Abstract
Transforming growth factor-β1 (TGF-β1) is secreted as part of an inactive complex consisting of the mature dimer, the TGF-β1 propeptide (latency-associated peptide (LAP)), and latent TGF-β-binding proteins. Using in vitro mutagenesis, we identified the regions of LAP that govern the cooperative assembly and stability of the latent TGF-β1 complex. Initially, hydrophobic LAP residues (Ile53, Leu54, Leu57, and Leu59), which form a contiguous epitope on one surface of an amphipathic α-helix, interact with mature TGF-β1 to form the small latent complex. TGF-β1 binding is predicted to alter LAP conformation, exposing ionic residues (Arg45, Arg50, Lys56, and Arg58) on the other side of the α-helix, which form the binding site for latent TGF-β-binding proteins. The stability of the resultant large latent complex is dependent upon covalent dimerization of LAP, which is facilitated by key residues (Phe198, Asp199, Val200, Leu208, Phe217, and Leu219) at the dimer interface. Significantly, genetic mutations in LAP (e.g. R218H) that cause the rare bone disorder Camurati-Engelmann disease disrupted dimerization and reduced the stability of the latent TGF-β1 complex.
Keywords: Extracellular Matrix Proteins, Protein Folding, Protein-Protein Interactions, Signal Transduction, Transforming Growth Factor Beta (TGFbeta), Camurati-Engelmann Disease, Extracellular Matrix Proteins, Latent TGF-β, Binding Protein, TGF-β, Prodomain, Latency-associated Peptide
Introduction
Transforming growth factor-β1 (TGF-β1)2 is the prototypical member of a large family of structurally related proteins, with well documented roles in cellular proliferation, differentiation, apoptosis, adhesion, and extracellular matrix deposition (1, 2). Like other family members, TGF-β1 is synthesized as a large precursor molecule consisting of an N-terminal prodomain (latency-associated peptide (LAP)) and a C-terminal mature domain. Within this precursor, LAP acts as a multifunctional peptide capable of regulating the crucial roles TGF-β1 plays throughout development and in the maintenance of tissue homeostasis in adult life (3–5).
Initially, hydrophobic residues near the N terminus of LAP bind to mature TGF-β1 (6), and this interaction is necessary to maintain the molecule in a conformation competent for dimerization. Two precursors are then covalently linked at sites within both the mature growth factor and LAP to form the small latent complex (SLC) (7–10). The SLC can be cleaved by proprotein convertases (11, 12), but LAP remains non-covalently associated with the mature TGF-β1 dimer (13, 14). During the secretory process, LAP also interacts covalently with latent TGF-β-binding proteins (LTBPs) to form the large latent complex (LLC), and it is in this form that TGF-β1 is secreted from the cell (7, 15). In the absence of LTBPs, the reactive cysteine (Cys33) within LAP forms an incorrectly paired disulfide bond with a free cysteine in mature TGF-β1, ensuring the SLC is secreted in an inactive form (10).
Extracellularly, LAP confers latency to TGF-β1 by shielding the type II receptor-binding epitope on the outer convex surface of the mature dimer (16, 17). Lacking additional secreted antagonists, this is the major point of regulation of TGF-β1 biological activity. Interestingly, of the 33 TGF-β ligands, only TGF-β1, -β2, -β3, myostatin, and GDF-11 (growth and differentiation factor-11) bind their propeptides with high enough affinity to confer latency (16, 18–21). For other family members (e.g. activins and bone morphogenetic proteins), affinity for receptors is greater than affinity for propeptides, and they are secreted in an “active” form (6, 22).
LTBPs associate with LAP via signature 8-Cys domains, which are unique to these proteins and the structurally related molecules, fibrillin-1 and fibrillin-2 (23–25). Once secreted, LTBPs target latent TGF-β1 to fibrillin microfibrils within the extracellular matrix. TGF-β1 signaling is dependant upon liberation of the mature ligand from the LLC, a process mediated by activators, such as thrombospondin-1 and integrins, that bind to specific residues in LAP (54LSKL and 244RGD, respectively) (4, 26–28). By altering the conformation of LAP, these activators permit TGF-β1 to engage its signaling receptors.
Perturbation of the extracellular control of TGF-β1 signaling has been implicated in many human diseases, including Marfan syndrome, geleophysic dysplasia, Loeys-Dietz syndrome, and Camurati-Engelmann disease (CED) (29–34). Instructively, missense mutations in LAP underlie the pathology observed in the rare bone disorder, CED. These mutations lead to up-regulation of TGF-β1 signaling via increased ligand activation or secretion of the mutant protein (35, 36). Interestingly, most of the identified CED mutations are in the vicinity of the cysteine residues (Cys223 and Cys225) required for covalent dimerization of LAP (10).
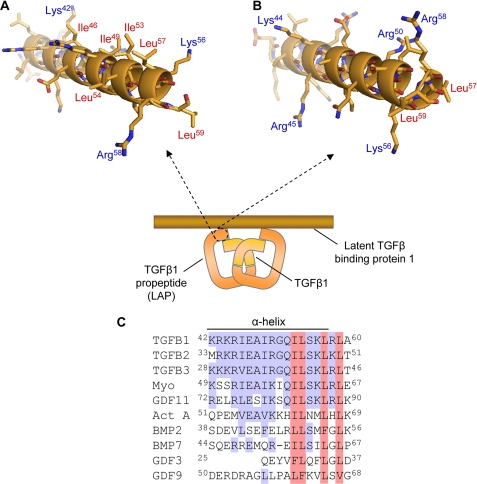
The numerous studies that have elucidated the mechanism of latent TGF-β1 assembly, secretion, matrix deposition, and activation have highlighted three functionally important regions in LAP: (i) a TGF-β1 binding site at the N terminus; (ii) an LTBP-1 binding epitope in the vicinity of Cys33; and (iii) a LAP dimerization interface at the C terminus (Fig. 1). Together, these sites coordinate the assembly and stability of the large latent TGF-β1 complex. In this study, using in vitro mutagenesis and functional analyses, we have identified the residues in LAP that mediate these functions. Importantly, despite low sequence homology (∼15%) between propeptides of TGF-β ligands, these structurally significant regions are broadly conserved.
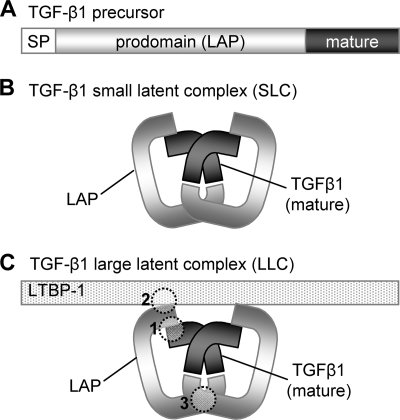
FIGURE 1.
Formation of the large latent TGF-β1 complex. TGF-β1 is synthesized from a larger precursor protein, comprising a signal peptide (SP), LAP, and a C-terminal mature domain (A). During assembly, two TGF-β1 precursors are covalently linked at sites within both the mature growth factor and LAP to form the SLC (B). The SLC then binds to LTBPs to form the LLC (C). Studies have indicated that the assembly and stability of the LLC is governed by three key regions in LAP (1–3).
EXPERIMENTAL PROCEDURES
Production of TGF-β1 Mutants
Mutations were introduced in full-length TGF-β1 using the QuikChange Lightning site-directed mutagenesis kit (Stratagene, La Jolla, CA). A pCDNA3.1 (Invitrogen) vector containing full-length human TGF-β1 served as the template in these reactions. For each construct, the mutated region was confirmed by DNA sequencing. Wild type and mutant TGF-β1 were produced by transient transfection in HEK-293T cells using Lipofectamine 2000 (Invitrogen). Briefly, HEK-293T cells were plated at 8.0 × 105 cells/well in a 6-well plate. Wild type or mutant TGF-β1 DNA (5 μg) was combined with Lipofectamine according to the manufacturer's instructions. After a 20-min incubation, DNA· Lipofectamine complexes were added directly to the plated cells and incubated in serum-free Opti-MEM medium (Invitrogen) for a further 48 h at 37 °C in 5% CO2.
The transfected HEK-293T culture medium and cell lysates for the TGF-β1 mutants were assessed by Western blotting. At 48 h post-transfection, the culture medium was removed and concentrated 50-fold using Nanosep microconcentration devices with a 10 kDa molecular mass cut-off (Pall Life Sciences). The cells were lysed in 1% Triton X-100 in phosphate-buffered saline (pH 7.4). Non-reduced samples were loaded onto 10% SDS-PAGE and subjected to Western blotting. After electrophoresis, samples were transferred onto ECL Hybond membranes (GE Healthcare). TGF-β1 was detected using an R&D Systems TGF-β1 monoclonal antibody (MAB240, R&D Systems (Minneapolis, MI)).
TGF-β1 ELISA
A TGF-β1 ELISA (R&D Systems) was employed to measure the amount of TGF-β1 in HEK-293T conditioned medium. Conditioned medium was diluted in 1.4% bovine serum albumin in phosphate-buffered saline (pH 7.4) prior to the assay. The sensitivity of the assay was 2 pg/ml.
Assessment of the Stability of TGF-β1 Latent Complexes
To assess TGF-β1 latent complex stability, a time course activation assay was developed. In order to release the mature growth factor from the latent complex, conditioned medium containing TGF-β1 variants was incubated with 1 m HCl for 10 min at room temperature and then neutralized with 1.2 m NaOH in 0.5 m HEPES. Neutralized samples were incubated at room temperature for 0.25, 0.5, 1, 2, and 4 h prior to assay. Active and total levels were measured for each time point in the TGF-β1 ELISA, as described above. The ability of wild type LAP to reassociate with mature TGF-β1 relative to CED variants was analyzed using one-phase decay parameters in the Prism Program (version 5; GraphPad Software (San Diego, CA)).
Production of Wild Type and Mutant LAP
Wild type and mutant LAP were generated by PCR (antisense primer 5′-CTAGGAATTCCTATTTGTCGTCGTCGTCTTTGTAGTCGGAGCTTTGCAGATGCTGGGCCCTCTC-3′) and cloned into the HindIII and EcoRI sites of a pCDNA3.1+ vector (Invitrogen). The primers incorporated a 3′ FLAG tag. LAP variants were produced by transient transfection in HEK-293T cells using Lipofectamine (Invitrogen) as described above. LAP expression was analyzed by Western blotting using the FLAG M2 monoclonal antibody (Sigma). The membranes were then probed with a sheep anti-mouse serum conjugated to the fluorescent dye IR800 (Rockland Immunochemicals (Gilbertsville, PA)). The membranes were scanned on an infrared imaging system (Odyssey IR imaging scanner; Li-Cor Biosciences (Lincoln, NE)), and relative LAP concentrations were determined.
TGF-β1 in Vitro Bioassay
LAP variants were assessed for their ability to suppress TGF-β1 bioactivity in a luciferase assay. Briefly, HEK-293T cells were plated in poly-d-lysine-coated 48-well plates at 75,000 cells/well. After a 24-h incubation, cells were transfected with the TGF-β1-responsive A3-Lux reporter construct (37) and FAST2 transcription factor (37) as described previously (38). Each well was transfected with A3-Lux (25 ng), FAST-2 (50 ng), and empty vector (425 ng of pcDNA3.1+) using Lipofectamine according to the manufacturer's protocol (Invitrogen). At 24 h post-transfection, cells were treated with 100 pm TGF-β1 (Peprotech) and increasing doses of conditioned medium containing either wild type or mutant LAP. After a 16-h incubation, cells were harvested in solubilization buffer (1% Triton X-100, 25 mm glycylglycine, pH 7.8, 15 mm MgSO4, 4 mm EGTA, and 1 mm dithiothreitol), and the TGF-β1-induced luciferase activity was determined.
TGF-β1-LTBP-1 Binding Experiments
A vector expressing the cDNA for the long isoform of LTBP-1 was kindly provided by Dr. Mark Gibson (University of Adelaide, Australia). PCR was used to isolate the 8-Cys domain of LTBP-1 and its two flanking epidermal growth factor domains (EGF-12 and EGF-13) (39). The generated PCR product (LTBP-1 trunc) served as a template in subsequent rounds of PCR, to enable the incorporation of a rat serum albumin leader sequence at the N terminus of LTBP-1 trunc, and three hemagglutinin epitope tags (YPYDVPDYA) at the C terminus.
To assess LTBP-1 interactions with TGF-β1 variants, the LTBP-1 trunc construct was co-expressed with equivalent amounts of either wild type or mutant TGF-β1 variants in HEK-293T cells by transient transfection (as described above). Binding of the TGF-β1 variants to LTBP-1 trunc was determined by Western blot analysis. Conditioned medium was concentrated 50-fold using Nanosep microconcentration devices with a 10 kDa molecular mass cut-off (Pall Life Sciences). Non-reduced samples were loaded onto 5% SDS-PAGE and analyzed by Western blot. The LTBP-1 fragment was detected using an anti-hemagglutinin high affinity monoclonal antibody (Roche Applied Science). TGF-β1 LAP was detected using LAP monoclonal antibody MAB246 (R&D Systems). Expression of TGF-β1 was determined using a TGF-β1 monoclonal antibody (MAB240, R&D systems), specific for the mature domain of TGF-β1.
Statistical Analysis
Significance (p < 0.05 or p < 0.01) was determined using one-way Student's t tests for independent groups. All error bars shown represent S.D.
RESULTS
Hydrophobic Residues at the N Terminus of LAP Govern Formation of the Small Latent TGF-β1 Complex
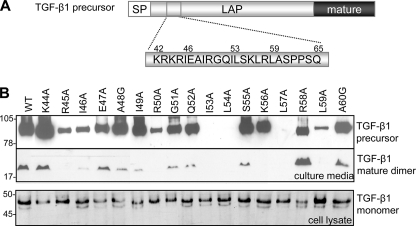
Previously, we identified a hydrophobic motif within the prodomain of inhibin A that governs the biosynthesis and secretion of the mature ligand (6). Conservation of the hydrophobic residues in the prodomains of other TGF-β ligands suggested that a similar assembly mechanism may exist across the family. For TGF-β1, the identified motif lies within a stretch of 24 amino acids (Lys42–Gln65) that is highly conserved between the TGF-β isoforms and has previously been implicated in binding the mature growth factor (4, 14). All residues throughout this region were substituted with alanine using in vitro mutagenesis (Fig. 2A). Western blot analysis on a panel of these TGF-β1 variants indicated that conditioned medium from cells transfected with wild type TGF-β1 contained both mature (25 kDa) and precursor (95 kDa) TGF-β1 forms (Fig. 2B). As anticipated, mutation of residues within the hydrophobic motif (Ile53, Leu54, Leu57, and Leu59) as well as Ile46 resulted in a significant reduction (>70%) in the amount of total TGF-β1 produced and secreted (Fig. 2B and Table 1). An analysis of lysates from the transfected cells indicated that the decrease in TGF-β1 production was not due to a loss of TGF-β1 precursor expression because it was present at similar levels for all variants tested (Fig. 2B, bottom).
FIGURE 2.
Effects of LAP mutations on TGF-β1 biosynthesis. Target residues in TGF-β1 LAP were substituted with alanine using in vitro mutagenesis (A). To determine the effects of amino acid substitutions on TGF-β1 production, conditioned media (top panels) and cell lysate (bottom panel) from HEK-293T cells transfected with either wild type (WT; lane 1) or mutant TGF-β1 were analyzed by Western blot (B). Samples were detected with a TGF-β1 mAb, specific for the mature domain. The 95-kDa TGF-β1 precursor and 25-kDa mature dimer are noted. SP, signal peptide.
TABLE 1.
Effect of LAP mutations on TGF-β1 biosynthesis
TGF-β1 levels were determined in the ELISA for each variant in triplicate; values represent mean ± S.D. ND, not determined.
| TGF-β1 variant | Active TGF-β1 | Total TGF-β1 | Percentage active |
|---|---|---|---|
| ng/ml | ng/ml | % | |
| Wild type | 2.42 ± 0.29 | 218.5 ± 12.8 | 1.11 ± 0.13 |
| K42A | 1.14 ± 0.16 | 121.7 ± 3.9a | 0.94 ± 0.13 |
| K43A | 2.60 ± 0.14 | 264.8 ± 2.7 | 0.98 ± 0.05 |
| K44A | 2.14 ± 0.07 | 141.9 ± 5.0b | 1.51 ± 0.05b |
| R45A | 1.64 ± 0.14 | 65.6 ± 2.2a | 2.50 ± 0.21a |
| I46A | 0.63 ± 0.15 | 58.4 ± 0.8a | 1.09 ± 0.26 |
| E47A | 1.55 ± 0.16 | 194.4 ± 6.8b | 0.80 ± 0.08 |
| A48G | 1.14 ± 0.21 | 109.9 ± 7.5a | 1.04 ± 0.19 |
| I49A | 0.64 ± 0.16 | 102.8 ± 4.1a | 0.62 ± 0.15 |
| R50Ac | 1.23 | 70.2 | 1.75 |
| G51A | 1.55 ± 0.16 | 180.5 ± 1.7b | 0.86 ± 0.09 |
| Q52A | 1.46 ± 0.29 | 125.4 ± 7.2b | 1.16 ± 0.23 |
| I53A | <0.6 | <0.6 | ND |
| L54A | <0.6 | <0.6 | ND |
| S55A | 2.10 ± 0.08 | 108.8 ± 6.6a | 1.93 ± 0.07a |
| K56A | 3.54 ± 0.09 | 10.4 ± 0.30a | 33.8 ± 0.82a |
| L57A | <0.6 | 1.75 ± 0.09a | ND |
| R58A | 4.42 ± 0.30 | 279.8 ± 9.8b | 1.58 ± 0.11 |
| L59A | 1.35 ± 0.09 | 64.6 ± 6.7a | 2.08 ± 0.14b |
| A60G | 1.65 ± 0.18 | 236.4 ± 21.7 | 0.70 ± 0.08 |
| S61A | 1.70 ± 0.00 | 259.7 ± 9.3b | 0.65 ± 0.00b |
| P62A | 1.80 ± 0.23 | 210.7 ± 6.4 | 0.85 ± 0.11 |
| P63A | 1.85 ± 0.26 | 83.5 ± 2.4b | 2.21 ± 0.31b |
| S64A | 2.15 ± 0.65 | 261.8 ± 7.0b | 0.82 ± 0.25 |
| Q65A | 2.45 ± 0.40 | 221.0 ± 6.6 | 1.11 ± 0.18 |
a p < 0.01.
b p < 0.05.
c n = 2.
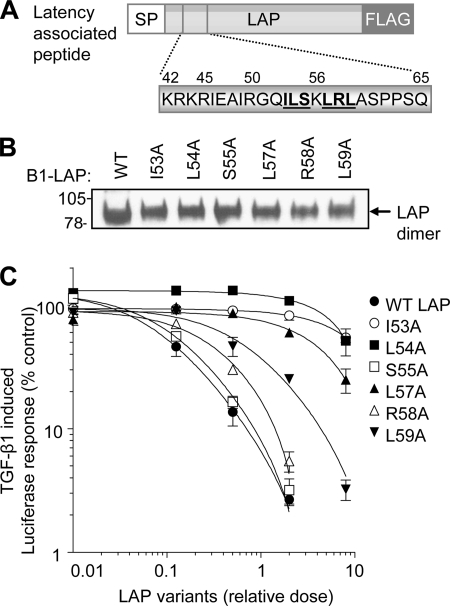
TGF-β1 is secreted as a latent complex, consisting of the mature growth factor non-covalently associated with LAP. We next sought to establish whether the hydrophobic LAP residues (Ile46, Ile53, Leu54, Leu57, and Leu59) important for assembly also confer latency. HEK-293T cells were transfected with a TGF-β1-responsive luciferase reporter and treated with recombinant TGF-β1 and increasing doses of conditioned medium containing either wild-type or mutant LAP (Fig. 3, A and B). Wild-type LAP was able to suppress the TGF-β1-induced luciferase response in a dose-dependant manner (Fig. 3C). In contrast, the LAP variants carrying mutations in the hydrophobic motif had a reduced ability to suppress TGF-β1 signaling (Fig. 3C). Indeed, the I53A and L54A mutants displayed an 80-fold reduction in affinity for mature TGF-β1 compared with wild type LAP. These results extend previous findings (4, 6, 14) that demonstrated that the N terminus of LAP interacted with the TGF-β1 dimer to mediate growth factor assembly, secretion, and latency.
FIGURE 3.
Analysis of the interaction between LAP and mature TGF-β1. Wild type and mutant LAP variants (boldface type and underlined) were generated by site-directed mutagenesis (A), and relative expression levels in transfected HEK-293T cells were determined by Western blot using the FLAG M2 mAb (B). The ability of wild type (WT) and mutant LAP variants to bind mature TGF-β1 and block TGF-β1 signaling was assessed in an in vitro bioassay (C). SP, signal peptide. Error bars, S.D.
Ionic Residues at the N Terminus of LAP Govern Formation of the Large Latent TGF-β1 Complex
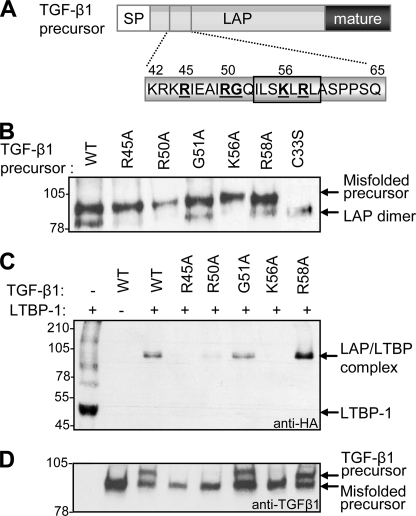
Our site-directed mutagenesis strategy also identified ionic LAP residues (Arg45, Arg50, Lys56, and Arg58) in the vicinity of the hydrophobic motif that affected TGF-β1 production (Fig. 2B and Table 1). Cells transfected with the R58A variant secreted higher levels (∼22%) of total TGF-β1, suggesting that this mutation improved LAP-mature interactions and subsequent growth factor processing. In contrast, cells transfected with the R45A, R50A, and K56A variants produced significantly lower levels (<65%) of total TGF-β1 (Fig. 2B and Table 1). The K56A variant was particularly interesting in that cells secreted wild type levels of precursor in the virtual absence of mature growth factor. Previous studies have indicated that, within the SLC, LAP covalently associates with LTBP-1, and, in the absence of LTBPs, secreted SLC contains incorrectly paired disulfide bonds (10). For this reason, we looked more closely at the composition of the precursor forms secreted by cells following transfection with the TGF-β1 variants (Fig. 4B). As a positive control, cells were transfected with mutant TGF-β1(C33S), which is unable to covalently link with LTBP, ensuring correct folding of the SLC (40). For the C33S variant, the SLC dissociated into its constituent components upon SDS-PAGE (as evidenced by the high levels of LAP dimer and the absence of TGF-β1 precursor) (Fig. 4B, lane 7). Strikingly, nearly all of the SLC secreted by cells transfected with the R45A, R50A, and K56A variants was misfolded (Fig. 4B, lanes 2, 3, and 5). This was partly attributable to the low levels of endogenous LTBPs in HEK-293T cells because >50% of the SLC produced by cells transfected with wild type TGF-β1 was also misfolded (Fig. 4B, lane 1). However, a significant proportion of wild type TGF-β1 was able to fold correctly, given the levels of LAP dimer evident following SDS-PAGE.
FIGURE 4.
Amino acids involved in the formation of the large latent TGF-β1 complex. Target residues in TGF-β1 LAP (boldface type, underlined) were substituted with alanine using in vitro mutagenesis (the hydrophobic residues identified to be important for LAP-mature interactions are boxed) (A). The effect of mutating these ionic residues on formation of the SLC was determined by Western blot using an anti-LAP antibody (B). The inability of the R45A, R50A, and K56A variants to fold correctly suggested that they were compromised in their ability to bind LTBPs. To verify this, cells were transfected with wild type (WT) or mutant TGF-β1 variants together with a truncated form of LTBP-1. The formation of the LLC was monitored by Western blot using an anti-hemagglutinin (anti-HA) mAb, specific for LTBP-1 (C). Expression of the TGF-β variants was verified by Western blot using a TGF-β1 mAb (D).
Chen et al. (39) recently showed that LAP binds to ionic residues (Glu1022, Asp1029, Asp1034, Asp1056, and Glu1059) in the third 8-Cys domain of LTBP-1, resulting in a disulfide exchange between a pair of cysteines in LTBP-1 and Cys33 in LAP. We therefore postulated that the identified cationic LAP residues (Arg45, Arg50, Lys56, and Arg58) may mediate non-covalent interactions with LTBPs. To verify this, wild type and mutant TGF-β1 variants were co-transfected with a truncated fragment of LTBP-1 (LTBP-1 trunc). The conditioned medium from transfected cells was analyzed by Western blot using antibodies specific for LTBP-1 trunc (anti-HA) (Fig. 4C). When expressed alone, LTBP-1 trunc was secreted as a 50-kDa polypeptide (Fig. 4C, lane 1); however, in the presence of latent TGF-β1, LTBP-1 trunc formed a covalent complex with LAP, which was evident at 100 kDa (Fig. 4C, lane 3). In line with its positive effect on TGF-β1 production, the R58A variant consistently bound LTBP-1 with high affinity (Fig. 4C, lane 8). In contrast, alanine substitution of Arg45, Arg50, and Lys56 in LAP disrupted covalent complex formation with LTBP-1 (Fig. 4C, lanes 4, 5, and 7). Western blot using antibodies specific for TGF-β1 (Fig. 4D) revealed that the absence of LAP·LTBP complex formation was not due to a loss of TGF-β1 expression.
LAP Residues That Mediate Assembly of the Large Latent Complex Form Part of an α-Helix
The binding epitopes for mature TGF-β1 and LTBP-1 overlap near the N terminus of LAP (Arg45–Leu59). Protein structure prediction (QuickPhyre, Imperial College, London) indicated that this region of LAP encompasses an α-helix, and modeling (PyMOL Molecular Graphics System) predicted that the helical domain was amphipathic (Fig. 5). Hydrophobic LAP residues (Ile46 Ile53, Leu54, Leu57, and Leu59), which bind to TGF-β1, formed a contiguous epitope on one surface of the α-helix, whereas ionic residues (Arg45, Arg50, Lys56, and Arg58) that interact with LTBP-1 are mostly clustered on the opposite surface (Fig. 5, A and B). Interestingly, sequence alignment indicated that the α-helical region is almost completely conserved in the prodomains of other latent human TGF-β superfamily members (TGF-β2, TGF-β3, myostatin, and GDF-11) (Fig. 5C).
FIGURE 5.
LAP residues (Met38–Leu57) form an amphipathic α-helix. The N terminus of LAP is predicted to form an amphipathic α-helix (A). In this model, hydrophobic residues Ile46, Ile53, Leu54, Leu57, and Leu59 present along one face of the helix, to enable LAP interactions with the mature TGF-β1 dimer. Charged residues, Arg45, Arg50, Lys56, and Arg58, that mediate LAP interactions with LTBP-1, are mostly clustered on the other side of the helix (B). Sequence alignment of the prodomains of TGF-β ligands highlighting the conservation of residues involved in binding to mature ligands (red) and LTBP-1 (blue) (C).
LAP Residues Distant from the N-terminal α-Helix Affect Stability of the Latent TGF-β1 Complex
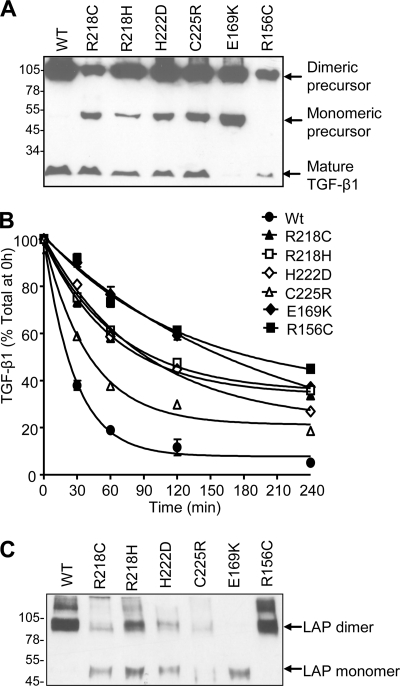
Missense mutations in LAP increase activation of TGF-β1 and cause the rare bone disorder, CED (35, 41). Cells transfected with the CED mutants R156C and E169K expressed substantially lower levels of total TGF-β1 (29 and 7 ng/ml, respectively) compared with wild type (118 ng/ml), whereas the R218C, R218H, H222D, and C225R variants produced slightly lower levels of total TGF-β1 (Fig. 6 A and Table 2). Each of the missense mutations, however, resulted in higher levels of active TGF-β1 in the conditioned medium compared with controls (Table 2). We sought to understand the mechanism for the decreased latency of these TGF-β1 variants. Conditioned medium from transfected cells was treated with acid to release mature TGF-β1 from the SLC and then neutralized. The ability of LAP to reassociate with TGF-β1 was monitored over time by TGF-β1 ELISA (Fig. 6B). The data were analyzed using one-phase decay parameters in the Prism program. Based on this, the half-life (i.e. time taken for 50% of mature TGF-β ligand to reassociate with LAP) was calculated. In contrast to wild type LAP (half-life of 19 min) (Fig. 6B), the CED LAP variants were compromised in their ability to bind mature TGF-β1, requiring longer periods of incubation for the SLC to reform (half-life ranging from 28 min for C225R to 138 min for E169K) (Fig. 6B). These results indicate that the CED mutations disrupt the interaction between LAP and TGF-β1 and subsequent stability of the latent complex.
FIGURE 6.
Analysis of the effect of CED mutations on TGF-β1 biosynthesis and latent complex stability. Conditioned medium from cells transfected with wild type (WT) or CED variants of TGF-β1 was initially analyzed by Western blot using a TGF-β1 mAb (A). Subsequently, medium was treated with acid to release mature TGF-β1 from the SLC and then neutralized. The ability of LAP to reassociate with TGF-β1 was monitored over time by a TGF-β1 ELISA (B). The presence of monomeric TGF-β1 precursor in the medium of cells transfected with the CED variants (A) suggested that these mutations affect LAP dimerization. To address this possibility, wild type and CED mutant LAP variants were generated and expressed transiently in HEK-293T cells (C). Error bars, S.D.
TABLE 2.
Effect of CED-causing and dimer interface mutations on TGF-β1 production and latent complex stability
TGF-β1 levels were determined in the ELISA for each variant in triplicate; values represent mean ± S.D. ND, not determined.
| TGF-β1 variant | Active TGF-β1 | Total TGF-β1 | Percentage active |
|---|---|---|---|
| ng/ml | ng/ml | % | |
| CED-causing TGF-β1 variants | |||
| Wild type | 1.26 ± 0.11 | 118.5 ± 2.3 | 1.07 ± 0.09 |
| R156C | 1.08 ± 0.00 | 28.9 ± 0.60a | 3.71 ± 0.00a |
| E169K | 1.26 ± 0.17 | 6.83 ± 0.09a | 18.50 ± 2.43a |
| R218C | 7.05 ± 0.16 | 70.4 ± 0.52a | 10.01 ± 0.22 |
| R218H | 4.85 ± 0.18 | 86.1 ± 1.7a | 5.64 ± 0.21a |
| H222D | 5.23 ± 0.23 | 68.7 ± 1.7a | 7.61 ± 0.33a |
| C225R | 3.29 ± 0.13 | 97.0 ± 5.8a | 3.39 ± 0.13a |
| Dimer interface TGF-β1 variants | |||
| Wild type | 3.03 ± 0.12 | 171.5 ± 1.6 | 1.77 ± 0.07 |
| W195A | 2.17 ± 0.24 | 107.5 ± 5.3a | 2.02 ± 0.23 |
| F198A | 1.10 ± 0.09 | 45.8 ± 0.82a | 2.41 ± 0.20b |
| D199A | 0.84 ± 0.08 | 43.2 ± 1.5a | 1.94 ± 0.19 |
| V200A | 1.02 ± 0.05 | 90.5 ± 1.2a | 1.13 ± 0.05a |
| G203A | 1.64 ± 0.04 | 168.0 ± 5.2 | 0.98 ± 0.03a |
| Q207A | 2.12 ± 0.11 | 141.4 ± 15.2 | 1.50 ± 0.08b |
| W208A | 1.62 ± 0.08 | 96.5 ± 2.8a | 1.68 ± 0.08 |
| L209A | 2.15 ± 0.08 | 101.2 ± 1.7a | 2.12 ± 0.07b |
| S210A | 2.05 ± 0.16 | 150.6 ± 4.2b | 1.36 ± 0.10b |
| F217A | <0.6 | 12.14 ± 0.18a | ND |
| L219A | 2.65 ± 0.16 | 129.5 ± 3.8a | 2.05 ± 0.12b |
a p < 0.01.
b p < 0.05.
Interestingly, four of the identified mutations lie within a short stretch of 8 residues (Arg218–Cys225), which encompasses the cysteine residues (Cys223 and Cys225) involved in LAP dimerization (10). The presence of monomeric TGF-β1 precursor in the medium of cells transfected with the CED variants (Fig. 6A) suggested that these mutations affect LAP dimerization. To address this, wild type and CED mutant LAP variants were generated and expressed transiently in HEK-293T cells. Wild type LAP and the R156C mutant were expressed in dimeric form (95 kDa); however, the other LAP variants were expressed poorly and as a mixture of monomer and dimer (Fig. 6C). Thus, most CED mutations disrupt the covalent dimerization of LAP, which, in the context of full-length TGF-β1, results in a more rapid dissociation of the small latent complex into its constituent components.
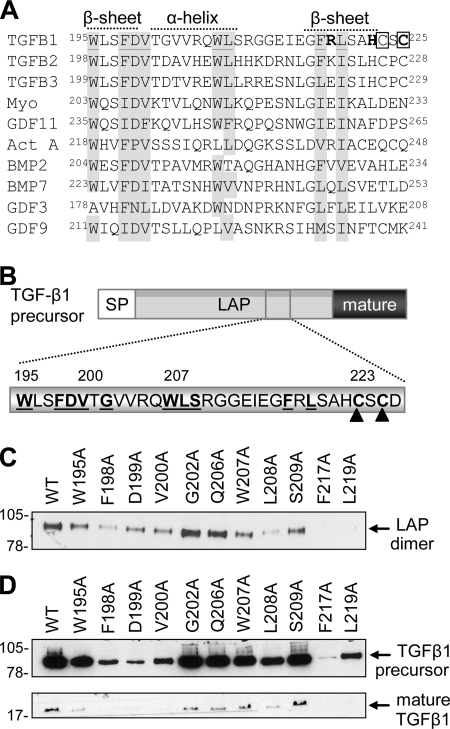
A Hydrophobic Domain at the C Terminus of LAP Mediates Dimerization
We analyzed the C-terminal region of LAP to identify residues that form the dimer interface. Sequence alignment of the TGF-β isoforms (Fig. 7A) identified a hydrophobic domain (Trp196–Leu219) that is highly conserved and neighbors the cysteine residues (Cys223 and Cys225) involved in interchain disulfide bond formation. Alanine mutants were generated across this region (Fig. 7B) and assessed for their effects on LAP dimerization (Fig. 7C) and TGF-β1 production (Fig. 7D and Table 2). Western blot analysis indicated that Phe198, Asp199, Val200, Leu208, Phe217, and Leu219 were critical for LAP dimerization, whereas Trp195 and Trp207 were peripherally involved (Fig. 7C). When these mutations were incorporated into full-length TGF-β1 constructs, they reduced mature TGF-β1 expression in line with their effects on LAP dimerization (Fig. 7D and Table 2). Mutation of non-conserved residues within this region, Gly202, Gln206, and Ser210, had little effect on TGF-β1 production or LAP biosynthesis (Fig. 7, C and D). Interestingly, the hydrophobic residues involved in LAP dimerization are conserved across the TGF-β superfamily (Fig. 7A), suggesting that prodomain dimerization is a common phenomenon.
FIGURE 7.
Characterization of the LAP dimerization interface. Sequence alignment of the prodomains of TGF-β ligands highlighting conserved residues near the C terminus (shaded gray) (A). In addition, the genetic mutations in LAP associated with CED are highlighted (boldface type), as are the cysteine residues that form interchain disulfide bonds (boxed). Conserved residues (boldface type, underlined) were mutated to alanine by in vitro mutagenesis (B), and their effect on LAP dimerization (C) and TGF-β1 production (D) was determined by Western blot analysis using the FLAG M2 mAb or a TGF-β1 mAb, respectively. SP, signal peptide. WT, wild type.
DISCUSSION
TGF-β1 is secreted as part of an LLC consisting of the mature growth factor, the TGF-β1 prodomain (LAP), and a molecule of LTBP (7, 15). After secretion, the LLC binds to the extracellular matrix via the N-terminal domain of LTBP, and this interaction is essential for latent TGF-β activation (25, 26). The absence of LTBPs leads to cardiac (Ltbp1L−/−), craniofacial (Ltbp3−/−), and pulmonary and myocardial (Ltbp4−/−) defects, which highlights the importance of these multifunctional proteins in normal TGF-β signaling (42–46). Furthermore, disruption of the covalent association between LAP and LTBPs in vivo (Tgfb1C33S/C33S mice) recapitulates most aspects of the Tgfb1−/− mouse phenotype, including multiorgan inflammation, lack of skin Langerhans cells, and a shortened life span (40). Thus, formation of the LLC is critical for regulating TGF-β bioavailability.
In this study, we characterized the residues in LAP that coordinate the formation and stability of the large latent TGF-β1 complex. Using in vitro mutagenesis, we identified a hydrophobic domain at the N terminus of LAP (Ile53–Leu59) that is essential for formation of the SLC. This domain encompasses the 54LSKL motif that was previously shown to be important for LAP induction of latency and binding of the TGF-β1-activating molecule, thrombospondin-1 (4, 47). LAP residues Ile53, Leu54, Leu57, and Leu59 mediate high affinity binding to TGF-β1, which shields the type II receptor binding sites in the mature growth factor and maintains TGF-β1 in a latent form. The 53ILSKLRL59 domain is almost completely conserved in the propeptides of other latent human TGF-β superfamily members (TGF-β2, TGF-β3, myostatin, and GDF-11), with the only divergence being the substitution of the arginine for a lysine in TGF-β2. Thus, the 53ILSKLRL59 domain probably represents the optimal high affinity binding site for mature TGF-β ligands, and propeptides that contain this sequence remain complexed with their respective growth factors, even in the presence of signaling receptors. In contrast, although the hydrophobic nature of this domain is maintained in the other 28 TGF-β ligands (6), they invariably lack one or both of the ionic residues corresponding to Lys56 and Arg58 of LAP. Thus, variation of these key ionic residues presumably reduces the affinity of activin, bone morphogenetic protein, and growth and differentiation factor propeptides for their mature growth factors and ensures that these ligands are secreted in an “active” form.
Within the large latent complex, LAP and LTBPs are linked by covalent bonds between Cys33 in each of two LAP chains and a pair of cysteine residues in LTBP (24, 25). Chen et al. (39) identified the five negatively charged residues in LTBP-1 (Glu1022, Asp1029, Asp1034, Asp1056, and Glu1059) that mediate initial non-covalent interactions with LAP. In the current study, we identified the corresponding positively charged residues on LAP (Arg45, Arg50, Lys56, and Arg58) that form the binding epitope for LTBP-1. These results explain the previous findings of Young and Murphy-Ulrich (4), who showed that Lys56 is essential for TGF-β1 complex formation and secretion. Interestingly, LAP could only associate with LTBP-1 in the presence of mature TGF-β1 (data not shown), suggesting that the LLC assembles cooperatively through recruitment of LTBPs to the SLC. In support, an earlier study showed that extensive conformational changes occurred in LAP upon formation of the SLC (48). The identified LTBP-1 binding motif on LAP is highly conserved among latent TGF-β ligands (i.e. Arg45 and Lys56 (TGF-β1 numbering) are invariant, Arg50 is substituted for a Lys in myostatin and GDF-11, and Arg58 is a Lys in TGF-β2). The importance of LTBPs as extracellular regulators of TGF-β activity has been well described (25, 49–51); however, little is known about interactions between LTBPs and other superfamily members. A recent study indicated that LTBP-3 associates non-covalently with latent myostatin and that this interaction sequesters myostatin within the extracellular matrix (52). Our data would indicate that this interaction is mediated by ionic residues in the N-terminal region of the myostatin propeptide. Based on sequence conservation, latent GDF-11 would also be anticipated to bind LTBPs via its propeptide. The LTBP-1 binding motif is poorly conserved among other TGF-β ligands, and, although LTBP-1 has been shown to synergize with activin and nodal to induce mesoderm in Xenopus (53), it is unlikely that this activity is mediated via a direct interaction.
The juxtaposition of mature TGF-β1 and LTBP-1 binding sites on LAP led to an analysis of the secondary structure of this region of the molecule. Protein structure prediction indicated that LAP residues (Met38–Leu57) form an amphipathic α-helix. The helix probably orientates such that key hydrophobic residues (Ile53, Leu54, and Leu57) contact residues in the “finger” domains of the mature dimer to facilitate ligand folding and complex formation, as previously observed for inhibin and activin (6). The exposed cationic surface of the α-helix would then be predicted to contact anionic residues in LTBPs and mediate covalent bond formation. Disruption of LAP binding to mature TGF-β1 by mutation of Ile53, Leu54, or Leu57 inhibited formation of the SLC. In contrast, TGF-β1 variants (R45A, R50A, and K56A) unable to bind LTBP-1 formed the SLC, but it was misfolded due to an aberrant covalent bond between Cys33 of LAP and a cysteine in the mature growth factor. Together, these results highlight the importance of the N-terminal α-helix of LAP in coordinating the assembly of the large latent TGF-β1 complex. Secondary structure analysis of the N-terminal region of all other TGF-β prodomains indicated that an α-helix is likely to be present. The conserved hydrophobic surface of these helices would govern the assembly and secretion of the mature growth factors, whereas the opposite exposed surface could be involved in interactions with accessory proteins. In support, preliminary studies in our laboratory suggest that this region of the BMP-7 (bone morphogenetic protein-7) prodomain mediates interactions with fibrillin (54, 55), an LTBP-related extracellular matrix protein required for elastic fiber formation.
Surprisingly, the N-terminal α-helix alone is not sufficient to maintain stability of the latent TGF-β1 complex. Brunner et al. (10) showed that mutation of Cys223 and Cys225 involved in LAP dimerization resulted in the secretion of bioactive TGF-β1. Moreover, naturally occurring mutations that involve these cysteines (C225R) or surrounding residues (R218C, R218H, and H222D) lead to increased TGF-β1 signaling and cause the rare bone disorder CED (29, 41). We showed that these CED mutations disrupted the covalent dimerization of LAP, which, in the context of full-length TGF-β1, resulted in a more rapid dissociation of the small latent complex into its constituent components. Interestingly, the TGF-β isoforms are the only family members in which prodomains are covalently linked. It is postulated that this adaptation was necessary to increase the affinity of LAP for mature TGF-β1 above that of the TGF-β receptor complex. Myostatin and GDF-11, which have significantly lower affinity for their signaling receptors than the TGF-β isoforms (20, 56, 57) remain associated with their prodomains even in the absence of covalent bonds.
Given the importance of LAP dimerization for TGF-β1 latency, we sought to identify the residues that form the dimer interface and thereby facilitate covalent bond formation. Within the vicinity of Cys223 and Cys225, protein structure analysis predicted the presence of two β-strands (Trp195–Val200 and Gly216–His222) separated by an α-helix (Thr201–Ser209). Because these structural elements often mediate protein interactions, we performed site-directed mutagenesis across this region and showed that Phe198, Asp199, Val200, Leu208, Phe217, and Leu219 are essential for LAP dimerization. The characterization of the LAP dimer interface enhances our understanding of the molecular etiology of Camurati-Engelmann disease, particularly in relation to Arg218, which lies between two critical hydrophobic residues and is a CED mutation hot spot (representing 60% of all mutations) (58). Interestingly, the residues involved in LAP dimerization are highly conserved across the TGF-β superfamily, suggesting that prodomain dimerization is a common phenomenon required for growth factor assembly. In support, mutations across this region in GDF-9 (V216M and T238A) and BMP-15 (A180T, N196K, and R206H) have been associated with premature ovarian failure (59–61).
Our comprehensive structure-function analysis of the TGF-β1 prodomain has identified two distinct regions that coordinate assembly of the latent complex. Residues at the N terminus of LAP mediate formation of the large latent complex by binding to TGF-β1 (Ile53–Leu59) and LTBPs (Arg45, Arg50, Lys56, and Arg58). At the opposing C terminus, hydrophobic residues (Trp195–Cys225) promote LAP dimerization. Importantly, despite low sequence homology (∼15%) between prodomains of TGF-β ligands, these structurally significant regions are broadly conserved. Future studies will seek to understand how mutations in LAP (Y81H, R156C, and E169K) outside the regions defined in this study affect the assembly, secretion, and activation of TGF-β1 and cause Camurati-Engelmann disease.
Acknowledgment
We thank Dr. Mark Gibson for providing the LTBP-1 construct.
This work was supported by National Health and Medical Research Council of Australia Project 494804 (to C. A. H.) and Program Grant 241000 (to K. L. W. and D. M. R.) and by the Victorian Government's Operational Infrastructure Support Program.
- TGF
- transforming growth factor
- LAP
- latency-associated peptide
- LTBP
- latent TGF-β binding protein
- LLC
- large latent complex
- SLC
- small latent complex
- CED
- Camurati-Engelmann disease
- mAb
- monoclonal antibody
- HEK-293T
- human renal epithelial cell
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Taipale J., Saharinen J., Keski-Oja J. (1998) Adv. Cancer Res. 75, 87–134 [DOI] [PubMed] [Google Scholar]
- 2.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 3.Gordon K. J., Blobe G. C. (2008) Biochim. Biophys. Acta 1782, 197–228 [DOI] [PubMed] [Google Scholar]
- 4.Young G. D., Murphy-Ullrich J. E. (2004) J. Biol. Chem. 279, 38032–38039 [DOI] [PubMed] [Google Scholar]
- 5.Derynck R., Akhurst R. J., Balmain A. (2001) Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 6.Walton K. L., Makanji Y., Wilce M. C., Chan K. L., Robertson D. M., Harrison C. A. (2009) J. Biol. Chem. 284, 9311–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazono K., Hellman U., Wernstedt C., Heldin C. H. (1988) J. Biol. Chem. 263, 6407–6415 [PubMed] [Google Scholar]
- 8.Assoian R. K., Komoriya A., Meyers C. A., Miller D. M., Sporn M. B. (1983) J. Biol. Chem. 258, 7155–7160 [PubMed] [Google Scholar]
- 9.Frolik C. A., Dart L. L., Meyers C. A., Smith D. M., Sporn M. B. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 3676–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner A. M., Marquardt H., Malacko A. R., Lioubin M. N., Purchio A. F. (1989) J. Biol. Chem. 264, 13660–13664 [PubMed] [Google Scholar]
- 11.Dubois C. M., Laprise M. H., Blanchette F., Gentry L. E., Leduc R. (1995) J. Biol. Chem. 270, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 12.Gentry L. E., Lioubin M. N., Purchio A. F., Marquardt H. (1988) Mol. Cell Biol. 8, 4162–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentry L. E., Nash B. W. (1990) Biochemistry 29, 6851–6857 [DOI] [PubMed] [Google Scholar]
- 14.Sha X., Yang L., Gentry L. E. (1991) J. Cell Biol. 114, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakefield L. M., Smith D. M., Flanders K. C., Sporn M. B. (1988) J. Biol. Chem. 263, 7646–7654 [PubMed] [Google Scholar]
- 16.Böttinger E. P., Factor V. M., Tsang M. L., Weatherbee J. A., Kopp J. B., Qian S. W., Wakefield L. M., Roberts A. B., Thorgeirsson S. S., Sporn M. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5877–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Crescenzo G., Grothe S., Zwaagstra J., Tsang M., O'Connor-McCourt M. D. (2001) J. Biol. Chem. 276, 29632–29643 [DOI] [PubMed] [Google Scholar]
- 18.Ge G., Hopkins D. R., Ho W. B., Greenspan D. S. (2005) Mol. Cell Biol. 25, 5846–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill J. J., Davies M. V., Pearson A. A., Wang J. H., Hewick R. M., Wolfman N. M., Qiu Y. (2002) J. Biol. Chem. 277, 40735–40741 [DOI] [PubMed] [Google Scholar]
- 20.Lee S. J., McPherron A. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thies R. S., Chen T., Davies M. V., Tomkinson K. N., Pearson A. A., Shakey Q. A., Wolfman N. M. (2001) Growth Factors 18, 251–259 [DOI] [PubMed] [Google Scholar]
- 22.Sengle G., Ono R. N., Lyons K. M., Bächinger H. P., Sakai L. Y. (2008) J. Mol. Biol. 381, 1025–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rifkin D. B. (2005) J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
- 24.Gleizes P. E., Beavis R. C., Mazzieri R., Shen B., Rifkin D. B. (1996) J. Biol. Chem. 271, 29891–29896 [DOI] [PubMed] [Google Scholar]
- 25.Saharinen J., Taipale J., Keski-Oja J. (1996) EMBO J. 15, 245–253 [PMC free article] [PubMed] [Google Scholar]
- 26.Annes J. P., Chen Y., Munger J. S., Rifkin D. B. (2004) J. Cell Biol. 165, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z., Mu Z., Dabovic B., Jurukovski V., Yu D., Sung J., Xiong X., Munger J. S. (2007) J. Cell Biol. 176, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young G. D., Murphy-Ullrich J. E. (2004) J. Biol. Chem. 279, 47633–47642 [DOI] [PubMed] [Google Scholar]
- 29.Janssens K., Gershoni-Baruch R., Guañabens N., Migone N., Ralston S., Bonduelle M., Lissens W., Van Maldergem L., Vanhoenacker F., Verbruggen L., Van Hul W. (2000) Nat. Genet. 26, 273–275 [DOI] [PubMed] [Google Scholar]
- 30.Dietz H. C., Cutting G. R., Pyeritz R. E., Maslen C. L., Sakai L. Y., Corson G. M., Puffenberger E. G., Hamosh A., Nanthakumar E. J., Curristin S. M. (1991) Nature 352, 337–339 [DOI] [PubMed] [Google Scholar]
- 31.Le Goff C., Morice-Picard F., Dagoneau N., Wang L. W., Perrot C., Crow Y. J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., Ge G., Greenspan D. S., Bonnet D., Le Merrer M., Munnich A., Apte S. S., Cormier-Daire V. (2008) Nat. Genet. 40, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeys B. L., Chen J., Neptune E. R., Judge D. P., Podowski M., Holm T., Meyers J., Leitch C. C., Katsanis N., Sharifi N., Xu F. L., Myers L. A., Spevak P. J., Cameron D. E., De Backer J., Hellemans J., Chen Y., Davis E. C., Webb C. L., Kress W., Coucke P., Rifkin D. B., De Paepe A. M., Dietz H. C. (2005) Nat. Genet. 37, 275–281 [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita A., Saito T., Tomita H., Makita Y., Yoshida K., Ghadami M., Yamada K., Kondo S., Ikegawa S., Nishimura G., Fukushima Y., Nakagomi T., Saito H., Sugimoto T., Kamegaya M., Hisa K., Murray J. C., Taniguchi N., Niikawa N., Yoshiura K. (2000) Nat. Genet. 26, 19–20 [DOI] [PubMed] [Google Scholar]
- 34.Wu S., Liang S., Yan Y., Wang Y., Li F., Deng Y., Huang W., Yuan W., Luo N., Zhu C., Wang Y., Li Y., Liu M., Wu X. (2007) Bone 40, 1630–1634 [DOI] [PubMed] [Google Scholar]
- 35.Janssens K., ten Dijke P., Ralston S. H., Bergmann C., Van Hul W. (2003) J. Biol. Chem. 278, 7718–7724 [DOI] [PubMed] [Google Scholar]
- 36.Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T. R., Peng X., Hu J., Feng X., Van Hul W., Wan M., Cao X. (2009) Nat. Med. 15, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F., Pouponnot C., Massagué J. (1997) Genes Dev. 11, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y. T., Liu J. J., Luo Y. E. C., Haltiwanger R. S., Abate-Shen C., Shen M. M. (2002) Mol. Cell Biol. 22, 4439–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Ali T., Todorovic V., O'leary J. M., Kristina Downing A., Rifkin D. B. (2005) J. Mol. Biol. 345, 175–186 [DOI] [PubMed] [Google Scholar]
- 40.Yoshinaga K., Obata H., Jurukovski V., Mazzieri R., Chen Y., Zilberberg L., Huso D., Melamed J., Prijatelj P., Todorovic V., Dabovic B., Rifkin D. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18758–18763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito T., Kinoshita A., Yoshiura K., Makita Y., Wakui K., Honke K., Niikawa N., Taniguchi N. (2001) J. Biol. Chem. 276, 11469–11472 [DOI] [PubMed] [Google Scholar]
- 42.Dabovic B., Chen Y., Colarossi C., Obata H., Zambuto L., Perle M. A., Rifkin D. B. (2002) J. Cell Biol. 156, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dabovic B., Chen Y., Colarossi C., Zambuto L., Obata H., Rifkin D. B. (2002) J. Endocrinol. 175, 129–141 [DOI] [PubMed] [Google Scholar]
- 44.Dabovic B., Levasseur R., Zambuto L., Chen Y., Karsenty G., Rifkin D. B. (2005) Bone 37, 25–31 [DOI] [PubMed] [Google Scholar]
- 45.Drews F., Knöbel S., Moser M., Muhlack K. G., Mohren S., Stoll C., Bosio A., Gressner A. M., Weiskirchen R. (2008) Biochim. Biophys. Acta 1783, 34–48 [DOI] [PubMed] [Google Scholar]
- 46.Sterner-Kock A., Thorey I. S., Koli K., Wempe F., Otte J., Bangsow T., Kuhlmeier K., Kirchner T., Jin S., Keski-Oja J., von Melchner H. (2002) Genes Dev. 16, 2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribeiro S. M., Poczatek M., Schultz-Cherry S., Villain M., Murphy-Ullrich J. E. (1999) J. Biol. Chem. 274, 13586–13593 [DOI] [PubMed] [Google Scholar]
- 48.McMahon G. A., Dignam J. D., Gentry L. E. (1996) Biochem. J. 313, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dallas S. L., Miyazono K., Skerry T. M., Mundy G. R., Bonewald L. F. (1995) J. Cell Biol. 131, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taipale J., Miyazono K., Heldin C. H., Keski-Oja J. (1994) J. Cell Biol. 124, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taipale J., Saharinen J., Hedman K., Keski-Oja J. (1996) J. Histochem. Cytochem. 44, 875–889 [DOI] [PubMed] [Google Scholar]
- 52.Anderson S. B., Goldberg A. L., Whitman M. (2008) J. Biol. Chem. 283, 7027–7035 [DOI] [PubMed] [Google Scholar]
- 53.Altmann C. R., Chang C., Muñoz-Sanjuán I., Bell E., Heke M., Rifkin D. B., Brivanlou A. H. (2002) Dev. Biol. 248, 118–127 [DOI] [PubMed] [Google Scholar]
- 54.Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. (2005) J. Biol. Chem. 280, 27970–27980 [DOI] [PubMed] [Google Scholar]
- 55.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin H. Y., Moustakas A., Knaus P., Wells R. G., Henis Y. I., Lodish H. F. (1995) J. Biol. Chem. 270, 2747–2754 [DOI] [PubMed] [Google Scholar]
- 57.Lin H. Y., Wang X. F., Ng-Eaton E., Weinberg R. A., Lodish H. F. (1992) Cell 68, 775–785 [DOI] [PubMed] [Google Scholar]
- 58.Janssens K., Vanhoenacker F., Bonduelle M., Verbruggen L., Van Maldergem L., Ralston S., Guañabens N., Migone N., Wientroub S., Divizia M. T., Bergmann C., Bennett C., Simsek S., Melançon S., Cundy T., Van Hul W. (2006) J. Med. Genet. 43, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixit H., Rao L. K., Padmalatha V., Kanakavalli M., Deenadayal M., Gupta N., Chakravarty B., Singh L. (2005) Menopause 12, 749–754 [DOI] [PubMed] [Google Scholar]
- 60.Dixit H., Rao L. K., Padmalatha V. V., Kanakavalli M., Deenadayal M., Gupta N., Chakrabarty B., Singh L. (2006) Hum. Genet. 119, 408–415 [DOI] [PubMed] [Google Scholar]
- 61.Zhao H., Qin Y., Kovanci E., Simpson J. L., Chen Z. J., Rajkovic A. (2007) Fertil. Steril. 88, 1474–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]