Abstract
The S6 kinases (S6Ks) have been linked to a number of cellular processes, including translation, insulin metabolism, cell survival, and RNA splicing. Signaling via the phosphotidylinositol 3-kinase and mammalian target of rapamycin (mTOR) pathways is critical in regulating the activity and subcellular localization of S6Ks. To date, nuclear functions of both S6K isoforms, S6K1 and S6K2, are not well understood. To better understand S6K nuclear roles, we employed affinity purification of S6Ks from nuclear preparations followed by mass spectrometry analysis for the identification of novel binding partners. In this study, we report that in contrast to S6K1, the S6K2 isoform specifically associates with a number of RNA-binding proteins, including heterogeneous ribonucleoproteins (hnRNPs). We focused on studying the mechanism and physiological relevance of the S6K2 interaction with hnRNP F/H. Interestingly, the S6K2-hnRNP F/H interaction was not affected by mitogenic stimulation, whereas mTOR binding to hnRNP F/H was induced by serum stimulation. In addition, we define a new role of hnRNP F in driving cell proliferation, which could be partially attenuated by rapamycin treatment. S6K2-driven cell proliferation, on the other hand, could be blocked by small interfering RNA-mediated down-regulation of hnRNP F. These results demonstrate that the specific interaction between mTOR and S6K2 with hnRNPs is implicated in the regulation of cell proliferation.
Keywords: Cell/Intracellular Processing, Nucleus, Phosphorylation/Kinases/Serine-Threonine, Protein/Binding/RNA, RNA/Ribonuclear Protein RNP, Signal Transduction
Introduction
Regulation of cellular processes by extracellular stimuli, such as growth factors and nutrients, is an essential process in cell size and cell cycle control. The mammalian target of rapamycin (mTOR)2 is a key regulator of cellular signaling and is highly dependent on the presence of nutrients, as well as growth factor-stimulated signaling from the phosphotidylinositol 3-kinase (PI3K) pathway. Activation of PI3K by a number of growth factor receptors leads to increased levels of phosphatidylinositol 3,4,5-triphosphate (1), a secondary messenger that recruits downstream kinases, such as 3-phosphoinositide-dependent kinase 1 (PDK1), to the plasma membrane (2). Membrane-associated PDK1 in turn phosphorylates and activates Akt at the T loop (3, 4), whereas activated Akt phosphorylation of tuberous sclerosis complex 2 (TSC2) blocks its inhibitory role in mTOR signaling (5, 6). TSC2 forms a heterodimer with TSC1, which as a complex acts as a GTPase activator toward the small GTPase, Rheb (Ras homolog enriched in brain) (7, 8). Rheb associates directly with the catalytic domain of mTOR and activates it by antagonizing FKBP38, the proposed endogenous inhibitor of mTOR (9). Activated mTOR with its interacting partner protein Raptor, as part of the mTOR complex I (mTORC1) is able to phosphorylate S6Ks (at Thr389 in S6K1 and Thr388 in S6K2) (10). This early phosphorylation event is required for subsequent phosphorylation by PDK1 of Thr229 and Thr228 in the activation loops of S6K1 and S6K2, respectively (11). Phosphorylation by both mTORC1 and PDK1 leads to S6K activation, resulting in full activation of S6Ks and phosphorylation of its downstream targets.
mTOR and its downstream effector S6 kinase (S6K) have been methodically demonstrated to regulate cell size in both mouse (12) and Drosophila (13) knockout studies. More recently, it was also demonstrated that the mTOR kinase and S6K2 have the ability to regulate cell proliferation and cell cycle progression (15, 16). Most studies on S6K regulation and function focused on S6K1, establishing that S6K1 is able to regulate a number of key cellular processes, including insulin metabolism (17, 18), cell survival (19, 20), translation (21–24), and more recently RNA splicing (25). Inactive S6K1 was reported to localize on the translational preinitiation complex through its interaction with eukaryotic initiation factor 3. Upon stimulation, mTOR arrives at the preinitiation complex leading to phosphorylation and activation of S6K1. As such, S6K1 is part of the cytoplasmic translational preinitiation complex (24).
Of the two isoforms of S6Ks, S6K2 harbors a C-terminal nuclear localization sequence that is absent in S6K1 (26–28), which indicates a distinct nuclear role of S6K2 from that of S6K1. Nuclear/cytoplasmic shuttling of S6Ks could be regulated by upstream kinases. The protein kinase CK2 phosphorylates S6K1 at Ser17, and this signaling event induces nuclear export of S6K1 (29). Similarly, phosphorylation of Ser486 located within the C-terminal nuclear localization sequence of S6K2 by protein kinase C leads to cytoplasmic retention of S6K2 (30). Known S6K functions in the nucleus include phosphorylation of the cAMP response element modulator CREMτ (31) and, more importantly, regulation of cap-dependent RNA splicing through S6K1 interaction with the SKAR (S6K1 Aly, Ref-like target) protein (25).
Deregulation of S6K signaling has been linked to various human pathologies, including cancer and diabetes. Gene amplification and overexpression at mRNA and protein levels were reported for both S6K1 and S6K2 in various malignancies. Interestingly, overexpression and higher nuclear localization of S6K2 were observed in human breast cancer (14) and also were linked to pro-survival and chemoresistance signaling in small cell lung cancer (20).
Heterogeneous ribonucleoproteins (hnRNPs) were first identified as a large group of chromatin-associated RNA-binding proteins. Early studies identified six core hnRNPs that indiscriminately associate with nascent RNA polymerase II transcripts and are packaged to form hnRNP particles (32, 33). Subsequently, it was further determined that the hnRNP-hnRNA interaction is far more complex than previously envisaged by the isolation of more hnRNPs associating with RNA, along with the observation that each individual hnRNP contains RNA-binding motifs that exhibit different RNA sequence binding preferences (34). Although first suggested to function as a large macromolecular complex, more recent reports on hnRNPs have been limited to individual hnRNP effects, including RNA splicing, internal ribosome entry site translation, RNA polymerase II regulation, and telomeric regulation (for review see Ref. 35).
Although hnRNPs F and H are two hnRNPs with high structural homology, they are expressed from distinct genes (36). Both hnRNPs F and H have three RNA recognition motifs with binding preferences of GGGA (37) or DGGGD (where D represents a U, G, or A nucleotide) motifs (38). In addition, hnRNP F/H have an extensive glycine-rich domain near the C terminus, which may facilitate hetero- or homodimerization between hnRNP F and H (39). hnRNP F/H are able to catalyze splicing events in both viral (40–43) and nonviral transcripts (44–46) and have a possible role in transcription through their association with RNA polymerase II (46) and the TATA-binding protein (47).
Here, we present for the first time a novel interactive and functional nuclear complex of the signaling kinases, mTOR, and S6K2 with heterogeneous ribonucleoproteins. First, affinity co-immunoprecipitation followed by mass spectrometry allowed us to identify a number of RNA-binding proteins associating with nuclear S6K2. We further demonstrated that S6K2 and mTOR-Raptor specifically associate with several hnRNPs as part of a macromolecular complex in vivo. The interaction of hnRNP F/H with mTOR, but not S6K2, was inducible by the addition of fetal bovine serum, which could be abolished by pretreatment of cells with the mTOR inhibitor, rapamycin. In the course of this study, we defined a new functional role for hnRNP F in the regulation of cell proliferation. Overexpression of hnRNP F, but not hnRNP H, promotes cell proliferation, with a reverse effect observed in siRNA knockdown of hnRNP F. Furthermore, hnRNP F-driven cell proliferation was observed to be mTOR-dependent because the addition of rapamycin, which blocks mTOR association with hnRNP F, had an inhibitory effect on the hnRNP F-driven cell proliferation. These results suggest that both S6K2 and mTOR function in the nucleus along with hnRNP F to regulate cell proliferation via a mechanism that is yet to be determined.
EXPERIMENTAL PROCEDURES
Materials Used for Cell Treatment
Rapamycin (25 nm), LY294002 (20 μm), and PD 098059 (10 μm) were obtained from LC Laboratories. Cisplatin was purchased from Sigma.
Antibodies and Sera
Anti-hnRNP F/H (sc-32310), anti-hnRNP C1/C2 (sc-32308), and anti-tubulin (H-235) antibodies were acquired from Santa Cruz Biotechnologies. Anti-hnRNP U antibodies (R6278) and anti-actin β antibodies (AC15) were acquired from Sigma-Aldrich. Anti-lamin (2032) antibody was purchased from Cell Signaling Technologies.
Generation of Tetracycline-inducible S6K Cell Lines
p70S6K1 and p54S6K2 cDNA sequences with the N-terminal EE (Glu-Glu) tags were cloned into pcDNA4/TO-inducible plasmids followed by transfection using Exgen 500 transfection reagent (Fermentas) into a tetracycline-regulated expression system for mammalian cells (T-Rex system; Invitrogen). EE-p70S6K1- and EE-p54S6K2-expressing Hek293 T-Rex cells (termed EE-p70S6K1Hek293 and EE-p54S6K2Hek293, respectively) were selected in Dulbecco's modified Eagle's medium (DMEM; PAA Laboratories) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone), 5 μg/ml blasticidin (Sigma), and 100 μg/ml Zeocin (Sigma). EE tag monoclonal antibody is commercially available and commonly used in immunoprecipitation or immunoblot studies. Induction of exogenous protein expression was achieved by the addition of tetracycline at 2 μg/ml working volume 12 h prior to analysis.
Generation of Myc-hnRNP F and Myc-hnRNP H Stable Cell Lines
Full-length cDNA clones of human hnRNP F (IRATp970C01109D) and hnRNP H (IRAUp969E013D) were obtained from ImaGenes. hnRNP F with a Myc tag sequence introduced at the N terminus of the coding region was amplified by PCR) using the forward primer 5′-ATCTCTAGAAATGGAACAGAAGCTTATTTCCGAAGAGGATCTGATGCTGGGCCCTGAGGGA-3′ and the reverse primer 5′-ATCGGATCCCTAGTCATAGCCACCCAT-3′. Similarly, hnRNP H with an N-terminal Myc tag sequence was amplified by PCR with the forward primer 5′-ATCTCTAGAATGGAACAGAAGCTTATTTCCGAAGAGGATCTGATGATGTTGGGC-3′ and the reverse primer 5′-ATCGGATCCCTATGCAATGTTTGATTG-3′. The amplified sequences were digested with XbaI and BamHI restriction endonucleases (Fermentas) and cloned into pcDNA 3.1 (Invitrogen) mammalian expression vectors. Myc-hnRNP F and Myc-hnRNP H clones were transfected into Hek293 cells and selected by 800 μg/ml G-418 addition into cell culture growth medium (DMEM, 10% FBS). The resultant cell lines were termed Myc-hnRNPFHek293 and Myc-hnRNPHHek293.
Cell Culture
Hek293, MCF-7, A549, EE-p70S6K1Hek293, and EE-p54S6K2Hek293 cells were grown as a monolayer in DMEM supplemented with 10% (v/v) FBS and 5% (v/v) streptomycin/penicillin mix (PAA), in a humidified atmosphere of 10% CO2 at 37 °C. Myc-hnRNPFHek293 and Myc-hnRNPHHek293 cells were maintained as with previously described cell lines with the addition of G-418 to the growth medium at 200 μg/ml final concentration. For studies or the responses to serum stimulation, the cells were grown to 60% confluency, washed once with 1× phosphate-buffered solution, and fed with serum-free culture medium for 24 h prior to addition of FBS at a final 10% (v/v) concentration for the stated time points.
Cell Lysis and Fractionation
Total cell lysis was achieved by addition of lysis buffer (20 mm Tris-HCl, pH 7.5, 1% Triton X-100, 150 mm NaCl, 5 mm EDTA, 50 mm NaF, and protease inhibitor mixture; Roche Applied Science) to harvested cells on ice for 30 min, followed by centrifugation at 14,000 rpm for 30 min at 4 °C on a bench top centrifuge to pellet cellular debris. In cases where nuclear fractions was required, cultured cells were trypsinized and washed once with 1× phosphate-buffered solution, followed by resuspension in ice-cold hypotonic buffer (20 mm HEPES, 0.5 mm dithiothreitol, protease inhibitor mixture added, pH 7.9) for 15 min. 10% Nonidet P-40 was added after 15 min at a concentration of 40 μl/ml and vortexed for 10 s followed by centrifugation at 800 × g for 1 min, 4 °C. The resulting supernatant (cytoplasmic and cellular membrane fraction) was removed and saved separately on ice. The pelleted nuclei were washed once with hypotonic buffer supplemented with 240 μl/ml of 10% Nonidet P-40 with nuclei pelleted by centrifugation at 800 × g for 1 min, 4 °C. The nuclei were further washed and pelleted in hypotonic buffer alone. Ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 1% Triton X-100, 150 mm NaCl, 5 mm EDTA, 50 mm NaF, protease inhibitor mixture) was added to lyse nuclear pellet for 30 min at 4 °C, followed by centrifugation at top speed for 30 min at 4 °C on a bench top centrifuge to remove nuclear debris.
Immunoprecipitation
Immunoprecipitation of specific proteins was carried out using appropriate antibodies specific for the target protein. The antibodies were preincubated with protein A-Sepharose beads (Amersham Biosciences) for 2 h. Protein sample was added, and incubation was carried out at 4 °C on a rotating wheel overnight. Antibody-bounded beads were centrifuged at 450 × g on a bench top centrifuge for 2 min. The beads were washed with lysis buffer three times prior to SDS-PAGE analysis. In studies involving elution of EE tag antibody-bound nuclear complexes, protein A-Sepharose beads bound to EE antibody were washed three times with lysis buffer. An equal volume of lysis buffer containing 0.3 μg/μl EE peptide was added to the 50% antibody-protein A-Sepharose slurry. This was left to stand for 10 min at room temperature, with gentle agitation every 2 min to allow mixing. The suspension was centrifuged at 450 × g for 2 min on a bench-top centrifuge. The resultant supernatant (S6K sample and EE peptide) was collected for analysis. The elution process was repeated two more times.
Mass Spectrometric Analysis
Excised bands were digested in Trypsin, and the resulting peptides were processed for the identification by mass spectrometry. 75-μm-inner diameter nanospray tips (New Objective) were used; the ion spray voltage was set to 2800 V, the ion source gas was set at 10, and the interface heater temperature was 150 °C scanning across the mass range of m/z 400–1600 over 1.12 s in an enhanced MS scan. An enhanced resolution scan was then performed in 0.79 s. Information-dependent acquisition was employed for enhanced product ion scans (MS/MS) to fragment the four most abundant multiply charged ions from each enhanced resolution scan that had counts exceeding 90,000. Ions were excluded after four fragmentations for 2 min, and MS/MS fragments in the m/z 100–1600 range were acquired.
MS and MS/MS data were extracted and searched against the NCBInr 20080210 data base, Homo sapiens taxonomy (127924 sequences) on a Mascot in-house server version 2.2.04 (Matrix Science) using Mascot Daemon version 2.2.2 (Matrix Science). The search parameters were peptide and fragment tolerances of 1 and 0.8 atomic mass unit, respectively, with one missed cleavage and carboamidomethylation of cysteines as a fixed modification and oxidation of methionine, protein N-terminally acetylated, Gln > pyro-Glu as variable modifications. Matches with a significance p threshold of <0.05 were reported for consideration for follow-up work.
Size Exclusion Chromatography
A Superose 6 HR size exclusion chromatography column (GE Healthcare) was used to separate different protein complexes obtained from MCF-7 cell lysates. 2 mg of total protein in 500 μl of lysis buffer were filtered through 0.2-μm syringe filters (Millipore) and then loaded onto the column. Elution was carried out in lysis buffer in 24 fractions of 1 ml.
Protein molecular weights were estimated by comparison with five standards (Bio-Rad), and dextran blue was used to determine the column void volume. The standards used gave elution volumes of 12.8, 15.9, 17.5, 18.8, and 21.5 ml for globular proteins of 670, 158, 49, 17, and 1.35 kDa, respectively.
siRNA Knockdown of hnRNP F and hnRNP H (Cisplatin Treatment)
The cells were plated in 0.5 ml of medium (24 wells) without antibiotics so as to achieve a confluency of 50% on the next day. On the following day, 35 nmol of specific siRNA was mixed in serum- and antibiotics-free medium. At the same time, 3 μl of Oligofectamine (Invitrogen) was added to 12 μl of serum and antibiotics-free medium and incubated for 5 min at room temperature. The mixture of siRNA and medium was then added to the Oligofectamine mix and left to stand for 20 min at room temperature to allow transfection complexes to form. The resultant mix was added dropwise to cells. 6 h post-transfection, the transfection mix containing medium was replaced with normal growth medium. 48 h later, the cells were treated with cisplatin at the stated different concentrations. After a further 48-h period, the cells were fixed in 4% paraformaldehyde for 10 min before staining with 0.075% crystal violet. Following three washes in distilled H2O, the plates were dried, and the crystal violet precipitates were solubilized in 10% acetic acid prior to optical density measurements at 590 nm.
Generation of GST-hnRNP F Domain Mutants
To express generate GST fusion constructs of hnRNP F, we used human hnRNP F (IRATp970C01109D) as a template and amplified various fragments of hnRNP F using Pfu polymerase and specific sets of primers. Amplified fragments were cloned into pGEX 4T3 expression plasmid, and the resulting constructs were transformed Rosetta (DE3) pLysS competent cells. The expression of GST fusion proteins was induced by isopropyl β-d-thiogalactopyranoside, and recombinant proteins were purified on glutathione-Sepharose beads.
RESULTS
S6K2 Associates with RNA-binding Proteins in the Nucleus
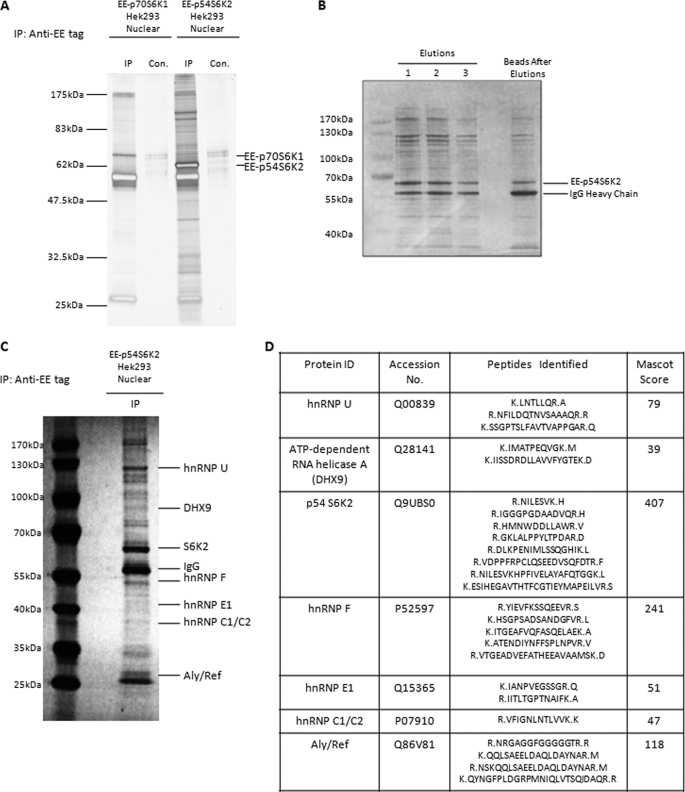
Although a cytoplasmic S6K1 complex regulating protein synthesis was reported (24), nuclear complexes and functions of S6K1 or S6K2 remain to be investigated. To identify novel binding partners of both S6K1 and S6K2 in the nucleus, we utilized an affinity purification approach of overexpressed S6Ks coupled with mass spectrometric identification. Tetracycline-inducible T-Rex Hek293 cell lines were generated for EE-tagged p70S6K1 and p54S6K2 overexpression. The expression of EE-p70S6K1or EE-p54S6K2 was induced by the addition of tetracycline for 12 h, followed by nuclear fractionation to enrich cellular nuclei. Antibodies against the EE tag were used to isolate EE-p70S6K1 and EE-p54S6K2 together with associating binding partners from the respective cell lines followed by SDS-PAGE and silver staining (Fig. 1A). Surprisingly, immunoprecipitation of nuclear EE-p54S6K2 but not nuclear EE-p70S6K1 co-precipitated a number of associated proteins, indicating that S6K2 has distinct nuclear functions from S6K1. These interactions were shown to be S6K2-specific, because neither control precipitations using protein A-Sepharose beads alone nor the immunoprecipitation of EE-p70S6K1 using an identical anti-EE antibody co-precipitate the distinctive pattern of binding proteins observed with S6K2. Furthermore, S6K2 and its nuclear associated proteins could be eluted from the nuclear immunocomplex by the addition of competitive EE peptide (Fig. 1B), demonstrating that associated proteins specifically associate with EE-p54S6K2 and not with the protein A-Sepharose beads.
FIGURE 1.
Mass spectrometric identification of RNA-binding proteins associated with S6K2. A, multiple nuclear proteins associate with EE-p54S6K2 but not EE-p70S6K1. Nuclear fractions of EE-p70S6K1 and EE-p54S6K2 Hek293 cells, respectively, were isolated and subjected to affinity purification using antibodies against EE tag. Isolated protein complexes were analyzed by SDS-PAGE followed by silver staining. Control precipitations were carried out in the absence of antibodies. B, addition of EE peptide to EE-p70S6K2 nuclear precipitates elutes bound S6K2 and S6K2 interacting proteins. Three elutions (10 mg of EE peptide in 30 μl of lysis buffer) were made in sequence (lanes 1–3), with lane 1 being the first elution, followed by lane 2 and lane 3. The beads left after elution were also analyzed as a control. C and D, RNA-associated proteins were identified by NanoLC-MS/MS to interact with nuclear EE-p54S6K2. Matches with a significance p threshold of <0.05 are shown in D). IP, immunoprecipitation; Con., control.
Protein bands co-precipitated with nuclear EE-p54S6K2 were excised from the SDS-PAGE gel and analyzed by NanoLC-MS/MS. hnRNPs U, F, E1, and C1/C2, along with ATP-dependent RNA helicase A and Aly/Ref, were identified as potential S6K2 nuclear associating partners (Fig. 1, C and D), demonstrating that a panel of RNA-binding proteins are able to associate with nuclear S6K2.
hnRNPs Associate with S6K2 and mTOR-Raptor in Vivo
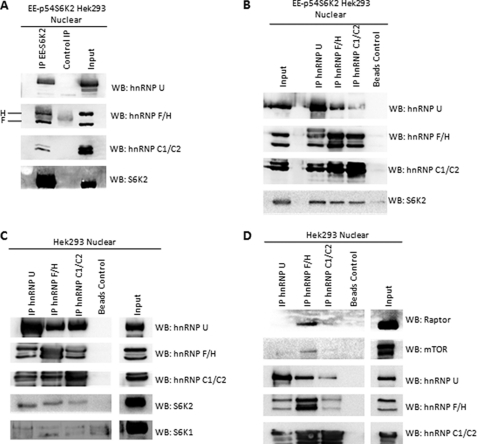
To further establish the interactions identified by mass spectrometry, we investigated the specificity of the S6K2-hnRNP interaction by Western blot analysis. Immunoprecipitation of nuclear EE-p54S6K2 from EE-p54S6K2Hek293 cells specifically co-precipitates hnRNP U, hnRNP F, and hnRNP C1/C2. In addition, by using an antibody that detects both hnRNP F and H, we found that hnRNP H, an hnRNP F-related protein, was also found to co-precipitate with nuclear EE-p54S6K2 (Fig. 2A). These interactions were further confirmed by reciprocal immunoprecipitation of hnRNPs from EE-p54S6K2Hek293 nuclear fractions, followed by Western blotting with S6K2 antibody (Fig. 2B). Antibodies against hnRNPs were also applied in Hek293 nuclear fractions and specifically co-precipitated endogenous S6K2 but not S6K1, further demonstrating that S6K2 (and not S6K1) interacts with hnRNPs (Fig. 2C). The mTOR kinase is the S6K1/S6K2 upstream regulator and is essential for signaling to S6K-mediated processes. We hypothesized that if S6K2 was found to interact with hnRNPs, the mTOR kinase may also interact with hnRNPs. Indeed, immunoprecipitation of hnRNP F/H, but not hnRNP U and C1/C2 from Hek293 nuclear lysates, co-purified endogenous mTOR. Furthermore, Raptor, the substrate-presenting molecule for mTOR complex 1, also co-precipitated with hnRNP F/H (Fig. 2D), indicating that the endogenous mTOR complex 1 along with S6K2 is able to associate with hnRNPs in vivo.
FIGURE 2.
S6K2 and the mTOR-Raptor complex associate with hnRNPs U, F/H, and C1/C2. A, hnRNPs U, F/H, and C1/C2 co-precipitate with nuclear EE-p54S6K2. Exponentially growing EE-p54S6K2 cells were harvested followed by nuclear fractionation. Nuclear immunoprecipitations (IP) were achieved with antibodies against the EE tag of overexpressed S6K2. Control purification was achieved by incubating equal amount of nuclear fraction with protein A-Sepharose beads bound to an unrelated antibody. B, antibodies against hnRNPs U, F/H, and C1/C2 were used, respectively, to co-precipitate EE-p54S6K2 using nuclear fractions as in A. C, hnRNPs co-precipitate endogenous S6K2. Hek293 nuclear lysates was used along with antibodies specific to individual hnRNPs. Western blots (WB) of co-precipitated proteins revealed that S6K2, but not S6K1, co-precipitates with hnRNPs. D, mTOR and Raptor associate with hnRNPs. Antibodies specific to hnRNPs were used to purify immunocomplexes in Hek293 nuclear fractions. Antibodies against mTOR and Raptor were used in a Western blot to determine association.
S6K2, mTOR, and hnRNP Form a Multimeric Protein Complex in the Nucleus
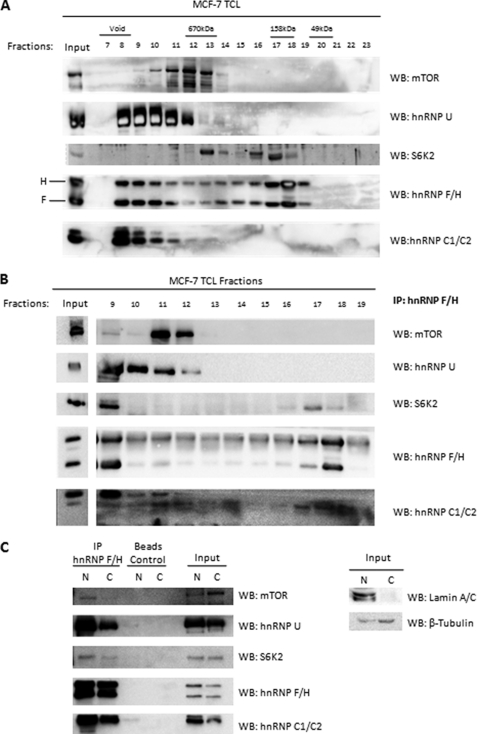
Because S6K2 and mTOR-Raptor were demonstrated to be hnRNP-interacting proteins, whereas hnRNPs were previously suggested to be part of a multi-protein complex, we sought to determine whether S6K2 and mTOR form a complex with hnRNPs in the cell nucleus. Total cell lysate from exponentially growing MCF-7 cells was analyzed by size exclusion chromatography to separate protein complexes (Fig. 3A). hnRNPs U and C1/C2, along with mTOR were observed to elute in high molecular weight fractions, whereas S6K2 was localized predominantly in low molecular weight fractions. Interestingly, hnRNP F/H was distributed throughout all of the fractions analyzed, with peaks in both high and low molecular weight fractions. The co-localization of hnRNPs, mTOR, and S6K2 was found in high molecular weight fractions (fractions 9–13), whereas S6K2 and hnRNP F/H was observed in fractions corresponding to low molecular weight standards (fractions 15–18).
FIGURE 3.
S6K2, mTOR, and hnRNPs are part of a multimeric complex in the cell nucleus. A, mTOR and S6K2 co-localize with hnRNPs in size exclusion chromatography fractions. MCF-7 breast carcinomas cells were grown exponentially and lysed. Total cell lysate was applied into a Superose 6 size exclusion chromatography column. Complexes and proteins were subsequently eluted according to size into 1-ml fractions. 40 μl of each fraction was loaded into a SDS-PAGE gel and analyzed by Western blot (WB) analysis. B, S6K2 and mTOR co-precipitate with hnRNPs within individual fractions. 500 μl of each fraction (fractions 9–19) was subjected to immunoprecipitation (IP) with antibody against hnRNP F/H. Co-precipitated hnRNPs, mTOR, and S6K2 were analyzed by Western blotting. C, the hnRNP-mTOR-S6K2 complex is predominantly nuclear. Exponentially growing Hek293 cells were separated into nuclear and cytoplasmic fractions as described under “Experimental Procedures.” hnRNP F/H was immunoprecipitated from nuclear and cytoplasmic fractions, respectively, followed by analysis of complex components by Western blotting.
To further elucidate the complex components of the hnRNP, mTOR, and S6K2 interactions, antibodies against hnRNPs F/H were used to isolate protein complexes from fractions 9–19, respectively (Fig. 3B). Three distinct complexes were observed to co-precipitate with hnRNP F/H. First, a complex involving all hnRNPs, mTOR, and S6K2 was observed (Fig. 3B, fraction 9) in high molecular weight size exclusion fractions. Second, a complex of mTOR and hnRNPs but not S6K2 was also observed (Fig. 3B, fractions 11 and 12). Finally, a low molecular weight complex of S6K2 and hnRNP F/H was present in fractions 17 and 18. Together, these results indicate that mTOR and S6K2 are able to form complexes with hnRNPs of differing protein compositions.
Subcellular fractionation followed by immunoprecipitation of hnRNP F/H indicated that the hnRNP-mTOR-S6K2 complex is predominantly localized in the nucleus because the mTOR and S6K2 interaction with immunoprecipitated hnRNP F/H was significantly lower in the cytoplasm. This result is in contrast to nuclear hnRNP F/H immunoprecipitations, where all components of the hnRNP-mTOR-S6K2 complex were clearly present (Fig. 3C).
mTOR, but Not S6K2, Interaction with hnRNP F/H Is Regulated by Serum
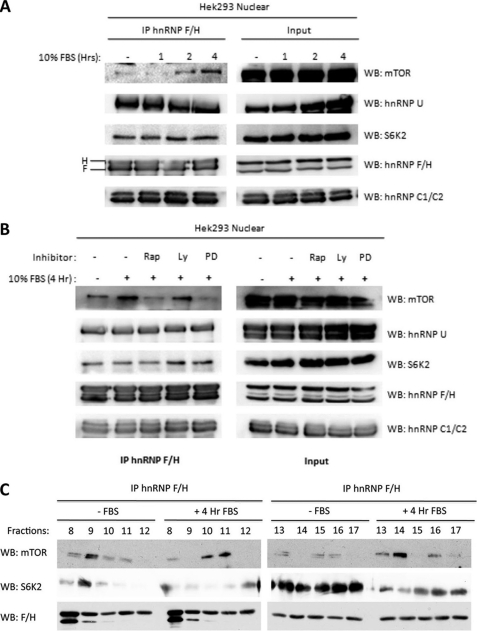
Activation of the mTOR/S6K pathway is highly dependent on the presence of nutrients and growth factors. To investigate whether the hnRNP-mTOR-S6K2 complex formation could be regulated by growth factors, we serum-starved Hek293 cells for 24 h, followed by the addition of FBS for various time points prior to cellular nuclear fractionation. Growth of Hek293 cells in serum-starved conditions reduced mTOR, but not S6K2, interaction with hnRNPs F/H (Fig. 4A). Following the addition of FBS, an increase in mTOR interaction with hnRNP F/H was observed in a time-dependent manner. Similar to S6K2, hnRNPs U and C1/C2 interaction with hnRNP F/H were not affected by serum deprivation or readdition, indicating constitutive and growth factor-independent interaction of S6K2 with hnRNPs. In addition, the FBS-induced mTOR-hnRNP F/H interaction could be attenuated in the presence of inhibitors of the mTORC1 pathway. The addition of rapamycin, a direct inhibitor of mTORC1 function, prior to serum stimulation of Hek293 cells blocked serum-induced mTOR association with hnRNP F/H (Fig. 4B). Similarly, PD 098059, the MAPK pathway inhibitor, and to a lesser extent LY 294002, which inhibits PI3K, reduced serum-induced association of mTOR and hnRNP F/H, indicating that upstream signaling pathways that are essential for mTOR regulation are important in the association of hnRNP F/H and mTOR. We further investigated the effect of serum starvation and stimulation on the mTOR and S6K2 association with hnRNP F/H by gel filtration followed by the analysis of immune complexes by Western blotting. We found that serum starvation or stimulation of MCF-7 cells did not observably alter the distribution of hnRNP F/H, mTOR, and S6K2 when gel filtration fractions of whole cell lysates were analyzed by immunoblotting with corresponding antibodies (supplemental Fig. S1). However, when hnRNP F/H was immunoprecipitated from these fractions and immune complexes examined by Western blotting, we observed that the mTOR and S6K2 interaction with hnRNP F/H redistributed to different gel filtration fractions, indicating a reorganization of complex integrity (Fig. 4C).
FIGURE 4.
Interaction of hnRNPs with mTOR but not S6K2 is regulated by growth factors. A, interaction of mTOR, but not S6K2, is regulated by serum withdrawal and stimulation. Hek293 cells were grown to 70% confluency followed by serum withdrawal for 24 h. Serum was reintroduced at a final concentration of 10% (v/v) for 1, 2, and 4 h, respectively. Nuclear fractions were obtained, and each sample was used in affinity purification of hnRNP F/H. B, interaction of hnRNPs with mTOR, but not S6K2, were inhibited by specific PI3K and mTOR pathway inhibitors. Hek293 cells were grown to 70% confluency and serum-starved for 24 h. Prior to serum stimulation, the starved cells were treated with rapamycin (Rap, 50 nm), LY 294002 (LY, 20 mm), and PD 098059 (PD, 10 mm) for 30 min. After 4 h of serum stimulation, the cells were harvested and nuclear fractionated. hnRNP F/H was affinity-purified from nuclear fractions, and the co-precipitated proteins were analyzed. C, distribution of the mTOR-S6K2-hnRNP F complex integrity in gel filtration in response to serum withdrawal and readdition. Gel filtration-derived fractions of serum-starved MCF7 cell lysates were incubated with antibodies against hnRNP F/H, followed by Western blot (WB) analysis of S6K2 and mTOR. IP, immunoprecipitation.
hnRNP F Regulates Cell Proliferation
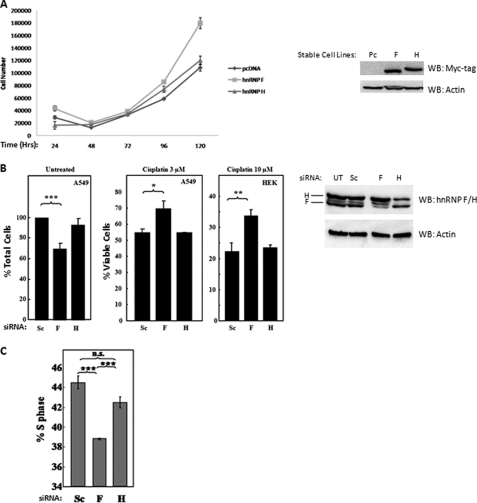
As mentioned prior, both S6K2 and mTOR have been reported to be involved in the regulation of cell proliferation (15, 16). We therefore reasoned that if S6K2 and mTOR are present in complex with hnRNPs, with inducible interactions between mTOR and hnRNP F/H, it is possible that hnRNPs, such as hnRNP F and H might be involved in the regulation of cellular growth and proliferation. To investigate this possibility, hnRNP F and hnRNP H were cloned, respectively, into pcDNA3.1 vectors. A Myc tag sequence was inserted in the N terminus of hnRNP F and hnRNP H to identify overexpressed proteins. Stable cell lines were constructed from the Myc-hnRNP F and Myc-hnRNP H plasmids in Hek293 cells. The Hek293 stable cell lines, Myc-hnRNPFHek293 and Myc-hnRNPHHek293, were investigated along with pcDNA3.1 stable Hek293 controls in a cell proliferation assay. Briefly, 25,000 cells were plated in 24-well plates, followed by analysis of cell number over 120 h of exponential growth (Fig. 5A). In contrast to Myc-hnRNP H and pcDNA3.1 Hek293 controls, stable cell lines overexpression of Myc-hnRNP F proliferate significantly faster. To further confirm the role of hnRNP F in cell proliferation, we used siRNA targeting hnRNP F or hnRNP H to reduce the target proteins in A549 cells (Fig. 5B, left panel). In support of overexpression studies, siRNA knockdown of hnRNP F, but not H, reduces the proliferation of A549 cells 3 days post-transfection, as compared with scrambled controls. In addition, siRNA-mediated silencing of hnRNP F in A549 (Fig. 5B, center panel) or Hek293 (Fig. 5B, right panel) cells cultured in the presence of cisplatin, a chemotherapy drug that targets actively proliferating cells, was observed to exhibit higher survival rates as compared with scrambled controls, further demonstrating that deletion of hnRNP F protein expression reduces cell proliferation. The reduction of cell proliferation in hnRNP F siRNA-treated cells could also be observed at the biochemical level. Hek293 cells were treated with either scrambled siRNA, siRNA targeting hnRNP F, or siRNA targeting hnRNP H. siRNA-treated cells were pulsed with BrdUrd for 2 h, followed by analysis by fluorescence-activated cell sorter (Fig. 5C). Hek293 cells treated with hnRNP F targeting siRNA incorporated significantly less BrdUrd in the S phase, as compared with hnRNP H siRNA or scrambled treated controls, indicating a lower DNA replication rate in hnRNP F-deficient Hek293 cells. These experiments taken together clearly indicate a proliferation regulatory role of the hnRNP F protein.
FIGURE 5.
hnRNP F drives cell proliferation. A, stable expression of exogenous Myc-hnRNP F drives cell proliferation. 25,000 cells from Myc-hnRNP F, Myc-hnRNP H, and pcDNA (Pc) control expressing stable cell lines (right panel) were plated, respectively, in FBS-containing DMEM. The cell numbers were determined by a Casy cell counter every 24 h to determine total cell numbers (left panel). B, siRNA knockdown of hnRNP F but not H reduces cell number and increases survival to cisplatin treatment. The cells were transfected with siRNA for the different targets at 35 nm (right panel). 48 h later, the cells were further treated with cisplatin at different concentrations. 48 h post-treatment, the cells were fixed in 4% paraformaldehyde for 10 min before staining with 0.075% crystal violet. Following three washes in distilled H2O, the plates were dried, and the crystal violet precipitates were solubilized in 10% acetic acid prior to optical density measurements at 590 nm (left panel). Sc, scrambled; UT, untransfected; F, hnRNP F; H, hnRNP H. C, siRNA knockdown of hnRNP F decreases percentage of cells in S phase. The cells were transfected with siRNA specific for either hnRNP F or hnRNP H at 35 nm. 48 h post-transfection, cells were incubated with 5-bromo-2′-deoxyuridine 5′-triphosphate for 2 h followed by fixation in 70% ethanol and analysis by fluorescence-activated cell sorter. WB, Western blot.
Interdependence of mTOR, S6K2, and hnRNP F in the Regulation of Cell Proliferation
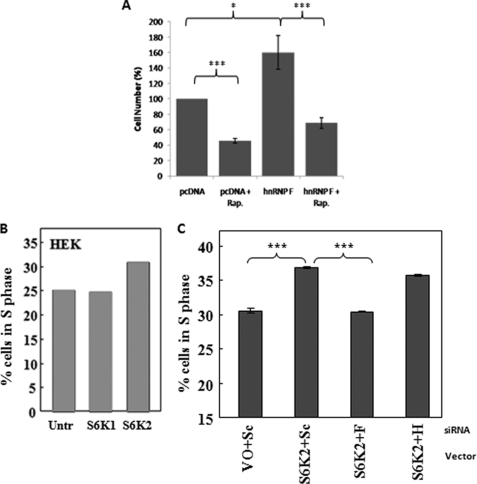
Because rapamycin was able to block the mTOR and hnRNP F interaction, we sought to determine whether this interaction is essential for hnRNP F-regulated cell proliferation. Myc-hnRNPFHek293 or pcDNA3.1 control stable cell lines were plated as before and allowed to proliferate for 48 h. Rapamycin was introduced into the cell growth medium at this point, and the cells were allowed to grow for a further 72 h in the presence or absence of rapamycin. As expected, mock-treated Myc-hnRNPFHek293 cells were able to proliferate faster as compared with control cells. Also not surprisingly, the addition of rapamycin blocked cell proliferation of pcDNA3.1 control cells. More importantly, the addition of rapamycin to Myc-hnRNPFHek293 cells inhibited hnRNP F-induced cell proliferation to levels similar to Rapamycin-treated control cells (Fig. 6A), demonstrating that hnRNP F-driven cell proliferation is partially dependent on the mTOR kinase.
FIGURE 6.
Regulation of cell proliferation by the mTOR-S6K2-hnRNP F complex. A, rapamycin blocks hnRNP F-dependent cell proliferation. Myc-hnRNP F and pcDNA control cells were plated at equal cell numbers and allowed to grow exponentially in 10% FBS-supplemented DMEM for 48 h. Rapamycin was added at a final concentration of 25 nm, and the cells were further allowed to proliferate for another 72 h. Total cell numbers were analyzed by Casy cell counter. The error margins represent the standard errors of the mean. *, p < 0.05; ***, = p < 0.05. B, overexpression of S6K2 drives cell proliferation. Hek293 cells were transfected with a mammalian expression vector encoding either S6K1 or S6K2 or left untransfected (Untr). C, siRNA down-regulation of hnRNP F blocks S6K2-driven cell proliferation. Hek293 cells were co-transfected with plasmid DNA encoding S6K2 or the corresponding empty vector (VO), and either scramble oligonucleotides (Sc) or siRNA targeting hnRNP F or H. B and C, 48 h later, the resulting cells were pulsed in the presence of BrdUrd and analyzed by flow cytometry. The results shown are representative of at least three independent experiments where 10,000 cells were analyzed. The error margins represent the standard errors of the mean. ***, p < 0.05.
S6K2 has been previously reported to drive cell proliferation (15). This effect is also observed here, where overexpression of S6K2 but not S6K1 in Hek293 cells increased the number of cells in the S phase in a BrdUrd incorporation assay (Fig. 6B). Here, we found that the addition of hnRNP F-targeting siRNA, but not hnRNP H siRNA nor scrambled controls, to S6K2-overexpressing cells abrogated the increase in the number of cells in S phase because of S6K2 overexpression (Fig. 6C), demonstrating that hnRNP F is required for S6K2-regulated cell proliferation. Taken together, these results suggest that the nuclear interactions of mTOR, S6K2, and hnRNP F are involved in the regulation of cell proliferation.
DISCUSSION
Since the discovery of S6K1 as the phosphorylating kinase for the ribosomal protein rS6 (48–50), our knowledge of the S6 kinases in cellular growth processes has been well elucidated. However, most studies reported thus far have focused on the S6K1 isoform, with less understanding of the role of S6K2. Deletion of S6K1 in mice, contrary to past expectations, did not abrogate the phosphorylation of rS6 (51), which inevitably led to the identification of S6K2 (26, 51–53). The presence of a C-terminal nuclear localization sequence in S6K2 and other structural differences between the two S6K isoforms indicate that S6K2 has a potential role independent of its S6K1 isoform (27, 28). This has partly been elucidated, because S6K2 but not S6K1 has been shown to be localized to centrosomes in cells (54). In addition, it has also been reported that S6K2 but not S6K1 is able to regulate cell survival (20). Similarly, S6K1 but not S6K2 is involved with intron-containing RNA splicing through its association with SKAR (25). As such, certain roles of S6K1 and S6K2 have been established to be distinct from one another.
Here, we present the evidence for a novel multi-protein complex in the nucleus comprising of mTOR, S6K2, and a number of nuclear proteins. We demonstrated that of the two S6K isoforms, S6K2 is involved in association with the hnRNP complex, with the specificity of identified interactions confirmed using both overexpressed and endogenous proteins in immunoprecipitation and Western blot experiments. In addition, we also observed that the mTOR kinase, together with its interacting protein Raptor, associates with hnRNPs. Of the two mTOR complexes (mTORC1 and mTORC2), it is the mTORC1 complex that harbors the Raptor-associated protein. It is also the mTOR complex that is involved in phosphorylation and regulation of the S6 kinases. Thus, identification of these interactions may indicate regulation of S6K2 in the nucleus involving association with hnRNPs. mTOR has been previously linked to another RNA-associating protein, the splicing factor SF2/ASF (55). Together, the work presented in this paper and that reported by Karni et al. (55) demonstrate potential multiple nuclear roles of mTOR.
Because hnRNP F was found in a functional complex with S6K2 and mTOR in the regulation of cell proliferation, we attempted to further characterize hnRNP F interaction with S6K2 at the molecular level. To answer this question, we have generated a panel of GST fusion proteins, containing the full-length and various domains of hnRNP F (supplemental Fig. S2A). GST-hnRNP F fusion proteins were affinity-purified on glutathione-Sepharose (supplemental Fig. S2B) and used in a GST pull-down assay with either bacterially expressed S6K2 or EE-p54S6K2 extracted from S6K2 overexpressed cell lysates. We were unable to co-precipitate S6K2 in both experimental setups. This was an unanticipated finding, because we expected to detect the interaction in either experimental set-up. We would like to note that a number of RNA-binding proteins, including hnRNP F, have also been recently identified in complex with S6K2 by a tandem affinity purification approach.3 Taking this into account, we decided to test whether the S6K2 and hnRNP F interaction might be dependent on the intact hnRNP-RNA complex. To test this hypothesis, Hek293 cell lysates were incubated with or without RNase A prior to hnRNP F/H immunoprecipitation. When the immune complexes were immunoblotted with anti-S6K2 antibodies, we observed a marked reduction in S6K2 co-immunoprecipitation with hnRNP F/H in cell lysates pretreated with RNase A. Pretreatment with RNase A had also reduced the levels of hnRNP U and hnRNP C1/C2 in immune complexes with hnRNP F/H (supplemental Fig. S3). These data suggest that the interaction between S6K2 and hnRNPs might occur when hnRNPs are in complex with hnRNAs. Interestingly, mTOR interaction with hnRNP F/H was increased in RNase A-treated samples, indicating a RNA-independent association of mTOR with hnRNP F/H (supplemental Fig. S3).
Our experiments indicate that the observed associations would exist as a complex, because immunoprecipitation of either hnRNP U, F/H, or C1/C2 led to co-precipitation of the other investigated hnRNPs. Furthermore, it has been reported that hnRNPs exist as a large protein complex tethered to RNA (34). To investigate this possibility, we utilized size exclusion chromatography along with immunoprecipitation of hnRNP F/H from derived chromatography fractions. Three distinct complexes were observed in these experiments. Immunoprecipitation of hnRNP F/H from fractions 9–19 indicated a large macromolecular complex consisting of mTOR, S6K2, and hnRNPs in fraction 9; an intermediate complex of mTOR and hnRNPs in fractions 11 and 12; and a low molecular weight complex of hnRNPs F/H, C1/C2, and S6K2. The different compositions of these complexes indicate dynamic interactions between these complexes.
Factors that determine the composition of the dynamic hnRNP-mTOR-S6K2 complexes were further investigated in this report. The complex comprised of S6K2, mTOR, and hnRNP F/H occurred primarily in the nucleus, indicating nuclear roles of both kinases associated with hnRNPs. Furthermore, we have also shown that this complex is regulated by upstream signaling pathways previously reported to be essential for mTOR and S6K2 activation and function. Serum stimulation promoted mTOR binding to the hnRNP complex, which is shown here to include a constitutively associated S6K2, but not S6K1. This is further demonstrated by the observation that inhibitors targeting upstream regulatory signaling pathways of the mTOR kinase (and as such S6K) could inhibit the serum-induced association of mTOR with hnRNP F/H. Rapamycin, a direct inhibitor of mTOR, achieves inhibition by first associating with FKBP12. Together with FKBP12, rapamycin binds to the FKBP-rapamycin binding domain and inhibits mTOR activation (56). The addition of rapamycin to cells led to the inhibition of mTOR association with hnRNP F/H. mTOR activation is also dependent on the PI3K and the MAPK pathway. PI3K activation of the Akt kinase results in phosphorylation and inhibition of TSC2 (5), the upstream antagonizer of the mTOR function. On the other hand, p90 ribosomal S6 kinase, a S6K-related kinase downstream of MAPK, is able to phosphorylate and promote Raptor association with mTOR (16). LY 294002 and PD 098059, inhibitors of PI3K and MAPK pathways, respectively, as of rapamycin, reduced mTOR interaction with hnRNP F/H. In contrast, S6K2 interaction with hnRNPs was not regulated by serum withdrawal or addition, indicating differential regulation of S6K2 and mTOR interaction with the hnRNPs.
It is possible that mitogen-induced association of mTOR with the hnRNP-S6K2 complex results in phosphorylation and subsequent activation of S6K2. In an active state, S6K2 might phosphorylate novel substrates, including hnRNPs. Certainly, ongoing studies in our laboratory indicate that hnRNP U and C1/C2 are in vitro substrates for S6K2 (results not shown).
What is the function of the mTOR-S6K2-hnRNP F complex? Both S6K2 (15) and mTOR (16) have been reported to regulate cell proliferation. We therefore hypothesized that hnRNPs may be involved in cell proliferation regulation through its interaction with S6K2 and mTOR. Indeed, overexpression of hnRNP F is able to drive cell proliferation, thus identifying a new physiological role for this hnRNP. The role of hnRNP F in cell proliferation is further established in siRNA knockout studies. hnRNP F siRNA-treated cells both proliferate at a slower rate and incorporate less BrdUrd as compared with control cells. Surprisingly, neither overexpression nor siRNA knockdown of hnRNP H is able to regulate cell proliferation. Although hnRNP F/H were reported to form heterodimers, with similar functional effects in splicing control (57), we have demonstrated that in the case of cell proliferation, only hnRNP F seems to be involved in this regulation. Because rapamycin is able to inhibit the mTOR-hnRNP F/H interaction, we used rapamycin in an hnRNP F overexpression cell proliferation assay to investigate whether the disruption of the mTOR-hnRNP F association would attenuate hnRNP F-driven cell proliferation. Growth of hnRNP F-overexpressing cells in the presence of rapamycin reduced cell proliferation, indicating that the mTOR-hnRNP F interaction is essential for the effect of hnRNP F on cell proliferation. In extension of this observation, we asked whether hnRNP F would be involved in S6K2-driven cell proliferation. Indeed, siRNA knockdown of hnRNP F in S6K2-overexpressing cells blocked S6K2-driven cell proliferation.
In summary, our results define a novel role for S6K2 and mTOR by formation of a complex with hnRNPs. This complex is predominantly nuclear and is sensitive to regulators of the mTOR kinase. We have also identified a novel role for hnRNP F in cell proliferation, which is dependent on its interaction with the mTOR kinase. In view of this, it is now clear that both S6K 1 and 2 are able to perform distinct roles in the cell nucleus. S6K1 was reported to be associated with SKAR (S6K Aly, Ref-like) protein. SKAR in turn recruits S6K1 to the exon junction complex on RNA, whereby S6K1 was able to regulate RNA splicing (25). S6K2, as reported in this study, is able to interact with hnRNPs in the cell nucleus. Together with mTOR, this complex is shown here to be involved in the regulation of cell proliferation. We propose that the roles of mTOR and S6K2 in cell proliferation are dependent on their association with hnRNP F and could also involve other, related hnRNPs.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
O. Pardo, unpublished observations.
- mTOR
- mammalian target of rapamycin
- S6K
- S6 kinase
- hnRNP
- heterogeneous ribonucleoprotein
- PI3K
- phosphotidylinositol 3-kinase
- siRNA
- small interfering RNA
- PDK
- 3-phosphoinositide-dependent kinase
- TSC
- tuberous sclerosis complex
- mTORC
- mTOR complex
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- GST
- glutathione S-transferase
- MAPK
- mitogen-activated protein kinase
- BrdUrd
- bromodeoxyuridine
- FKBP
- FK506-binding protein.
REFERENCES
- 1.Auger K. R., Serunian L. A., Soltoff S. P., Libby P., Cantley L. C. (1989) Cell 57, 167–175 [DOI] [PubMed] [Google Scholar]
- 2.Anderson K. E., Coadwell J., Stephens L. R., Hawkins P. T. (1998) Curr. Biol. 8, 684–691 [DOI] [PubMed] [Google Scholar]
- 3.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 4.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 5.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 6.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 7.Inoki K., Li Y., Xu T., Guan K. L. (2003) Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. (2003) Nat. Cell Biol. 5, 578–581 [DOI] [PubMed] [Google Scholar]
- 9.Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 10.Lee G., Chung J. (2007) Biochem. Biophys. Res. Commun. 357, 1154–1159 [DOI] [PubMed] [Google Scholar]
- 11.Pullen N., Thomas G. (1997) FEBS Lett. 410, 78–82 [DOI] [PubMed] [Google Scholar]
- 12.Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. (2004) Mol. Cell. Biol. 24, 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montagne J., Stewart M. J., Stocker H., Hafen E., Kozma S. C., Thomas G. (1999) Science 285, 2126–2129 [DOI] [PubMed] [Google Scholar]
- 14.Filonenko V. V., Tytarenko R., Azatjan S. K., Savinska L. O., Gaydar Y. A., Gout I. T., Usenko V. S., Lyzogubov V. V. (2004) Exp. Oncol. 26, 294–299 [PubMed] [Google Scholar]
- 15.Cruz R., Hedden L., Boyer M., Kharas M. G., Fruman D. A., Lee-Fruman K. K. (2005) J. Leukocyte Biol. 78, 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 17.Shah O. J., Wang Z., Hunter T. (2004) Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 18.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 19.Harada H., Andersen J. S., Mann M., Terada N., Korsmeyer S. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardo O. E., Wellbrock C., Khanzada U. K., Aubert M., Arozarena I., Davidson S., Bowen F., Parker P. J., Filonenko V. V., Gout I. T., Sebire N., Marais R., Downward J., Seckl M. J. (2006) EMBO J. 25, 3078–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. (2001) EMBO J. 20, 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raught B., Peiretti F., Gingras A. C., Livingstone M., Shahbazian D., Mayeur G. L., Polakiewicz R. D., Sonenberg N., Hershey J. W. (2004) EMBO J. 23, 1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorrello N. V., Peschiaroli A., Guardavaccaro D., Colburn N. H., Sherman N. E., Pagano M. (2006) Science 314, 467–471 [DOI] [PubMed] [Google Scholar]
- 24.Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 25.Ma X. M., Yoon S. O., Richardson C. J., Jülich K., Blenis J. (2008) Cell 133, 303–313 [DOI] [PubMed] [Google Scholar]
- 26.Gout I., Minami T., Hara K., Tsujishita Y., Filonenko V., Waterfield M. D., Yonezawa K. (1998) J. Biol. Chem. 273, 30061–30064 [DOI] [PubMed] [Google Scholar]
- 27.Reinhard C., Thomas G., Kozma S. C. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4052–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coffer P. J., Woodgett J. R. (1994) Biochem. Biophys. Res. Commun. 198, 780–786 [DOI] [PubMed] [Google Scholar]
- 29.Panasyuk G., Nemazanyy I., Zhyvoloup A., Bretner M., Litchfield D. W., Filonenko V., Gout I. T. (2006) J. Biol. Chem. 281, 31188–31201 [DOI] [PubMed] [Google Scholar]
- 30.Valovka T., Verdier F., Cramer R., Zhyvoloup A., Fenton T., Rebholz H., Wang M. L., Gzhegotsky M., Lutsyk A., Matsuka G., Filonenko V., Wang L., Proud C. G., Parker P. J., Gout I. T. (2003) Mol. Cell. Biol. 23, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Groot R. P., Ballou L. M., Sassone-Corsi P. (1994) Cell 79, 81–91 [DOI] [PubMed] [Google Scholar]
- 32.Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. (1977) Cell 11, 127–138 [DOI] [PubMed] [Google Scholar]
- 33.Dreyfuss G., Choi Y. D., Adam S. A. (1984) Mol. Cell. Biol. 4, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piñol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. (1988) Genes Dev. 2, 215–227 [DOI] [PubMed] [Google Scholar]
- 35.Carpenter B., MacKay C., Alnabulsi A., MacKay M., Telfer C., Melvin W. T., Murray G. I. (2006) Biochim. Biophys. Acta 1765, 85–100 [DOI] [PubMed] [Google Scholar]
- 36.Matunis M. J., Xing J., Dreyfuss G. (1994) Nucleic Acids Res. 22, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caputi M., Zahler A. M. (2001) J. Biol. Chem. 276, 43850–43859 [DOI] [PubMed] [Google Scholar]
- 38.Schaub M. C., Lopez S. R., Caputi M. (2007) J. Biol. Chem. 282, 13617–13626 [DOI] [PubMed] [Google Scholar]
- 39.Chou M. Y., Rooke N., Turck C. W., Black D. L. (1999) Mol. Cell. Biol. 19, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fogel B. L., McNally M. T. (2000) J. Biol. Chem. 275, 32371–32378 [DOI] [PubMed] [Google Scholar]
- 41.Jacquenet S., Méreau A., Bilodeau P. S., Damier L., Stoltzfus C. M., Branlant C. (2001) J. Biol. Chem. 276, 40464–40475 [DOI] [PubMed] [Google Scholar]
- 42.Oberg D., Fay J., Lambkin H., Schwartz S. (2005) J. Virol. 79, 9254–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNally L. M., Yee L., McNally M. T. (2006) J. Biol. Chem. 281, 2478–2488 [DOI] [PubMed] [Google Scholar]
- 44.Garneau D., Revil T., Fisette J. F., Chabot B. (2005) J. Biol. Chem. 280, 22641–22650 [DOI] [PubMed] [Google Scholar]
- 45.Crawford J. B., Patton J. G. (2006) Mol. Cell. Biol. 26, 8791–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida T., Kokura K., Makino Y., Ossipow V., Tamura T. (1999) Genes Cells 4, 707–719 [DOI] [PubMed] [Google Scholar]
- 47.Yoshida T., Makino Y., Tamura T. (1999) FEBS Lett. 457, 251–254 [DOI] [PubMed] [Google Scholar]
- 48.Jenö P., Ballou L. M., Novak-Hofer I., Thomas G. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee P., Ahmad M. F., Grove J. R., Kozlosky C., Price D. J., Avruch J. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8550–8554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozma S. C., Ferrari S., Bassand P., Siegmann M., Totty N., Thomas G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7365–7369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shima H., Pende M., Chen Y., Fumagalli S., Thomas G., Kozma S. C. (1998) EMBO J. 17, 6649–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koh H., Jee K., Lee B., Kim J., Kim D., Yun Y. H., Kim J. W., Choi H. S., Chung J. (1999) Oncogene 18, 5115–5119 [DOI] [PubMed] [Google Scholar]
- 53.Lee-Fruman K. K., Kuo C. J., Lippincott J., Terada N., Blenis J. (1999) Oncogene 18, 5108–5114 [DOI] [PubMed] [Google Scholar]
- 54.Rossi R., Pester J. M., McDowell M., Soza S., Montecucco A., Lee-Fruman K. K. (2007) FEBS Lett. 581, 4058–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karni R., Hippo Y., Lowe S. W., Krainer A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15323–15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oshiro N., Yoshino K., Hidayat S., Tokunaga C., Hara K., Eguchi S., Avruch J., Yonezawa K. (2004) Genes Cells 9, 359–366 [DOI] [PubMed] [Google Scholar]
- 57.Balcazar N., Sathyamurthy A., Elghazi L., Gould A., Weiss A., Shiojima I., Walsh K., Bernal-Mizrachi E. (2009) J. Biol. Chem. 284, 7832–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.