Abstract
In this study we aimed to identify small molecules with high affinity involved in the allosteric regulation of LVIS553, a MarR member from Lactobacillus brevis ATCC367. Using high throughput screening, novobiocin was found to specifically bind LVIS553 with a KD = 33.8 ± 2.9 μm consistent with a biologically relevant ligand. Structure guided site-directed mutagenesis identified Lys9 as a key residue in novobiocin recognition. The results found in vitro were correlated in vivo. An increased tolerance to the antibiotic was observed when LVIS553 and the downstream putative transport protein LVIS552 were either expressed in a low copy plasmid in L. brevis or as a single copy chromosomal insertion in Bacillus subtilis. We provide evidence that LVIS553 is involved in the specific regulation of a new mechanism of tolerance to novobiocin.
Keywords: Antibiotics, Bacterial Transcription, Drug Resistance, Multidrug Transporters, Transcription Factors, Lactobacillus, Novobiocin, High Throughput Screening, Small Molecule
Introduction
The MarR family of transcriptional regulators is a diverse group of small proteins (120–207 amino acids long) that share similar structural scaffold (1). Conversely, analysis of the amino acid sequence alignment of well characterized MarR members, such as MarR (PDB code 1JGS), OhrR (PDB code 1Z91), SlyA (PDB code 1LJ9), and EmrR (PDB code 2GXG) revealed that very few amino acids are conserved (2). Consequently, the main focus of several previous reports has been to further understand the contribution of conserved structures to DNA binding and the key roles of individual amino acids. As a result, the location of the MarR DNA binding domain was previously determined to reside within amino acids 61–121 (3). Further reports have shown that mutations on residues allocated in this area have both structural and functional consequences that decrease DNA binding as shown for MarR (3), SlyA in Salmonella (4), and MdtR (YusO) in Bacillus subtilis (5). To date, however, we are far from understanding in depth the molecular mechanisms behind the structural modifications associated with ligand binding.
Several factors have contributed to the uncertainties that remain regarding ligand binding. First and foremost, the ability to identify specific amino acids and their associated functional involvement has been hindered by the overlapping nature of both the DNA and ligand binding motifs. In addition, attempts to examine the mechanisms in detail have been further hampered due to the overall lack of physiologically relevant ligands. Annotations for the majority of family members are usually based on in silico data without direct physiological evidence. The founding member of this family is MarR from Escherichia coli (6, 7), which has been biochemically, structurally, and phenotypically characterized. MarR binds salicylate and other phenolic compounds such as plumbagin, 2,4-dinitrophenol, and menadione in the low millimolar range (0.5–2 mm) (8, 9).
Many orthologs have been reported thereafter and binding to salicylate has been claimed (10), although in most cases, their binding affinities are too low to be considered biologically or biochemically relevant. Other well characterized members of this family such as EmrR (11) and HucR bind small molecules in the low micromolar range (2.0–15.0 and 11.6 μm, respectively) (12, 13). Accordingly, we are particularly interested in identifying alternative ligands, elucidating the mechanisms by which small molecules regulate the formation and release of transcriptional regulators from the cognate DNA sequence, and discussing the molecular results under the light of its physiological consequences.
Due to their involvement in multidrug resistance and tolerance to highly toxic compounds, MarR members have been predominately studied in pathogenic microorganisms, although they are found throughout many bacterial and archaeal groups (14). It is particularly interesting to determine how commensal bacteria such as Lactobacillus are able to tolerate and survive antibiotics as a means of persistence in the intestinal tract or as a competitive advantage over other commensal members. Lactobacilli are in general, very resistant and capable of tolerating a wide variety of stress conditions (for a review, see Lorca and Font de Valdez (15)). Lactobacillus brevis, in particular, has received a lot of attention due to its role in spoilage of beer (16), which involves induction of efflux pumps as a mechanism of hop resistance (17, 18). By comparative genomics we determined that L. brevis has the highest number of transport proteins (13%) involved in the uptake and efflux of drugs and toxic compounds (19, 20). Collectively, these characteristics make L. brevis a good model for understanding how commensal bacteria respond to environmental stressors such as antibiotics, via mechanisms in which transcriptional regulators may be involved.
In this study we used LVIS553 as a model protein to locate and examine high affinity small molecules involved in the allosteric regulation of a MarR member. The identified molecule, novobiocin, was found to specifically bind LVIS553 in the low micromolar range. We provide in vivo evidence that LVIS553 is involved in the regulation of a new mechanism of tolerance to novobiocin.
EXPERIMENTAL PROCEDURES
Bacterial Strains
L. brevis ATCC367 was obtained from the American Type Culture Collection (Manassas, VA). Lactobacillus strains were grown at 37 °C in MRS broth (Difco Laboratories, Detroit, MI). E. coli DH5α cells, used to carry and propagate all vectors, were grown in Luria-Bertani medium (Difco). B. subtilis M168 was used for heterologous gene expression. Growth was performed in LB medium at 37 °C under aerobic conditions. When appropriate, medium was supplemented with erythromycin (7.5 μg/ml) (for L. brevis), kanamycin (5 μg/ml) (for B. subtilis), or ampicillin (100 μg/ml) (for E. coli). All antibiotics and chemicals were purchased from Sigma.
DNA Manipulations and Gene Cloning
Standard methods were used for site-directed mutagenesis, chromosomal DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation (21). Plasmids were isolated using spin miniprep kits (Qiagen, Valencia, CA) and PCR products were purified using QIAquick purification kits (Qiagen). Mutagenesis was performed using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA).
For protein expression and purification, the LVIS553 gene was amplified from L. brevis ATCC367 chromosomal DNA by PCR. The primers utilized are described in Table 1. The plasmid p15TV-L (GenBank accession EF456736) obtained from the Structural Genomics Consortium (SGC, Toronto) was employed as a vector. This construct also provides a N-terminal hexahistidine tag, separated from the protein by a tobacco etch virus protease recognition site (ENLYFQ↓GS).
TABLE 1.
Oligonucleotides used in this work
| Primer | To LVIS553 ATG | Oligonucleotide sequence (5′ → 3′) |
|---|---|---|
| Protein purification | ||
| LB45 Fw | …+1… | ttgtatttccagggcatgactcaacccattacctttaataaatatattga |
| LB45 Rv | …+438… | caagcttcgtcatcatcagttcctttcttgtcgctgttga |
| EMSA | ||
| PLvis552 Fwb | …+724… | aaaactcatggcgcagaaag |
| PLvis552 Rv | …+489… | aacaacttgcggccatttac |
| PLvis553 Fwb | …+24… | attaaaggtaatgggttgagtcat |
| PLvis553 Rv | …−273… | gtgcggacctcctttgaat |
| PLvis553-Int Fwb | …+203… | aagtcacgagcgaccagact |
| PLvis553-Int Rv | …−134… | gaagacccatttgcgttgaa |
| PLvis554 Fwb | …−107… | atcgttttcaacgcaaatgg |
| PLvis554 Rv | …−411… | gccttaacaccaagggtttg |
| Site-directed mutagenesis | ||
| Lvis553_K9A Fw | …+7… | caacccattacctttaatgcatatattgccagtatttac |
| Lvis553_K9A Rv | …+45… | gtaaatactggcaatatatgcattaaaggtaatgggttg |
| Lvis553_R16A Fw | …+28… | tatattgccagtatttacgcccaatctaagcatgacttc |
| Lvis553_R16A Rv | …+66… | gaagtcatgcttagattgggcgtaaatactggcaatata |
| Cloning into pRV610 | ||
| Lvis552–553 KpnI Fw | …+1152… | ggggtaccgaatcagtctggctgggaagc |
| Lvis552–553 XbaI Rv | …−272… | gctctagagcggacctcctttgaattttc |
| Competition assay | ||
| Lvis553 protection Fw | …−18… | aaaaaatagtggataagtccattaaattct |
| Lvis553 protection Rv | …−47… | agaatttaatggacttatccactatttttt |
| DNase I footprinting | ||
| Lvis553fprint_FAM Rv | …+203… | aagtcacgagcgaccagact |
| Lvis553fprint_VIC Fw | …−273… | gtgcggacctcctttgaat |
| Quantitative reverse transcription-PCR | ||
| Lvis552 Fw | …+550… | tcgccaatttctagcattcc |
| Lvis552 Rv | …+718… | caccgaaaataacggaagga |
| Lvis553 Fw | …+4… | actcaacccattacctttaat |
| Lvis553 Rv | …+435… | gttcctttcttgtcgctgttg |
| rpoD Fw | attcccgttcatatggtgga | |
| rpoD Rv | gaaccttttccgttgccata | |
a Italics show the extra bases added to the 5′ end for the ligation independent cloning using the BD-infusion CF Dry-Down PCR cloning kit (BD Biosciences).
b Biotin labeled.
c Underlines indicate the restriction sites.
Cloning in the pRV610 plasmid (22) was performed amplifying LVIS552-LVIS553 (including the complete PLVIS553) by PCR, using the primers described in Table 1. Clones were confirmed by sequencing using universal M13 primers.
For heterologous gene expression of L. brevis genes in B. subtilis, LVIS552-LVIS553 was amplified using PCR, cut with KpnI and XbaI restriction enzymes, and cloned in pSac-Kan (23). Recombinant clones were obtained in E. coli DH5α and subsequently sequenced. Transformation of B. subtilis was performed by natural competence. Integration in the sacA gene was verified by the inability to utilize sucrose as a carbon source on minimal medium (24).
Protein Purification
Protein purification was carried out as previously described (25). Briefly, the His-tagged fusion proteins were overexpressed in E. coli BL21-Star(DE3) cells (Stratagene). The cells were grown in LB at 37 °C to an A600 ∼0.6 and expression induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. After addition of isopropyl 1-thio-β-d-galactopyranoside, the cells were incubated with shaking at 15 °C overnight. The cells were harvested, resuspended in binding buffer (500 mm NaCl, 5% glycerol, 50 mm HEPES, pH 7.5, 5 mm imidazole), flash frozen in liquid N2, and stored at −70 °C. The thawed cells were lysed and passed through a French Press after the addition of 0.5% Nonidet P-40 and 1 mm each of phenylmethylsulfonyl fluoride and benzamidine. The lysate was clarified by centrifugation (30 min at 17,000 × g) and applied to a metal chelate affinity column charged with Ni2+. After the column was washed and the protein was eluted from the column in elution buffer (binding buffer with 500 mm imidazole). The His6 tag was then cleaved from the protein by treatment with recombinant His-tagged tobacco etch virus protease. The cleaved protein was then resolved from the cleaved His tag and the His-tagged protease by passing the mixture through a second Ni2+ column. The purified proteins were dialyzed against 10 mm HEPES, pH 7.5, 500 mm NaCl, and concentrated using a BioMax concentrator (Millipore, Billerica, MA).
Size Exclusion Chromatography
A 100-μl protein sample contained 10 mm HEPES, pH 7.5, 500 mm NaCl, 25 μm LVIS553, and where indicated, 100 μm novobiocin. Following a 20-min incubation on ice, samples were injected onto a Superose 12 10/300 GL gel filtration column (Amersham Biosciences) installed on an Äkta system (Amersham Biosciences) equilibrated with 10 mm HEPES, pH 7.5, 500 mm NaCl. Filtration was performed at 4 °C at a flow rate of 0.5 ml/min and the protein concentration was monitored by measuring the absorbance at 280 nm. Blue dextran 2000 was used to determine the void volume. A mixture of protein molecular mass standards, containing β-amylase (200 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa), was applied to the column under similar conditions. The elution volumes and molecular masses of the protein standards were used to generate a standard curve from which the apparent molecular mass was determined.
Electrophoretic Mobility Shift Assays (EMSA)2
EMSA for LVIS553 was performed using protein purified and concentrated according to the procedures described above. Fragments for the PLVIS552, PLVIS553, and PLVIS554 promoter regions (Table 1) were generated by PCR using biotin-prelabeled (5′-end) primers, then purified using QIAquick spin columns (Qiagen). Incubation mixtures for EMSA (20 μl) contained 2.5 nm of a 5′-labeled DNA fragment, 50 mm Tris-HCl, pH 7.5, 150 mm KCl, 10 mm MgCl, 0.01% Triton X-100, 50 ng/μl of poly(dI-dC) nonspecific competitor DNA, purified LVIS553 protein (0–100 nm), and ligand (0–1 mm) when indicated.
After incubation for 20 min at 37 °C, samples were separated on 6% acrylamide-bisacrylamide nondenaturing gels in 0.5× Tris borate-EDTA buffer, pH 8.3 (TBE). Electrophoresis was performed at 100 V using ice-cold 0.5× TBE as a running buffer. DNA was then transferred from the polyacrylamide gel to the Biodyne B Positive Nylon Membrane (Pierce) by electroblotting at 380 mA for 40 min in 0.5× TBE. Transferred DNA was cross-linked for 15 min using a UV cross-linker equipped with 312-nm bulbs. Biotin-labeled DNA was detected using a horseradish peroxidase/Super Signal Detection System (Pierce). Membranes were exposed to Kodak x-ray film.
For EMSA competition assays, oligonucleotides corresponding to both strands of protected regions in the footprint assay (Table 1) were synthesized. Annealing was carried out by mixing equimolar amounts (at a concentration of 100 μm) of each complementary oligonucleotide in 0.25 m Tris-HCl, pH 8.0. The mixture was incubated at 95 °C for 5 min and then chilled on ice.
DNase I Footprinting
DNase I footprint assay was carried out as described previously by Zianni et al. (26) in the Plant and Microbe Genomics Facility at Ohio State University, Columbus, OH. Briefly, 5′-VIC or 5′-FAM-labeled probes were generated by PCR using the primers listed in Table 1. The reaction mixture containing 750 ng of labeled probe, 14 μg of LVIS553, 50 mm Tris-HCl, pH 7.5, 150 mm KCl, 12.5 mm MgCl2, 0.5 mm CaCl2, 0.01% Triton X-100, 25 ng/μl of poly(dI-dC)-nonspecific competitor DNA, and 0.006 units of DNase I (New England Biolabs, Ipswich, MA) was incubated for 20 min at 37 °C. The reaction was terminated with 10 mm EDTA, pH 8.0. The mixture was heat inactivated at 72 °C for 10 min. The DNA fragments were purified with phenol/chloroform/isoamyl alcohol (ratio 25:24:1) and precipitated with ethanol. A nondigested fragment was used for sequencing reactions with the Thermo Sequence dye primer manual cycle sequencing kit (U. S. Biochemical Corp.). The digested DNA and sequencing reaction products were analyzed with a 3730 DNA analyzer and the protected regions were identified with GeneMarker (Soft genetics).
Quantitative Reverse Transcription-PCR Studies
Bacterial cells were cultured in MRS broth with novobiocin (0.5 or 1 μm) or coumermycin A1 (0.1 or 0.5 μm) when required. The cells were collected by centrifugation at 4 °C when A600 ∼0.5. Total RNA was subsequently isolated with RiboPureTM-Bacteria (Ambion, Austin, TX) in accordance with the manufacturer's protocol. cDNAs were synthesized with the SuperscriptTM first-strand synthesis kit (Invitrogen) in accordance with the manufacturer's instructions and stored at −80 °C prior to use. Real time quantitative PCR was carried out on the iCycler IQTM apparatus (Bio-Rad) using Platinum® SYBR® Green qPCR SuperMix for iCycler (Invitrogen) in accordance with the manufacturer's recommended protocol. Quantitative reverse transcription-PCR primers LVIS552 and LVIS553 are described in detail in Table 1. The rpoD gene was used as an internal control.
Small Molecule Screening by Differential Scanning Fluorimetry
Purified LVIS553 protein was screened against a library of 160 intracellular compounds (27) at a final concentration of 100 μm or against the Prestwick chemical library of 1152 compounds (Prestwick Chemical, France) at a final concentration of 1.3 μg/ml using fluorometry as previously described (27, 28). LVIS553 was diluted to a final concentration of 10 μm in 100 mm HEPES, pH 7.5, 150 mm NaCl. 25-μl aliquots of a protein solution containing the chemical compounds were placed in duplicate into 96-well plates (Bio-Rad) and heated from 25 to 80 °C at the rate of 1 °C per minute. A real time PCR device (iCycler IQTM, Bio-Rad) was used to monitor protein unfolding by an increase in the fluorescence of the fluorophor SYPRO Orange (Invitrogen). Fluorescence intensities were plotted against temperature for each sample well and transition curves were fitted using the Boltzmann equation using Origin 8 software (Northampton, MA). The midpoint of each transition was calculated and compared with the midpoint calculated for the reference sample. If the difference between them was greater than 2.0 °C, the corresponding compound was considered to be a “hit” and the experiment was repeated to confirm the effect in a dose-dependent manner.
Isothermal Titration Calorimetry
Measurements were performed on a VP-Microcalorimeter (MicroCal, Northampton, MA) at 30 °C. The protein was thoroughly dialyzed against 10 mm HEPES, pH 7.5, and 500 mm NaCl. A solution of novobiocin (1 mm) was directly prepared in dialysis buffer. Each titration involved a series of 4-μl injections of effector molecule into the protein solution. The mean enthalpies measured from injection of the ligand in the buffer were subtracted from raw titration data prior to data analysis with ORIGIN software (MicroCal). Titration curves were fitted by a nonlinear least squares method to a function for the binding of a ligand to a macromolecule (29). From the curve thus fitted, the parameters ΔH (reaction enthalpy), KA (binding constant, KA = 1/KD), and n (reaction stoichiometry) were determined. From the values of KA and ΔH, the changes in free energy (ΔG) and entropy (ΔS) were calculated with the equation: ΔG = −RT lnKA = ΔH − TΔS, where R is the universal molar gas constant and T is the absolute temperature.
RESULTS
LVIS553 Operon Structure and Binding Site
In L. brevis the MarR family (COG1846) is the most abundant and the most diverse group with 22 proteins or 15.4% of all transcription factors. These data correlate well with the high amount of drug efflux systems (13% of the total transport proteins) found in the genomes of Lactobacillus (19, 20). In this report, we chose LVIS553 as our model protein for the first step in unveiling small regulatory molecules for this diverse group of proteins.
To determine the binding site for LVIS553, the genomic environment was analyzed (Fig. 1). LVIS553 is encoded in the minus strand and LVIS554 is encoded downstream on the plus strand. This protein is annotated as a putative Na+-driven multidrug efflux pump and classified using the Transport Classification Data base (30) within the multi-antimicrobial extrusion (MATE) family, TCDB 2.A.66.1. Upstream of LVIS553 on the minus strand is LVIS552, which encodes for a protein with four predicted transmembrane domains. No characterized homologs of LVIS552 were identified on either TCDB or non-redundant databases. The genomic environment of LVIS552-LVIS553, however, was found to be conserved in Lactobacillus plantarum, but not in other Lactobacillus. Interestingly, the only two other genomes where this putative operon is conserved is in Ruminococcus obeum and Blautia hydrogenotrophica. LVIS554 shares a 64% identity at the protein level with the gene lp_1386, which is located in a different genomic environment than lp_0816 and lp_0817 (homologs to LVIS552 and LVIS553, respectively), suggesting that they do not belong to the same transcriptional unit.
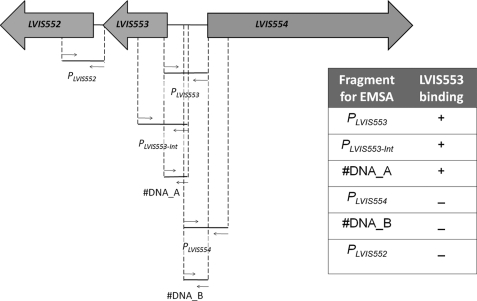
FIGURE 1.
Identification of the LVIS553 binding site. The genomic environment of LVIS553 was extracted and analyzed to identify putative binding regions. Two membrane proteins were encoded upstream and downstream of LVIS553. Different primer combinations (Table 1) were used to determine the smallest DNA binding region for LVIS553. EMSA results are summarized in the inset. +, positive binding of LVIS553 using 10 nm pure protein; −, negative binding using up to 500 nm purified LVIS553 protein.
Based on the vast literature available in which local transcriptional regulators are usually encoded upstream or downstream of the regulated genes, we amplified by PCR two fragments: the first includes the intergenic region between LVIS552 and LVIS553 (PLVIS552) and the second harbors the LVIS553-LVIS554 (PLVIS553) (Fig. 1). The protein-DNA interaction was tested by EMSA. We found that LVIS553 binds to PLVIS553 but not to PLVIS552 (Fig. 1). To narrow down the binding site we tested different combinations of primers within PLVIS553 including the promoter-like region upstream of LVIS554. The smallest region that binds LVIS553 was found within 130 bp of the predicted start codon of LVIS553.
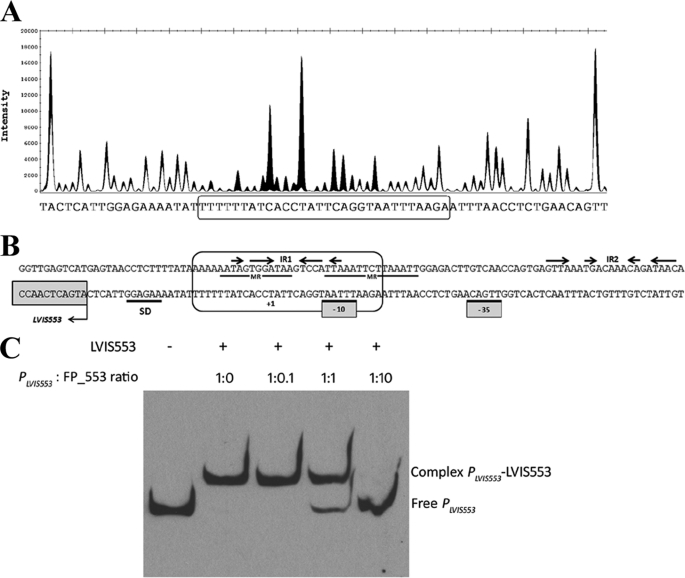
To determine the binding site of LVIS553, a DNase I footprinting assay was performed. We detected a protected region of 30 nucleotides located from base +13 to base −17 of both the plus and minus strands according to the +1 position of the predicted transcription start site (Fig. 2A). LVIS553 appears to protect a region that contains the predicted −10 region of PLVIS553 as well as the hypothetical transcription start site (Fig. 2B). This protected region includes one imperfect inverted repeat (5′-TAaTGGActtaTCCAcTA-3′) flanked on both sides by AT-rich sequences. The protected sequence also has two short mirror sequences, 5′-TTATCcaCTATT-3′ and 5′-AATTTAAgAATTTAA-3′, that partially overlap the inverted repeats.
FIGURE 2.
Analysis of the interaction between LVIS553 with the PLVIS553 promoter. A, DNase I footprinting. The electropherogram shows a fragment of the digested probe in the absence (black) or presence (white) of LVIS553 highlighting the protected region. The reaction mixture was treated as described under “Experimental Procedures” using as probe the primers shown under Table 1. The nucleotide sequence protected by LVIS553 is shown in the bottom of panel A and boxed in B. B, analysis of PLVIS553 promoter. Predicted Shine-Dalgarno sequence and −10 and −35 of the PLVIS553 are underlined. The protected regions of both plus and minus strands are indicated in a circled box. IR, inverted repeats; MR, mirror region. C, competition EMSA. Labeled PLVIS553 promoter with 10 nm LVIS553 were mixed with increasing concentrations of unlabeled double-stranded FP-553 (30 bp-sequence identified by DNase I footprinting).
To confirm whether this protected region is the binding site for LVIS553, a competition assay was performed with the small, unlabeled double-stranded DNA of 30 bp, identified by DNase I footprinting. The LVIS553-PLVIS553 interaction was incubated with increasing concentrations of this unlabeled DNA (FP-553). A 50% disruption of the LVIS553·PLVIS553 complex was found at 1:1 ratio (PLVIS553 DNA:FP-553 DNA) with a complete disruption at 1:10 ratio (Fig. 2C).
Screening of the Small Molecule Library for Binding to LVIS553
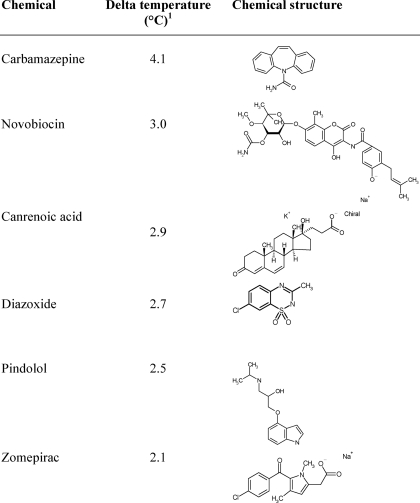
The expression of LVIS553 from L. brevis ATCC367 produced a soluble polypeptide (see “Experimental Procedures” for details) with an apparent molecular mass of 17.2 kDa and a yield of 12 mg/liters. LVIS553 was tested for binding against 158 metabolic scaffolds (31) and the Prestwick library by high-throughput protein stability assay, using fluorometry (27, 32). First, the thermal melt conditions for LVIS553 were established, to generate interpretable unfolding data (described under “Experimental procedures”). The midpoint transition was established at 56.6 °C. The compounds that induced a shift in the midpoint transition temperature (ΔTm) of LVIS553 by more than 2.0 °C were considered hits. We found that LVIS553 showed an increase in Tm with carbamazepide (ΔTm = 4.1 °C), novobiocin (ΔTm = 3.0 °C), canrenoic acid (ΔTm = 2.9 °C), diazoxide (ΔTm = 2.7 °C), pindolol (ΔTm = 2.5 °C), and zomepirac (ΔTm = 2.1 °C) (Table 2). These results were confirmed by analyzing the dose dependence. This was accomplished by using increasing concentrations (0–1 mm) of each chemical. Interestingly, a common chemical scaffold was not identified among all hit molecules.
TABLE 2.
Stabilization effect of ligand binding
The thermal stabilization of each protein by 0.1 μm ligand was evaluated using fluorometry.
1 Delta temperature was calculated as the difference in the transition temperature between the protein in the absence and presence of a given ligand.
Effect of Putative Hits on DNA Binding in Vitro
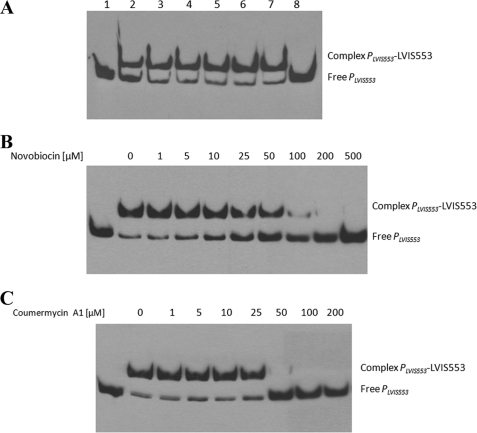
The PLVIS553 fragment was used to assess the effect of ligands on the protein-DNA interaction. Carbamazepine, canrenoic acid, diazoxide, zomepirac, and pindolol at 100 μm or 1 mm (Fig. 3A) had no effect on the binding ability of LVIS553 with PLVIS553. According to EMSA results, novobiocin was the only ligand identified from screening that caused dissociation of LVIS553 from PLVIS553 (Fig. 3A) in a concentration-dependent manner (Fig. 3B).
FIGURE 3.
Effect of different small molecules on LVIS553 binding to PLVIS553. EMSA results using 2.5 nm biotin-labeled PLVIS553 and 10 nm LVIS553 with different inducer molecules (A) or with increasing concentrations of novobiocin (B) or coumermycin A1 (C). No protein was added to the first lane. The full binding conditions are described under “Experimental Procedures.” In A, lane 1 shows the migration of the target DNA fragment; lanes 2–7, LVIS553 at 10 nm. Carbamezepine (lane 3), canrenoic acid (lane 4), diazoxide (lane 5), pindolol (lane 6), zomepirac (lane 7), and novobiocin (lane 8) were added at a concentration of 100 μm.
Using the structure of novobiocin as a model (33), we attempted to determine the smallest chemical scaffold that elicited an effect on LVIS553 using EMSA. We found that none of the coumarin-related molecules tested (coumarin, esculin, esculetin, umbelliferone, and scopoletin) had an effect on the stability of the LVIS553·PLVIS553 complex when tested at 1 mm concentration (data not shown). Coumermycin A1 is a structurally related antibiotic that is also active on DNA gyrase (33). We determined the effect of coumermycin A1 on the binding of LVIS553 to PLVIS553 (Fig. 3C). EMSA experiments showed that coumermycin A1 is able to disrupt the DNA-protein complex at lower concentrations (50 μm) than novobiocin (100 μm).
Modulation of LVIS553 Activity in Vivo
The effect of novobiocin and coumermycin A1 on LVIS553 activity was assessed by measuring the expression of LVIS553 and the downstream gene LVIS552. Cells were grown in MRS broth in the presence or absence of increasing concentrations of novobiocin (1 or 5 μm) or coumermycin A1 (0.1 or 0.5 μm). The difference in concentrations used for novobiocin and coumermycin A1 was due to higher sensitivity of L. brevis to the latter chemical. Novobiocin increased the expression of LVIS553 ∼2000-fold at 5 μm, whereas LVIS552 was induced at a lower level (2-fold). Coumermycin A1 was found to be a weak inducer of LVIS553 (24-fold) and had no effect on the expression of LVIS552 (supplemental Fig. S1). The difference in expression between LVIS552 and LVIS553 is intriguing because transcriptional terminators could not be identified in the intergenic region. These results indicate that although coumermycin A1 has an apparent higher affinity for LVIS553 in vitro, as shown on EMSA, the in vivo assays indicate that novobiocin might be the biologically relevant ligand for this transcriptional regulator.
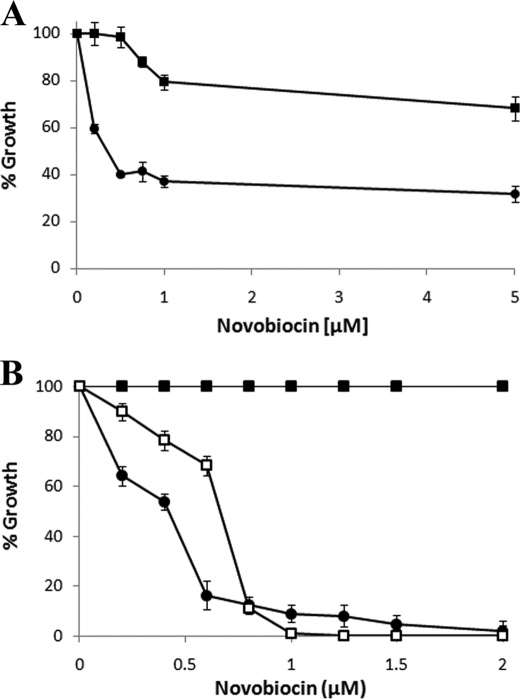
To test this hypothesis, LVIS552 and LVIS553 genes (including the complete PLVIS553) were cloned in the low copy vector pRV610 generating the pLVIS552_553 plasmid. The rationale being that if L. brevis ATCC 367 harbors a plasmid resulting in higher numbers of the transporter protein that mediate antibiotic efflux, an increased tolerance to novobiocin will be observed. L. brevis-pRV610 (L. brevis with empty plasmid as control) cells were grown with increasing concentrations of novobiocin (0.1–5 μm). An initial growth inhibition, indicated by a 60% decrease in the optical density with respect to a culture without novobiocin, was observed at 0.5 μm (Fig. 4A). This residual tolerance was not inhibited by concentrations up to 5 μm. Interestingly, L. brevis-pLVIS552_553 showed an increased tolerance (80% of the initial density) with 5 μm novobiocin (Fig. 4A). These results suggest that the putative transport protein LVIS552 is involved in the efflux of novobiocin. The residual tolerance observed at 0.5 μm in L. brevis-pRV610 could be explained by the activation of multiple unspecific drug efflux systems known to be abundant in the L. brevis genome (20).
FIGURE 4.
Heterologous expression of LVIS552 and LVIS553 in L. brevis and B. subtilis. A, L. brevis cells were grown with increasing concentrations of novobiocin in the presence (closed squares) or absence (closed circles) of LVIS552-LVIS553 cloned in pRV610. B, growth of B. subtilis with a chromosomal insertion of LVIS552-LVIS553 in the sacA locus (closed squares), empty controls (kanamycin cassette in the sacA locus, closed circles), and LVIS552-LVIS553 (K9A) mutant (open squares). Growth was expressed as a percentage of the OD of cells grown without novobiocin.
Characterization of Novobiocin-LVIS553 Binding
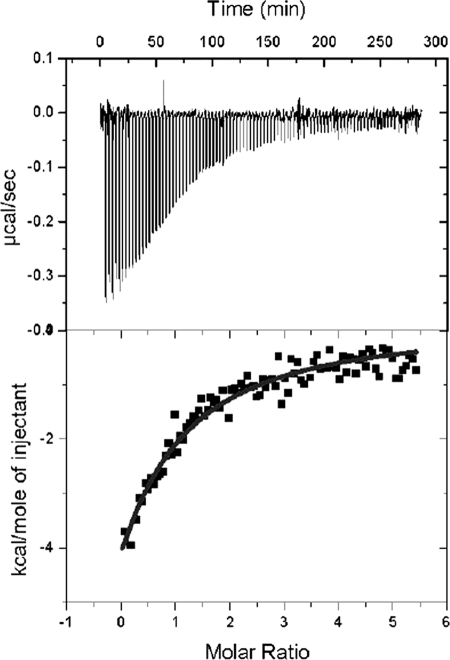
To further characterize the LVIS553 interactions with the newly identified effector molecule, the thermodynamic properties of LVIS553 interactions with novobiocin were determined using isothermal titration calorimetry. The titration of LVIS553 followed an exothermal heat change profile giving rise to a hyperbolic binding curve (Fig. 5). Data were fitted with Origin software using the “one set of sites model.” The derived thermodynamic parameters from the calorimetric titration of LVIS553 with novobiocin are as follows: KD = 33.8 ± 2.9, n = 2, KA = 2.7 × E4 m−1, ΔH = −3.4 ± 0.3 kcal/mol, TΔS = 50.3 kcal/mol, ΔG = −506.5 kcal/mol. In agreement with the results obtained through EMSA, the LVIS553 dissociation constant (KD) for novobiocin is in the low micromolar range (33.8 ± 2.9 μm). Stoichiometry of the reaction is 2, which is consistent with the available crystallographic data on MTH313 from Methanothermobacter thermautotrophicus ΔH (2).
FIGURE 5.
Isothermal titration calorimetric data for the binding of novobiocin to LVIS553. Heat changes (upper panel) and integrated peak areas (lower panel) for the injection of a series of 4-μl aliquots of 1 mm ligand in a solution of 40 μm protein. Protein solutions at 40 μm in 10 mm HEPES, pH 7.5, and 500 mm NaCl were titrated with 1 mm solution of effector. Experiments were carried out at 30 °C.
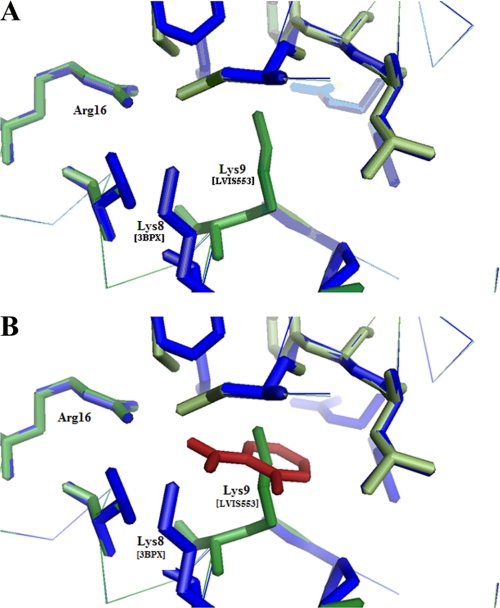
Although LVIS553 shares low sequence identity (21.6%) with MTH313, in silico modeling at Swiss-Model (34, 35) in the automated mode retrieved PDB code 3BPX (MTH313 with salicylate) as the best hit (E-value = 8.4e-19) (supplemental Fig. S2A). In MTH313, two distinctive binding sites were determined for salicylate. In salicylate binding pocket 1 (SAL1) (2) LysB8 and ArgB16 make distinct contacts (ionic interactions or hydrogen bonds) with the salicylate molecule. These 2 amino acids are found at the junction of the dimerization domain and the DNA binding pocket (2). LysB8 and ArgB16 of MTH313 are conserved in LVIS553 (Lys9 and Arg16) (Fig. 6 and supplemental Fig. S2B). Interestingly, the predicted model of LVIS553 revealed that the orientation of the Lys9 side chain is twisted toward the center of the binding pocket (Fig. 6). Amino acids with hydrophobic characteristics were also found in the MTH313 SAL1 pocket, which are also conserved in LVIS553. Based on the location of these residues, it was proposed that the binding of a salicylate molecule to MTH313 induces a structural change in the DNA binding motif of each monomer, resulting in the twisting out of the DNA binding lobes, decreasing the binding to DNA (2). In accordance with this model LVIS553 behaves as a dimer in solution with an apparent molecular mass of 34 kDa as determined by fast protein liquid chromatography separation (data not shown). The addition of different concentrations of novobiocin (10, 100, or 1000 μm) to either the reaction mixture or running buffer did not affect the oligomeric state of LVIS553, suggesting that a similar mechanism to MTH313 might be involved in molecular regulation by novobiocin.
FIGURE 6.
Close view (6-Å radius) of the SAL1 binding site of MTH313 (PDB code 3BPX) (2) aligned with the predicted model for LVIS553. A, model of the LVIS553 structure (in green) constructed using MTH313 (in blue) as a template. B, salicylate from MTH313 (in red) is displayed.
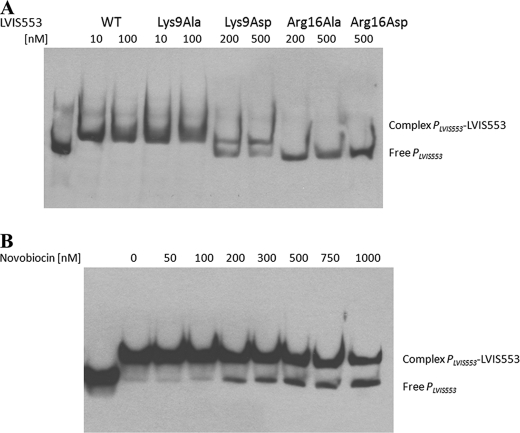
An unbiased BLAST search (36) identified a large number (43) of homologs to LVIS553, with partial sequence alignment in which Arg16 was conserved but Lys9 was not (supplemental Fig. S2B). Based on these results it was hypothesized that Arg16 might be involved in DNA binding, whereas Lys9 might be involved in signal recognition. This was tested by changing Lys9 and Arg16 to either a small amino acid (alanine) or an acidic amino acid (aspartic acid). All these proteins were expressed in soluble form and tested by EMSA experiments. Mutations in Arg16 (either to alanine or aspartic acid) rendered this protein unable to bind DNA, confirming that this amino acid is essential to DNA binding (Fig. 7A). Similarly, the mutant containing a drastic shift in the amino acid charge, LVIS553 K9D, also resulted in decreased affinity for DNA (Fig. 7A) and no effect of novobiocin was observed (data not shown).
FIGURE 7.
Distinctive functions of Lys9 and Arg16 residues identified by EMSA. A, DNA binding properties of LVIS553 substitution mutants. WT, wild type protein. B, effect of novobiocin on the stability of LVIS553-K9A·PLVIS553 complex.
LVIS553 K9D showed binding of PLVIS553 to the DNA fragment at 10 nm (as the wild type). Concentrations of novobiocin up to 1 mm had a very slight effect on the LVIS553 K9D·PLVIS553 complex (Fig. 7B), suggesting that Lys9 is the residue involved in binding to novobiocin. The residual ligand binding activity observed might be mediated by weak interactions with other residues in the binding cavity.
The effect of the LVIS553 K9D mutant was tested in vivo using B. subtilis as a surrogate, due to technical difficulties associated with the genetic manipulation of L. brevis ATCC367. The genes LVIS552-LVIS553 (or its mutant variant K9D) were cloned in pSac-Kan and integrated in the sacA gene of B. subtilis M168. The tolerance to novobiocin was increased by 10-fold in B. subtilis LVIS552-LVIS553 when compared with B. subtilis sacA::Kanr (empty vector integrated at the sacA locus as control), with a minimal inhibitory concentration of 5 and 0.5 μm, respectively. The high tolerance to novobiocin conferred by LVIS552 was reverted when LVIS553 was mutated in Lys9 (Fig. 4B). These results confirmed the role of Lys9 as the amino acid involved in the specific binding of novobiocin.
DISCUSSION
In this report we took an unbiased approach, using high throughput screening of small molecules library, to identify novobiocin as the specific effector molecule for LVIS553. In silico modeling of LVIS553 identified MTH313 (PDB code 3BPX) (2) as the closest structural homolog. MTH313 is one of the few available structures with its ligand as salicylate, which shed light into the mechanism of interaction of the small molecule/protein. Interestingly, the physiologically relevant SAL1 is found in the interface of the dimerization domain and DNA binding domain, making it difficult to discriminate residues involved in DNA binding, ligand binding, or both. A detailed analysis performed in SlyA identified several amino acids within the winged-helix region, as well as two hydrophobic residues (Leu12 and Leu126) within α1 and α6 that are required for DNA binding (4). Using structure-guided mutagenesis, we aimed to identify residues involved in ligand binding within the SAL1 cavity. Arg16 is highly conserved among different MarR members. We found that an alanine substitution (LVIS553 R16A) resulted in impaired DNA binding, suggesting a role in sequence recognition.
On the contrary, Lys9 is only conserved in homologs close to LVIS553, indicating that Lys9 may be the regulatory residue involved in the specific binding of novobiocin. LVIS553 K9A showed similar affinity for DNA as the wild type strain, whereas no further response to novobiocin was detected. Although it is speculated that the SAL1 cavity has the potential to bind a wide variety of small molecules, LVIS553 only bound novobiocin or the closely related molecule coumermycin A1. The determined affinity constants for novobiocin were in the low micromolar range, consistent with a molecule having physiological relevance. These in vitro results were correlated with the in vivo induction in gene expression of LVIS553 and its downstream gene LVIS552, a putative membrane protein.
The antibacterial property of novobiocin is well established in the literature. It targets the DNA gyrase activity that translates into pleiotropic effects on the cell, affecting DNA replication, DNA repair, recombination, and ultimately cell growth (37). The novobiocin biosynthetic cluster of Streptomyces caeruleus is positively regulated by NovG and NovE (38). NovG is an unusual transcription factor with a predicted central helix-turn-helix DNA binding domain that specifically recognizes the sequence GTTCRACTG(N)11CRGTYGAAC (39). The effect of novobiocin on NovG binding properties, if any, has not yet been reported.
We identified the DNA binding sequence for LVIS553 using footprinting assay. The protected region is 30 nucleotides long and located on the minus strand, upstream of LVIS553, overlapping the predicted −10 region of the LVIS553 promoter. It includes a 7-bp imperfect palindrome (5′-TAaTGGActtaTCCAcTA-3′) flanked on both sides by AT-rich sequences. A competitive EMSA with the 30-nucleotide fragment confirmed the specificity of this region. A similar arrangement of the binding sequence has been described for two other MarR members, MepR (40) and OhrR (41).
The identification of new mechanisms of tolerance to antibiotics is very relevant. Novobiocin in particular, has been suggested as a treatment for Gram-positive infections in penicillin-resistant strains (42). Different mechanisms of tolerance to novobiocin have been described including mutations on the DNA gyrase (33) and the activation of multidrug efflux pumps (5). In L. brevis, the expression in trans of LVIS552-LVIS553 results in an increased tolerance to novobiocin, indicating that the putative membrane protein LVIS552 is involved in detoxification of novobiocin. These results were confirmed using B. subtilis as a surrogate strain. We revealed that the heterologous expression of LVIS552 increased the tolerance to novobiocin 4-fold. In B. subtilis, the multidrug efflux transporter MdtP (YusP) was associated with increased tolerance to several antibiotics, including fusidic acid, novobiocin, streptomycin, and actinomycin D (5). Similarly to LVIS552, the expression of MdtP is regulated by MdtR (YusO), a MarR member encoded upstream of the multidrug efflux transporter (YusP). Kim et al. (5) reported that mutations in the regulatory protein MdtR (R83K or A67T) impaired DNA binding and resulted in a 2-fold increase in tolerance to streptomycin, actinomycin D, and novobiocin, as well as a 5-fold increase in tolerance to fusidic acid. Interestingly, EMSA experiments with MdtR revealed binding affinities in the millimolar range for novobiocin and fusidic acid, questioning their biological significance.
In conclusion, we provide evidence that LVIS553 is involved in the specific regulation of a new mechanism of tolerance to novobiocin. In contrast to the other MarR members that respond to “preconceived” ligands (i.e. salycilate) that have been studied, we provide in vitro data that correlates with in vivo responses. The identification of key residues involved in specific binding of small molecules, contributes not only to the growing body of knowledge currently needed to decipher the intricate mechanism of interactions between the signal molecules and MarR, but may also serve as a starting point for the design of innovative therapeutics in the near future.
Supplementary Material
Acknowledgments
We thank Asma Sayed Abdelgeliel Mahmou and Tenisha Wilson for technical help. We acknowledge Dr. Joanna Long, Department of Biochemistry and Molecular Biology, for the use of isothermal titration calorimetry equipment. We thank Dr. Claudio Gonzalez for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R03AI078001 from the NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- EMSA
- electrophoretic mobility shift assay
- SAL1
- salicylate binding pocket 1.
REFERENCES
- 1.Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. (2001) Nat. Struct. Biol. 8, 710–714 [DOI] [PubMed] [Google Scholar]
- 2.Saridakis V., Shahinas D., Xu X., Christendat D. (2008) J. Mol. Biol. 377, 655–667 [DOI] [PubMed] [Google Scholar]
- 3.Alekshun M. N., Kim Y. S., Levy S. B. (2000) Mol. Microbiol. 35, 1394–1404 [DOI] [PubMed] [Google Scholar]
- 4.Okada N., Oi Y., Takeda-Shitaka M., Kanou K., Umeyama H., Haneda T., Miki T., Hosoya S., Danbara H. (2007) Microbiology 153, 548–560 [DOI] [PubMed] [Google Scholar]
- 5.Kim J. Y., Inaoka T., Hirooka K., Matsuoka H., Murata M., Ohki R., Adachi Y., Fujita Y., Ochi K. (2009) J. Bacteriol. 191, 3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seoane A. S., Levy S. B. (1995) J. Bacteriol. 177, 3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin R. G., Rosner J. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alekshun M. N., Levy S. B. (1999) J. Bacteriol. 181, 4669–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L., Fang J., Wei Y. (2009) Biochemistry 48, 2099–2108 [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson S. P., Grove A. (2006) Curr. Issues Mol. Biol. 8, 51–62 [PubMed] [Google Scholar]
- 11.Brooun A., Tomashek J. J., Lewis K. (1999) J. Bacteriol. 181, 5131–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson S. P., Grove A. (2004) J. Biol. Chem. 279, 51442–51450 [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson S. P., Grove A. (2005) J. Mol. Biol. 350, 617–630 [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Rueda E., Collado-Vides J., Segovia L. (2004) Comput. Biol. Chem. 28, 341–350 [DOI] [PubMed] [Google Scholar]
- 15.Lorca G. L., Font de Valdez G. (2009) in Lactobacillus Molecular Biology: From Genomics to Probiotics (Ljungh A., Wadström T. eds) pp. 115–137, Caister Academic Press, Norfolk, UK [Google Scholar]
- 16.Simpson W. J., Fernandez J. L. (1992) Lett. Appl. Microbiol. 14, 13–16 [Google Scholar]
- 17.Sakamoto K., Margolles A., van Veen H. W., Konings W. N. (2001) J. Bacteriol. 183, 5371–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K., Iijima K., Ozaki K., Yamashita H. (2005) Appl. Environ. Microbiol. 71, 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova K., Slesarev A., Wolf Y., Sorokin A., Mirkin B., Koonin E., Pavlov A., Pavlova N., Karamychev V., Polouchine N., Shakhova V., Grigoriev I., Lou Y., Rohksar D., Lucas S., Huang K., Goodstein D. M., Hawkins T., Plengvidhya V., Welker D., Hughes J., Goh Y., Benson A., Baldwin K., Lee J. H., Díaz-Muñiz I., Dosti B., Smeianov V., Wechter W., Barabote R., Lorca G., Altermann E., Barrangou R., Ganesan B., Xie Y., Rawsthorne H., Tamir D., Parker C., Breidt F., Broadbent J., Hutkins R., O'Sullivan D., Steele J., Unlu G., Saier M., Klaenhammer T., Richardson P., Kozyavkin S., Weimer B., Mills D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorca G. L., Barabote R. D., Zlotopolski V., Tran C., Winnen B., Hvorup R. N., Stonestrom A. J., Nguyen E., Huang L. W., Kim D. S., Saier M. H., Jr. (2007) Biochim. Biophys. Acta 1768, 1342–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 22.Crutz-Le Coq A. M., Zagorec M. (2008) Plasmid 60, 212–220 [DOI] [PubMed] [Google Scholar]
- 23.Middleton R., Hofmeister A. (2004) Plasmid 51, 238–245 [DOI] [PubMed] [Google Scholar]
- 24.Petit-Glatron M. F., Chambert R. (1992) J. Gen. Microbiol. 138, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 25.Lorca G. L., Ezersky A., Lunin V. V., Walker J. R., Altamentova S., Evdokimova E., Vedadi M., Bochkarev A., Savchenko A. (2007) J. Biol. Chem. 282, 16476–16491 [DOI] [PubMed] [Google Scholar]
- 26.Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. (2006) J. Biomol. Tech. 17, 103–113 [PMC free article] [PubMed] [Google Scholar]
- 27.Vedadi M., Niesen F. H., Allali-Hassani A., Fedorov O. Y., Finerty P. J., Jr., Wasney G. A., Yeung R., Arrowsmith C., Ball L. J., Berglund H., Hui R., Marsden B. D., Nordlund P., Sundstrom M., Weigelt J., Edwards A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15835–15840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niesen F. H., Berglund H., Vedadi M. (2007) Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 29.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 30.Saier M. H., Jr., Yen M. R., Noto K., Tamang D. G., Elkan C. (2008) Nucleic Acids Res. 37, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobeli I., Thornton J. M. (2006) BioEssays 28, 534–545 [DOI] [PubMed] [Google Scholar]
- 32.Senisterra G. A., Markin E., Yamazaki K., Hui R., Vedadi M., Awrey D. E. (2006) J. Biomol. Screen 11, 940–948 [DOI] [PubMed] [Google Scholar]
- 33.Hooper D. C., Wolfson J. S., McHugh G. L., Winters M. B., Swartz M. N. (1982) Antimicrob. Agents Chemother. 22, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 36.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson A. P., Maxwell A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11232–11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dangel V., Eustáquio A. S., Gust B., Heide L. (2008) Arch. Microbiol. 190, 509–519 [DOI] [PubMed] [Google Scholar]
- 39.Eustáquio A. S., Li S. M., Heide L. (2005) Microbiology 151, 1949–1961 [DOI] [PubMed] [Google Scholar]
- 40.Kumaraswami M., Schuman J. T., Seo S. M., Kaatz G. W., Brennan R. G. (2009) Nucleic Acids Res. 37, 1211–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005) Mol. Cell. 20, 131–141 [DOI] [PubMed] [Google Scholar]
- 42.Gombert M. E., Aulicino T. M. (1984) Antimicrob. Agents Chemother. 26, 933–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.