Abstract
Caspase-8 is a cysteine protease activated by membrane-bound receptors at the cytosolic face of the cell membrane, initiating the extrinsic pathway of apoptosis. Caspase-8 activation relies on recruitment of inactive monomeric zymogens to activated receptor complexes, where they produce a fully active enzyme composed of two catalytic domains. Although in vitro studies using drug-mediated affinity systems or kosmotropic salts to drive dimerization have indicated that uncleaved caspase-8 can be readily activated by dimerization alone, in vivo results using mouse models have reached the opposite conclusion. Furthermore, in addition to interdomain autoprocessing, caspase-8 can be cleaved by activated executioner caspases, and reports of whether this cleavage event can lead to activation of caspase-8 have been conflicting. Here, we address these questions by carrying out studies of the activation characteristics of caspase-8 mutants bearing prohibitive mutations at the interdomain cleavage sites both in vitro and in cell lines lacking endogenous caspase-8, and we find that elimination of these cleavage sites precludes caspase-8 activation by prodomain-driven dimerization. We then further explore the consequences of interdomain cleavage of caspase-8 by adapting the tobacco etch virus protease to create a system in which both the cleavage and the dimerization of caspase-8 can be independently controlled in living cells. We find that unlike the executioner caspases, which are readily activated by interdomain cleavage alone, neither dimerization nor cleavage of caspase-8 alone is sufficient to activate caspase-8 or induce apoptosis and that only the coordinated dimerization and cleavage of the zymogen produce efficient activation in vitro and apoptosis in cellular systems.
Keywords: Apoptosis, Caspase, Death Protease, Protease, Protein Processing, TEV Protease
Introduction
Caspases are a family of cysteine proteases that can be broadly divided by function into the inflammatory caspases and the apoptotic caspases (reviewed in Refs. 1, 2). Although the former group, which includes caspase-1, -4, and -5, is involved in the innate immune response through the cleavage-induced maturation of pro-inflammatory cytokines, the majority of the mammalian caspases are involved in the initiation and execution of apoptosis.
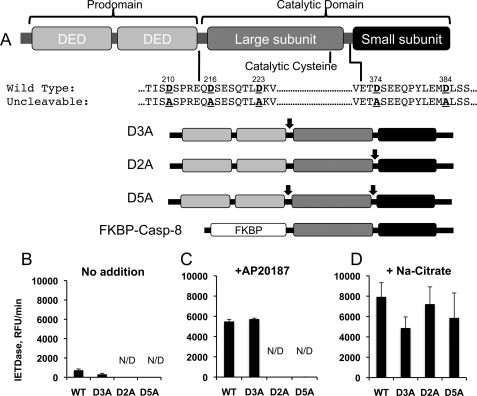
The apoptotic caspases, caspase-2, -3, and -6–10, can be further divided based on protein architecture and mode of activation. Caspase-2 and -8–10 are termed the “initiator” or “apical” caspases and are composed of a long N-terminal prodomain containing protein-protein interaction motifs and a C-terminal catalytic domain, often composed of an ∼20-kDa large subunit containing the active site cysteine and an ∼10-kDa small subunit (3). The initiator caspases are present in the cytosol of nonapoptotic cells as inactive monomers and are activated by recruitment to large protein complexes where they are induced to dimerize by protein-protein interactions with their prodomains. Once dimerized, initiator caspases undergo a series of autocleavage events at aspartic acid residues in their interdomain linker regions (4–6). Caspase-8 is first cleaved at one of two aspartic acid residues (Asp-374 or -384) between its large and small subunits and subsequently at one of three such residues (Asp-210, -216, or -223) between the large subunit and prodomain (Fig. 1A) (7). The resulting fully active enzyme is made up of a dimer of catalytic domains, each composed of a large and a small subunit.
FIGURE 1.
In vitro activation assays reveal discrepancies between prodomain-induced and salt-induced dimerization of caspase-8. A, schematic representation of the cleavage mutants of human caspase-8 employed in this study. Prohibitive aspartate-to-alanine substitutions were introduced at the sites previously reported to undergo autocleavage, as indicated by arrows. Prohibitive mutations were introduced between the prodomain and the large subunit (D3A mutant), between the large and small subunits (D2A mutant), or in both regions (D5A mutant). Numbers refer to the mutated Asp residue, counting from the N terminus of human caspase-8. In addition to the indicated mutants of full-length caspase-8, similar mutations were introduced into a version of caspase-8 in which the prodomain was replaced with a modified FKBP-inducible dimerization domain. DED, death effector domain. B, FKBP-Caspase-8 bearing the indicated mutations was purified, and activity was assayed using the fluorogenic substrate Ac-IETD-afc. C and D, these constructs were activated by incubation with 1 m sodium citrate (C) or via addition of the dimerizer drug AP20187 (D), and fluorogenic substrate activity was assayed using AcIETD-afc. N/D, not detected.
The executioner caspases, caspase-3, -6, and -7, share the large and small subunit architecture of the initiator caspases but lack long prodomains. Their mechanism of activation is also distinct from that of the initiators. The executioner caspases are present in the cytosol of nonapoptotic cells as inactive dimers and are activated by cleavage of the linker region between the large and small subunits by activated initiator caspases. This cleavage event causes intramolecular rearrangements and produces a fully active caspase enzyme (8).
Caspase-8 is the initiator caspase responsible for initiation of the extrinsic pathway of apoptosis in mammals. Caspase-8 is activated when the monomeric zymogen is recruited to the death-inducing signaling complex (DISC)4 formed at the cytoplasmic tail of activated receptors of the TNF family through homotypic interactions with the death effector domains in the prodomain of the zymogen (9). The outcome of receptor-mediated caspase-8 activation depends upon the cellular context; in some cell types (termed type I cells), caspase-8 can initiate apoptosis by directly cleaving and activating executioner caspases, although in other cell types (type II) it must engage the mitochondrial pathway of apoptosis by cleaving the Bcl-2 family member Bid to efficiently drive cell death (10). This difference is due to variations in the effects of the inhibitor of apoptosis proteins, which block executioner caspase function unless derepressed by proteins released from the mitochondria (11, 12).
The role of cleavage in caspase-8 activation is complex. Early studies using drug-inducible dimerization indicated that although mutation of interdomain cleavage sites reduced enzymatic activity upon dimerization, it did not eliminate it, and that uncleavable mutants of caspase-8 could be productively activated in vitro (7). Later work by ourselves and others relied upon kosmotropic salts to induce dimerization of caspase-8 in vitro. Insolubility of the prodomain confounds the production of full-length recombinant caspase-8, but recombinant proteins containing the large and small subunits can be induced to dimerize by the addition of 1 m sodium citrate. Production of such proteins with either prohibitive mutations at the interdomain cleavage sites or with thrombin cleavage sites at these positions revealed that noncleavable caspase-8 could be readily activated by kosmotropic salts (13). Under these in vitro conditions, it was determined that interdomain cleavage served to stabilize the dimer but was not required for activation.
However, these data are at odds with in vivo studies. Creation of a mouse expressing a caspase-8 mutant with a prohibitive mutation at the autocleavage site between the large and small subunits revealed that cells or tissues from this mouse were highly resistant to apoptosis induced by Fas ligation. These results indicate that noncleavable caspase-8 cannot support extrinsic apoptosis (14). Recent studies in which the DISC was reconstituted in vitro using purified recombinant proteins also support this idea; in these studies noncleavable caspase-8 could not be productively activated by DISC formation (15).
The effect of caspase-8 cleavage alone has also been the subject of some controversy. Although the in vitro work carried out by ourselves and others has supported the idea that cleavage alone is not sufficient to activate caspase-8 (13), studies in cell lines have come to the opposite conclusion (16, 17).
To accurately define the effects of both dimerization and cleavage of caspase-8 in living cells, we sought a system in which each event could be independently controlled. The regulated homodimerization system presents an ideal and well characterized system to regulate dimerization (18–20). Briefly, this system includes modified FKBP-12 protein domains, and a modified version of the cell-permeable drug FK1012, which binds tightly to these domains and induces their dimerization. This system has been used to study the activation of initiator caspases in the past (7, 21, 22). To study the effects of caspase cleavage, we adapted the tobacco etch virus (TEV) protease for use in mammalian cells. The TEV protease has been used extensively in vitro for removal of affinity tags and works under pH and temperature ranges that would indicate compatibility with in vivo applications (23, 24). TEV protease also has the advantage of a large consensus cleavage site, so its expression would not be expected to cause off-target effects (25, 26). Using these tools, we discovered that neither dimerization nor cleavage of caspase-8 zymogens, in the absence of the other process, is sufficient to activate the proteolytic activity of the enzyme under cellular conditions.
EXPERIMENTAL PROCEDURES
Cell Lines and DNA Constructs
HeLa and 293A cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, glutamine, and antibiotics. NB7 and Jurkat cells and cell lines were maintained in similarly supplemented RPMI 1640 medium.
The indicated mutations were inserted into human caspase-8, caspase-3, or caspase-7 using the QuikChange mutagenesis kit (Stratagene). Transient expression studies employing full-length caspases were carried out using indicated human caspases or mutants thereof in a pCDNA3.1 vector. Stable expression of caspase-8 was accomplished by cloning full-length caspase-8 into the pBabe-Puro retroviral vector, upstream of the T2A ribosomal skipping sequence followed by enhanced GFP. Stable expressers were obtained by retroviral transduction of these vectors followed by two rounds of sorting for GFP-positive cells. FKBP-caspase-8 constructs were obtained by cloning the region of caspase-8 corresponding to amino acid 206 to the C terminus into the SpeI site of the pC4-FV1E vector (ARIAD Pharmaceuticals). A 4-glycine linker was added between the FKBP domain and the caspase domain. Transient expression studies using FKBP-caspase-8 were carried out using this vector. In vitro work was accomplished by subcloning the FKBP-caspase-8 genes into the pET28b vector.
TEV protease was initially obtained as the S219V variant (Addgene, pRK793); this gene was subsequently cloned into pCDNA3.1. The optimized version of TEV protease, corresponding to the amino acid sequence of the S219V protease, was synthesized at our request by BioNexus, Inc., and cloned into pCDNA3.1 with an N-terminal V5 epitope tag.
Purification of Recombinant Protein
FKBP-caspase-8 mutants were expressed in Escherichia coli BL21(DE3) as N-terminal His constructs. Upon induction with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, cells were grown at 25 °C for 4 h and purified on nickel-affinity chromatography, followed by further purification on the Mono Q anion exchange Sepharose using 50 mm Tris, pH 8, NaCl buffer. TEV protease was purified as described previously (24). Proteins were stored at −80 °C.
Caspase Activation and Activity Assays
For caspase assays, FKBP-caspase-8 mutants were diluted to 10–50 nm into caspase buffer (10 mm Pipes, pH 7.2, 0.1 m NaCl, 1 mm EDTA, 10% sucrose, 0.05% CHAPS, 5 mm dithiothreitol), followed by addition of homodimerizer drug AP21087 in stoichiometric concentrations. For activation with kosmotropes, assay buffer contained 30 mm Tris-HCl, pH 7.4, 1 m sodium citrate, 5 mm dithiothreitol, 0.05% CHAPS, carefully adjusted to pH 7.4. Mixtures were incubated at 25 °C for 30–45 min, and the activity was determined at 30 °C by using 100 μm Ac-IETD-afc (27).
Cleavage with TEV protease was done by incubating FKBP-caspase-8 mutants (1.5 μm) with TEV protease (0.25 μm) for 5 h at room temperature in a buffer containing 50 mm Tris, 0.1 m NaCl, 5 mm dithiothreitol, pH 8. Similar caspase-7 to TEV protease ratios were used for caspase-7(TEV) cleavage. Reaction was stopped with 3× SDS-PAGE buffer, and proteins were resolved in 8–18% SDS-PAGE, followed by GelCode blue staining.
Antibodies
Monoclonal anti-human caspase-8 (C15) was a kind gift from Dr. Marcus Peter. Anti-hemagglutinin was purchased from Sigma (H9658); anti-caspase-3 was from Santa Cruz Biotechnology (H277); anti-actin was from ICN (C4); anti-V5 was from Invitrogen; anti-HSP90 was from Assay Designs; anti-Bid was from BD Biosciences; anti-caspase-7 was from Cell Signaling Technologies; and anti-poly(ADP-ribose) polymerase was from BD Biosciences.
Transfections and Biotin-Val-Ala-Asp-Fluoromethyl Ketone (b-VAD) Pulldown
HeLa and 293A cells were transfected with Lipofectamine 2000 (Invitrogen), and NB7 cells were transfected with the Nanojuice transfection reagent (EMD Bioscience) according to the manufacturers' instructions. Medium was changed 3–4 h post-transfection. In all cases, transfections were balanced using empty vector such that each sample from a given experiment received the same total quantity of DNA.
b-VAD pulldown was accomplished by transfecting cells for 16 h as described using 200 ng of caspase-8 vector/60-mm plate and then preincubating with b-VAD-fmk (50 μm) for 60 min prior to addition of TNF. Following TNF treatment (100 ng/ml TNF, 2 μg/ml cycloheximide) for 24 h, cells were lysed using RIPA buffer, and cleared lysates were incubated overnight with high capacity Neutravidin resin (Thermo-Fisher). Resin was washed three times in RIPA buffer and one time in phosphate-buffered saline, and bound proteins were eluted by boiling in SDS-PAGE loading buffer.
RESULTS
Intact Interdomain Cleavage Site Is Required for Prodomain-mediated Activation of Caspase-8 in Vitro
Previous studies have come to conflicting conclusions regarding the requirement for caspase-8 zymogen autocleavage during activation of caspase-8. Productive activation of noncleavable caspase-8 by kosmotropic salts or drug-driven dimerization has been described (7, 13), although other reports find that expression of full-length noncleavable caspase-8 in cells fails to sensitize them to extrinsic apoptosis (14). In an effort to reconcile these differences, we compared FKBP-driven dimerization to kosmotrope-driven dimerization using a battery of caspase-8 mutants carrying prohibitive mutations in the interdomain cleavage sites (Fig. 1A). The ARGENT inducible dimerization system (ARIAD Pharmaceuticals) employs modified FKBP protein domains and a drug, AP20187, that induces their dimerization. FKBP-caspase-8 constructs were expressed in bacteria and affinity-purified (supplemental Fig. S1), and productive homodimerization upon addition of dimerizer was confirmed by gel filtration (data not shown).
In agreement with previous findings (28), caspase-8 constructs with an intact autocleavage site between the large and small subunits (WT or D3A mutants) underwent spontaneous cleavage (supplemental Fig. S1) and partial activation (Fig. 1B) when expressed in bacteria, whereas those carrying prohibitive mutations (D2A and D5A) in the large/small subunit linker region did not. Also consistent with our previous observations, upon addition of kosmotropic salt, all constructs exhibited a robust increase in enzymatic activity (Fig. 1C). However, to our surprise, and in contrast to previously published results (7), when our constructs were dimerized by addition of the dimerization drug AP20187, robust activation was observed only in the constructs with permissive interdomain cleavage sites between the large and small subunits (Fig. 1D); indeed, under these conditions no detectable enzymatic activity was observed upon dimerization of the D2A and D5A constructs, both of which carry prohibitive mutations in the large/small subunit linker region. Given the more physiological nature of prodomain-driven dimerization as compared with kosmotropic salt-mediated dimerization, this finding led us to hypothesize that interdomain cleavage is required for caspase-8 activation in vivo.
Cleavage of Caspase-8 between the Large and Small Subunits Is Necessary for Efficient Induction of Apoptosis via the Extrinsic Pathway
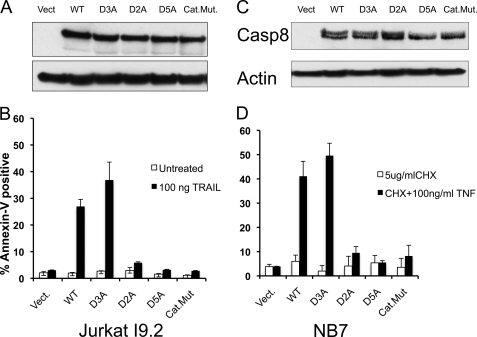
In an effort to further explore the dual roles of dimerization and cleavage in caspase-8 activation in a more physiological setting, we stably reconstituted human cell lines lacking endogenous caspase-8 with full-length caspase-8 bearing mutations described in Fig. 1A. To mitigate the selection problems often associated with stable expression of pro-apoptotic proteins such as caspases, we employed constructs expressing caspase-8 upstream of a 2A “ribosomal skipping” sequence, followed by GFP (29). In this manner GFP expression absolutely requires productive translation of caspase-8, allowing precise sorting of stable clones to yield cell lines expressing consistent, homogeneous, and nonlethal levels of caspase-8.
The caspase-8-deficient human cell lines NB7 or Jurkat I9.2 were retrovirally transduced with vectors encoding the previously mentioned mutants of caspase-8 and sorted three times each by FACS to achieve homogeneous GFP expression. Expression levels were confirmed by Western blotting for caspase-8 (Fig. 2, A and C). Jurkat I9.2 or NB7 cell lines were then treated with TRAIL or TNF, respectively, and apoptosis was measured by annexin-V binding and FACS analysis (Fig. 2, B and D). Consistent with our results using drug-inducible dimerization, apoptosis was efficiently induced only by constructs carrying permissive cleavage sites between the large and small subunits (WT and D3A). Mutation of these sites (D2A and D5A) effectively blocked apoptotic induction via the extrinsic pathway.
FIGURE 2.
Elimination of the interdomain autocleavage sites of caspase-8 abrogates its apoptosis inducing activity. Caspase-8-deficient I9.2 Jurkat cells (A and B) or the caspase-8-deficient neuroblastoma cell line NB7 (C and D) were stably transduced with full-length human caspase-8 or the indicated mutants. Equal expression of each mutant, as well as absence of endogenous caspase-8, was confirmed by Western blot for caspase-8 (A and C). Jurkat cells were treated with TRAIL (100 ng/ml) for 6 h, although NB7 cells were treated with 100 ng/ml TNFα + 5 μg/ml cycloheximide for 6 h. Following these treatments, cells were stained with annexin-V-allophycocyanin, and apoptosis was assessed by FACS. Vect., vector; Cat.Mut., catalytic mutant; CHX, cycloheximide.
Studying Caspase Activation Using Inducible Cleavage
The results presented above indicate that permissive interdomain cleavage sites are required for activation of caspase-8 by drug-mediated dimerization, as well as for efficient induction of apoptosis by the extrinsic apoptotic pathway. However, such sites are apparently dispensable for kosmotrope-mediated caspase-8 activation. To further explore the role of interdomain cleavage in initiator caspase activation, we sought a system in which cleavage and dimerization of caspase-8 could be precisely controlled in living cells. To achieve this, we modified the auto-processing sites within caspase-8 to sites that could be cleaved by an exogenous protease. The TEV protease was an ideal candidate for this because its stringent substrate preference and relatively low enzymatic activity make it an exquisitely selective protease. It has long been used for removal of affinity tags during recombinant protein purification and therefore has well defined catalytic parameters and works well under physiological pH and temperature conditions.
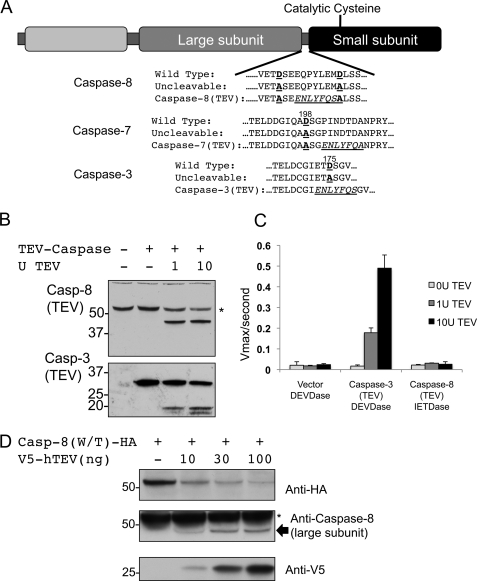
To develop an inducible cleavage system, we first introduced mutations into caspase-8 that changed its interdomain autocleavage sites to a TEV protease cleavage site. All permissive aspartic acid residues were mutated to alanine residues to eliminate the possibility of autocleavage or cleavage by other caspases, and the TEV consensus sequence ENLYFQS was introduced in the interdomain linker region (Fig. 3A). These mutations were introduced into both full-length caspase-8 and versions of caspase-8 in which the prodomain was replaced by the FKBP dimerization domain, as illustrated in Fig. 1. We found that upon purification of the FKBP-caspase-8 constructs carrying a TEV site, no autocleavage was observed, indicating that autoprocessing of these constructs was indeed eliminated by the mutations introduced (supplemental Fig. S1). Furthermore, these FKBP-caspase-8 constructs carrying TEV cleavage sites behaved similarly to the noncleavable FKBP-caspase-8 constructs when activated by AP20187- or kosmotrope-mediated dimerization; they were not activated by AP20187 but displayed robust activation in kosmotrope (supplemental Fig. S2). We also made TEV-cleavable versions of the executioner caspases, caspase-3 and caspase-7 (Fig. 3A).
FIGURE 3.
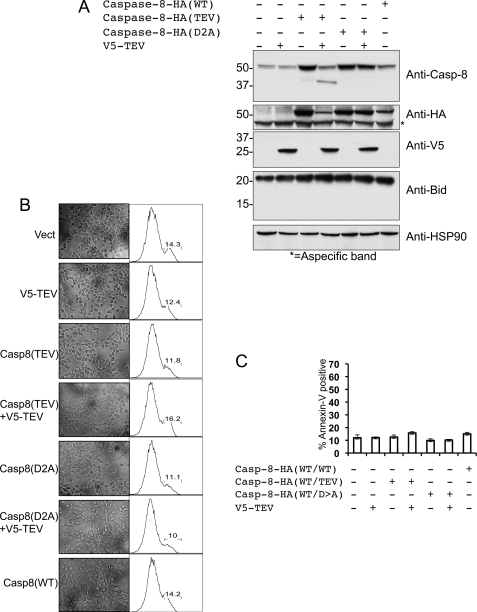
Creation of an inducibly cleavable caspase system by adaptation of the TEV protease. A, summary of mutations introduced into caspase-8, as well as into the executioner caspases caspase-3 and caspase-7. Cleaved aspartate residues or their alanine substitutes are depicted in red, and introduced TEV cleavage sites are italicized and underlined. Numbers indicate the mutated Asp residues, beginning from the N terminus of the human protein. RFU, relative fluorescent units. B, lysates from MCF-7 cells transfected with caspase-8(TEV) or caspase-3(TEV) were treated for 3 h with the indicated quantities of recombinant TEV protease, and effective cleavage was assessed by Western blot. C, caspase activity was assayed in lysates from MCF-7 cells transfected with vector, caspase-8(TEV), or caspase-3(TEV) and then treated with recombinant TEV protease as in B. The fluorogenic substrate Ac-DEVD-afc was used to assess caspase-3 activity, although Ac-IETC-afc was used for caspase-8. D, cleavage of caspase-8(TEV) via co-expression with optimized V5-TEV. 293A cells were co-transfected with caspase-8 bearing a C-terminal hemagglutinin (HA) tag and increasing amounts of V5-TEV, and cleavage was assessed by Western blot. Blotting for hemagglutinin reveals disappearance of full-length caspase-8(TEV)-hemagglutinin, whereas blotting with the C15 monoclonal caspase-8 antibody, which recognizes an epitope in the large subunit, reveals appearance of cleaved caspase-8. Vector was included in each transfection mix such that each sample received the same total quantity of DNA. * denotes residual signal from endogenous caspase-8.
Current models of caspase activation hold that inactive executioner caspases are present in the cytosol as inactive dimers that are activated by cleavage by initiator caspases, whereas initiator caspase zymogens are monomeric and are activated by dimerization (8); a corollary of this model is that cleavage alone is sufficient to activate executioner, but not initiator, caspases. As an initial test of our TEV-cleavable caspase system, we expressed TEV-cleavable caspase-3 or TEV-cleavable caspase-8 in 293T cells. Lysates from these cells were treated with increasing amounts of recombinant TEV, and cleavage was confirmed by Western blot (Fig. 3B). Caspase activity was then measured using an appropriate fluorogenic substrate as follows: Ac-DEVD-afc for caspase-3 and Ac-IETD-afc for caspase-8 (Fig. 3C). We observed robust dose-dependent induction of caspase-3 activity upon TEV addition but no such activation of caspase-8; TEV protease alone showed no activity on either DEVD-afc (Fig. 3C) or IETD-afc (data not shown). Together, these results both validate our TEV-cleavable caspase constructs and confirm that interdomain cleavage is sufficient to activate executioner, but not initiator, caspases.
To specifically cleave individual caspases in living cells using TEV protease, we next sought to adapt TEV protease to expression in mammalian cells. Transfection of mammalian cells with a gene encoding a V5-tagged version of naturally occurring TEV protease (C-terminally truncated to increase activity) yielded only limited protein expression and activity (supplemental Fig. S3). We suspected that nonoptimal codon usage in the viral version of the gene could be limiting its expression in mammalian cells. We therefore synthesized a version of the protease with codon usage optimized for expression in mammalian cells, and we found that this version expressed at much higher levels and readily cleaved a version of caspase-8 in which the interdomain linker region was mutated to a TEV protease consensus sequence (supplemental Fig. S3). Importantly, no cell death was observed upon expression of TEV protease alone in any of the cell types used in this study (data not shown). We next co-transfected TEV-cleavable caspase-8 and increasing amounts of human-optimized TEV protease into 293A cells (Fig. 3D). This optimized TEV protease efficiently cleaved caspase-8 in a dose-dependent manner, validating the in cellulo use of this system to study caspase cleavage.
Interdomain Cleavage of Caspase-7 Is Sufficient to Induce Apoptosis
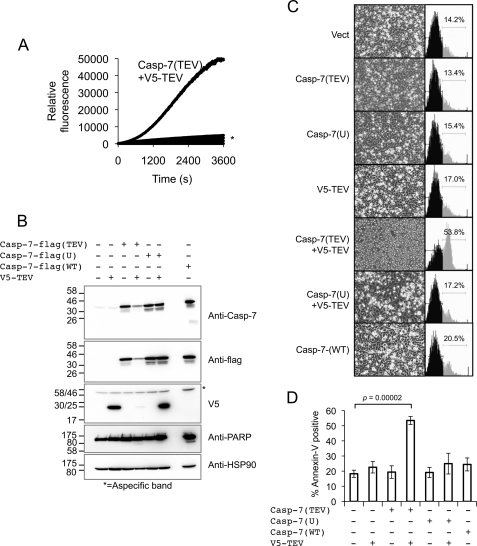
We next sought to use our cell-based TEV protease system to explore the consequences of interdomain cleavage of executioner and initiator caspases in living cells. Because caspase-7 was found to be more readily cleaved than caspase-3 by human-optimized TEV upon co-expression in cultured cells, we chose that executioner caspase for further study. We first validated our TEV-cleavable caspase-7 construct in vitro. Incubation of recombinantly expressed purified His6-tagged procaspase-7(TEV) with recombinant TEV protease resulted in robust activation (Fig. 4A). Importantly, TEV protease alone had no activity on the caspase substrate used (Ac-DEVD-Afc) and was unable to activate wild-type procaspase-7.
FIGURE 4.
In cellulo cleavage of caspase-7 by TEV protease results in cell death. A, His-tagged recombinant caspase-7(TEV) was incubated with TEV protease, and caspase activity was assessed immediately after TEV protease addition using the fluorogenic substrate Ac-DEVD-afc. * denotes overlaid control traces including the following: buffer alone, caspase-7(TEV) alone, TEV protease alone, caspase-7 (WT), and caspase-7(WT) + TEV protease. B, 293A cells were transfected with the indicated pCDNA3.1 vectors, and lysates were immunoblotted as indicated 48 h post-transfection. HSP-90 is used as a loading control. PARP, poly(ADP-ribose) polymerase. C, cells transfected as in B were stained with annexin V-PE and analyzed by FACS. Representative FACS histograms are presented. Inverted microscope images of corresponding cells are shown to confirm the morphologic events associated with apoptosis. D, quantification of annexin-V positivity in cells transfected as indicated. This figure is representative of 3–4 independent experiments.
To demonstrate that TEV protease can activate a caspase and induce cell death in mammalian cells, V5-tagged TEV protease was co-expressed with C-terminally FLAG-tagged caspase-7 in which the interdomain cleavage site was mutated to a TEV site (caspase-7(TEV)) and protein expression was assessed by immunoblotting. Caspase-7(TEV) was readily processed upon co-expression with V5-TEV in 293A cells (note reduction in full-length caspase, see Fig. 4B). Processing was accompanied by some cleavage of the caspase-7 death substrate poly(ADP-ribose) polymerase. Conversely, less or no processed poly(ADP-ribose) polymerase was observed in single DNA-transfected samples or when TEV protease was co-expressed with uncleavable caspase-7 D198A mutant (caspase-7(U)).
Co-expression of caspase-7(TEV) and TEV protease was expected to induce rapid apoptosis of co-transfected cells, making visualization of TEV protease and cleaved caspase-7 by Western blot difficult in these cells (Fig. 4B, 4th lane). To ensure that co-expression of TEV protease and caspase-7(TEV) was indeed causing productive caspase-7 cleavage, we repeated this transfection in the presence of the pancaspase inhibitor benzyloxycarbonyl-VAD-fmk (supplemental Fig. S4). When death was blocked by benzyloxycarbonyl-VAD, both TEV expression and cleaved caspase-7 were readily visible (supplemental Fig. S4).
Cleavage of caspase-7(TEV) resulted in morphologically distinguishable apoptosis as observed by light microscopy (Fig. 4C). To better quantify the ability of TEV protease-activated caspase-7(TEV) to induce death, we performed annexin-V-PE staining of transfected cells. Co-expression of caspase-7(TEV) and TEV protease led to greater rates of apoptosis than did any of the controls tested (Fig. 4, C and D). As a means of comparison, and in the experimental conditions used, transfection of WT caspase-7 alone resulted in no significant increase in cell death. Annexin-V staining as a measure of apoptosis of transfected cells under the indicated conditions was quantified (Fig. 4D). Taken together, these results show that TEV protease can induce cell death via the processing of an engineered caspase, thus validating our approach.
Interdomain Cleavage of Caspase-8 in the Absence of a Dimerization Signal Does Not Induce Apoptosis
We next sought to extend this approach to caspase-8. Although the findings presented above indicate that caspase-8 cleavage is indeed necessary for its activation in cells, the question of whether cleavage is sufficient to activate in this context has been the subject of some debate. Our earlier in vitro studies (13) have indicated that unlike the executioner caspases (see Figs. 3C and 4A), caspase-8 cleavage is not sufficient to induce its proteolytic activity. However, earlier reports (16, 17) claimed that cleavage of caspase-8 was indeed sufficient to induce its proteolytic activity in the absence of receptor-driven dimerization. We therefore studied the effects of specific cleavage of caspase-8 zymogen in living cells. Uncleavable or TEV-cleavable caspase-8 was transiently expressed in 293A cells, in a manner analogous to that previously employed for caspase-7 (Fig. 5A, compare with Fig. 4B). Cleavage of caspase-8(TEV), but not of WT caspase-8, was readily observed upon co-expression with TEV protease. However, blotting for the caspase-8 substrate Bid revealed no observable Bid cleavage in the presence of cleaved caspase-8. Furthermore, microscopic observation or FACS analysis of these cells revealed that, unlike the case with caspase-7, TEV-mediated cleavage of caspase-8 did not induce apoptosis in these cells (Fig. 5B). Annexin-V reactivity induced by TEV-mediated caspase-8 cleavage in living cells was also negative (Fig. 5C). We therefore conclude that caspase-8 cleavage is not sufficient to induce its activity or to drive apoptosis.
FIGURE 5.
In cellulo cleavage of caspase-8 by TEV protease does not result in cell death. A, 293A cells were transfected with the indicated pCDNA3.1 vectors (Vect), and lysates were immunoblotted as indicated 48 h post-transfection. Caspase-8 was detected using the C15 monoclonal antibody, which recognizes an epitope in the large subunit. HSP-90 is used as a loading control. B, cells transfected as in A were stained with annexin V-PE and analyzed by FACS. Representative FACS histograms are presented. Inspection of cells via light microscopy revealed no discernible apoptotic phenotype in any sample (data not shown). C, quantification of annexin-V positivity in cells transfected as indicated. This figure is representative of three independent experiments.
Both Cleavage and Dimerization Are Necessary for Caspase-8 Activation in Vivo
Our results indicate that dimerization in the absence of interdomain cleavage is not sufficient to activate caspase-8; permissive autocleavage sites in the interdomain region of caspase-8 are required for dimerization-induced caspase-8 activation in vitro, as well as for death receptor-induced activation of caspase-8 and subsequent apoptosis in living cells. Furthermore, our TEV-cleavable system demonstrated that, unlike the executioner caspases, specific cleavage of caspase-8 does not lead to its activation nor does it drive apoptosis. We therefore sought to unify these findings by investigating the possibility that both dimerization and interdomain cleavage are required to activate caspase-8 and drive apoptosis.
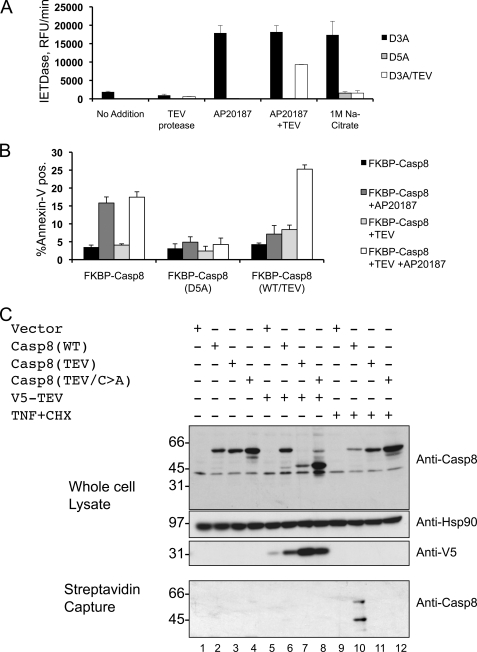
We utilized purified FKBP-Caspase-8 proteins carrying TEV cleavage sites in the interdomain linker regions (supplemental Fig. S5). Because their mutations render them resistant to autocleavage, these proteins did not demonstrate significant activation upon drug-induced dimerization in the absence of TEV protease (Fig. 6A and supplemental Table 1). Consistent with our previous results and previously published findings (13), TEV treatment alone did not activate these constructs to appreciable levels. However, treatment with TEV protease followed by drug-induced dimerization yielded potent activation, demonstrating that both dimerization and cleavage were required for caspase-8 activation in vitro under physiologically relevant conditions (Fig. 6A and supplemental Table 1).
FIGURE 6.
Both cleavage and dimerization are required for caspase-8 activation in vitro and for apoptosis in cellulo. A, FKBP-Caspase-8, or the indicated mutants thereof, were produced in bacteria and purified (see supplemental Fig. S4). Following purification, proteins were incubated with recombinant TEV protease, the dimerizer AP20187, or 1 m sodium citrate in the indicated combinations, and proteolytic activity was assayed using the fluorogenic substrate AcIETD-afc. RFU, relative fluorescent units. B, HeLa cells were transfected with FKBP-Caspase-8 or the indicated mutants thereof, along with either V5-TEV protease in pCDNA3.1 vector, or empty vector such that each sample received the same total quantity of DNA. Enhanced GFP was included to mark transfected cells. Twenty four hours post-transfection, cells were treated with AP20187 or vehicle as indicated, and a further 6 h later cells were harvested and stained with annexin-V-APC. Cells were analyzed by FACS; graphs represent transfected cells that were also annexin-V-positive. The graph represents quantification of three independent experiments. C, caspase-8-deficient NB7 cells were transfected with the indicated full-length caspase-8 and TEV protease vectors. Twenty four hours post-transfection, biotinylated-VAD-fmk was added, and cells were then treated with 2 μg/ml cycloheximide (CHX) plus 100 ng/ml TNF as indicated. Cells were then lysed, and lysates were incubated with streptavidin beads to trap biotin-labeled proteins. Inputs and captured proteins were then analyzed by Western blot. Caspase-8 was detected using the C15 antibody.
We next sought to extend these findings to living cells. Constructs encoding wild type, noncleavable, or TEV-cleavable versions of FKBP-caspase-8 were transfected into HeLa cells in the presence or absence of TEV protease. Dimerizer was then added, and apoptosis was assessed by annexin staining and FACS. No variation in apoptotic response was observed in the presence or absence of TEV protease for wild-type or noncleavable caspase-8 upon addition of dimerizer. Wild-type FKBP-caspase-8 readily induced apoptosis upon addition of dimerizer, whereas noncleavable FKBP-caspase-8 did not (Fig. 6B). Dimerization of TEV-cleavable FKBP-caspase-8 in the absence of TEV protease also failed to induce apoptosis, as did co-expression of this construct with TEV protease in the absence of dimerizer. However, when TEV was co-expressed with this construct and dimerizer was subsequently added, apoptosis was induced with efficiency comparable with wild-type FKBP-caspase-8.
Finally, we sought to determine the activation requirements of full-length caspase-8 in living cells. For this experiment, we utilized the biotin-labeled pan-caspase inhibitor b-VAD. This activity-based probe binds irreversibly to the catalytic cysteine of active caspases, and the inclusion of a biotin moiety allows subsequent affinity purification of b-VAD-bound proteins to determine which, if any, caspases are active under specific cellular conditions (30, 31). Caspase-8-deficient NB7 cells were transfected with wild-type or TEV-cleavable caspase-8 constructs. These constructs were either co-expressed with TEV protease or activated by TNF treatment, and active caspases were trapped using b-VAD in each case (Fig. 6C). Consistent with our earlier findings, cleavage of caspase-8 by TEV in the absence of a dimerization signal did not lead to activation or b-VAD binding (Fig. 6C, lane 8). Likewise, TNF treatment of cells expressing autoprocessing-prohibitive mutants of caspase-8 did not result in b-VAD binding (Fig. 6C, lane 11). Receptor-driven dimerization of caspase-8 was only able to induce its activity in the presence of a permissive interdomain cleavage site (Fig. 6C, lane 10).
DISCUSSION
The findings presented in this study indicate that both dimerization and interdomain cleavage of caspase-8 are necessary for apoptosis induction under physiological conditions. Although autocleavage has previously been shown to stabilize dimeric caspase-8, it was thought that dimerization of caspase-8 zymogens was synonymous with activation, with cleavage optimizing proteolytic activity and stabilizing the enzyme (5–7, 13). Here, we show that cleavage, although not sufficient to activate caspase-8, is nonetheless necessary. Additionally, previous reports have found that cleavage of caspase-8 alone can lead to its activation (16, 17). We find that cleavage in the absence of dimerization does not produce an active enzyme nor does it drive apoptosis.
Many studies have shown that dimerization of caspase-8 zymogens can activate caspase-8 proteolytic activity and induce apoptosis. Apoptotic caspase-8 activation in cells leads to its interdomain autoproteolytic processing; however, the role and requirement of this cleavage event have been the subject of some debate. In vitro work using purified FKBP-caspase-8 driven to dimerize with the FKBP-binding drug AP20187 indicated that mutation of the interdomain autoprocessing sites of caspase-8 reduced but did not abrogate its activation following dimerizer addition; these same studies found that transfection of these constructs into HeLa cells followed a similar pattern, with elimination of the interdomain cleavage sites reducing, but not eliminating, apoptosis upon dimerizer addition (7). Additional in vitro work has supported the idea that caspase-8 dimerization, in the absence of interdomain processing, can lead to its activation. Previously published work from ourselves and others used purified recombinant caspase-8 lacking a prodomain and induced this construct to dimerize by addition of kosmotropic salts (5, 13). These studies also found that dimerization in the absence of interdomain processing yielded reduced but still significant proteolytic activity, with interdomain processing serving to stabilize the active dimer.
The mechanism proposed by these biochemical studies is not entirely supported by work performed in vivo. Most significantly, when caspase-8-deficient mice were reconstituted with a caspase-8 construct that cannot undergo interdomain processing, cells and tissues from the resulting mice were resistant to stimuli that induce apoptosis via caspase-8 (14). In addition, very recent work has confirmed the apparent requirement for intact interdomain processing sites for induction of apoptosis by caspase-8, by reconstituting caspase-8-deficient Jurkat cells with various noncleavable mutants of caspase-8 (15).
The findings presented here support the idea that interdomain cleavage of caspase-8 is required for efficient activation and induction of apoptosis under physiological conditions. We found that purified FKBP-Caspase-8 that could not undergo interdomain cleavage was indeed able to be activated by addition of kosmotropic salts, as reported previously. However, upon dimerization via the FKBP domain, which is predicted to more faithfully mimic physiological dimerization by activated receptors, elimination of interdomain processing sites rendered caspase-8 nonactivatable, most likely due to the inability of noncleavable caspase-8 to form a stable dimeric enzyme. We suggest that dimerization induced by buffer conditions, such as high kosmotropic salt levels, drives caspase zymogens into sustained dimeric conformations by acting on the entire dimer interface, although prodomain-driven dimerization merely brings the zymogens into proximity and requires cleavage and molecular rearrangement to “lock in” the dimeric conformation.
Our findings conflict with those presented by others who used similar FKBP-caspase-8 constructs but found that noncleavable FKBP-caspase-8 activity could be induced by addition of dimerizer drug in vitro and that the same construct could drive apoptosis in cells, albeit to a lesser extent than the cleavable version. Because differences in protein purification conditions or concentrations of purified protein used in activity assays can differ, we sought to determine whether noncleavable caspase-8 could be productively activated by prodomain-driven dimerization in a more physiological context. We found that reconstitution of either caspase-8-deficient Jurkat or NB7 cell lines with noncleavable caspase-8 yielded cells insensitive to TRAIL or TNF treatment, respectively. These findings precisely mirror our in vitro activity results obtained using purified protein, confirming the requirement for interdomain processing to drive apoptosis in vivo. Given the manifestly nonphysiological nature of high kosmotropic salt conditions, and the finding that caspase-8-deficient cells reconstituted with noncleavable caspase-8 fail to undergo apoptosis in response to death ligand binding, we can also conclude that kosmotropic salt-mediated dimerization does not faithfully mimic physiological caspase-8 activation, which absolutely requires interdomain processing.
Another persistent question concerns the effect of caspase-8 cleavage at its interdomain autoprocessing sites by other enzymes. Both caspase-6 and granzyme-B have been found to effectively cleave caspase-8 at these sites, and reports have held that this cleavage is sufficient to induce caspase-8 activity (16, 17). This question is not one of mere idle interest, because active caspase-8 is able to activate executioner caspases; if the converse were also true, significant feedback amplification of the apoptotic signal would be expected to result. However, this seemingly simple premise has been difficult to test in practice, because of the limited utility of inhibitors purportedly specific to single caspases (32) and the self-amplifying nature of the executioner caspases. Our earlier work in vitro demonstrated that interdomain processing of caspase-8 zymogen was not sufficient to induce its proteolytic activity (13); however, the effect of such cleavage events in the cellular context warranted further study. We therefore adapted a viral protease, TEV protease, for use in mammalian cells. We found that codon optimization was required to achieve useful expression and activity levels in cells. As a proof of concept, we produced the executioner caspases, caspase-3 and caspase-7, with TEV protease cleavage sites, and as expected, we found that cleavage of these caspases with TEV protease was sufficient to activate their proteolytic activity in vitro and to drive apoptosis in cellulo. However, this was not the case with caspase-8; interdomain cleavage of this caspase by TEV protease induced neither activation in vitro nor apoptosis in cells. This finding differs from those of Sohn et al. (16) and Murphy et al. (17). In the study by Murphy et al. (17), caspase-8 was cleaved in lysates by addition of recombinant caspase-6 or granzyme-B, then purified by immunoprecipitation, and tested for activity; and it is possible that in this protocol immunoprecipitation itself induces proximity, and therefore activity, of cleaved caspase-8 zymogens. Sohn et al. (16) found activation of caspase-8 following etoposide-induced DNA damage as measured by affinity tag binding and concluded that this activation was the result of cleavage in the absence of a dimerization signal. However, work from our laboratory found that etoposide treatment induces Fas expression and subsequent caspase-8 activation via the well defined death receptor pathway (30).
A major advantage of the TEV protease system is its stringent specificity. The ideal TEV protease cleavage site is 7 amino acids in length and does not occur naturally in the human proteome; off-target effects are therefore expected to be minimal, and indeed cells tolerate TEV protease expression with no perceivable ill effects. Co-expression of TEV-cleavable proteins with the TEV protease therefore allows analysis of the effects of specific proteolytic cleavage events and will be an exceptionally useful tool in dissecting proteolysis-mediated pathways in vivo. The caspase activation cascade provides an excellent example of this utility. Over 700 putative caspase substrates have been described previously (33), but discerning which of these cleavage events actually influences cell fate is challenging. Because executioner caspase activation leads to a terminal cell fate, apoptosis, there is not expected to be evolutionary pressure against the presence of caspase cleavage sites. This has led to numerous reports correlating the cleavage of a given substrate to a specific feature of apoptosis, but due to the massive morphological changes associated with apoptosis, mutation of a putative caspase cleavage site to show its relevance can be difficult within the context of the dying cell. Using optimized TEV protease, it will be possible to introduce TEV consensus sites into such substrates and induce their cleavage in otherwise healthy cells. Although this work serves as a proof of concept, the potential utility of this system goes well beyond the results presented here.
The results discussed above indicate that neither dimerization nor cleavage alone, in the absence of the other, is sufficient to activate caspase-8 to drive apoptosis. We sought to prove this formally through the combination of FKBP-driven inducible dimerization and TEV-mediated inducible cleavage. We first purified TEV-cleavable FKBP-caspase-8 constructs, and as expected we found that neither dimerization nor cleavage alone induced proteolytic activity in these proteins. However, TEV cleavage followed by drug-induced dimerization did induce robust catalytic activity. We next moved this system into HeLa cells, using co-expression of our human-optimized TEV construct to induce cleavage, with results similar to those obtained in vitro; only when both cleavage and dimerization were induced did robust apoptosis result.
As a final test of this model, we transfected caspase-8-deficient NB7 cells with constructs encoding wild-type or TEV-cleavable caspase-8, and we found that TEV cleavage in the absence of dimerization, or TNF treatment of cells expressing noncleavable caspase-8, failed to induce caspase-8 activity measured by b-VAD-fmk labeling. Only dimerization of cleavage-competent caspase-8 led to activation. Close examination of this result reveals a puzzling wrinkle, whereas only cleavable caspase-8 labels with b-VAD-fmk, both full-length and cleaved caspase-8 appear in the streptavidin capture (Fig. 6C). Although this indicates that noncleaved caspase-8 can acquire activity, cells in which the TEV-cleavable construct (which is resistant to autoprocessing) was present showed no such labeling. This finding could indicate that cleavage of one member of a dimer is sufficient to induce b-VAD labeling of both members; indeed, this idea agrees with recently published results (15). Although other models are possible, the parsimonious explanation of the dual requirement for dimerization and cleavage is that dimerization activates caspase-8 but that the dimer formed at the DISC is transient and unstable. Only auto-proteolysis (which would be favored due to juxtaposition of caspase-8 in the DISC) and possibly cleavage of other DISC-associated proteins occur. This autoproteolysis stabilizes the caspase-8 dimer to allow sustained activity with cleavage and activation of downstream executioner caspases. This activation/stabilization model is essentially compatible with most published reports on caspase activation mechanisms.
The requirement for interdomain processing for activation of the initiator caspase-9 differs markedly from that of caspase-8. A number of publications indicate that caspase-9 bearing prohibitive mutations at the interdomain processing sites between the large and small subunits can be robustly activated in vitro by addition of purified Apaf-1 and cytochrome c (34–36). Furthermore, expression of noncleavable caspase-9 in cell lines is able to support robust activation of caspases following cytochrome c release, again indicating that interdomain processing of caspase-9 is dispensable for its activation (35). The best evidence that requirement for interdomain cleavage during activation is fundamentally different between caspase-8 and caspase-9 comes from studies using “caspase-9/8,” a fusion protein composed of a caspase-9 prodomain fused to caspase-8 catalytic domains. Both caspase-9 and caspase-9/8 can be readily activated by in vitro apoptosome formation, confirming that both caspase-8 and caspase-9 share a requirement for dimerization of inactive zymogens during activation (4). However, although noncleavable caspase-9 is still able to be robustly activated, caspase-9/8 containing prohibitive mutations at the interdomain cleavage sites of the caspase-8 catalytic domains is not activated by apoptosome formation in vitro. In light of the results presented here, this finding further confirms that unlike caspase-9, caspase-8 absolutely requires interdomain processing during activation.
The data presented here show that cleavage of caspase-8 is required for induction of apoptosis and that noncleavable caspase-8 cannot be activated by physiological homodimerization, nor can it drive apoptosis induced by the extrinsic pathway. These findings are in agreement with those generated by introducing noncleavable caspase-8 into a caspase-8-deficient mouse; tissues and cells from that animal are resistant to extrinsic apoptosis (14). However, an additional and extremely provocative finding of that study was that introduction of noncleavable caspase-8 nonetheless rescued the severe developmental defects and embryonic lethality caused by caspase-8 deficiency. These results indicate that uncleaved caspase-8 plays a fundamental role in nonapoptotic pathways required for development. A requirement for caspase-8 during T-cell proliferation has also been described, and recent work has indicated that caspase-8-deficient T-cells proliferate normally but die at high rates via the recently described “programmed necrosis” pathway (37, 38). This pathway is dependent on the receptor-interacting protein kinases RIPK1 and RIPK3. Intriguingly, both of these proteins contain caspase-8 cleavage sites (39, 40), and this has led to the hypothesis that limited autocleavage-independent activation of caspase-8 could be required to inactivate the receptor-interacting protein kinases following T-cell activation. This pathway could also play a role in development, explaining the developmental rescue observed upon re-introduction of noncleavable caspase-8. However, the biochemical mechanisms by which the cleavage-independent, nonapoptotic activity of caspase-8 is brought to bear remain elusive. Knockouts of the adaptor protein FADD or the caspase-like protein FLIP share the developmental phenotype of the caspase-8 knock-out, leading to the hypothesis that formation of caspase-8/FLIP heterodimers could be important for the nonapoptotic activity of caspase-8. However, biochemical analyses of these heterodimeric species indicate that they form more readily, are more active, and can drive apoptosis with higher efficiency than caspase-8 homodimers.
Supplementary Material
Acknowledgments
We thank ARIAD Pharmaceuticals for providing the dimerization system, Dr. Andrei Osterman for providing TEV plasmid for bacterial expression, and Scott Snipas for outstanding technical assistance. We also thank Pat Fitzgerald for invaluable advice and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants AI47891 (to D. R. G.), AI44828 (to D. R. G.), and CA69381 (to D. R. G. and G. S. S.). This work was also supported in part by Canadian Institutes of Health Research and Cancer Research Society operating grants (to J.-B. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table 1.
- DISC
- death-inducing signaling complex
- TNF
- tumor necrosis factor
- FACS
- fluorescence-activated cell sorter
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- Pipes
- 1,4-piperazinediethanesulfonic acid
- TEV
- tobacco etch virus
- b-
- biotin-
- fmk
- fluoromethyl ketone
- GFP
- green fluorescent protein
- WT
- wild type
- FKBP
- FK506-binding protein
- afc
- 7-amino-4-trifluoromethylcoumarin.
REFERENCES
- 1.Fuentes-Prior P., Salvesen G. S. (2004) Biochem. J. 384, 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pop C., Salvesen G. S. (2009) J. Biol. Chem. 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson D. W. (1999) Cell Death Differ. 6, 1028–1042 [DOI] [PubMed] [Google Scholar]
- 4.Pop C., Timmer J., Sperandio S., Salvesen G. S. (2006) Mol. Cell 22, 269–275 [DOI] [PubMed] [Google Scholar]
- 5.Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. (2003) Mol. Cell 11, 529–541 [DOI] [PubMed] [Google Scholar]
- 6.Donepudi M., Mac Sweeney A., Briand C., Grütter M. G. (2003) Mol. Cell 11, 543–549 [DOI] [PubMed] [Google Scholar]
- 7.Chang D. W., Xing Z., Capacio V. L., Peter M. E., Yang X. (2003) EMBO J. 22, 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riedl S. J., Shi Y. (2004) Nat. Rev. Mol. Cell Biol. 5, 897–907 [DOI] [PubMed] [Google Scholar]
- 9.Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K. M., Krammer P. H., Peter M. E. (1998) EMBO J. 17, 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samraj A. K., Keil E., Ueffing N., Schulze-Osthoff K., Schmitz I. (2006) J. Biol. Chem. 281, 29652–29659 [DOI] [PubMed] [Google Scholar]
- 11.Jost P. J., Grabow S., Gray D., McKenzie M. D., Nachbur U., Huang D. C., Bouillet P., Thomas H. E., Borner C., Silke J., Strasser A., Kaufmann T. (2009) Nature 460, 1035–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer S. L., Gaudet S., Albeck J. G., Burke J. M., Sorger P. K. (2009) Nature 459, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pop C., Fitzgerald P., Green D. R., Salvesen G. S. (2007) Biochemistry 46, 4398–4407 [DOI] [PubMed] [Google Scholar]
- 14.Kang T. B., Oh G. S., Scandella E., Bolinger B., Ludewig B., Kovalenko A., Wallach D. (2008) J. Immunol. 181, 2522–2532 [DOI] [PubMed] [Google Scholar]
- 15.Hughes M. A., Harper N., Butterworth M., Cain K., Cohen G. M., MacFarlane M. (2009) Mol. Cell 35, 265–279 [DOI] [PubMed] [Google Scholar]
- 16.Sohn D., Schulze-Osthoff K., Jänicke R. U. (2005) J. Biol. Chem. 280, 5267–5273 [DOI] [PubMed] [Google Scholar]
- 17.Murphy B. M., Creagh E. M., Martin S. J. (2004) J. Biol. Chem. 279, 36916–36922 [DOI] [PubMed] [Google Scholar]
- 18.Crabtree G. R., Schreiber S. L. (1996) Trends Biochem. Sci. 21, 418–422 [DOI] [PubMed] [Google Scholar]
- 19.Spencer D. M., Belshaw P. J., Chen L., Ho S. N., Randazzo F., Crabtree G. R., Schreiber S. L. (1996) Curr. Biol. 6, 839–847 [DOI] [PubMed] [Google Scholar]
- 20.Clackson T., Yang W., Rozamus L. W., Hatada M., Amara J. F., Rollins C. T., Stevenson L. F., Magari S. R., Wood S. A., Courage N. L., Lu X., Cerasoli F., Jr., Gilman M., Holt D. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10437–10442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan L., Freeman K. W., Khan T., Pham E., Spencer D. M. (1999) Hum. Gene Ther. 10, 2273–2285 [DOI] [PubMed] [Google Scholar]
- 22.Muzio M., Stockwell B. R., Stennicke H. R., Salvesen G. S., Dixit V. M. (1998) J. Biol. Chem. 273, 2926–2930 [DOI] [PubMed] [Google Scholar]
- 23.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 24.Kapust R. B., Waugh D. S. (2000) Protein Expr. Purif. 19, 312–318 [DOI] [PubMed] [Google Scholar]
- 25.Phan J., Zdanov A., Evdokimov A. G., Tropea J. E., Peters H. K., 3rd, Kapust R. B., Li M., Wlodawer A., Waugh D. S. (2002) J. Biol. Chem. 277, 50564–50572 [DOI] [PubMed] [Google Scholar]
- 26.Kapust R. B., Tözsér J., Copeland T. D., Waugh D. S. (2002) Biochem. Biophys. Res. Commun. 294, 949–955 [DOI] [PubMed] [Google Scholar]
- 27.Denault J. B., Salvesen G. S. (2008) Methods Mol. Biol. 414, 191–220 [DOI] [PubMed] [Google Scholar]
- 28.Denault J. B., Salvesen G. S. (2003) Curr. Protoc. Protein Sci. Unit 21, 13 [DOI] [PubMed] [Google Scholar]
- 29.Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., Vanin E. F., Vignali D. A. (2004) Nat. Biotechnol. 22, 589–594 [DOI] [PubMed] [Google Scholar]
- 30.Tu S., McStay G. P., Boucher L. M., Mak T., Beere H. M., Green D. R. (2006) Nat. Cell Biol. 8, 72–77 [DOI] [PubMed] [Google Scholar]
- 31.Denault J. B., Salvesen G. S. (2003) J. Biol. Chem. 278, 34042–34050 [DOI] [PubMed] [Google Scholar]
- 32.McStay G. P., Salvesen G. S., Green D. R. (2008) Cell Death Differ. 15, 322–331 [DOI] [PubMed] [Google Scholar]
- 33.Lüthi A. U., Martin S. J. (2007) Cell Death Differ. 14, 641–650 [DOI] [PubMed] [Google Scholar]
- 34.Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. (1999) J. Biol. Chem. 274, 8359–8362 [DOI] [PubMed] [Google Scholar]
- 36.Denault J. B., Eckelman B. P., Shin H., Pop C., Salvesen G. S. (2007) Biochem. J. 405, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ch'en I. L., Beisner D. R., Degterev A., Lynch C., Yuan J., Hoffmann A., Hedrick S. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17463–17468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su H., Bidère N., Zheng L., Cubre A., Sakai K., Dale J., Salmena L., Hakem R., Straus S., Lenardo M. (2005) Science 307, 1465–1468 [DOI] [PubMed] [Google Scholar]
- 39.Feng S., Yang Y., Mei Y., Ma L., Zhu D. E., Hoti N., Castanares M., Wu M. (2007) Cell. Signal. 19, 2056–2067 [DOI] [PubMed] [Google Scholar]
- 40.Rébé C., Cathelin S., Launay S., Filomenko R., Prévotat L., L'Ollivier C., Gyan E., Micheau O., Grant S., Dubart-Kupperschmitt A., Fontenay M., Solary E. (2007) Blood 109, 1442–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.