Abstract
Calsequestrin undergoes dynamic polymerization with increasing calcium concentration by front-to-front dimerization and back-to-back packing, forming wire-shaped structures. A recent finding that point mutation R33Q leads to lethal catecholaminergic polymorphic ventricular tachycardia (CPVT) implies a crucial role for the N terminus. In this study, we demonstrate that this mutation resides in a highly conserved alternately charged residue cluster (DGKDR; cluster 1) in the N-terminal end of calsequestrin. We further show that this cluster configures itself as a ring system and that the dipolar arrangement within the cluster brings about a critical conformational flip of Lys31-Asp32 essential for dimer stabilization by formation of a H-bond network. We additionally show that Ca2+-induced calsequestrin aggregation is nonlinear and reversible and can regain the native conformation by Ca2+ chelation with EGTA. This study suggests that cluster 1 works as a molecular switch and governs the bidirectional transition between the CASQ2 monomer and dimer. We further demonstrate that mutations disrupting the alternating charge pattern of the cluster, including R33Q, impair Ca2+-CASQ2 interaction, leading to altered polymerization-depolymerization dynamics. This study provides new mechanistic insight into the functional effects of the R33Q mutation and its potential role in CPVT.
Keywords: Calcium, Calcium-binding Proteins, Circular Dichroism (CD), Molecular Dynamics, Sarcoplasmic Reticulum, Arrhythmia, Calsequestrin, Heart
Introduction
Cardiac muscle contraction is initiated by release of Ca2+ from the sarcoplasmic reticulum (SR).4 The reuptake of Ca2+ by the sarco/endoplasmic reticulum Ca2+-ATPase pump into the SR results in muscle relaxation and restoration of the store (1). This reuptake is facilitated by buffering of Ca2+ inside the SR so that free [Ca2+] is maintained at a low level, allowing sarco/endoplasmic reticulum Ca2+-ATPase-mediated Ca2+ uptake against a gradient (2–4). Ca2+ buffering inside the SR lumen is accomplished mainly by a high capacity (40–50 Ca2+ ions/molecule), moderate affinity Ca2+-binding protein called calsequestrin (CASQ2, the cardiac isoform) (5, 6). Because of CASQ2, the concentration of total stored Ca2+ can reach up to 20 mm, whereas free [Ca2+] remains at ∼1 mm (7). CASQ2 interacts with the ryanodine receptor via the junctional SR proteins triadin and junctin, keeping high concentrations of Ca2+ close to the site of release, and also modulates the gating behavior of the ryanodine receptor (8–13).
Electron microscopic studies indicate that CASQ2 molecules are packaged as long wires beneath the junctional face of the SR of cardiomyocytes (14, 15). Based on crystal structures, it has been suggested that CASQ2 undergoes polymerization first by front-to-front dimerization and that two such dimers establish back-to-back contact, making a tetramer (11, 16–18). In this way, a long chain of highly ordered linear polymer is expected to be formed as the Ca2+ concentration continues to increase, resembling the wire-shaped structures observed by electron microscopy. It has also been suggested that CASQ polymers can depolymerize when free [Ca2+] is lowered, although the exact molecular mechanism fast enough to be physiologically relevant is not known (18–23). Each CASQ monomer is a mixed α-β protein composed of three domains formed by five β-strands sandwiched by four α-helices (11, 17). Each domain consists of a hydrophobic core with the acidic residues on the surface. This makes the surface electrostatic potential highly electronegative, and it can be neutralized only by cations (17).
The role of CASQ2 in Ca2+ release has gained further attention due to the finding that mutations in or absence of CASQ2 causes catecholaminergic polymorphic ventricular tachycardia (CPVT) in humans (13, 24–28). CPVT is a familial arrhythmogenic disorder characterized by episodes of syncope and sudden death primarily by exposure to emotional or physical stress in the absence of structural alterations of the heart. Both in vitro and in vivo studies have attempted to define the causative mechanism for the reported arrhythmia with limited success. Among the CPVT mutations, R33Q (corresponds to R14Q once the signal sequence is cleaved in the mature protein) is located in the extended N-terminal arm, which is inserted into the partner monomer in the crystal structure. In this case, a neutral residue, glutamine, replaces the completely conserved and strongly basic Arg33 residue. Recent studies by Terentyev et al. (29) suggested that the R33Q mutation has lost the ability to inhibit ryanodine receptor-2, but other studies (16, 30, 31) showed that the R33Q mutation modifies its ability to sense Ca2+ and Ca2+-buffering capacity. However, the molecular basis of how the mutation affects CASQ2 polymerization is not understood.
Our hypothesis is that the charged amino acid clusters at the N terminus are important for front-to-front dimerization and that mutations that disrupt this charge pattern would impede CASQ2 polymerization. Therefore, the major goal of this study was to determine the role of charged clusters in the N-terminal region. We specifically studied how the CPVT mutation R33Q affects CASQ2 function/polymerization. We demonstrate that an alternately charged residue cluster works as a molecular switch and governs the bidirectional transition between CASQ2 monomer and dimer. Our study further shows that the CPVT mutation R33Q impairs Ca2+-CASQ2 interaction, leading to altered polymerization-depolymerization dynamics.
EXPERIMENTAL PROCEDURES
Multiple Alignment of Calsequestrin from Different Organisms
Protein-protein BLAST was performed using the NCBI Database (www.ncbi.nlm.nih.gov/BLAST) using the rat CASQ2 sequence as the template. Sequences from vertebrates with e-values <6 × e−106 were selected. Multiple sequence alignment was performed with ClustalW 2.0.11 using the EMBL-EBI Database (www.ebi.ac.uk/clustalw) together with the skeletal CASQ (CASQ1) isoform of mouse and human and CASQ from Ciona intestinalis (sea squirt) and Caenorhabditis elegans. In mouse CASQ2, the first 19 residues act as a signal sequence, and thus, in the mature protein, the first amino acid starts from amino acid 20 of the immature protein.
Mutagenesis and Purification of Mutant CASQ2
Rat wild-type (WT) CASQ2 was cloned into the pET21a vector (Novagen) between NdeI and XhoI restriction sites. The CASQ2 mutants were generated using a site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. Five different kinds of mutants were generated: D29A-D32A, K31A-R33A, R33Q, E39A, and K40A-K43A. In addition, deletion mutants in multiples of five amino acids (up to 20 amino acids) from the N-terminal end of the mature protein were generated, Δ5CASQ2, Δ10CASQ2, Δ15CASQ2, and Δ20CASQ2. The primers used for the generation of mutants are listed in Table 1. The proteins were expressed in bacterial cells and purified using a previously published method (32). The conditions for overexpression and purification were individually optimized for each mutant to ensure maximum yield of the protein. The protein concentration was estimated using Bradford reagent (Bio-Rad) following the supplier's protocol using bovine serum albumin as a standard.
TABLE 1.
Primers used for site-directed mutagenesis and systematic N-terminal deletions
| Mutation | Primer sequence |
|---|---|
| R33QFP | 5′-CAAGTGGTCAGCCTTTCTGAGAAG-3′ |
| R33QRP | 5′-CTTCTCAGAAAGGCTGACCACTTG-3′ |
| D29A-D32AFP | 5′-CACGTACGCTGGGAAGGCCCGAGTGGTCAG-3′ |
| D29A-D32ARP | 5′-CTGACCACTCGGGCCTTCCCAGCGTACGTG-3′ |
| K31A-R33AFP | 5′-CACGTACGATGGGGCGGACGCAGTGGTCAGC-3′ |
| K31A-R33ARP | 5′-GCTGACCACTGCGTCCGCCCCATCGTACGTG-3′ |
| E39AFP | 5′-GTGGTCAGCCTTTCTGCGAAGAACTTGAAGCAAGTG-3′ |
| E39ARP | 5′-CACTTGCTTCAAGTTCTTCGCAGAAAGGCTGACCAC-3′ |
| K40A-K43AFP | 5′-CAGTCTTTCTGAGGCAAACTTGGCGCAAGTGTTGA-3′ |
| K40A-K43ARP | 5′-TCAACACTTGCGCCAAGTTTGCCTCAGAAAGACTG-3′ |
| Δ5CASQ2FP | 5′-GGAATTCCATATGTTGCCGACGTACGATGGGAAGGACC-3′ |
| Δ10CASQ2FP | 5′-GGAATTCCATATGGGTAAGGACCGAGTGGTCAGCCT-3′ |
| Δ15CASQ2FP | 5′-GGAATTCCATATGGGTCAGCCTTTCTGAGAAGAACTTG-3′ |
| Δ20CASQ2FP | 5′-GGAATTCCATATGAAGAACTTGAAGCAAGTGTTGAAGA-3′ |
| CASQRP | 5′-CCGCTCGAGTTCATCGTCATCATCGTCATCATCAC-3′ |
Circular Dichroism Spectroscopy
CD spectra were acquired using an Aviv spectropolarimeter with a temperature-controlled cell holder. Far-UV CD spectra were collected with 10 μm protein in 5 mm NaH2PO4 buffer (pH 7.4) containing 25 mm NaCl. CD scans were recorded using a quartz cell with a path length of 1 mm, a response time of 2 s, a scan speed of 10 nm/min, and a bandwidth of 1.0 nm. Two scans were accumulated and averaged for each spectrum after base-line subtraction. Unless stated otherwise, all of the spectra were recorded at 25 °C. The effect of temperature on the stability of CASQ2 folding was examined by incubating the protein at the indicated temperatures (20–80 °C) for 15 min and recording the spectra. Polymerization studies were carried out in 50 mm Tris-HCl (pH 7.5) containing 20 mm NaCl. The protein was mixed with CaCl2 (5 mm) in the above Tris-HCl buffer and incubated for 5 min before recording the spectra. After recording the spectra, Ca2+-bound CASQ was mixed with 1.5 mm EGTA and incubated for 5 min, and spectra were recorded. The addition of EGTA was repeated until the CD spectra overlapped with the native spectra.
Limited Proteolysis by Trypsin
Purified WT and mutant CASQ2 proteins were subjected to proteolysis by l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (New England Biolabs) in 50 mm Tris-HCl (pH 8.0) at 25 °C. The reactions were performed in the absence and presence of various concentration of CaCl2 (1–5 mm). Proteins were incubated in the reaction buffers for 20 min at 25 °C in a reaction volume of 200 μl before trypsin was added at a protease/CASQ2 ratio of 1:100 (w/w). The digestion was limited to 30 min, and the samples were quickly mixed with Laemmli sample buffer (Bio-Rad) and heated at 99 °C for 2 min. Equal amounts of protein from each sample were loaded and analyzed on a 12% SDS-polyacrylamide gel. The gels were stained with Coomassie Brilliant Blue R-250 staining solution (Bio-Rad), destained, and imaged.
Turbidimetric Measurements
Ca2+-induced aggregation of CASQ2 proteins was monitored in 50 mm Tris-HCl (pH 7.5) containing 20 mm NaCl by adding aliquots (2–10 μl) of CaCl2 stock solutions (0.1–2 m) to a solution of 2.5 μm CASQ2 at 25 ± 1 °C using a Genesys spectrophotometer. Ca2+ chelator was not added; therefore, low levels of Ca2+ might be present at the starting point. After Ca2+ addition, protein samples were incubated to equilibrate for 5 min, and then absorbance was recorded at 350 and 600 nm. The absorbance of samples was corrected by subtracting the absorbance of buffer alone at the same calcium concentration. For experiments involving EGTA-mediated chelation, WT CASQ and cluster 1 mutants were treated with 8 mm CaCl2. 500 μl of each sample was centrifuged at 20,000 × g for 1 h, and the soluble fraction was estimated for protein concentration using Bradford reagent as described above. From the protein quantity, the percentage of Ca2+-induced precipitation of CASQ2 was calculated and plotted against Ca2+ concentration. The concentration of CaCl2 at which 50% of CASQ2 protein had undergone precipitation (i.e. EC50 of Ca2+-induced aggregation) was calculated. 2–10 μl of EGTA (0.1–0.5 m) was added to the aggregated solution aliquots and allowed to equilibrate for 5 min, and the percentage of protein still in precipitation was calculated as described above. The concentration of EGTA necessary to resolubilize 50% of CASQ2 protein from precipitation (i.e. EC50 of EGTA-mediated resolubilization) was calculated.
Molecular Dynamics
The molecular dynamics studies were conducted (Schrödinger Inc., New York) to examine the effect of charge alteration in cluster 1 of CASQ2 using the crystal structures of the monomer form (Protein Data Bank code 2VAF) and dimer form (code 1SIJ) of CASQ2. Generally, the crystal structures have missing residues and lack hydrogen atoms; therefore, the crystal structures were improved to correct such discrepancies using the Protein Preparation module of the Schrödinger Suite and further manually verified employing the Builder module of Maestro. The corrected dimer structure was subjected to molecular dynamics simulation with the OPLS2005 force field in the presence of the GB/SA continuum water model. The dynamics simulations were carried out by heating the system to 300 K with 10-ps equilibration time. The production dynamics simulations were carried out for 500 ps with a step size of 1.5 fs at 300 K. A shake algorithm was used to constrain covalent bonds to hydrogen atoms. The five residues (DGKDR) of chain A of the dimer were selected to define the 15-Å radius for total freedom for residue movement. The next 5-Å circular selections were used for harmonic constraints of 100 newtons and beyond that provided a frozen parameter. The stable structure from dynamics was further subjected to global minimization using the OPLS force field until the iteration number reached 5000 or the energy difference between two consecutive iterations became <0.05 kJ/mol. Cluster 1 mutants were generated using the Mutate command of the Builder module of Maestro and were subjected to minimization as described above for further structural analysis and interpretation.
RESULTS
CPVT Mutation R33Q Resides in a Highly Conserved Charged Cluster
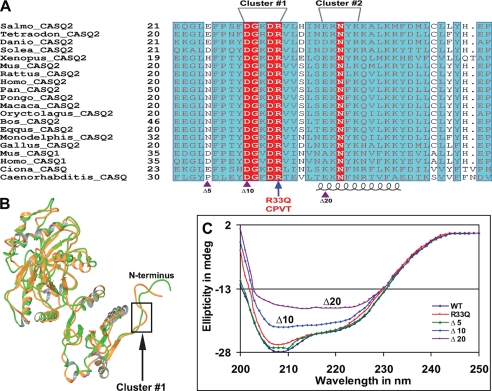
Multiple alignment of the N-terminal region of CASQ revealed that the N-terminal end of CASQ2 is highly conserved among various mammals (Fig. 1A). Of the first 40 residues, 36 amino acids are strictly conserved. The degree of conservation of residues decreased when sequences from other vertebrates were included in alignment. When vertebrate CASQ2 sequences were aligned, we found two alternately charged residue clusters (cluster 1, amino acids 29–33 (DGKDR); and cluster 2, amino acids 39–43 (EKNLK)) in the N-terminal domain that are involved in CASQ dimerization by insertion into the partner molecule and domain swapping. However, cluster 1 at amino acids 29–33 is evolutionarily much more conserved than cluster 2 and is even present in the skeletal CASQ1 isoform of mouse and man. It is also found in calsequestrin sequences of sea squirt and C. elegans. Interestingly, CPVT mutant R33Q is also part of cluster 1. These two clusters were selected for further analysis by charge neutralization mutagenesis and biophysical-biochemical methods to validate the significance of the N-terminal end in protein folding and Ca2+-induced structural changes. The complete alignment is shown in supplemental Fig. S1.
FIGURE 1.
The N terminus of CASQ is highly conserved and involved in protein folding. A, multiple sequence alignment of the N terminus. The number of amino acids before the mature protein is indicated to the left of each sequence. Identical residues in the alignment are indicated by white letters in red boxes, similar residues are shown as gray letters in cyan boxes, and a blue frame indicates similarity across groups. The alternately charged residue clusters 1 (DGKDR) and 2 (EKNLK) are labeled. The CPVT-related residue Arg33′ is located in cluster 1. The accession numbers for each sequence and the complete alignment are provided in the legend to supplemental Fig. S1. Gaps in alignment are represented as dots. Alignment was generated using the program ESPRIPT (42). Violet triangles at the bottom indicate the positions of deletion mutations: Δ5CASQ2 (Δ5), Δ10CASQ2 (Δ10), and Δ20CASQ2 (Δ20). B, superposition of the monomer and dimer forms of CASQ2. One of the major structural differences can be seen in the N terminus. The loop formed by cluster 1 (DGKDR) is highlighted by a black rectangle. C, far-UV CD spectra of different N-terminal deletion mutants. Deletion mutants Δ10CASQ2 and Δ20CASQ2 failed to fold, but mutant Δ5CASQ2 could fold to a conformation equivalent to that of the WT protein. mdeg, millidegrees.
The N-terminal End Is Required for Proper Folding of CASQ2
The major goal of this study was to determine the role of the extended N-terminal end in the proper folding of CASQ2. In addition, the presence of a CPVT mutation in the N terminus provided the rationale for further exploration of this region by mutagenesis. Additionally, upon superimposition of the dimer (Protein Data Bank code 1SIJ; green) and monomer (2VAF; orange) forms of CASQ2 (Fig. 1B), one of the major structural alterations is seen in the N terminus, although the overall three-dimensional architecture is similar. The cluster 1 DGKDR sequence forms a loop and is highlighted by the rectangle. The N-terminal deletions and point mutants listed in Table 1 were generated and purified. However, we were not able to overexpress Δ15CASQ2, and this mutant could not be purified. We employed CD spectroscopy to investigate the molecular folding pattern. The shorter deletion Δ5CASQ2 showed CD spectra that overlapped with those of the WT CASQ2 protein. On the other hand, the spectra of mutants Δ10CASQ2 and Δ20CASQ2 showed that deletion of N-terminal residues had a significant effect on the ellipticity/protein conformation. Deletion of the first 20 residues (Δ20CASQ2) had a drastic effect on protein folding, whereas mutant Δ10CASQ2 showed a modest reduction in ellipticity compared with WT CASQ2 (Fig. 1C). The spectra of the point mutants (R33Q, K31A-R33A, D29A-D32A, E39A, and K40A-K43A) closely resembled those of the WT CASQ2 protein, indicating that the point mutations do not affect protein folding at the single-molecule level (supplemental Fig. S2).
Thermal Stability of CASQ2 Is Reduced if the N-terminal Charged Cluster Is Altered
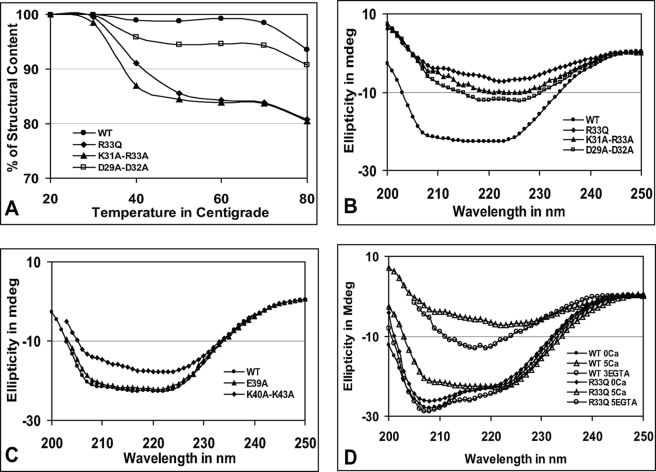
The CD spectra were recorded at different temperatures (20–80 °C) for each of the N-terminal cluster mutants. The secondary structural content of the mutant with an alteration in cluster 1 was lost more rapidly, whereas the overall spectral characteristics were not significantly altered at higher temperatures. The spectra of the WT protein and cluster 1 (including R33Q) versus cluster 2 mutants are shown in supplemental Fig. S3. Because the overall spectral characteristic was not altered, to better interpret the effect of mutation on the CASQ2 folding pattern at high temperature, the percentage of the secondary structural content was calculated. The ellipticity at 222 nm and 20 °C was taken as 100% for each protein. The results showed that the K31A-R33A and R33Q mutants behaved comparably. The R33Q and K31A-R33A mutants lost their secondary structural content significantly between 30 and 50 °C from 99 and 98% to 86 and 84%, respectively. The data for D29A-D32A were intermediate between those for the WT and mutant (K31A-R33A and R33Q) proteins (Fig. 2A).
FIGURE 2.
Mutations in cluster 1 affect CASQ stability and polymerization. A, comparison of secondary structural content in cluster 1 mutants. The cluster 1 mutants lose secondary structural content rapidly at high temperatures. (The actual CD spectra are shown in supplemental Fig. S3.) B and C, CD spectra of cluster 1 and 2 mutants, respectively, in the presence of 5 mm CaCl2. The CD spectra of R33Q, K31A-R33A, and D29A-D32A show highly reduced ellipticity, suggesting random aggregation. The K40A-K43A mutation had an intermediate effect, but the E39A mutation had no effect, and its CD spectrum is similar to that of the WT protein. D, polymerization and depolymerization of the WT protein and R33Q. The WT protein and mutant were treated with Ca2+ and chelated with EGTA as described under “Experimental Procedures.” WT CASQ regained the native conformation upon Ca2+ chelation by EGTA (∼3 mm), whereas R33Q failed to regain the native conformation, indicating that reversibility of CASQ2 polymerization is affected by the R33Q mutation. mdeg, millidegrees.
Calcium-induced Polymerization Is Impaired if the N-terminal Charged Cluster Is Altered
We employed CD spectroscopy, turbidimetric assay, and partial trypsinization to address how alteration of N-terminal charged clusters affects protein structure and function, including Ca2+-induced aggregation. First, Ca2+-induced aggregation of WT and mutant proteins was analyzed by CD spectroscopy. In the presence of 5 mm CaCl2, cluster 1 mutants R33Q, K31A-R33A, and D29A-D32A showed significantly different CD spectra with the least ellipticity, and the negative maxima around 208 nm were completely lost (Fig. 2B and supplemental Fig. S4). The CD spectral ellipticity for double mutant K40A-K43A in cluster 2 was intermediate; however, single mutant E39A in cluster 2 showed similar CD spectra compared with the WT protein (Fig. 2C) in the presence of CaCl2. After Ca2+-induced aggregation, the solution was subjected to chelation with EGTA. Interestingly, the WT protein regained its secondary structural content similar to its native conformation upon Ca2+ chelation by EGTA at ∼3 mm; however, the R33Q mutant could not regain CD ellipticity comparable with the native conformation even up to ∼5 mm EGTA (Fig. 2D). Point mutations E39A and K40A-K43A could regain their native conformation, but the K31A-R33A and D29A-D32A mutants failed to regain the native conformation upon Ca2+ chelation (supplemental Fig. S4).
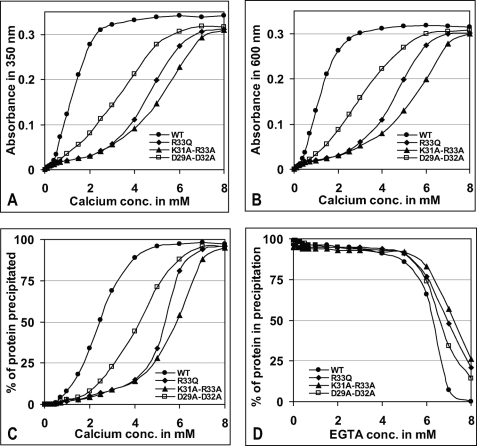
The effect of N-terminal mutation on Ca2+-induced CASQ2 aggregation was further investigated by turbidimetry. As shown in Fig. 3 (A and B), the WT protein became aggregated very rapidly and reached the highest achievable aggregation with 2.5 and 3.0 mm CaCl2 as measured by absorbance at 350 and 600 nm, respectively. 50% precipitation was achieved with 2.42 mm CaCl2 (Fig. 3C). In contrast, the least amounts of Ca2+ required for maximum aggregation of R33Q, K31A-R33A, and D29A-D32A (at 350 and 600 nm) were found to be 6.2, 7.1, and 5.7 mm and 6.4, 7.4, and 6.1 mm, respectively. 50% precipitation (EC50) for R33Q, K31A-R33A, and D29A-D32A was achieved at higher Ca2+ concentrations of 5.3, 5.9, and 4.11 mm, respectively. A closer visual examination revealed that the aggregates formed by the WT protein were needle-shaped crystalline structures as observed by others, whereas the R33Q mutant produced flake-shaped irregular structures (33, 34). Similarly irregular aggregates were observed for K31A-R33A, D29A-D31A, Δ10CASQ2, and Δ20CASQ2, whereas other mutants formed aggregates like the WT protein. When CASQ aggregate was subjected to Ca2+ chelation by EGTA, the WT protein became solubilized with a very low concentration of EGTA, whereas the cluster 1 mutant proteins required higher EGTA for resolubilization (Fig. 3D and supplemental Fig. S5). The EC50 values of Ca2+-induced aggregation and EGTA-mediated chelation are shown in Table 2.
FIGURE 3.
Conformational changes in cluster 1 mutants as analyzed by turbidimetric assay. Aggregation measured at 350 nm (A) and 600 nm (B) showed that cluster 1 mutants R33Q, K31A-R33A, and D29A-D32A are Ca2+-insensitive at low calcium concentrations. Shown is the percentage of protein precipitated due to Ca2+-induced aggregation (C) and Ca2+ chelation mediated by EGTA (D) as described under “Experimental Procedures.” Cluster 1 mutants required higher EGTA concentrations to become resolubilized. For EGTA-mediated Ca2+ chelation, the WT protein and cluster 1 mutants were first aggregated with 8 mm CaCl2. The replot of aggregation-disaggregation of each of the cluster 1 mutants is shown in supplemental Fig. S5.
TABLE 2.
Dimer stability and EC50 values for Ca2+-induced polymerization and EGTA-mediated chelation
Mutations were generated from the dimer form (Protein Data Bank code 1SIJ). The cluster 1 mutants are less stable and require higher CaCl2 and EGTA for bidirectional transition between monomer and polymer. The EC50 values were calculated from turbidimetric assay.
| Protein | Overall potential energy (MMFF94s) | EC50 of CaCl2 (for aggregation) | EC50 of EGTA (for resolubilization) |
|---|---|---|---|
| kcal/mol | mm | mm | |
| WT | −17,540.5 | 2.42 ± 0.15 | 2.45 ± 0.1 (after 5 mm CaCl2) |
| R33Q | −17,127.5 | 5.3 ± 0.2 | |
| D29A-D32A | −17,190.1 | 4.11 ± 0.15 | |
| K31A-R33A | −16,870.4 | 5.9 ± 0.3 |
Limited Proteolysis Shows That Mutations in Cluster 1 Affect CASQ Polymerization
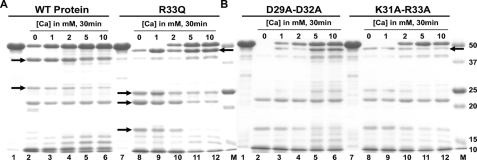
CASQ2 becomes highly resistant to trypsin upon Ca2+ binding; therefore, we determined the behavior of cluster 1 mutants upon partial digestion with trypsin. Interestingly, the patterns of tryptic fragments generated from the R33Q, K31A-R33A, and D29A-D32A mutants were different from that of the WT protein even in the absence of Ca2+, but they resembled each other in being highly sensitive to trypsin digestion, unlike the WT protein (Fig. 4, A and B). The differences in the tryptic fragments are highlighted by arrows. R33Q and K31A-R33A did not gain resistance to trypsinization at 1 mm CaCl2, although D29A-D32A became mildly resistant. At 2 mm CaCl2, R33Q and K31A-R33A started to gain some resistance, and D29A-D32A became more protected. However, at high Ca2+ concentrations (5–10 mm), all three mutants became significantly protected from trypsin digestion.
FIGURE 4.
Cluster 1 mutants have impaired Ca2+ response. The trypsin digestion pattern of cluster 1 mutants is different from that of the WT protein. A, lanes 1–6, untreated WT protein and incubation with 0, 1, 2, 5, and 10 mm CaCl2, respectively; lanes 7–12, untreated R33Q mutant protein and treatment with 0, 1, 2, 5, and 10 mm CaCl2, respectively. B, lanes 1–6, untreated D29A-D32A mutant protein and treatment with 0, 1, 2, 5, and 10 mm CaCl2, respectively; lanes 7–12, untreated K31A-R33A mutant protein and treatment with 0, 1, 2, 5, and 10 mm CaCl2, respectively. The M lanes are protein markers (Bio-Rad). The differences in the tryptic fragments are highlighted with black arrows.
Dynamic Dimerization Is Dependent on the Integrity of the Charge Pattern of Cluster 1
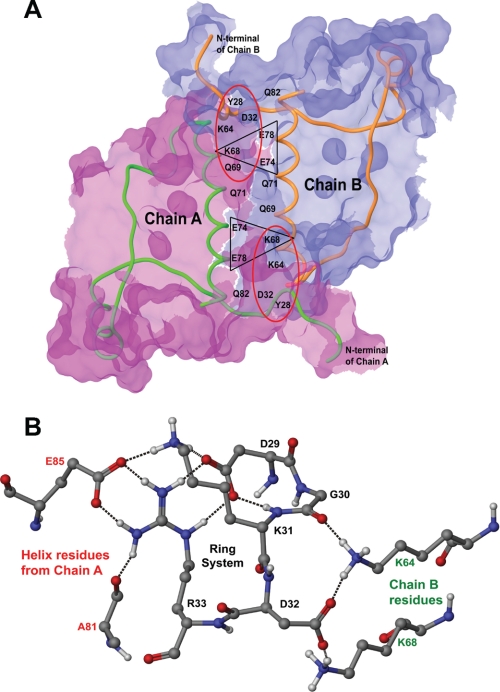
To further understand the role of cluster 1, we employed molecular dynamics simulation using three-dimensional coordinates of the CASQ2 structure. As shown in Fig. 1B, the highly conserved cluster 1 forms a loop. Further analysis revealed that the loop is stabilized by an intrachain H-bond between CPVT Arg33 and Asp29, giving the cluster 1 a conformation of a ring system. The location of the ring system in the dimer is shown in Fig. 5A. Detailed structural analysis following simulation revealed that a conformational flip of Asp32 to the exo-conformation in the dimer is important for dimerization (Fig. 5B). Associated with Asp32, there occurs another conformational flip of Lys31 in the reverse orientation with respect to Asp32. Asp32 remains in an endo-conformation in the monomer without a H-bond network. The arrangement of the ring system in the dimer and monomer is shown in Fig. 5 (C and D, respectively). Overall, the Lys31-Asp32 flipping brings the dipoles into close proximity (Glu74 and Glu78 of chain A and Lys68 of chain B and vice versa) to bridge them by the H-bond network, establishing loop-helix and helix-helix interactions, as shown in Fig. 6A. The side chain of the inward flipped Lys31 residue forms H-bonds with Asp29 and Glu85. The side chain of the outward flipped Asp32 residue forms a H-bond with helix residues Lys64 and Lys68 with its partner monomer. The H-bond network established by cluster 1 to stabilize the dimer conformation is shown in Fig. 6B.
FIGURE 5.
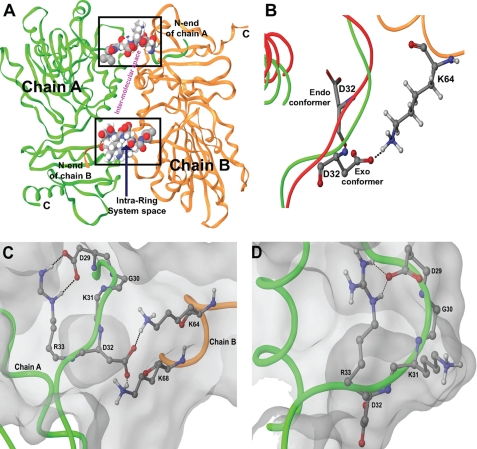
Organization of cluster 1 into a ring system to function as a molecular switch. The dipoles emanating from alternately charged residues in the ring system are arranged such that they can support the coordinated conformational flip of Lys31-Asp32, resulting in a H-bond network. Black dashes represent H-bonds. A, location of cluster 1 in the dimer. The cluster located at the interface of the dimer in the form of a ring system regulates formation of a H-bond network to stabilize dimer conformation. The intermolecular space (shown in pink) is the site of helix-loop and helix-helix interactions shown in detail in Fig. 6A. B, conformational flip of Asp32. Superimposition studies of the monomeric (red; endo-conformer) and dimeric (green; exo-conformer) structures suggested the conformational flipping of Asp32. Asp32 and Lys64 (of chain B, shown in orange) of the dimer are shown in ball and stick form, and Asp32 of the monomer is shown in tube form. C and D, comparison of the ring system in the dimer and monomer, respectively. The ring system in the monomer has Lys31 exposed outward and Asp32 oriented inward. Arg33 regulates the outward flip of Asp32 to interact with Lys64 and Lys68 of chain B. The flip of alternately charged residues Lys31 and Asp32 suggests its plausible role in Ca2+ sensing and polymerization.
FIGURE 6.
Stabilization of the CASQ dimer by the formation of a H-bond network. A, intermolecular surface of the dimer. The structure of the front-to-front dimer between chains A and B is stabilized through intermolecular H-bonding by loop-helix interactions of Asp32 with Lys68 and Lys64 (red ellipse) and helix-helix interactions of Lys68 with Glu74 and Glu78 (black triangle). The surface of chain A is shown in pink, and that of chain B is shown in blue. The interacting surface has overlapping pink and blue colors. B, the H-bond network formed by the DGKDR ring system in a dimer. Black dashes represent H-bonds. The conformational flip of Lys31-Asp32 brings together helices from chains A (in red) and B (in green) into proximity and is stabilized by the H-bond network.
DISCUSSION
Structural studies suggested that, upon Ca2+ binding, CASQ2 undergoes linear polymerization first by front-to-front dimerization and then by back-to-back packaging, generating needle-shaped polymers (11, 19). Although it was suggested that Ca2+-induced polymerization is nonlinear and might be reversible (17, 20, 35), the exact mechanism controlling the nonlinear polymerization-depolymerization in nanoseconds is not known. On the other hand, recent studies have indicated that the front-to-front dimer is the key step in overall CASQ2 function (16, 17). We hypothesized that the N-terminal end might be involved in dimer formation and stabilization. Therefore, the major goal of this study was to define the role of structural elements in the N-terminal end in dynamic polymerization. Our structural and computational studies revealed that the highly conserved alternately charged cluster (DGKDR) is the key for polymerization dynamics and that mutation R33Q disrupts the charge pattern of the cluster.
The N-terminal End Is Crucial for Protein Folding and Polymerization
It was suggested that the front-to-front dimer formation and stabilization may involve arm exchange and domain swapping involving the extended N-terminal end (16). Therefore, we investigated whether the extended N-terminal end is vital for CASQ2 folding. Interestingly, we found that N-terminal deletion mutants Δ10CASQ2 and Δ20CASQ2 cannot fold properly, indicating that the N-terminal end is required for correct folding of CASQ2. We predicted that the alternating charged clusters 1 (DGKDR) and 2 (EKNLK) might be involved in Ca2+-induced polymerization. Results showing that Δ5CASQ2 can fold and polymerize like the WT protein and that Δ10CASQ2 fails to fold suggest the importance of cluster 1 (ring system) residues. This is further highlighted by the CD data showing a reduction in the thermal stability of cluster 1 mutants. On the other hand, charge neutralization mutagenesis suggested that cluster 2, which is less conserved in evolution, is not critical.
Recent studies using a knock-in mouse model for R33Q suggested that mutation might affect protein stability and that a decrease in CASQ protein expression itself can contribute to CPVT disease phenotype (28, 30, 36). This and other studies showed that the mutant protein is more susceptible to proteolysis (30, 31). Our study further confirmed this finding, and our data show that R33Q fails to polymerize at low Ca2+ concentration (1 mm) when the WT protein has already reached lower order oligomeric states. In addition, we have demonstrated that mutations in cluster 1 make the proteins highly susceptible to trypsin digestion in the absence of Ca2+ and that mutants fail to gain resistance to trypsin digestion at 1 mm CaCl2.
Results from the turbidimetric experiment indicate that the R33Q, K31A-R33A, and D29A-D32A mutants do not aggregate at low Ca2+ concentrations of 1 and 2 mm but undergo a significant level of aggregation at ∼5 mm Ca2+, additionally supporting our proteolysis data. However, at high Ca2+ concentrations, R33Q does not exhibit needle-shaped crystalline structure as seen for the WT protein and appears to be formed by random aggregation rather than a properly oriented linear polymer (33, 34). The random aggregation could be promoted by the neutralization of surface charges by Ca2+.
Cluster 1 Charged Residues Are Essential for Ca2+-induced Aggregation of CASQ2 and Resolubilization
One of the most interesting findings of this study is that, following Ca2+-induced aggregation, the WT protein can be resolubilized to the native conformation upon EGTA-mediated Ca2+ chelation. Our findings further support the proposal of Park et al. (17) that CASQ2 polymerization is reversible and that polymers can depolymerize when [Ca2+] falls below a critical level. Ca2+ chelation with EGTA, which has a much higher affinity for Ca2+ compared with CASQ2, compels CASQ2 polymers to undergo depolymerization by giving off Ca2+. This mimics the physiological condition that occurs in the myocyte during Ca2+-induced Ca2+ release; CASQ2 can release a portion of the bound Ca2+ without undergoing depolymerization because polymerization and Ca2+ concentration are nonlinear. However, when maximal Ca2+ release is needed, CASQ2 polymers have to depolymerize to support Ca2+ release. The turbidimetric data showed that the cluster 1 mutants (R33Q, K31A-R33A, and D29A-D32A) require higher Ca2+ and EGTA to undergo aggregation and disaggregation, respectively (Fig. 3, Table 2, and supplemental Fig. S5), suggesting that mutations in cluster 1 cause random aggregation. This is also supported by the CD data showing that the mutants failed to regain their native conformation (although they became soluble with higher EGTA concentrations). These findings concur with the dynamics study and the idea that the charge or dipolar arrangement in cluster 1 (due to the presence of alternately charged residues) is essential for correct bidirectional navigation in the polymerization-depolymerization pathway. The sigmoidal curve observed for polymerization-depolymerization in our turbidimetric assays is qualitatively similar to the mathematical calculation, and this finding is in agreement with the recent proposal that Ca2+-induced CASQ2 polymerization is nonlinear (3, 35, 37).
Cluster 1 Works as a Molecular Switch, and the R33Q Mutation Disrupts the Formation of a Ring System
The molecular dynamics and structural studies suggest that cluster 1, which is conserved from C. elegans to human, is the key to dimerization. This cluster works as a molecular switch and configures into a ring system stabilized by a H-bond between Asp29 and Arg33. Gly30 is useful as a spacer and accounts for the DGKDR ring system being large enough to maintain the critical conformational flip of Lys31-Asp32 and therefore might be conserved although not directly involved in dipolar interactions. The conformational flip may be catalyzed by Ca2+ and controlled by Arg33, which is assisted by the dynamicity of companion residues in the ring system. The dipoles from alternately charged residues might sense [Ca2+]. Being positively charged, Lys31 will be repelled by Ca2+, and being negatively charged, Asp32 will be attracted to Ca2+, which seems to regulate the flip of Lys31-Asp32 and the formation of a H-bond network (Fig. 6B), stabilizing the dimer conformation. Again, the rotational flexibility of the ring system backbone is high, allowing a flip of ∼180°. This is why the mutant molecule loses the property of Ca2+-induced polymerization when the charge pattern in the cluster is altered. Moreover, introduction of a glutamate residue in place of Arg33 reduces the backbone flexibility, and the ring system is not stabilized because the hydrogen bond between amino acids 29 and 33 cannot be formed. This disrupts the ring system, which is necessary for the critical flip of Lys31-Asp32, leading to impairment of Ca2+-induced dynamics. The above interpretation is further supported by potential energy determination using energy minimization. The cluster 1 mutants have higher potential energy compared with the WT protein (Table 2), suggesting that mutant dimers are less stable. During polymerization, dimer formation is the key step; therefore, if the dimer is less stable, its ability to form a properly oriented polymer and to undergo rapid depolymerization is significantly reduced. This concurs with the proposal that Ca2+ regulation by calsequestrin involves interplay among protein folding, Ca2+ binding, and calsequestrin polymerization-depolymerization (16, 17).
To be physiologically relevant, the dynamic changes in CASQ2 polymerization-depolymerization should occur in the nano-microsecond range. This can be facilitated only by the presence of a molecular switch that allows swift transition between protein conformations in a calcium-dependent manner. This type of conformational change is facilitated by structural motifs, and these motifs often operate as switches to regulate biological events dynamically in the nanosecond range (38–41).
Conclusions
The observations made in this study show that the extended N-terminal end in general and the alternately charged residue cluster in particular are key for the Ca2+-induced aggregation and function of CASQ2. The results also show that CASQ2 polymerization is reversible and nonlinear. This fits well with the recent proposal that Ca2+ buffering inside the SR and Ca2+ release are nonlinear (35). In addition, our results demonstrate that the R33Q mutation disrupts the alternating charge pattern of the N-terminal DGKDR ring system and weakens the ability of CASQ2 to undergo dynamic polymerization-depolymerization. On the basis of these findings, we propose that, under increased physiological demands, the R33Q mutant fails to undergo the dynamic conformational interconversions necessary to cope with increased Ca2+ handling and that this, along with decreased protein levels, could contribute to CPVT.
Supplementary Material
Acknowledgments
We thank Barbara Heck for editorial assistance. N. C. B. thanks Professor Rita Bernhardt for mentorship in protein engineering.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL64014 (to M. P.). This work was also supported by a Fund for Improvement of Science and Technology Infrastructure grant from the Department of Science and Technology, Government of India (to A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- SR
- sarcoplasmic reticulum
- CPVT
- catecholaminergic polymorphic ventricular tachycardia
- WT
- wild-type.
REFERENCES
- 1.Bers D. M. (1997) Basic Res. Cardiol. 92, Suppl. 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 2.MacLennan D. H., Campbell K. P., Reithmeier R. A. F. (1983) in Calcium and Cell Function (Cheung W. Y. ed) Vol. IV, pp. 151–173, Academic Press, Inc., New York [Google Scholar]
- 3.Restrepo J. G., Weiss J. N., Karma A. (2008) Biophys. J. 95, 3767–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon T. R., Wang F., Puglisi J., Weber C., Bers D. M. (2004) Biophys. J. 87, 3351–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slupsky J. R., Ohnishi M., Carpenter M. R., Reithmeier R. A. (1987) Biochemistry 26, 6539–6544 [DOI] [PubMed] [Google Scholar]
- 6.MacLennan D. H., Wong P. T. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard N. A., Laver D. R., Dulhunty A. F. (2004) Prog. Biophys. Mol. Biol. 85, 33–69 [DOI] [PubMed] [Google Scholar]
- 8.Knollmann B. C. (2009) J. Physiol. 587, 3081–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzini-Armstrong C. (2009) Appl. Physiol. Nutr. Metab. 34, 323–327 [DOI] [PubMed] [Google Scholar]
- 10.Györke S., Terentyev D. (2008) Cardiovasc. Res. 77, 245–255 [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Trumble W. R., Liao H., Wesson C. R., Dunker A. K., Kang C. H. (1998) Nat. Struct. Biol. 5, 476–483 [DOI] [PubMed] [Google Scholar]
- 12.Franzini-Armstrong C., Protasi F., Ramesh V. (1999) Biophys. J. 77, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu N., Priori S. G. (2008) Cardiovasc. Res. 77, 293–301 [DOI] [PubMed] [Google Scholar]
- 14.Renken C., Hsieh C. E., Marko M., Rath B., Leith A., Wagenknecht T., Frank J., Mannella C. A. (2009) J. Struct. Biol. 165, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagenknecht T., Hsieh C. E., Rath B. K., Fleischer S., Marko M. (2002) Biophys. J. 83, 2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E., Youn B., Kemper L., Campbell C., Milting H., Varsanyi M., Kang C. (2007) J. Mol. Biol. 373, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 17.Park H., Park I. Y., Kim E., Youn B., Fields K., Dunker A. K., Kang C. (2004) J. Biol. Chem. 279, 18026–18033 [DOI] [PubMed] [Google Scholar]
- 18.Park H., Wu S., Dunker A. K., Kang C. (2003) J. Biol. Chem. 278, 16176–16182 [DOI] [PubMed] [Google Scholar]
- 19.Gatti G., Trifari S., Mesaeli N., Parker J. M., Michalak M., Meldolesi J. (2001) J. Cell Biol. 154, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Launikonis B. S., Zhou J., Royer L., Shannon T. R., Brum G., Ríos E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2982–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLennan D. H., Abu-Abed M., Kang C. (2002) J. Mol. Cell. Cardiol. 34, 897–918 [DOI] [PubMed] [Google Scholar]
- 22.Terentyev D., Kubalova Z., Valle G., Nori A., Vedamoorthyrao S., Terentyeva R., Viatchenko-Karpinski S., Bers D. M., Williams S. C., Volpe P., Gyorke S. (2008) Biophys. J. 95, 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L., Varsányi M., Dulhunty A. F., Beard N. A. (2006) Biophys. J. 91, 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blayney L. M., Lai F. A. (2009) Pharmacol. Ther. 123, 151–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.di Barletta M. R., Viatchenko-Karpinski S., Nori A., Memmi M., Terentyev D., Turcato F., Valle G., Rizzi N., Napolitano C., Gyorke S., Volpe P., Priori S. G. (2006) Circulation 114, 1012–1019 [DOI] [PubMed] [Google Scholar]
- 26.Scheinman M. M., Lam J. (2006) Annu. Rev. Med. 57, 473–484 [DOI] [PubMed] [Google Scholar]
- 27.Knollmann B. C., Chopra N., Hlaing T., Akin B., Yang T., Ettensohn K., Knollmann B. E., Horton K. D., Weissman N. J., Holinstat I., Zhang W., Roden D. M., Jones L. R., Franzini-Armstrong C., Pfeifer K. (2006) J. Clin. Invest. 116, 2510–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chopra N., Kannankeril P. J., Yang T., Hlaing T., Holinstat I., Ettensohn K., Pfeifer K., Akin B., Jones L. R., Franzini-Armstrong C., Knollmann B. C. (2007) Circ. Res. 101, 617–626 [DOI] [PubMed] [Google Scholar]
- 29.Terentyev D., Nori A., Santoro M., Viatchenko-Karpinski S., Kubalova Z., Gyorke I., Terentyeva R., Vedamoorthyrao S., Blom N. A., Valle G., Napolitano C., Williams S. C., Volpe P., Priori S. G., Gyorke S. (2006) Circ. Res. 98, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 30.Rizzi N., Liu N., Napolitano C., Nori A., Turcato F., Colombi B., Bicciato S., Arcelli D., Spedito A., Scelsi M., Villani L., Esposito G., Boncompagni S., Protasi F., Volpe P., Priori S. G. (2008) Circ. Res. 103, 298–306 [DOI] [PubMed] [Google Scholar]
- 31.Valle G., Galla D., Nori A., Priori S. G., Gyorke S., de Filippis V., Volpe P. (2008) Biochem. J. 413, 291–303 [DOI] [PubMed] [Google Scholar]
- 32.Kalyanasundaram A., Bal N. C., Franzini-Armstrong C., Knollmann B. C., Periasamy M. (2010) J. Biol. Chem. 285, 3076–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer A., Tanaka M., Ozawa T., Fleischer S. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4036–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams R. W., Beeler T. J. (1986) J. Biol. Chem. 261, 12408–12413 [PubMed] [Google Scholar]
- 35.Royer L., Ríos E. (2009) J. Physiol. 587, 3101–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin J., Valle G., Nani A., Nori A., Rizzi N., Priori S. G., Volpe P., Fill M. (2008) J. Gen. Physiol. 131, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pape P. C., Fénelon K., Lamboley C. R., Stachura D. (2007) J. Physiol. 581, 319–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonsor D. A., Hecht O., Vankemmelbeke M., Sharma A., Krachler A. M., Housden N. G., Lilly K. J., James R., Moore G. R., Kleanthous C. (2009) EMBO J. 28, 2846–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakrapani S., Auerbach A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J., Karplus M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11905–11910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra P., Socolich M., Wall M. A., Graves J., Wang Z., Ranganathan R. (2007) Cell 131, 80–92 [DOI] [PubMed] [Google Scholar]
- 42.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.