Abstract
Interferon regulatory factors (IRFs) are crucial for transcription during innate immune responses. We have previously shown that the tyrosine kinase c-Src enhances IRF-3-dependent transcription in response to viral double-stranded RNA. In this study, we show that c-Src has distinct roles in Toll-like receptor (TLR)-mediated activation of IRF-5 and IRF-3. Surprisingly, c-Src inhibition markedly enhanced IRF-5 activation after treatment with unmethylated CpG, while suppressing IRF-3 activation. Also, CpG-elicited interleukin-6 mRNA production was increased, whereas IP10 mRNA synthesis was reduced in cells deficient in c-Src. Interestingly, c-Src regulated TLR-stimulated induction of activating transcription factor 3 (ATF3), a transcriptional repressor. Depletion of ATF3 by small interfering RNA markedly enhanced interleukin-6 production after CpG treatment, whereas IP10 production was reduced. These results demonstrate functional specificity for c-Src in TLR-stimulated responses and suggest that c-Src modulation and ATF3 activity may contribute to differential regulation of IRF-3- versus IRF-5-mediated gene expression.
Keywords: Cytokines, Immunology/Innate Immunity, Immunology/Toll Receptors, Receptors/Toll-like, Signal Transduction, Transcription/Regulation, Tyrosine Kinase
Introduction
Innate immune mechanisms against microbial pathogens rely on membrane-associated or cytoplasmic pattern recognition receptors. Toll-like receptors (TLRs)2 and cytosolic RIG-I-like receptors recognize pathogens extra- or intracellularly by binding to specific microbial patterns occurring at the cell surface (TLRs), in endosomes (TLRs), or in the cytoplasm (RIG-I-like receptors) (1). TLRs recognize microbial lipids (TLR1, -2, -4, -6, and -10), viral or bacterial DNA (TLR9), or viral single- or double-stranded RNA (TLR3 and TLR7/8), although RIG-I-like helicases respond to viral single- or double-stranded RNA. The transcriptional program that is activated by pathogen exposure involves a plethora of molecular components, including cytokines, chemokines, and cell cycle/apoptosis regulatory proteins (2). In these transcriptional events, the biological roles and activation of transcription factors belonging to the interferon (IFN) regulatory factors (IRFs) and nuclear factor of κB (NF-κB) families have been extensively studied.
Engagement of TLRs leads to recruitment of Toll-interleukin-1 receptor (TIR) domain-containing adapter proteins and activation of signaling cascades that stimulate the IRF transcription factors. Previously, nine cellular IRF genes along with certain virus-encoded analogues of cellular IRFs have been identified (1). Activated IRFs act in conjunction with other transcription factors and cofactors to induce immune response genes, e.g. IFNs, chemokines, adhesion molecules, and apoptosis-regulating genes. Interestingly, IRF family members exhibit distinct functions in immune responses (1, 3). IRF-3 and IRF-7 are considered to be the antiviral IRF family members and, together with NF-κB, constitute critical regulators of type I IFNs (4). Activation of IRF-3 is brought about by TLR ligand binding, which induces activation of the IκB kinase-related kinase TBK1 (TANK-binding kinase 1) and phosphorylation of IRF-3 in the C-terminal domain (5). Studies using knock-out mice have shown that IRF-5 regulates production of several inflammatory cytokines and chemokines such as IL-6 and IL-12β, although its contribution to transcription of IFN genes is uncertain and appears to be cell type-dependent (6, 7). In contrast to IRF-3, the activation mechanism of IRF-5 is poorly understood. Nevertheless, it has been reported that IRF-5 acts in a trimolecular complex with the TIR domain-containing adapter MyD88 and TRAF6 and that IRF-5-IRAK1 association triggers IRF-5 ubiquitination and activation (8). The IRFs share significant homology in the N-terminal DNA binding domain, which binds to an IFN-stimulated response element in promoters (9). However, despite their similarities, IRFs exhibit distinct functional properties, and the molecular basis for their different roles is incompletely understood.
We have previously reported that the tyrosine kinase c-Src is required for TLR3-mediated IRF-3 activation in response to double-stranded RNA (10). In this study, we compared the role of c-Src in transcription of IRF-regulated genes and in activation of the IRF family members IRF-3 and IRF-5. We found that c-Src exerts a differential role in TLR-elicited IRF-3 and IRF-5 activation and gene expression. Our results show that c-Src-mediated differences in TLR-elicited gene expression may be attributed, at least in part, by the activating transcription factor/cAMP-responsive element-binding protein (ATF/CREB) family member ATF3.
EXPERIMENTAL PROCEDURES
Reagents
CpG ODN 1826 and synthetic double-stranded RNA (pIC) were purchased from Coley and Amersham Biosciences, respectively. LPS, derived from Escherichia coli strain 0111:B4, was purchased from Invivogen. Resiquimod (R-848) was purchased from GLSynthesis. PP2 and PP3 were purchased from Calbiochem. Antibodies to ATF3, ATF-2, and α-tubulin were from Santa Cruz Biotechnology. HA-tagged ATF3 and ATF2 were generously provided by Dr. T. Hai (Ohio State University). The Gal4-IRF3 and Gal4-IRF5 luciferase reporter constructs were gifts from Dr. Tom Maniatis (Harvard University, Cambridge, MA) and Paula M. Pitha (The John Hopkins University, Baltimore, MD), respectively. NF-κB-luc was from Stratagene.
Cell Culture
RAW 264.7 cell line, the SYF mouse embryonic fibroblasts from the triple Src knock-out mouse lacking src, yes, and fyn, and the added-back version of the SYF cells (with the wild-type c-src gene introduced (11)) were obtained from ATCC. All cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 20 mg/ml garamycin, and 2 mm l-glutamine at 37 °C in 8% CO2. Bone marrow-derived macrophages from C57BL6 mice were allowed to differentiate for 9 days in L929-containing medium.
RNA Interference
siRNA oligonucleotides targeting ATF3, TRAF3, or CAT were synthesized by Santa Cruz Biotechnology and Qiagen (CAT), respectively. siRNA duplexes were transfected into cells in 6-well plates for 48 h using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol. Experiments were repeated twice producing similar results. Representative results are shown.
Gal4-based IRF Reporter Assays
HEK293 or murine TLR9YFP HEK293 cells were seeded in 96-well plates and were transfected 24 h later with the indicated plasmids using GeneJuice (Novagen) following the manufacturer's protocol. Forty nanograms of each plasmid (Gal4-IRF-3, Gal4-IRF-5, Gal4-DBD, or a luciferase reporter gene containing the Gal4 upstream activation sequence, UAS(GAL)) were used. Reporter gene activity was measured using a luciferase assay system (Promega). Experiments were repeated 3–4 times. Representative data are shown.
RNA Analysis
Total RNA from cells seeded in 6-well plates was isolated by Qiagen RNeasy kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from RNA by using the cDNA synthesis kit (Bio-Rad). Quantitative real time-PCR was carried out by Chromo 4 using the iQ SYBR Green supermix (Bio-Rad), and data were normalized against glyceraldehyde-3-phosphate dehydrogenase or β-actin levels. Primer sequences are given in the supplemental material. Fold induction was calculated using the formula 2−ΔΔCT (12). In experiments using bone marrow-derived macrophages, the specificity of amplification was assessed for each sample by melting curve analysis. Relative quantification was performed using standard curve analysis. The quantification data are presented as a ratio of gene copy number per 100 copies of β-actin. The experiments were performed three to five times, and representative results are shown.
Immunoblotting
Immunoblotting was performed as described previously (13).
Chromatin Immunoprecipitation
RAW 264.7 cells were seeded at an approximate density of 2.5 × 107 cells/15-cm dish. After stimulation, chromatin immunoprecipitation with anti-ATF3 (Santa Cruz Biotechnology) was performed based on the protocol from Aas et al. (14). DNA was purified by using QIAquick (Qiagen) columns and eluted with 30 μl of Tris/EDTA. For detection of the proximal mouse il-6 promoter region, 2 μl of eluted DNA (from immunoprecipitation or input DNA) was used. Primers specific for the mouse il-6 promoter and conditions for the PCR have been described previously (15). PCR products were visualized on ethidium bromide gels.
RESULTS
Inhibition of c-Src Differentially Affects TLR-elicited IRF-3 and IRF-5 Activation
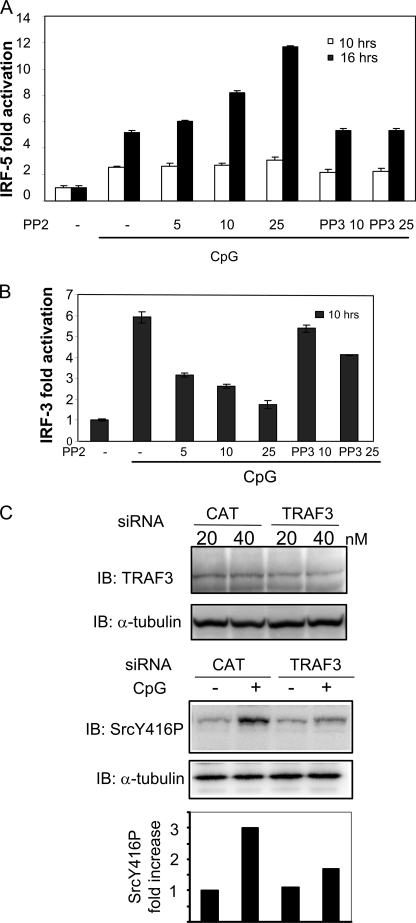
To compare the role of c-Src tyrosine kinase in IRF-3 and IRF-5 activation, we first examined if CpG stimulated activation of IRF-3 and IRF-5. HEK cells stably expressing yellow fluorescent protein-tagged mouse TLR9 were transfected with Gal4-based IRF-3 or IRF-5 luciferase reporters. CpG elicited activation of both IRF-3- and IRF-5-dependent luciferase activity (supplemental Fig. 1A). IRF-3 activation is correlated to phosphorylation of Ser396 (1). Hence, we also examined phosphorylation of IRF-3 after CpG treatment of TLR9-expressing HEK cells. As shown in supplemental Fig. 1B, we found that CpG increases phosphorylation at Ser396, thus suggesting that CpG activates endogenous IRF-3 in these cells. IRF-7 has previously been implicated in signaling emanating from TLR9 (9), whereas the role of IRF-3 CpG-stimulated signaling is less well understood. However, it is likely that IRF-3 is operative in CpG-elicited signaling in cells with low levels of IRF-7.
The existence of cytoplasmic DNA-binding receptors that induce type I IFNs has recently been suggested (16, 17). Therefore, we evaluated the importance of TLR9 expression for IRF-3 and IRF-5 activation. CpG did not induce significant IRF-3 or IRF-5 activation in non-TLR9-transfected HEK cells compared with TLR9-expressing HEK cells (supplemental Fig. 1C), thus suggesting that IRF-3 and IRF-5 activations in our cellular system are mediated mainly through TLR9. We have previously shown that the tyrosine kinase c-Src positively regulates IRF-3 activation (10). To elucidate the role of c-Src-dependent tyrosine phosphorylation in IRF-5 activation, HEK-TLR9YFP cells were transiently transfected with the Gal4-based IRF-5 reporter and pretreated with the c-Src family kinase inhibitor PP2 prior to CpG addition. Treatment with PP2 or its inactive analogue PP3 had no significant effect on IRF-5 activation after 10 h of treatment with CpG (Fig. 1A). In contrast, after prolonged treatment with CpG (16 h), we found that IRF-5 activation was significantly increased by PP2, displaying a dose-dependent effect of PP2 (Fig. 1A). The inactive PP2 analogue PP3 had no effect. Interestingly, PP2 had the opposite effect on IRF-3, attenuating IRF-3 activation after 16 h of treatment with CpG (Fig. 1B). We have previously found that c-Src positively regulates IRF-3 activation, acting downstream of TLR3 and RIG-I, and that c-Src associates with TRAF3 (9, 13). To examine the level at which c-Src acts in TLR9-mediated signaling, we treated RAW cells with TRAF3 or CAT siRNA as a nonspecific control. Transfection of TRAF3 siRNA partially down-regulated TRAF3 protein in RAW cells compared with the CAT siRNA nonspecific control (Fig. 1C, upper panel). Analysis by immunoblotting showed that CpG-stimulated activation of c-Src (as assessed by phosphorylation of its tyrosine 416) was markedly reduced in the presence of 40 nm TRAF3 siRNA (Fig. 1C, middle panel). The lack of complete reduction of c-Src activation may be due to incomplete knockdown of TRAF3 protein or that CpG-elicited c-Src activation is not solely dependent on TRAF3 but perhaps also relies on TRAF6. Collectively, these results suggest that TLR9-induced activation of c-Src is dependent on TRAF3.
FIGURE 1.
CpG induces TRAF3-dependent c-Src activation that differentially modulates activation of IRF-3 and IRF-5. HEK cells expressing yellow fluorescent protein-tagged moTLR9 were transfected with the Gal4-based IRF-5 (A) or IRF-3 (B) luciferase reporter constructs. After 24 h, cells were pretreated with various concentrations of PP2 or PP3 for 45 min prior to treatment with CpG (10 μg/ml). Luciferase activity was determined after 10 or 16 h of CpG stimulation. C, RAW 264.7 cells were transfected with 40 nm TRAF3 or CAT siRNA as a nonspecific control prior to treatment with CpG (50 min) and assessment of TRAF3 levels (upper panel). Phosphorylation of c-Src was determined by an antibody recognizing phosphorylation at Tyr416, and the blot was reprobed with α-tubulin as a loading control (middle panel). Band intensities of c-Src were quantified using the Kodak image analysis software and fold induction of phosphorylated c-Src relative to medium- and CAT-treated cells are shown (lower panel). IB, immunoblot.
In our previous report, we found that the Src family kinase inhibitor PP2 reduces IRF-3 activation after 6 h of pIC treatment (10). We next confirmed that PP2 reduced IRF-3 activation also after prolonged pIC treatment (16 h). Indeed, PP2 attenuated pIC-elicited IRF-3 activation after 16 h (supplemental Fig. 1D). Hence, PP2 reduces pIC- and CpG-stimulated IRF-3 activation after 6 and 16 h of treatment. Consistent with previous reports (3, 18), we found that pIC failed to induce activation of an IRF-5 reporter gene (data not shown), thus precluding studies of PP2 on pIC-elicited IRF-5 activation. This shows that c-Src inhibition differentially affects CpG-stimulated IRF-3 and IRF-5 activation and suggests that c-Src may exert a negative regulatory role on IRF-5 activation. Moreover, the effect of PP2 on IRF-5 required prolonged CpG treatment, which suggests that c-Src affects a delayed phase of CpG-mediated transcription.
Attenuation of c-Src Confers Distinct Effects on Endogenous Immune Modulators Induced by CpG
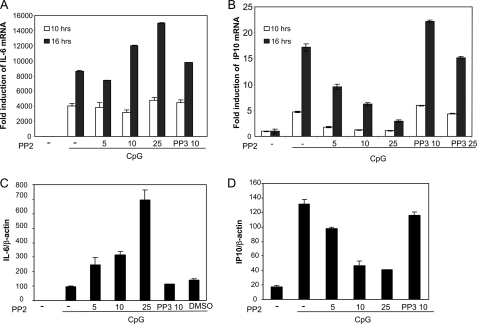
Studies performed in IRF-5 knock-out mice have shown that IRF-5 regulates IL-6 production in response to several TLR ligands (6). Moreover, IRF-3 has been suggested to be critical for TLR-elicited synthesis of the chemokine IP10 (19). To examine the effect of c-Src inhibition on CpG-stimulated transcription of endogenous immune components, we assessed mRNA levels of IL-6, IP10, and IRF-5 in RAW mouse macrophages. RAW cells were pretreated with PP2 prior to treatment with CpG for 10 or 16 h, followed by isolation of total RNA and analysis of IL-6 and IP10 expression by quantitative real time PCR (qRT-PCR) analysis. Increasing concentrations of PP2 augmented IL-6 mRNA levels after treatment with CpG for 16 h (Fig. 2A). Conversely, CpG-stimulated IP10 production was significantly reduced by the presence of PP2, showing a dose-dependent manner, although PP3 had no effect (Fig. 2B). We also examined these events in primary cells, using bone marrow-derived macrophages from C57BL6 mice. PP2 dose-dependently increased CpG-elicited IL-6 mRNA levels, while reducing IP10 synthesis in macrophages from C57BL6 mice (Fig. 2, C and D). Hence, these results show that c-Src inhibition augments CpG-elicited IL-6 transcription, while attenuating transcription of IP10 mRNA. These results may reflect that c-Src exerts a differential effect on IRF-3- and IRF-5-regulated immunomodulatory genes. In addition to their regulation by IRFs, the murine ip10 and il-6 promoters have been suggested to be regulated by the transcription factor NF-κB (20, 21), and the effect of PP2 could be mediated through NF-κB. To address this, TLR9- and TLR3-expressing HEK cells were transfected with a luciferase reporter containing four κB elements prior to treatment with PP2 and CpG or pIC. CpG and pIC effectively induced NF-κB activation after 10 and 16 h of treatment, but PP2 did not have any significant effect on NF-κB activation (supplemental Fig. 2). These results indicate that the effects of PP2 on IP10 and IL-6 transcription are independent of NF-κB.
FIGURE 2.
CpG-induced expression of IL-6 and IP10 mRNA is differentially affected by c-Src inhibition. RAW 264.7 mouse macrophages (A and B) or bone marrow-derived macrophages from C57BL6 (C and D) were pretreated with various concentrations of PP2, PP3, or DMSO prior to stimulation with CpG (10 μg/ml) for various times and isolation of total RNA. IL-6 (A and C) and IP10 (B and D) mRNA levels were determined by qRT-PCR and compared with glyceraldehyde-3-phosphate dehydrogenase (A and B) or β-actin (C and D) expression.
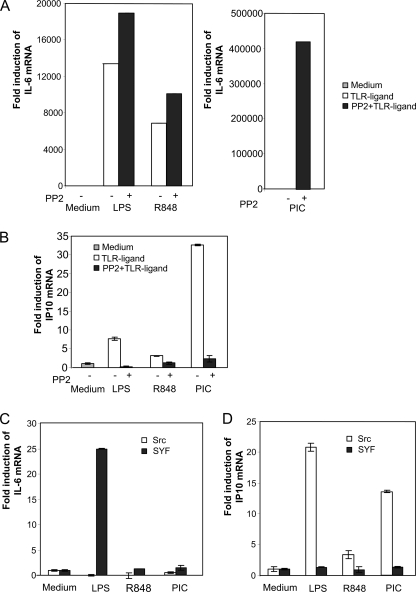
Effect of c-Src Inhibition on Gene Expression Stimulated by Various TLR Ligands
Different TLRs signal through distinct intracellular pathways utilizing individual cytoplasmic adapter proteins. Therefore, it was of interest to examine the role of c-Src on gene expression in response to different TLR ligands. RAW macrophages were pretreated with the c-Src chemical inhibitor PP2 prior to addition of the TLR ligands LPS (TLR4), R848 (TLR7/8), and pIC (TLR3), and expression of IL-6 and IP10 mRNA levels was assessed by qRT-PCR. The TLR stimuli applied induced expression of IL-6 and IP10 mRNA to different extents (Fig. 3, A and B). Importantly, c-Src inhibition resulted in enhanced IL-6 mRNA induction in response to LPS, R848, and pIC, although TLR-elicited IP10 transcription was attenuated. To confirm the effects of the c-Src pharmacological inhibitor, we examined TLR-induced transcription of IL-6 and IP10 mRNA in a c-src genetic knock-out model. For these experiments, we utilized embryonic fibroblasts derived from src/yes/fyn (SYF) triple knock-out mice and compared with transcription in SYF fibroblasts in which c-src has been stably introduced (denoted “c-Src” (11)). In c-Src-expressing cells, IP10 mRNA was induced by the TLR ligands LPS, R848, and pIC, although IL-6 mRNA was not induced after 16 h of stimulation (Fig. 3, C and D). In SYF cells (differing from c-Src cells in that they do not express c-Src), IL-6 mRNA induction in response to TLR ligands, in particular LPS, was enhanced (Fig. 3C). Conversely, TLR ligand-induced levels of IP10 were reduced in SYF cells lacking c-Src relative to SYF cells expressing c-Src (Fig. 3D). IRF-3 mRNA levels were not induced and were not significantly different in SYF and c-Src cells (data not shown). These results suggest that c-Src exerts similar roles downstream of MyD88-dependent and TIR domain-containing adapter-inducing interferon-β-dependent signaling pathways.
FIGURE 3.
Effect of c-Src inhibition on transcription of IL-6 and IP10 mRNA induced by various TLR ligands. RAW 264.7 cells were pretreated with 25 μm PP2 before addition of the TLR ligands LPS (50 ng/ml), R848 (100 ng/ml), and pIC (50 μg/ml). After 16 h, total RNA was isolated, and levels of IL-6 (A) and IP10 (B) mRNAs were determined by qRT-PCR and compared with glyceraldehyde-3-phosphate dehydrogenase mRNA levels. C and D, embryonal fibroblasts from c-Src-, Yes-, and Fyn-deficient mouse (SYF cells) or c-Src-expressing control cells (designated Src) were stimulated with TLR ligands for 16 h prior to isolation of total RNA and assessment of IL-6 (C) and IP10 (D) synthesis by qRT-PCR analysis.
TLR-elicited Induction of ATF3 Is Regulated by c-Src
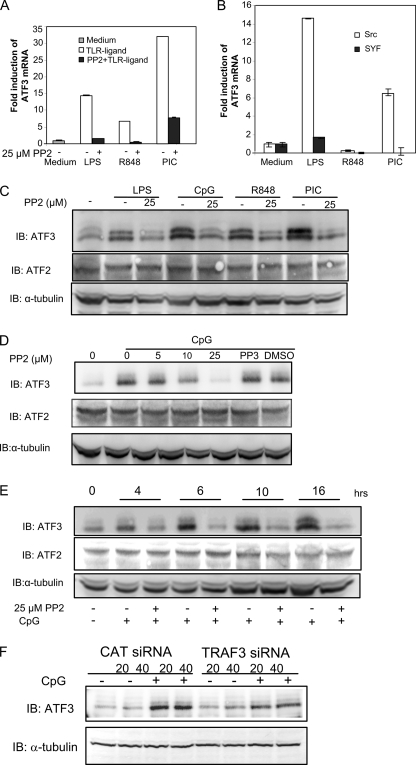
Our results indicated that c-Src may enhance transcription of IP10, while restraining transcription of IL-6 mRNA. TLR signaling has been shown to be negatively regulated by various means (22). Regarding IRF-5-mediated transcription, it was recently shown that IRF-4 negatively regulates IRF-5-dependent transcription through direct competition with IRF-5 for MyD88 association (23). However, we did not observe significant induction of IRF-4 mRNA after 2 or 16 h of TLR stimulation in RAW macrophages (supplemental Fig. 3A). Also, the presence of PP2 did not have a considerable effect on transcription of IRF-4 mRNA, while modestly affecting IRF-4 mRNA in the presence of CpG. Thus, IRF-4 does not appear to be implicated in differential TLR-elicited transcription after c-Src inhibition. Suppressors of cytokine signaling SOCS1 and SOCS3 have been shown to negatively regulate TLR signaling (24). We found that mRNA encoding SOCS3, but not SOCS1, was induced by LPS, R848, and CpG in RAW cells (supplemental Fig. 3B). The TLR-elicited induction of SOCS3 mRNA was significantly enhanced by the Src inhibitor PP2. However, we expected SOCS1 and SOCS3 to exert negative effects on TLR-elicited signaling and found it unlikely that the enhanced IL-6 expression was associated with increased levels of SOCS3. Recently, it was reported that activating transcription factor-3 (ATF3), a member of the ATF/CREB family of transcription factors, acts as a negative regulator of TLR signaling. In particular, ATF3 was shown to inhibit transcription of IL-12β and IL-6 (15, 25). To explore the potential role of ATF3 in c-Src-mediated enhancement of IL-6 transcription, we examined whether c-Src inhibition affected ATF3 mRNA expression after TLR treatment. Analysis by qRT-PCR showed that ATF3 transcription was induced by the TLR ligands LPS, R848, CpG, and pIC in RAW macrophages (Fig. 4A). Interestingly, addition of the c-Src pharmacological inhibitor PP2 abrogated TLR-elicited ATF3 mRNA induction (Fig. 4A). Thus, it is possible that reduced ATF3 levels upon c-Src inhibition are associated with enhanced IL-6 mRNA transcription. Next, we verified the results obtained by pharmacological inhibition using mouse embryonic fibroblasts in which the tyrosine kinases src, yes, and fyn have been deleted (“SYF” cells) and SYF cells in which c-src has been introduced by retroviral transduction (c-Src (11)). LPS, pIC, and CpG induced ATF3 transcription in SYF cells. However, TLR-elicited induction of ATF3 mRNA was abrogated in SYF cells compared with cells expressing c-Src (Fig. 4B), thus corroborating results obtained with the pharmacological Src inhibitor. To examine if altered ATF3 mRNA transcription translated into changes in ATF3 protein levels, RAW cells were pretreated with PP2 before stimulation with TLR ligands and assessment of ATF3 protein levels by immunoblotting. The TLR ligands LPS, CpG, R848, and pIC clearly stimulated ATF3 protein levels (Fig. 4C). Interestingly, TLR-elicited ATF3 protein levels were reduced upon c-Src inhibition (Fig. 4C). Next, we examined the dose-dependent effect of PP2 on CpG-elicited ATF3 and found that PP2 reduced ATF3 protein levels in a dose-dependent manner in RAW macrophages (Fig. 4D). Moreover, to relate ATF3 induction to the kinetics of enhanced IL-6 transcription, we examined the kinetics of ATF3 protein induction following addition of CpG. ATF3 protein was induced after 4 h of CpG treatment and was further increased after 6, 10, and 16 h of CpG stimulation (Fig. 4E). Importantly, protein levels of another member of this transcription factor family, ATF2 (which acts as a transcriptional enhancer (26)), remained unchanged after c-Src inhibition and treatment with TLR ligands.
FIGURE 4.
TLR-elicited induction of ATF3 mRNA and ATF3 protein are regulated by c-Src. A, RAW 264.7 cells were pretreated with 25 μm PP2 prior to treatment with TLR ligands for 16 h and assessment of mRNA levels of ATF3. B, mouse embryonic fibroblasts deficient for the Src family kinases Src, Yes, and Fyn (SYF) and c-Src-expressing control cells were treated with TLR ligands for 16 h before determination of ATF3 mRNA induction by qRT-PCR analysis. C–E, RAW 264.7 cells were pretreated with the indicated concentrations of PP2 or PP3 (25 μm PP2) prior to treatment with TLR ligands for 16 h (C and D) and immunoblotting (IB) for ATF3 or ATF2. The blots were reprobed with α-tubulin as a loading control. LPS (50 ng/ml), R848 (100 ng/ml), pIC (50 μg/ml), and CpG (10 μg/ml) are shown. F, RAW 264.7 cells were transfected with TRAF3 or CAT siRNA (20 or 40 nm) and treated with CpG (4 h) or not. Proteins were analyzed by immunoblotting for ATF3 or α-tubulin as a loading control.
We found that c-Src activation occurred downstream of TRAF3 (Fig. 1C). To examine if TRAF3 was implicated in CpG-stimulated ATF3 expression, we treated RAW cells with TRAF3 or CAT siRNA. Treatment with TRAF3 siRNA significantly reduced ATF3 induction stimulated by CpG compared with that in CAT siRNA-treated cells (Fig. 4F). Collectively, these results suggest that TRAF3-mediated c-Src activation contributes to TLR9-elicited ATF3 induction. This may suggest a specific role for ATF3 in modulation of c-Src-mediated TLR-dependent signaling.
ATF3 Interacts with IRF-5 and Mediates Enhanced CpG-stimulated IL-6 Production
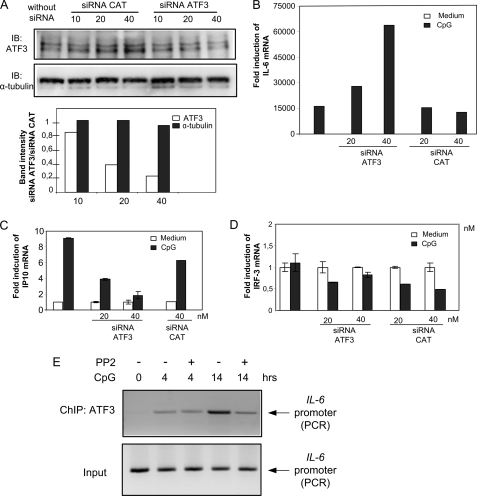
To investigate the functional role of ATF3 in TLR-elicited IL-6 and IP10 production, we used siRNA to down-regulate ATF3 mRNA. Immunoblotting confirmed that ATF3 protein levels were reduced to about 50% upon transfection of RAW cells with an ATF3 siRNA oligonucleotide (Fig. 5A). CAT siRNA used as a nonspecific control slightly increased ATF3 levels at the highest siRNA concentration. Next, we examined the effect of ATF3 siRNA in RAW cells and found that CpG-stimulated transcription of IL-6 was significantly enhanced in cells transfected with ATF3 siRNA compared with cells transfected with CAT siRNA (Fig. 5B). Conversely, CpG-stimulated IP10 mRNA was decreased by the presence of ATF3 siRNA relative to the presence of CAT siRNA (Fig. 5C). The molecular basis for reduced CpG-stimulated IP10 transcription upon ATF3 siRNA treatment is presently unknown. However, it is possible that ATF3 interacts or competes with protein(s) that could restrain IP10 production, e.g. similarly to inducible cAMP early repressor, which represses TLR-stimulated, ATF2-mediated expression of tumor necrosis factor (27). In such settings, reduced ATF3 levels could result in enhanced inhibition of CpG-stimulated IP10 transcription. Because IRF-3 mRNA is known to be largely constitutive in most cells, we examined IRF-3 mRNA levels after transfection of ATF3 siRNA. IRF-3 mRNA was not induced by CpG and was not influenced by the presence of ATF3 siRNA (Fig. 5D). Collectively, our results suggest that ATF3 is induced downstream of c-Src and negatively regulates CpG-elicited IL-6 production.
FIGURE 5.
ATF3 differentially regulates CpG-stimulated IL-6 and IP10 mRNA. A, RAW 264.7 cells were transfected with ATF3 siRNA or CAT siRNA as a nonspecific control (10, 20, or 40 nm) for 48 h. Lysates were analyzed by immunoblotting (IB) for ATF3 or α-tubulin as a control. Band intensities of ATF3 and α-tubulin were quantified using the Kodak image analysis software, and levels of ATF3 or α-tubulin in ATF3 siRNA-treated relative to CAT siRNA-treated cells are shown in the lower panel. B–D, RAW 264.7 cells were transfected with ATF3 siRNA or CAT siRNA as a nonspecific control (20 or 40 nm) for 48 h before addition of CpG (10 μg/ml). 16 h later, RNA was isolated, and IL-6 (B), IP10 (C), and IRF-3 (D) mRNA levels were determined by qRT-PCR. Data are expressed as fold induction relative to corresponding mRNA levels in medium-treated cells. E, RAW 264.7 cells were treated or not with 10 μg/ml CpG or 10 μm PP2 for 4 or 14 h. Chromatin immunoprecipitation assays were performed with anti-ATF3. DNA from chromatin immunoprecipitated (ChIP) or input fractions were measured by PCR amplification of IL-6 promoter sequences.
In an attempt to rationalize the differential effect of ATF3 on TLR-elicited genes, we used bioinformatics analysis to predict potential transcription factor-binding sites in putative promoter regions of IL-6, IRF-5, IP10, IL-12β, and IRF-3 genes. DNA sequences from −1500 to +200 bp around the transcriptional start sites of the corresponding promoters were scanned. Promoters of IL-6 and IL-12β were predicted to contain ATF and CREB consensus sites, whereas IP10 and IRF-3 promoters were not. To investigate if c-Src inhibition affected binding of ATF3 to the IL-6 promoter in vivo, we performed chromatin immunoprecipitation experiments. RAW cells were pretreated with PP2 or not prior to treatment with CpG for 4 or 14 h and precipitation of chromatin fragments using an ATF3 antibody. il-6 promoter DNA was amplified by PCR. We found that ATF3 was recruited to the il-6 promoter after 4 and 14 h of CpG treatment and that c-Src inhibition reduced recruitment of ATF3 after longer CpG treatments (Fig. 5E). This correlates to results showing that ATF3 is a negative regulator of IL-6 production at later phases of TLR engagement and shows a direct link between c-Src activity and CpG-induced recruitment of ATF3 to the IL-6 promoter.
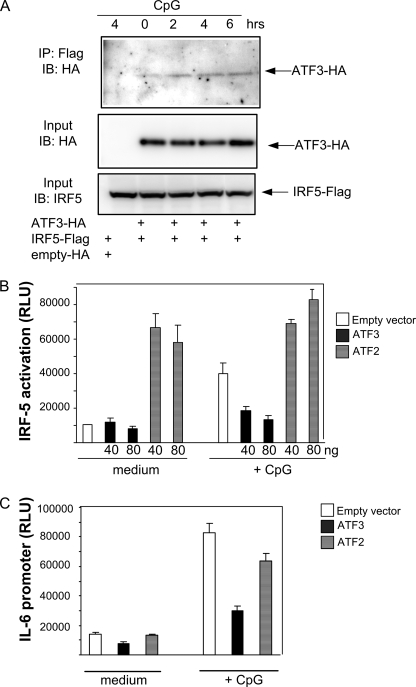
To further examine the relationship between ATF3- and IRF-5-mediated transcription, we examined whether ATF3 associated with IRF-5 by using coimmunoprecipitation. FLAG-tagged IRF-5 and HA-tagged ATF3 were transfected in TLR9-expressing HEK cells prior to treatment with CpG, precipitation with anti-FLAG antibody, and immunoblotting for HA. We found that ATF3 associated with IRF-5 in a CpG-dependent manner (Fig. 6A). Sendai virus, which is recognized by RIG-I in HEK cells (28), also stimulated interaction between ATF3 and IRF-5 (data not shown). Next, we investigated if overexpression of ATF3 affected IRF-5 activation or transcription from the IL-6 promoter. ATF3 expression alone had no effect on IRF-5 or IL-6 promoter activation, but it significantly reduced CpG-stimulated IRF-5 and IL-6 promoter activation (Fig. 6, B and C). Conversely, in the absence of stimuli, ATF2 expression activated IRF-5 in unstimulated cells. Collectively, these results suggest that ATF3 associates with IRF-5 and attenuates CpG-dependent IRF-5 activation and stimulation of the IL-6 promoter.
FIGURE 6.
ATF3 associates with IRF-5 and modulates CpG-stimulated IRF-5 and IL-6 promoter activation. A, HEK293 cells were cotransfected with IRF-5FLAG and HA-tagged ATF3, ATF2, or the empty vector. 24 h post-transfection, cells were treated with CpG (10 μg/ml). Lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblotting (IB) with anti-HA antibody. Membranes were reprobed with IRF-5 antibody. B and C, HEK293 cells were cotransfected with HA-tagged ATF3, ATF2, or the corresponding empty vector together with Gal4-IRF-5 reporter plasmids (B) or an IL-6 promoter reporter plasmid (C). 24 h post-transfection, cells were treated or not with CpG (10 μg/ml) and lysed before determination of luciferase activity. Amount of vector backbone DNA was kept constant in each transfection by adding empty vector.
DISCUSSION
The family of IRFs constitutes important transcription factors shaping the host immune response to pathogens. In this study, we have shown that the tyrosine kinase c-Src differentially regulates activation of IRF-3, IRF-5, and TLR-mediated gene expression. Inhibition of c-Src activity leads to reduced IRF-3 activation and IP10 production, while augmenting IRF-5 activation and IL-6 synthesis in response to TLR ligands. The stimulatory effect on IRF-5 activation and IL-6 production observed after c-Src inhibition occurred at later phases, being considerably enhanced after 16 h relative to 10 h of CpG treatment. This slow kinetics affecting late phases of TLR responses indicated that transcription and protein synthesis were involved. Hence, we proposed that c-Src inhibition modulated transcription of negative regulators restraining TLR signaling and IL-6 transcription. Therefore, we examined if expression of IRF-4, SOCS1, SOCS3, and ATF3 (previously identified as negative regulators of TLR signaling) were dependent on c-Src activity. IRF-4 was recently reported to inhibit MyD88-dependent TLR signaling by competing with IRF-5 for binding to MyD88, thus hampering downstream signaling (23). Negishi et al. (23) found that IL-6 expression (which depends on IRF-5 activation) was elevated in cells lacking IRF-4, and overexpression of IRF-4 in RAW cells reduced CpG-stimulated IL-6 production. However, we found that IRF-4 mRNA was largely unaffected by TLR stimulation/c-Src inhibition, and it is therefore unlikely that IRF-4 is responsible for the differential gene expression observed in our system. SOCS1 and SOCS3 have been shown to inhibit pathogen-elicited responses (22, 30). However, we did not observe changes in SOCS1 and SOCS3 expression that correlated with and could explain the enhanced IL-6 expression.
ATF3 is a member of the ATF/CREB family of transcription factors and binds to the ATF/cAMP-response element consensus sequence TGACGTCA (31). Transcription of ATF3 is induced by proinflammatory cytokines and stress signals, e.g. genotoxic and cell death-inducing agents. Interestingly, in contrast to other ATF family members, ATF3 has been shown to repress transcription from promoters with ATF/cAMP-response element-binding sites (32). We found that ATF3 was induced by CpG (within 2–4 h) and that TRAF3 and c-Src activity were required for ATF3 induction in response to TLR ligands. The kinetics of ATF3 protein expression showed that ATF3 levels were further elevated at prolonged treatments with CpG (Fig. 4E). Bioinformatics analysis of the 5′-regulatory regions of several immune modulatory genes showed that IL-6 and IL-12β promoters contain ATF or CREB sites to which ATF3 could bind. Hence, ATF3 may repress later phases of TLR-stimulated IL-6 production by binding to the ATF or CREB sites of its promoter. Indeed, recent studies using chromatin immunoprecipitation have shown that ATF3 binds to the IL-6 and IL-12β promoters in response to LPS treatment and to IL-4, IL-5, and IL-13 promoters in differentiated CD4+ T cells (15). Moreover, it was found that ATF3 is recruited to the IL-6 and IL-12β promoters more slowly than the NF-κB subunit Rel, reflecting the role of ATF3 in delayed phases of TLR signaling. We also found that CpG induces binding of ATF3 to the IL-6 promoter at 4 and 14 h of treatment and that c-Src inhibition reduces this recruitment (Fig. 5E). Gilchrist et al. (15) also reported that LPS induced association between nuclear ATF3 and the chromatin-modifying enzyme histone deacetylase. Therefore, ATF3 may repress transcription by inducing deacetylation of histones at the IL-6 promoter generating an inaccessible chromatin structure at the IL-6 promoter. Reduced ATF3 levels after c-Src inhibition may thus result in enhanced IL-6 expression due to changes in chromatin modification. Furthermore, we found that ectopic expression of ATF3 reduced IRF-5 activation in a Gal4-dependent reporter system (Fig. 6B) and that ATF3 associated with IRF-5, as assessed by coimmunoprecipitation (Fig. 6A). This suggests that ATF3 interacts directly with IRF-5, which may be sufficient to inhibit the transactivation ability of IRF-5 (independent of chromatin/DNA binding). Thus, this represents an alternative mechanism by which ATF3 may modulate IRF-5-dependent transcription. Although the subcellular localization of ATF3 has not been studied in detail, ATF3 is believed to reside mainly in the nucleus (33) and could interact with IRF-5 in the nucleus. To clarify the results obtained by us and others, a schematic model for the possible action of c-Src in IRF-3- and IRF-5-dependent transcription is depicted in Fig. 7. Interestingly, it has previously been shown that ATF and IRF proteins may associate with non-family member transcription factors, e.g. gadd153/Chop10 and bone morphogenetic proteins, respectively (34, 35). Specifically, ATF3 has been shown to heterodimerize with gadd153/Chop10, generating nonfunctional protein dimers (34). Likewise, ATF3 could bind to IRF-5 and suppress IRF-5 homodimerization and DNA binding, thus inhibiting IRF-5-dependent transcription. It would be interesting to study if c-Src inhibition (through altered ATF3 levels or other mechanisms) affected CpG-elicited activation of endogenous IRF-5. We attempted to study this through IRF-5 nuclear translocation, IRF-5 dimerization, or chromatin immunoprecipitation, but in our experimental settings commercial antibodies tested failed to specifically stain IRF-5 and discriminate between IRF-5 and IRF-3.
FIGURE 7.
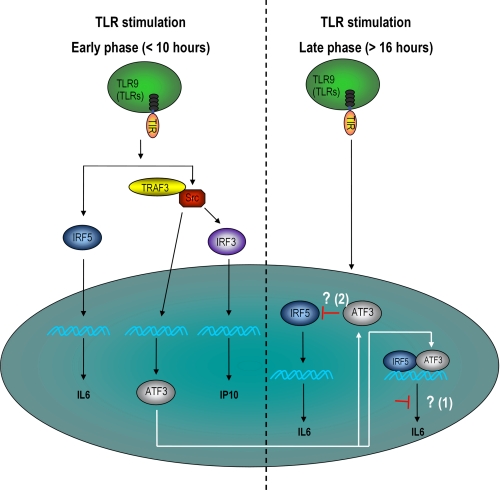
Schematic model illustrating the role of c-Src and ATF3 in TLR-mediated IRF activation and gene expression. During an early phase of TLR stimulation, c-Src positively affects IRF-3 activation, induction of IP10, and transcription of ATF3. In later phases of TLR stimulation, ATF3 represses TLR-dependent IL-6 expression by two possible mechanisms (indicated by white arrows) as follows: 1) by chromatin modification at the IL-6 promoter level, or 2) by direct interaction with IRF-5 and interference of IRF-5-mediated transcription.
ATF3-deficient mice were recently reported to exhibit increased protection against murine cytomegalovirus infection (29) and modulate hyper-responsiveness in asthma and airway inflammation. Hence, ATF3 mediates inflammatory responses by limiting expression of distinct cytokines in biologically relevant disease models. Interestingly, our results suggest that c-Src modulation of ATF3 differentially regulates IRF-3- and IRF-5-mediated gene expression, implicating c-Src in ATF3-mediated functions. In conclusion, our results identify a pathway by which c-Src contributes to differential IRF activation, thus affecting the dynamic transcriptional program stimulated by pathogen exposure and TLR activation.
Supplementary Material
This work was supported by the National Programme for Research in Functional Genomics in Norway and Mid-Norway (to M. W. A.), the Research Council of Norway (to M. W. A.), the Faculty of Medicine, Norwegian University of Science and Technology (to I. B. J. and M. W. A.), and the Cancer Fund at St. Olavs Hospital (to M. W. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Table SI, “Methods,” and additional references.
- TLR
- Toll-like receptor
- IFN
- interferon
- RIG-I
- retinoic-inducible gene
- ATF/CREB
- activating transcription factor/cAMP-responsive element-binding protein
- NF-κB
- nuclear factor of κB
- CAT
- chloramphenicol acetyltransferase
- IRF
- interferon regulatory factor
- qRT
- quantitative real time
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- LPS
- lipopolysaccharide
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Tamura T., Yanai H., Savitsky D., Taniguchi T. (2008) Annu. Rev. Immunol. 26, 535–584 [DOI] [PubMed] [Google Scholar]
- 2.Huang Q., Liu D., Majewski P., Schulte L. C., Korn J. M., Young R. A., Lander E. S., Hacohen N. (2001) Science 294, 870–875 [DOI] [PubMed] [Google Scholar]
- 3.Barnes B. J., Richards J., Mancl M., Hanash S., Beretta L., Pitha P. M. (2004) J. Biol. Chem. 279, 45194–45207 [DOI] [PubMed] [Google Scholar]
- 4.Panne D., Maniatis T., Harrison S. C. (2007) Cell 129, 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wathelet M. G., Lin C. H., Parekh B. S., Ronco L. V., Howley P. M., Maniatis T. (1998) Mol. Cell 1, 507–518 [DOI] [PubMed] [Google Scholar]
- 6.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W., Taniguchi T. (2005) Nature 434, 243–249 [DOI] [PubMed] [Google Scholar]
- 7.Paun A., Pitha P. M. (2007) Biochimie 89, 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balkhi M. Y., Fitzgerald K. A., Pitha P. M. (2008) Mol. Cell. Biol. 28, 7296–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K., Taniguchi T. (2006) Nat. Rev. Immunol. 6, 644–658 [DOI] [PubMed] [Google Scholar]
- 10.Johnsen I. B., Nguyen T. T., Ringdal M., Tryggestad A. M., Bakke O., Lien E., Espevik T., Anthonsen M. W. (2006) EMBO J. 25, 3335–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 13.Johnsen I. B., Nguyen T. T., Bergstroem B., Fitzgerald K. A., Anthonsen M. W. (2009) J. Biol. Chem. 284, 19122–19131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas P. A., Peña-Diaz J., Liabakk N. B., Krokan H. E., Skorpen F. (2009) DNA Repair 8, 822–833 [DOI] [PubMed] [Google Scholar]
- 15.Gilchrist M., Thorsson V., Li B., Rust A. G., Korb M., Roach J. C., Kennedy K., Hai T., Bolouri H., Aderem A. (2006) Nature 441, 173–178 [DOI] [PubMed] [Google Scholar]
- 16.Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Choi M. K., Ban T., Yanai H., Negishi H., Lu Y., Tamura T., Takaoka A., Nishikura K., Taniguchi T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoenemeyer A., Barnes B. J., Mancl M. E., Latz E., Goutagny N., Pitha P. M., Fitzgerald K. A., Golenbock D. T. (2005) J. Biol. Chem. 280, 17005–17012 [DOI] [PubMed] [Google Scholar]
- 19.Doyle S., Vaidya S., O'Connell R., Dadgostar H., Dempsey P., Wu T., Rao G., Sun R., Haberland M., Modlin R., Cheng G. (2002) Immunity 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 20.Melchjorsen J., Sørensen L. N., Paludan S. R. (2003) J. Leukocyte Biol. 74, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., Takeda K., Akira S. (2004) Nature 430, 218–222 [DOI] [PubMed] [Google Scholar]
- 22.Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 23.Negishi H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., Honda K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15989–15994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura A., Naka T., Kubo M. (2007) Nat. Rev. Immunol. 7, 454–465 [DOI] [PubMed] [Google Scholar]
- 25.Whitmore M. M., Iparraguirre A., Kubelka L., Weninger W., Hai T., Williams B. R. (2007) J. Immunol. 179, 3622–3630 [DOI] [PubMed] [Google Scholar]
- 26.Bhoumik A., Ronai Z. (2008) Cell Cycle 7, 2341–2345 [DOI] [PubMed] [Google Scholar]
- 27.Altmayr F., Jusek G., Holzmann B. (2010) J. Biol. Chem. 285, 3525–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K. A. (2005) J. Immunol. 175, 5260–5268 [DOI] [PubMed] [Google Scholar]
- 29.Rosenberger C. M., Clark A. E., Treuting P. M., Johnson C. D., Aderem A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pothlichet J., Chignard M., Si-Tahar M. (2008) J. Immunol. 180, 2034–2038 [DOI] [PubMed] [Google Scholar]
- 31.Hai T., Hartman M. G. (2001) Gene 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 32.Chen B. P., Liang G., Whelan J., Hai T. (1994) J. Biol. Chem. 269, 15819–15826 [PubMed] [Google Scholar]
- 33.Turchi L., Fareh M., Aberdam E., Kitajima S., Simpson F., Wicking C., Aberdam D., Virolle T. (2009) Cell Death. Differ. 16, 728–737 [DOI] [PubMed] [Google Scholar]
- 34.Chen B. P., Wolfgang C. D., Hai T. (1996) Mol. Cell. Biol. 16, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qing J., Liu C., Choy L., Wu R. Y., Pagano J. S., Derynck R. (2004) Mol. Cell. Biol. 24, 1411–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.