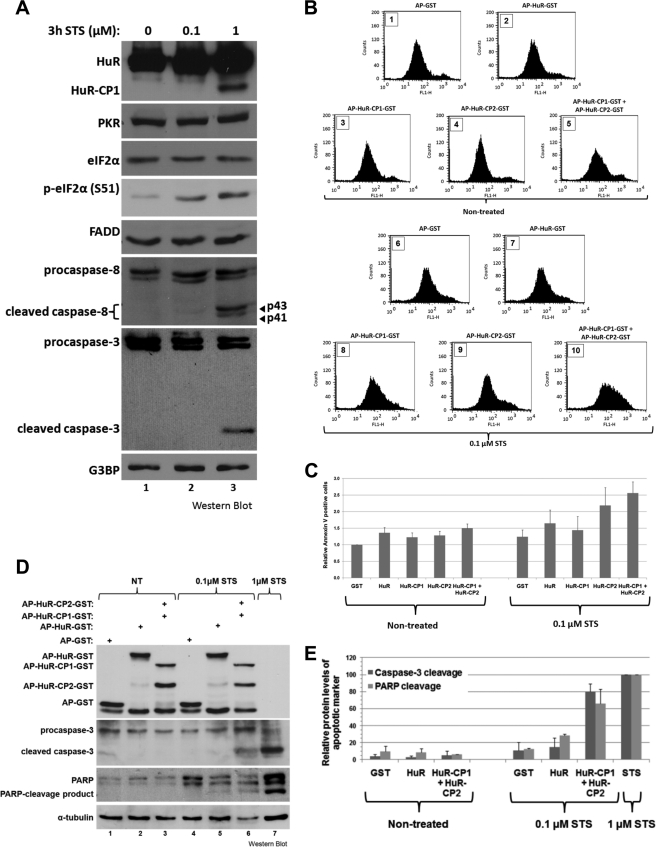
Having previously observed that preventing the cleavage of HuR delays apoptosis (9), we next wished to determine whether these CPs are sufficient to induce apoptosis under conditions where the PKR/FADD/caspase-8-mediated cleavage of HuR is not occurring. To have a tight control on the levels of expression, HuR and its CP isoforms were fused with the cell-permeable peptide AP (antennapedia), which enables the rapid and efficient uptake of recombinant proteins (9). HeLa cells, where caspase-8 and caspase-3 are not active (Fig. 4A, lane 1) were treated with AP-wtHuR, AP-HuR-CP1, AP-HuR-CP2, or both AP-HuR-CPs (Fig. 4, B, panels 2–5, and C). Cell death was evaluated by flow cytometry for these cells, counting Annexin V-positive cells (FL-1 channel), and no noticeable apoptosis was observed. This failure to engage apoptosis may be due to the absence of basal activity for factors acting downstream of the HuR-CPs when triggering cell death. We thus treated these cells with a nonlethal dose of STS, 0.1 μm, allowing us to evaluate the effect of the HuR-CPs in a setting where the caspase-8/caspase-3 pathway is not activated, and thus endogenous HuR is not cleaved (Fig. 4A, lane 2). At this 0.1 μm treatment, we also observed that HeLa cells underwent apoptosis in the presence of the two HuR-CPs more than with full-length HuR (Fig. 4B, compare panels 7 and 10). Additionally, although HuR-CP1 did not appear to cause apoptosis as analyzed by flow cytometry, addition of HuR-CP2 with 0.1 μm STS was sufficient to cause an increase in Annexin V-positive staining cells (Fig. 4B, panel 9). Physiologically, however, it would be expected that both cleavage products be generated simultaneously, so we assessed the consequence of the expression of both HuR-CPs. Additionally, to deliver the same quantity of AP/GST-conjugated proteins to the cells, the dose of each CP was reduced by half, so that all cells would be treated with a total dose of 150 nm conjugated proteins. With these modifications, the sensitization experiments were repeated on HeLa cells where lysates were prepared and analyzed by Western blotting, probing for the cleaved forms of both PARP and caspase-3, two well known markers of apoptosis (9) (Fig. 4, D and E). The two HuR-CPs again increased cell death substantially in the presence of 0.1 μm STS, even comparable in degree to a standard 3-h treatment with 1 μm STS (Fig. 4E), clearly demonstrating their ability to induce apoptosis when neither the cleavage of HuR, nor cell death, is engaged. Therefore, together these data argue that the PKR-mediated cleavage of HuR represents a key event that enables HuR to promote cell death by the generation of its proapoptotic cleavage products.
FIGURE 4.
HuR-CPs can induce apoptosis in the absence of caspase-8 activation. A, 0.1 μm STS is nonlethal and does not induce the cleavage of HuR. HeLa cells were treated, or not, with STS for 3 h at a dose of 0.1 or 1 μm. Total cell extracts were analyzed by Western blotting, looking at levels of PKR, eIF2α, p-eIF2α, FADD, caspase-8, caspase-3, HuR, and G3BP (loading control). The blot shown is representative from two independent experiments. B–E, HuR-CPs sensitize HeLa cells to apoptosis in conditions where endogenous HuR is not cleaved. B and C, HeLa cells were treated with 150 nm AP-conjugated GST, HuR-GST, HuR-CP1-GST, HuR-CP2-GST, or 150 nm each HuR-CP1-GST and HuR-CP2-GST for 1 h, and then treated, or not, with a 0.1 μm STS for 3 h. Cells were collected, incubated with Annexin V-FITC, and then analyzed by flow cytometry. Counts are the relative fluorescent intensity, and the FL-1 channel detects Annexin V-FITC. Flow cytometry data represent two independent experiments, with mean values graphed (C) and where the error bars represent the S.E. D and E, HeLa cells were treated as in B, except that 75 nm AP-HuR-CP1-GST and HuR-CP2-GST were used for combined treatment (instead of 150 nm each). Cells were then lysed and analyzed by Western blotting, incubating membranes with antibodies against PARP, caspase-3, GST, and α-tubulin (loading control). Band intensities of the Western blots were quantified, and mean values were graphed (E) with error bars representing the S.E. of two independent experiments, a representative sample of which is shown in D.
DISCUSSION
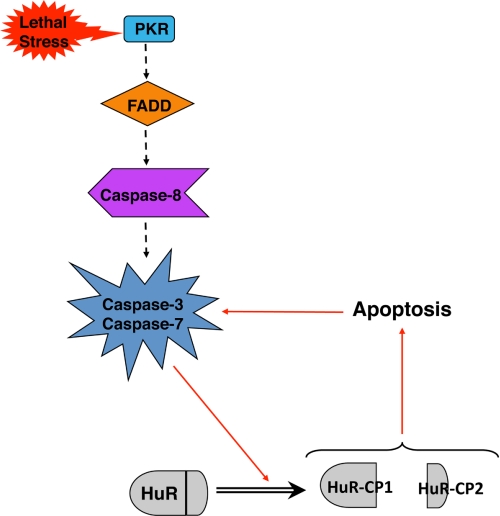
PKR has previously been described as being capable of triggering various downstream effects, ranging from the promotion of survival to the induction of apoptosis (26, 34). Here, we show that through its activation of FADD and caspase-8, PKR is also capable of triggering the cleavage of HuR. We demonstrated previously that this cleavage event is important for the proapoptotic activity of HuR, and we have determined here that these HuR-CPs are in fact capable of inducing cell death without the need for apoptotic activation. Therefore, we suggest a model whereby a lethal stress signals through PKR to promote the cleavage of HuR in a FADD/caspase-8-dependent manner, leading to cell death by apoptosis (Fig. 5). Intriguingly, our data also suggest that the implication of PKR in HuR cleavage is independent of its phosphorylation.
FIGURE 5.
Model illustrating the role of PKR/FADD/caspase-8 in mediating the proapoptotic cleavage of HuR. In this study, we found that in response to severe stress (here, 1 μm STS was used), HuR is cleaved in a manner dependent on the PKR/FADD/caspase-8 pathway, leading to the activation of caspases-3 and -7, which we previously showed to be necessary for the cleavage of HuR. The data obtained in this study as well as those published (9) have led us to suggest that the cleavage of HuR enables it to promote apoptosome-mediated cell death, acting as an amplification signal.
In our previous study where we identified the cleavage of HuR (9), we showed that this processing is due to the activities of caspases-3 and -7. We found here that caspase-8, through PKR and FADD, is necessary for the cleavage of HuR as well. Because it has been well established that caspase-8 may both directly process caspase-3 in an apoptosome-independent manner (19, 20) and also trigger mitochondrial cell death, we hypothesize that this caspase-8 activity is responsible for activating caspases-3 and -7 to produce HuR-CPs (Fig. 5). This argues that the HuR-CPs play a key role in apoptosis downstream of initial caspase-3 and -7 activation by amplifying/sustaining cytochrome c-mediated cell death. Thus, we speculate that through PKR/FADD/caspase-8, early levels of caspases are activated to generate HuR-CPs. This may also explain how, even in the absence of upstream components involved in cleaving HuR (Fig. 1 and 3), some levels of cleavage occur as apoptosis is still engaged, and caspases will eventually become active. Once generated, these CPs can then enhance the apoptotic response, possibly because HuR cleavage enables its ligand PHAPI to promote apoptosome activity (9). The possibility that HuR may differentially regulate other targets involved in apoptosis also exists, so the extent to which HuR can promote cell death upon cleavage may be greater than we believe. We have also seen that the two CPs may act differently in their ability to induce apoptosis (Fig. 4, B and C), so it would be particularly interesting to establish the mechanism(s) by which each cleavage product acts and may mediate cell death. Already we know that HuR-CP2, but not HuR-CP1, associates with PHAPI (9), and so further characterization of these CPs may provide important advancements toward the possibility of using them as apoptosis-inducing agents.
It is well established that HuR promotes cell growth and is involved in tumorigenesis (1, 35), but our data suggest that if HuR cleavage could be induced in these cases, it may instead revert to promoting cell death (Fig. 4). This also raises the possibility that in some chemoresistant cancer cells, HuR may not be cleaved. HuR has also been linked to numerous other cell activities and is known to regulate the expression of a variety of mRNA targets (1, 36). PKR is already known to govern cell outcome when activated. Surprisingly, in this study we have demonstrated that PKR, without being phosphorylated, employs the cleavage of HuR as one means of regulating cell fate. Given the broad scope of roles that HuR plays, this PKR-induced cleavage may prove to be a far reaching and powerful homeostatic mechanism for regulating cell biology, which clearly warrants further investigation.
Acknowledgments
We thank Drs. Rachid Mazroui and Sergio Di Marco for critical review of this manuscript, insightful discussion, and technical assistance and Cristina Gareau for critical review of the manuscript. We also thank N. Punjani for assistance with sensitization experiments.
This work was supported in part by National Cancer Institute of Canada (NCIC) Operating Grants 016247 and 018125 (to I.-E. G.).
- HuR
- human antigen R
- CP
- cleavage product
- PKR
- protein kinase RNA
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- wt
- wild type
- eIF2α
- eukaryotic translation initiation factor 2α
- MEF
- mouse embryonic fibroblast
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- G3BP
- Ras-GAP SH3 domain-binding protein
- ERK
- extracellular signal-regulated kinase
- STS
- staurosporine
- PARP
- poly(ADP-ribose)polymerase
- JNK
- c-Jun N-terminal kinase
- PP2A
- protein phosphatase 2A
- FADD
- Fas-associated protein with death domain
- PERK
- PKR-like endoplasmic reticulum kinase
- AP
- antennapedia
- MAPKAPK
- mitogen-activated protein kinase activated protein kinase.
REFERENCES
- 1.Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. (2008) Biol. Chem. 389, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Roretz C., Gallouzi I. E. (2008) J. Cell Biol. 181, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan X. C., Steitz J. A. (1998) EMBO J. 17, 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallouzi I. E., Steitz J. A. (2001) Science 294, 1895–1901 [DOI] [PubMed] [Google Scholar]
- 5.Abdelmohsen K., Lal A., Kim H. H., Gorospe M. (2007) Cell Cycle 6, 1288–1292 [DOI] [PubMed] [Google Scholar]
- 6.Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M. (2006) Mol. Cell Biol. 26, 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. (2005) EMBO J. 24, 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazan-Mamczarz K., Galbán S., López de Silanes I., Martindale J. L., Atasoy U., Keene J. D., Gorospe M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazroui R., Di Marco S., Clair E., von Roretz C., Tenenbaum S. A., Keene J. D., Saleh M., Gallouzi I. E. (2008) J. Cell Biol. 180, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beauchamp P., Nassif C., Hillock S., van der Giessen K., von Roretz C., Jasmin B. J., Gallouzi I.-E. (2010) Cell Death Differ., in press [DOI] [PubMed] [Google Scholar]
- 11.Brennan C. M., Gallouzi I. E., Steitz J. A. (2000) J. Cell Biol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Giessen K., Gallouzi I. E. (2007) Mol. Biol. Cell 18, 2619–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadeel B., Ottosson A., Pervaiz S. (2008) Cell Death Differ. 15, 443–452 [DOI] [PubMed] [Google Scholar]
- 14.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 15.Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., Wang X. (2003) Science 299, 223–226 [DOI] [PubMed] [Google Scholar]
- 16.Kim H. E., Jiang X., Du F., Wang X. (2008) Mol. Cell 30, 239–247 [DOI] [PubMed] [Google Scholar]
- 17.Schafer Z. T., Kornbluth S. (2006) Dev. Cell 10, 549–561 [DOI] [PubMed] [Google Scholar]
- 18.García M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. (2006) Microbiol. Mol. Biol. Rev. 70, 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García M. A., Meurs E. F., Esteban M. (2007) Biochimie 89, 799–811 [DOI] [PubMed] [Google Scholar]
- 20.Peters G. A., Dickerman B., Sen G. C. (2009) Biochemistry 48, 7441–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin X. M. (2000) Cell Res. 10, 161–167 [DOI] [PubMed] [Google Scholar]
- 22.van der Giessen K., Di-Marco S., Clair E., Gallouzi I. E. (2003) J. Biol. Chem. 278, 47119–47128 [DOI] [PubMed] [Google Scholar]
- 23.Gallouzi I. E., Brennan C. M., Stenberg M. G., Swanson M. S., Eversole A., Maizels N., Steitz J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. (2006) Mol. Biol. Cell 17, 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holcik M., Sonenberg N. (2005) Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 26.Donzé O., Deng J., Curran J., Sladek R., Picard D., Sonenberg N. (2004) EMBO J. 23, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z., Williams B. R. (2000) Mol. Cell. Biol. 20, 5285–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaishnav D., Jambal P., Reusch J. E., Pugazhenthi S. (2003) Biochem. Biophys. Res. Commun. 307, 855–860 [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Jiang M. S., Adams J. L., Lee J. C. (1999) Biochem. Biophys. Res. Commun. 263, 825–831 [DOI] [PubMed] [Google Scholar]
- 30.Casillas A. M., Amaral K., Chegini-Farahani S., Nel A. E. (1993) Biochem. J. 290, 545–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balachandran S., Kim C. N., Yeh W. C., Mak T. W., Bhalla K., Barber G. N. (1998) EMBO J. 17, 6888–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil J., Esteban M. (2000) Oncogene 19, 3665–3674 [DOI] [PubMed] [Google Scholar]
- 33.Medema J. P., Scaffidi C., Kischkel F. C., Shevchenko A., Mann M., Krammer P. H., Peter M. E. (1997) EMBO J. 16, 2794–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagus R., Joshi B., Barber G. N. (1999) Int. J. Biochem. Cell Biol. 31, 123–138 [DOI] [PubMed] [Google Scholar]
- 35.Gorospe M. (2003) Cell Cycle 2, 412–414 [PubMed] [Google Scholar]
- 36.Doller A., Pfeilschifter J., Eberhardt W. (2008) Cell. Signal. 20, 2165–2173 [DOI] [PubMed] [Google Scholar]