Abstract
NKG2D is an important activating receptor on lymphocytes. In human, it interacts with two groups of ligands: the major histocompatibility complex class I chain-related A/B (MICA/B) family and the UL-16 binding protein (ULBP) family, also known as retinoic acid early transcript (RAET1). MIC proteins are membrane-anchored, but all of the ULBP/RAET1 proteins, except for RAET1E and RAET1G, are glycosylphosphatidylinositol (GPI)-anchored. To address the reason for these differences we studied the association of RAET1G with the membrane. Using epitope-tagged RAET1G protein in conjunction with antibodies to different parts of the molecule and in pulse-chase experiments, we showed that the C terminus of the protein was cleaved soon after protein synthesis. Endoglycosidase H and peptide N-glycosidase treatment and cell surface immunoprecipitation indicated that most of the protein stayed in the endoplasmic reticulum, but some of the cleaved form was modified in the Golgi and transported to the cell surface. We examined the possibility of GPI anchoring of the protein in three ways: (i) Phosphatidylinositol (PI)-specific phospholipase C released the PI-linked form of the protein. (ii) The surface expression pattern of RAET1G decreased in cells defective in GPI anchoring through mutant GPI-amidase. (iii) Site-directed mutagenesis, to disrupt residues predicted to facilitate GPI-anchoring, resulted in diminished surface expression of RAET1G. Thus, a form of RAET1G is GPI-anchored, in line with most other ULBP/RAET1 family proteins. The cytoplasmic tail and transmembrane domains appear to result from gene duplication and frameshift mutation. Together with our previous results, our data suggest that RAET1G is regulated post-translationally to produce a GPI-anchored isoform.
Keywords: Glycolipids/Glycosylphosphatidylinositol Anchors, Immunology, Immunology/Cellular Response, Membrane/Proteins, Protein/Post-translational Modification, Protein/Processing, Natural Killer (NK) Cell, NKG2D
Introduction
NKG2D3 is a C-type lectin-like activating receptor, expressed on natural killer (NK) cell and T cells, that plays an important role in triggering cytotoxic activity. Remarkably, the receptor interacts with multiple different ligands. In humans, the ligands fall into two groups, namely the MHC class I chain-related A/B (MICA/B) family and the UL-16-binding protein (ULBP) or retinoic acid expressed transcript (RAET1) family (1–4). In mice, there are at least six NKG2D ligands; some of which (Rae1α-ϵ, H60c) are GPI-anchored, and the other three, H60a, H60b, and Mult1 (murine UL-16-binding protein-like transcript 1) are transmembrane proteins (5, 6).
MICA/B and ULBP/RAET1 family proteins differ in domain structure and share low amino acid sequence similarity. Both MIC family proteins, MICA and MICB, consist of three α domains, transmembrane domains, and cytoplasmic tail. In contrast, ULBP/RAET1 proteins have two α domains and, of the six molecules, four (ULBP1, ULBP2, ULBP3, and ULBP6 (RAET1L)), are GPI-anchored proteins (4, 7). The other two, ULBP4 (RAET1E) and ULBP5 (RAET1G) have been considered to have transmembrane domains (4, 8).
ULBP/RAET1 family proteins share different degrees of sequence similarity and may have arisen by a series of gene duplications (4). Some ULBP/RAET1 loci are recent duplicates. The extracellular domains of RAET1G and ULBP2 share 92% amino acid identity, for example (4). In addition to their sequence similarity, RAET1G and ULBP2 share similar expression patterns. Their transcripts are often coexpressed in cell lines and are induced by the same stimuli (9, 10). Therefore, it is striking that only two of the six ULBP/RAET1 proteins, RAET1E and RAET1G, span the cell membrane.
The cell surface expression of NKG2D ligands is tightly controlled and is regulated at multiple levels. First, the expression of NKG2D ligand transcript is generally absent in healthy cells but is up-regulated by cell stress signals such as viral infection (11), Toll-like receptor signaling (9, 12), and DNA damage (13). A second level of regulation has been proposed to act post-transcriptionally, mediated by microRNAs (14). Third, NKG2D ligand expression can be regulated at the post-translational level. For example, Nice et al. (15) reported post-translational regulation of Mult1 protein, which is considered as a functional ortholog of RAET1G in mice. The protein undergoes ubiquitination on lysine residues in the cytoplasmic tail and consequent lysosomal degradation, preventing its surface expression. On receipt of DNA damage signals, the ubiquitination of Mult1 protein was reduced, and the protein was stably expressed at the cell surface (15). RAET1G has a single lysine residue in the cytoplasmic tail, indicating that it is also a candidate for ubiquitination-mediated regulation.
Here, we examined the trafficking and post-translational modification of RAET1G in an effort to understand the role of its large C-terminal domain and how it might differ functionally from other NKG2D ligands. To approach this, we set out to clarify its mode of association with the cell membrane.
EXPERIMENTAL PROCEDURES
Cells and Plasmids
HT1080 and HeLa cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 units/ml penicillin. K562 cells were grown in RPMI 1640 medium containing 10% fetal bovine serum and 100 μg/ml streptomycin. K562 class K cell lines were a kind gift from Dr. Kanzawa and Prof. Kinoshita, Osaka University, Japan (16). The untagged, epitope-tagged, and GFP fusion constructs of RAET1G for transient transfection were created in the vector pcDNA3 (Invitrogen). Transient transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Stable cell lines were created with a lentiviral expression system (gift from Prof. Paul Lehner, University of Cambridge, UK) (17).
Antibodies and Reagents
Polyclonal anti-RAET1G-tail antiserum was previously described (18). Polyclonal and monoclonal anti-ULBP2 (AF1298 for Western blotting and immunoprecipitation, MAb1298 for flow cytometry), anti-MICB (AF1599 for Western blotting and immunoprecipitation, MAb1599 for flow cytometry) antibodies were purchased from R&D Systems. Anti-GFP antibody was from Abcam. Anti-V5 antibody (R960-25) was from Invitrogen. Isotype control mouse antibody (X0943), anti-goat (P0449), and mouse (P0447) Fc horseradish peroxidase-conjugated antibodies were purchased from Dako UK Ltd. Isotype control Fab antibody (MOR6391) and goat anti-human IgG F(ab′)2 horseradish peroxidase-conjugated antibody (0500-0099) were from AbD Serotec. Alexa Fluor 633 goat anti-mouse (A21053) and goat anti-human (A21091) antibodies were from Molecular Probes.
Preparation of Monoclonal Antibody
The monoclonal recombinant anti-RAET1G antibody (anti-RAET1G mAb) was generated by AbD Serotec, using the His-tagged extracellular domain of RAET1G as antigen of interest and His-tagged extracellular domain of closely related ULBP2 for negative selection. His-tagged extracellular domains of RAET1G and ULBP2 proteins were constructed in the vector pMW-H6 (a gift from Dr. A. Barrow), expressed in BL21 (DE3) pLysS competent cells (also from Dr. A. Barrow), purified using nickel-nitrilotriacetic acid-agarose (Qiagen) and refolded in 100 mm Tris-HCl (pH 8.0), 400 mm l-arginine hydrochloride, 2 mm EDTA, 5 mm reduced glutathione, 0.5 mm oxidized glutathione, and 0.1 mm PMSF, at 4 °C for 3 days. Each antibody had a 5-fold higher signal on ELISA detection of 5 μg/ml RAET1G, compared with ULBP2, when detected with 5 μg/ml antibody.
Endo H and PNGase Treatment
Cells were directly lysed in reducing SDS-PAGE sample buffer, boiled, and digested by Endo H or PNGase (New England Biolabs) for 30 min at 37 °C and subjected to Western blot analysis.
PI-PLC Treatment
Cells were washed with PBS and stripped with 10 mm EDTA in PBS. After washing with PBS twice, cells were incubated with 1 unit/ml PI-PLC (Sigma) in PBS for 30 min at 4 °C. The cells were washed with ice-cold PBS containing 1% bovine serum albumin and subjected to FACS analysis or centrifuged with 13,000 rpm for 15 min at 4 °C, and the resulting supernatant and pellet were subjected to Western blot analysis.
Western Blotting
Equal numbers of viable cells were lysed into reducing SDS-PAGE sample buffer, boiled, and separated by SDS-PAGE. Western blotting was performed using goat anti-MICB (AF1599) or goat anti-ULBP2 (AF1298) antibody (R&D Systems) or anti-RAET1G mAb described.
Pulse-Chase
Cells were harvested, washed in PBS, and then starved for 1 h at 37 °C in methionine/cysteine-free RPMI 1640 medium (Sigma) supplemented with 2 mmol/liter glutamine, 5% dialyzed fetal calf serum, and 10 mmol/liter HEPES. Cells were labeled with 1 mCi of [35S]methionine and [35S]cysteine Pro-mix (Amersham Biosciences; GE Healthcare)/107 cells for 0–60 min at 37 °C. Cells were chased in culture medium supplemented with excess unlabeled cysteine/methionine for 0–120 min, harvested, washed in ice-cold PBS, and then subjected to immunoprecipitation.
Immunoprecipitation
Equal numbers of viable cells were lysed in RIPA buffer (0.1% SDS, 0.5% DOC, 1% Nonidet P-40, 150 mm NaCl, and 50 mm Tris-HCl (pH 8.0)) containing protein inhibitor mixture tablet (Roche Diagnostics) and 10 mm PMSF for 30 min at 4 °C, and the resulting supernatant was incubated with antibody described and protein G-Sepharose 4B (GE Healthcare) with incubation overnight at 4 °C. After the beads were washed five times with RIPA buffer, proteins were eluted with SDS sample buffer by boiling for 5 min. Total protein and isolated labeled proteins were analyzed by Western blotting.
Flow Cytometry
Cells were washed with PBS and stripped off in 10 mm EDTA in PBS. After washing in ice-cold PBS containing 1% bovine serum albumin, cells were stained with mouse anti-ULBP2 (MAb1298), mouse anti-MICB (MAb1599) (R&D Systems) or anti-RAET1G antibody for 1 h at 4 °C, followed by goat anti-mouse Alexa Fluor 633 conjugate (A21053; Molecular Probes). As the stable cell lines were roughly 70% GFP-positive, GFP-negative cells were gated out in analysis. Data were then collected using BD FACSCalibur. All experiments were repeated at least twice.
Site-directed Mutagenesis
Mutation of RAET1G was generated by oligonucleotide-directed mutagenesis using primers 5′-CACCCACCATGCCCTCAGGCAC-3′ and 5′-GTGCTCCTGCACTTGGCTCC-3′ and KOD DNA polymerase (Novagen).
Cell Surface Immunoprecipitation
For cell surface immunoprecipitation, the cells were washed three times with ice-cold PBS then incubated for 1 h with anti-RAET1G mAb in ice-cold PBS. After washing three times with ice-cold PBS, the cells were lysed in RIPA buffer containing protein inhibitor mixture tablet (Roche Diagnostics) and 10 mm PMSF, and the resulting supernatant was incubated with anti-V5 agarose (A7345; Sigma) overnight at 4 °C. After the beads were washed five times with RIPA buffer, bound proteins were eluted with SDS sample buffer by boiling for 5 min. Total protein and isolated labeled proteins were analyzed by Western blotting. The experiments were independently repeated twice.
RESULTS
Detection of RAET1G Protein with Antibodies against the Extracellular and Cytoplasmic Domains
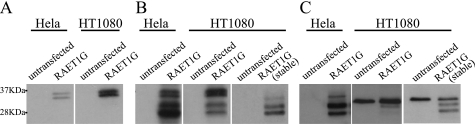
We compared expression patterns of RAET1G on Western blots of both HeLa and HT1080 cell lines using untagged, recombinant RAET1G, with three different reagents for detection. Reagent 1 was polyclonal anti-RAET1G-tail antiserum, which was raised using peptides corresponding to part of the cytoplasmic tail (18). Reagent 2 was anti-ULBP2 antibody, which recognizes both RAET1G and ULBP2, because their extracellular domains are almost identical to each other (18). Reagent 3 is an antibody specific to the extracellular portion of RAET1G (anti-RAET1G mAb), as described below.
The anti-RAET1G-tail antiserum detected bands of ∼37 kDa, corresponding to the predicted molecular mass of RAET1G (Fig. 1A). Anti-ULBP2 antibody detected two additional bands of ∼28 and ∼32 kDa (Fig. 1B). No bands were detected in lysates of untransfected cells.
FIGURE 1.
Expression pattern of RAET1G protein. Lysates of HeLa and HT1080 cells transfected with RAET1G were subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-RAET1G-tail antiserum raised with peptides corresponding to part of the cytoplasmic tail of the protein (A), polyclonal anti-ULBP2 antibody (AF1298), (B) or monoclonal recombinant anti-RAET1G antibody generated using the extracellular domain of the protein (anti-RAET1G mAb) (C). The band in untransfected HT1080 cells (C) is nonspecific. The anti-RAET1G mAb detected the doublet band only weakly in HT1080. The reason for this is not clear. It may be partly due to conversion of the high molecular mass form to smaller products (as in B, right panel, for example). The intensity of the nonspecific band makes longer exposure of these blots unfeasible.
To avoid any cross-reaction with endogenous NKG2D ligands, such as ULBP2 and RAET1L, we made specific monoclonal recombinant antibody against the RAET1G using phage display technology. Bacterially expressed recombinant RAET1G and ULBP protein were produced as antigens. We screened for positive binding to RAET1G while eliminating any antibodies that bound recombinant ULBP2 in a negative selection step. In Western blot and FACS analysis this anti-RAET1G antibody specifically recognized recombinant RAET1G protein transfected into Chinese hamster ovary cells, but not ULBP2 or RAET1L (supplemental Fig. S1). Using this RAET1G-specific reagent, we confirmed RAET1G expression in the transfected HeLa and HT1080 cell lines. The antibody clearly detected bands of ∼28 and ∼32 kDa, in patterns that were similar to those detected by anti-ULBP2 antibody (Fig. 1C).
Cleavage of RAET1G Protein Occurs Soon after Protein Synthesis
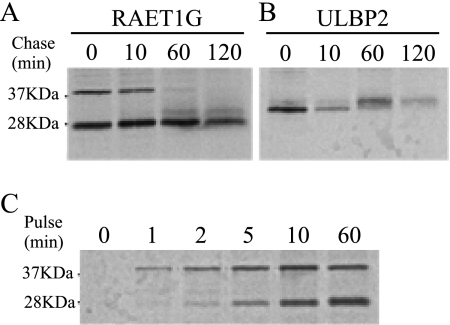
To characterize the RAET1G bands further, we performed pulse-chase experiments. Samples from cells transduced with RAET1G or ULBP2 were pulsed for 20 min and chased for 0–120 min before the proteins were immunoprecipitated. In the cells transduced with RAET1G, a ∼37 kDa band, corresponding to the full-length protein, appeared from the beginning of the 0-min chase and disappeared in 60 min (Fig. 2A). Another band of ∼28 kDa was also detected at the 0-min chase, and a further one of ∼32 kDa appeared after a 60-min chase. In the cells transduced with ULBP2, an initial band became weaker after 10 min, corresponding to the appearance of a higher band, consistent with modification of the ULBP2 protein (Fig. 2B).
FIGURE 2.
Time course of the cleavage of RAET1G protein. A and B, HT1080 cells transduced with RAET1G (A) or ULBP2 (B) were pulse-labeled for 20 min and chased for 0–120 min, followed by immunoprecipitation of RAET1G or ULBP2 with polyclonal anti-ULBP2 antibody (AF1298) and electrophoresis on 15% acrylamide gels. C, HT1080 cells were transduced with RAET1G pulse-labeled for 0–60 min, followed by immunoprecipitation of RAET1G and electrophoresis on 15% acrylamide gels.
To follow the timing of the RAET1G band of ∼28 kDa more closely, a shorter pulse time was applied, this time without chase. The ∼37 kDa band appeared from the beginning of the pulse, and the ∼28 kDa band appeared later and became gradually more intense (Fig. 2C).
These data suggest that the ∼28 kDa RAET1G band is derived from cleavage of the ∼37 kDa band and that the cleavage occurs soon after protein synthesis. They also suggest that the ∼32 kDa band might be derived from modification of the 28 kDa band.
C terminus of RAET1G Protein Is Cleaved
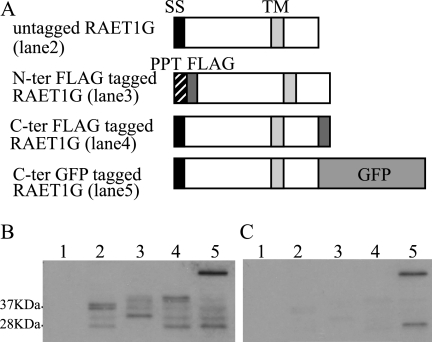
To confirm the pulse-chase results, we made epitope-tagged RAET1G constructs. RAET1G proteins were fused downstream of a generic ER leader sequence and a FLAG tag, or upstream of a FLAG, or enhanced GFP, tag (Fig. 3A). Anti-ULBP2 antibody, which recognizes the extracellular domain of RAET1G, detected ∼37 kDa bands in the lane containing untagged protein and slightly higher bands in lanes with the N-terminal and C-terminal FLAG-tagged RAET1G. A much higher band was observed in the C-terminal GFP-tagged lane (Fig. 3B). These data confirmed expression of full-length RAET1G with its cytoplasmic tail. In addition to bands representing the full-length proteins, bands of ∼28 and ∼32 kDa were detected in lanes containing untagged RAET1G, the C-terminal FLAG-tagged RAET1G, and the C-terminal enhanced GFP-tagged molecule. Slightly higher bands were seen in the lane with an N-terminal FLAG tag, suggesting that the C terminus of RAET1G might be cleaved.
FIGURE 3.
Cleavage of cytoplasmic tail of RAET1G protein. A, schematic representation of epitope-tagged RAET1G constructs is shown. RAET1G was cloned downstream of a generic ER leader sequence and a FLAG tag, upstream of a FLAG tag or GFP. B and C, lysates of HT1080 cells transduced with RAET1G or epitope-tagged RAET1G were collected and subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 antibody (AF1298) (B) or with anti-GFP antibody (C). Lane 1, untransfected; lane 2, transfected with untagged; lane 3, with N-terminal FLAG-tagged; lane 4, transfected with C-terminal FLAG-tagged; and lane 5, transfected with C-terminal GFP-tagged RAET1G.
With the anti-GFP antibody, in addition to the higher band corresponding to full-length RAET1G-GFP, a band of ∼27 kDa was observed, corresponding to the molecular mass of GFP (Fig. 3C). This supports the idea that the C terminus of RAET1G protein is removed. Similar results were obtained with the other cell lines, including HeLa, MelJuSo, and HCA7 (data not shown).
Transport of RAET1G after Cleavage
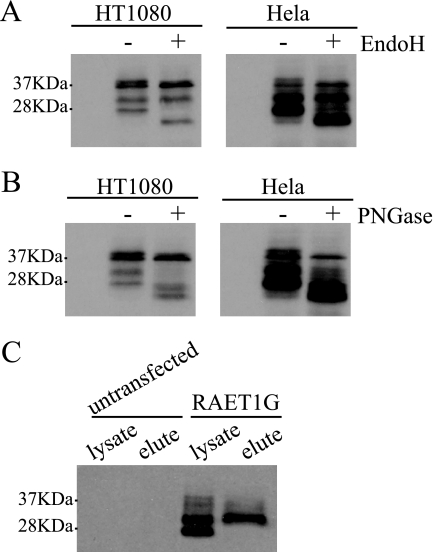
To determine the intracellular localization of the four forms of RAET1G, lysates of cells transduced with RAET1G were treated with two glycosidases, Endo H and PNGase F, and protein was detected by Western blotting (Fig. 4, A and B). Endo H does not cleave processed N-linked sugars, but PNGase F does, so differential sensitivity to these two glycosidases can be interpreted to establish transit to the Golgi. The upper band of ∼37 kDa and the lower band of ∼28 kDa were sensitive to both Endo H and PNGase F, suggesting that they were glycosylated but remained in the ER. The lower band of the ∼37 kDa doublet was resistant to both Endo H and PNGase F, indicating that this is immature protein. The ∼32 kDa band was resistant to Endo H but sensitive to PNGase F, suggesting that this form might be glycosylated and transported, at least to the Golgi. These data are consistent with modification of the RAET1G protein in the Golgi after cleavage of its C-terminal region.
FIGURE 4.
Cellular localization of each form of RAET1G protein. A and B, lysates of HT1080 and HeLa cells transfected with RAET1G were digested by Endo H (A) or PNGase (B) and subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 antibody (AF1298). C, HT1080 cells transduced with RAET1G were surface-stained with monoclonal anti-RAET1G antibody (anti-RAET1G mAb) and after washing, the surface proteins were lysed, immunoprecipitated, and subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 antibody (AF1298).
Cell surface immunoprecipitation was performed to determine which forms of the protein are expressed in this location. Cells transduced with RAET1G were stained with anti-RAET1G antibody (anti-RAET1G mAb), and the bound cell surface proteins were immunoprecipitated and detected in Western blots. Only the ∼32 kDa band was clearly detected in the eluate sample (Fig. 4C), suggesting that this form is transported to the cell surface after modification in the Golgi.
GPI Anchoring of RAET1G
Only RAET1E and RAET1G have been reported as transmembrane proteins, among the ULBP family proteins, although ULBP2 and RAET1L share high similarity to RAET1G. We checked for GPI-anchoring motifs in RAET1G using website software, PredGPI. It was suggested, with only 17% probability, that RAET1G was GPI-anchored. Previous reports of mutational analysis of the C-terminal GPI signal sequences of several proteins have demonstrated that the length of the hydrophobic region and the spacer sequence between the GPI-anchoring site and the hydrophobic region are important for GPI anchoring (19), and we found that these features were conserved in RAET1G, ULBP2, and RAET1L. Because we established that RAET1G protein is cleaved at its C terminus, we examined sequences of the products of protein cleavage. In this case, PredGPI indicated with 100% probability that RAET1G protein is GPI-anchored by a serine residue (Ser216) that is conserved among the three proteins.
PI-PLC Treatment Released RAET1G Protein
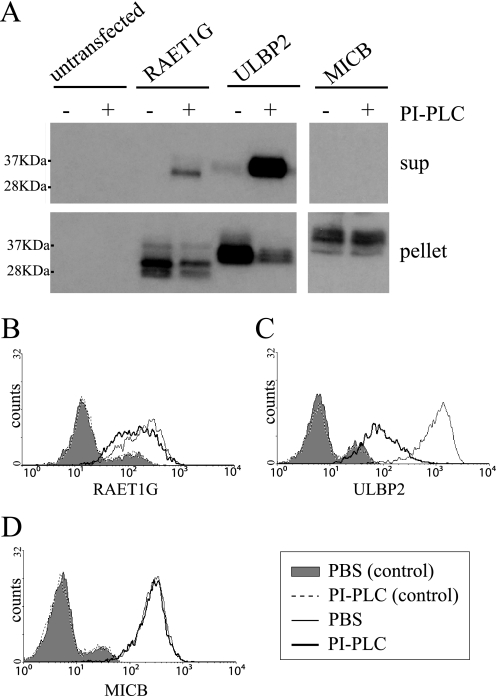
To test experimentally whether RAET1G protein is GPI-anchored, three experiments were performed. First, we checked by Western blotting whether the protein is released by PI-PLC treatment. GPI-anchored protein ULBP2 was used as a positive control and transmembrane-associated MICB as a negative control. A band of released RAET1G protein was detected in the supernatant of only PI-PLC-treated cells, which suggests that at least some RAET1G is GPI-anchored (Fig. 5A). A ULBP2 band was also seen, but no band for MICB, in the supernatant of PI-PLC treated cells. Similar data were obtained for HeLa cells (not shown).
FIGURE 5.
RAET1G protein is released by PI-PLC treatment. A, HT1080 cells transduced with RAET1G, ULBP2, and MICB were treated with PI-PLC and centrifuged. Supernatant and pellet were collected for SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 (AF1298) and anti-MICB (AF1599) antibody. B–D, HT1080 cells transduced with RAET1G (B), ULBP2 (C), and MICB (D) were treated with PI-PLC and washed with PBS, and the level of surface RAET1G was assessed by FACS using monoclonal anti-RAET1G (anti-RAET1G mAb), anti-ULBP2 (MAb1298), and anti-MICB (MAb1599) antibody in the FL4 channel. Shaded area and black dashed lines represent isotype controls.
To confirm these findings, expression on the surface of cells was examined in FACS. Surface expression of RAET1G was reduced slightly after PI-PLC treatment (Fig. 5B). The surface expression of ULBP2 was dramatically reduced, but PI-PLC treatment did not affect the surface level of MICB. These experiments show that at least some RAET1G is GPI-linked to the cell surface. The slight reduction in cell surface expression of RAET1G is in contrast to the other data (e.g. Fig. 6C), which indicate that a substantial fraction of the molecules are GPI-anchored. The reason for this discrepancy is not known. The apparent small shift after PI-PLC treatment for RAET1G may relate to its low initial level on the cell surface. Alternatively, RAET1G may be a poor substrate for PI-PLC at the cell surface.
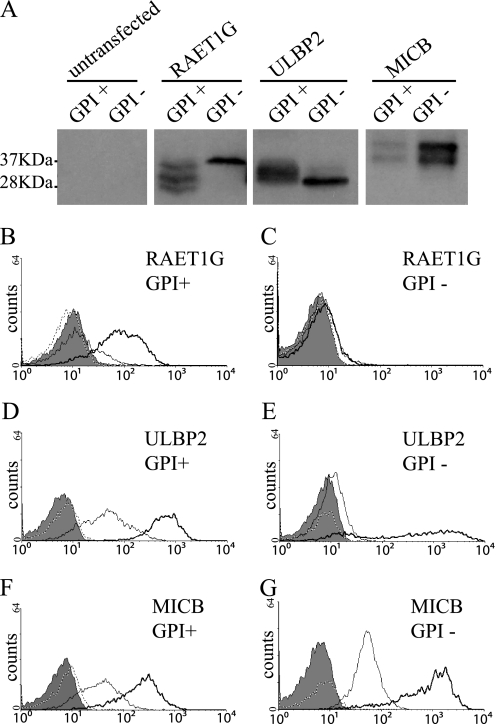
FIGURE 6.
RAET1G expression in GPI-biosynthetic mutant cells. A, lysates of K562 and K562 class K cell lines transduced with RAET1G, ULBP2, or MICB were subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 (AF1298) or anti-MICB (AF1599) antibody. B–G, surface expression level of RAET1G (B and C), ULBP2 (D and E), or MICB (F and G) in K562 (B, D, and F) and K562 class K (C, E, and G) cell lines transduced with RAET1G, ULBP2, or MICB were assessed by FACS using monoclonal anti-RAET1G (anti-RAET1G mAb), anti-ULBP2 (MAb1298), or anti-MICB (MAb1599) antibody in the FL4 channel. Shaded area and dashed line represent isotype controls in untransfected and transduced cells, respectively. Solid black line and bold black line represent each antibody staining in untransfected and transduced cells, respectively.
Reduced Surface Expression of RAET1G in GPI-negative Cells
As an alternative way of examining GPI anchoring of RAET1G, we monitored expression of the protein in a GPI biosynthetic mutant. We chose a K562 cell line, K562 class K, which has defects in one of the major enzymes of GPI biosynthesis, GPI amidase (16). Three bands were detected of ∼28 kDa, ∼32 kDa, and ∼37 kDa in normal, GPI-positive, K562 cells. The ∼28 kDa and ∼32 kDa bands disappeared in the GPI-negative cells, and the intensity of the 37 kDa band increased (Fig. 6A). The upper band of ULBP2, corresponding to the GPI-anchored form, was detected in GPI-positive cells but not in GPI-negative cells. No difference in MICB expression was detected between GPI-positive and GPI-negative cells, although total band intensity was changed. Furthermore, the surface expression of RAET1G was dramatically reduced in the GPI-negative, compared with the normal GPI-positive, cells (Fig. 6B). The surface expression of ULBP2 was also reduced in GPI-negative cells, but MICB expression on the cell surface was high in both GPI-positive and GPI-negative cells.
GPI Anchoring of RAET1G Protein
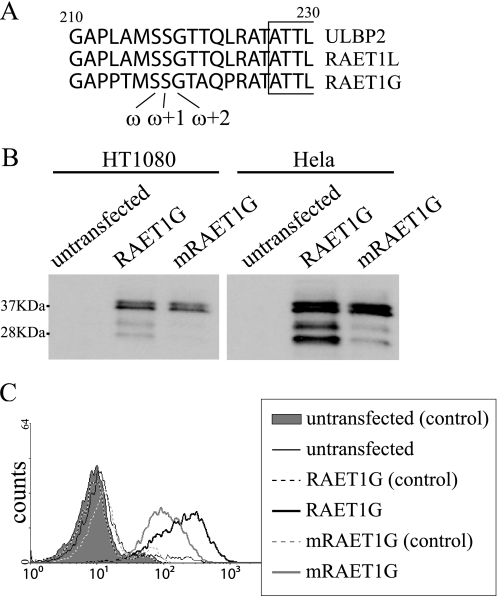
To determine whether RAET1G is directly GPI-anchored, we mutated GPI anchor residues in the molecule. PredGPI indicated that Ser216 in both ULBP2 and RAET1L is a ω site, meaning that it is the residue most likely to be directly associated with GPI anchor. In addition to Ser216, the adjacent two residues are conserved among RAET1G, ULBP2, and RAET1L as these sequences all have Ser217 in the ω+1 site and Gly218 in the ω+2 site (Fig. 7A). We made RAET1G protein in which the serine residue in the ω site was exchanged for a proline, a residue that has never been reported to be used as a ω site (20). In Western blots, the intensity of the ∼28 kDa and ∼32 kDa bands decreased, although the intensity of the ∼37 kDa bands did not change (Fig. 7B). In addition, surface expression by FACS of RAET1G protein, mutated in the GPI anchor, was lower than that of normal RAET1G (Fig. 7C). Similar results were obtained in HeLa cells (data not shown).
FIGURE 7.
Expression of RAET1G mutated in predicted GPI-anchoring position. A, amino acid alignment of RAET1G, ULBP2, and RAET1L proteins around the predicted GPI-anchoring position is shown. Conserved residues are shown in bold. B, lysates of HT1080 cells transfected with wild-type RAET1G (RAET1G) and RAET1G mutated in GPI-anchoring position (mRAET1G) were subjected to SDS-PAGE (15% gel) separation and Western blotting with polyclonal anti-ULBP2 antibody (AF1298). C, HT1080 cells transduced with wild-type RAET1G (RAET1G) and RAET1G mutated in GPI-anchoring position (mRAET1G) were assessed by FACS using anti-RAET1G mAb in FL4 channel. Shaded area and black and gray dashed lines represent isotype controls.
DISCUSSION
NKG2D ligands are critical activators of NK cells and subsets of T cells. Their variation in different species suggests that they are under selection driven by immune evasion. Mice have at least six different NKG2D ligands, some of which are transmembrane molecules and the others GPI-anchored (5, 6). Similarly, human MIC proteins, which are not present in mouse, span the membrane, but ULBP1–3 depend on GPI attachment. RAET1G is predicted to span the cell membrane and to contain a long cytoplasmic tail. We now show that the arrangement of this molecule is more complex and that it undergoes post-translational cleavage and modification before being transported to the cell surface.
Our data suggest that RAET1G undergoes GPI anchoring, in line with most of the other ULBP family proteins, and the ∼32 kDa band corresponds to the functional GPI-anchored form (supplemental Fig. 2). The immediate appearance of the ∼37 kDa band and its subsequent replacement by ∼28 kDa and ∼32 kDa bands in the pulse-chase suggest that RAET1G might be first translated as the full-length form and then cleaved just after protein synthesis. The data using epitope-tagged RAET1G called for cleavage of the C terminus of the protein. The disappearance of both ∼28 kDa and ∼32 kDa bands in Western blots, using a GPI-anchoring mutant defective in GPI-transamidase, suggests that the cleavage and GPI anchoring might occur simultaneously by the enzyme located in the ER. The pulse-chase data showing that the ∼32 kDa band appeared after the ∼28 kDa band, coupled with its resistance of Endo H digestion, suggest that the ∼32 kDa GPI-anchored RAET1G form might be transported to the Golgi, where it could be modified further. The FACS data showing the reduced cell surface expression of RAET1G in the GPI-anchoring mutant cell line support the data. Furthermore, the cell surface immunoprecipitation data suggest that this form might be transported to the cell surface.
The extracellular domain of RAET1G protein shares high similarity with those domains of ULBP2 and RAET1L proteins. It has been reported that RAET1G and ULBP2 transcripts are generally coexpressed in different cell lines and tissues (9, 10). The long cytoplasmic tail is a specific feature of the RAET1G protein. However, we showed here that the cytoplasmic tail of RAET1G is cleaved and that at least a proportion of the protein works as a GPI-anchored protein. What then is the functional difference between RAET1G and ULBP2? One hypothesis is that their expression might be regulated differently. RAET1G might mainly use post-translational regulation to respond to stimuli rapidly, whereas ULBP2 could mainly use transcriptional regulation. Indeed, the mode of expression of RAET1G at the cell surface might be regulated by transport from the ER to the Golgi or by modification in the Golgi. This arrangement could reflect the need to express this NKG2D ligand rapidly at the surface of the cell in response to infection. Most RAET1G protein might reside inside the cell in normal conditions, and environmental stimuli such as viral infection might trigger release to the cell surface. This hypothesis could explain the reason for the lower expression of RAET1G on the cell surface compared with ULBP2 and MICB. The slower glycosylation of RAET1G, compared with ULBP2, in our pulse-chase data is consistent with this. Further investigation is needed to examine what kind of environmental stimuli might regulate the cell surface expression of RAET1G post-translationally.
An alternative hypothesis is that RAET1G functions as a decoy in viral infection. RAET1G protein interacts weakly with NKG2D compared with ULBP2 (7, 10). RAET1G protein might sequester viral proteins that target ULBP2, for example, although it binds UL-16 very weakly if at all. It is also possible that NKG2D is not the primary receptor for RAET1G and that it serves as a ligand for other immunoreceptor molecules.
It has been reported that the cytoplasmic tail of Mult1 protein, which is proposed to be a functional ortholog of RAET1G in mice, undergoes ubiquitination-mediated regulation (15). However, on activation of cellular stress pathways by DNA-damaging agents the ubiquitin-mediated turnover of Mult1 was reduced, and the protein was expressed at the cell surface. RAET1G has been considered to a candidate to be regulated by similar mechanism (15), although our data suggest that the C terminus of RAET1G is proteolytically cleaved. A question arises concerning the function of the cytoplasmic tail of RAET1G. The tail does not share any motif or similarity to other proteins, although its transmembrane and hydrophobic regions were similar to those of the other ULBPs (supplemental Fig. S3). There is no splice junction in the vicinity, indicating that the cytoplasmic tail of RAET1G might have been derived by duplication of ULBP2, followed by a frameshift in the hydrophobic region.
Our experiments all suggest that RAET1G protein undergoes GPI anchoring, like most other ULBP/RAET1 family proteins. However, the antiserum against the cytoplasmic tail detected protein in some tissue, including cancer cells (18). One interpretation of all the data is that RAET1G protein might exist as both isoforms, GPI-anchored and transmembrane. Slight reduction of cell surface expression of RAET1G by PI-PLC treatment is consistent with this hypothesis. In fact, the ratio between the prospective transmembrane form (∼37 kDa band) and the GPI-anchored form (∼32 kDa band) varies depending on cell types and conditions (data not shown). Therefore, it is possible that the cleavage and processing of RAET1G could be altered on receipt of cellular stress signals. There are other reports of two isoforms of some proteins regulated post-transcriptionally (21). Expression of both transmembrane and GPI-linked forms of NKG2D ligands could be a host strategy to evade subversion by viruses as two alternative coupling mechanisms would be more difficult to target. It might be a feature of other NKG2D ligands that has not been explored in detail yet. Although we detected the tail of RAET1G in tissue staining, our experiments would not distinguish whether the tail was attached to the surface protein. The antiserum could also recognize the C-terminal cleavage product in the cytoplasm.
In vivo expression of RAET1G has been difficult to follow due to the lack of specific antibody. Because of the high similarity of the extracellular domain to ULBP2 and RAET1L, antiserum against the cytoplasmic tail (anti-RAET1G-tail antiserum) was used previously to detect the protein (18). However, our data in this paper suggest that the antiserum might not be able to trace the entire expression of all RAET1G protein because of cleavage of the cytoplasmic tail. Specific anti-RAET1G antibody will be crucial to analyze the function of this protein in vivo. We attempted to detect endogenous expression of the protein in several nontransfected cell lines using the recombinant monoclonal antibody (anti-RAET1G mAb), but no significant protein expression was observed by Western blotting or FACS in cell lines expressing RAET1G transcripts (data not shown). Levels of protein expression of RAET1G in the cell lines might not be sufficient to be detected by the antibody in Western blotting or FACS, and expression might only be increased on receipt of certain signals, such as viral infection or DNA damage. There are other reports demonstrating that different cells and tissues express NKG2D ligand transcripts but lack corresponding protein expression (22–26). Clearly, NKG2D ligand expression is subject to complex regulation at several levels: the transcriptional level, the post-transcriptional level by microRNAs (14), and at post-translational level, for example the processing of RAET1G as we have reported here and the S-acylation of MICA (27). It is also clear that these mechanisms vary between NKG2D ligands and may vary between cell types, presenting a considerable challenge to understand the NKG2D immune recognition system fully.
Expression of RAET1G and RAET1E is subject to alternative splicing to produce soluble proteins (8, 18, 28, 29), and soluble ULBP2 is released by metalloprotease in tumor cells (30). It will be interesting to examine whether soluble forms of RAET1G are detected by anti-RAET1G antibody in ELISAs. This issue needs further investigation, using other cell lines, tissues, and conditions to analyze the protein in vivo. It will also be crucial to assess whether RAET1G trafficking and post-translational processing is altered in specific cell types or when cells are subjected to stress such as viral infection or DNA damage. RAET1G expression has been detected in gut epithelial cell layers (8, 18). These cells are highly specialized and form polarized cell layers allowing interface with the gut lumen on one side and lymphocytes on the other. MICA has been shown to have specific trafficking properties in these cell types (31), and it will be interesting to assess RAET1G trafficking and modification in this cell type.
Supplementary Material
Acknowledgments
We thank Dr. N. Kanzawa and Prof. T. Kinoshita for cell lines, Prof. P. Lehner for plasmid, Dr. A. P. Kelly for helpful advice, and Dr. L. H. Boyle for help with the pulse-chase experiment.
This work was supported by a grant from Cancer Research United Kingdom.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- NKG2D
- natural killer group 2 member D
- NK
- natural killer
- MIC
- major histocompatibility complex class I chain-related protein
- ULBP
- UL-16-binding protein
- RAET1
- retinoic acid expressed transcript 1 protein
- GPI
- glycosylphosphatidylinositol
- Mult1
- murine UL-16-binding protein-like transcript 1
- GFP
- green fluorescent protein
- MAb
- monoclonal antibody
- Endo H
- endoglycosidase H
- PNGase
- peptide N-glycosidase
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorting
- RIPA
- radioimmune precipitation assay buffer
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Bahram S., Bresnahan M., Geraghty D. E., Spies T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6259–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leelayuwat C., Townend D. C., Degli-Esposti M. A., Abraham L. J., Dawkins R. L. (1994) Immunogenetics 40, 339–351 [DOI] [PubMed] [Google Scholar]
- 3.Cosman D., Müllberg J., Sutherland C. L., Chin W., Armitage R., Fanslow W., Kubin M., Chalupny N. J. (2001) Immunity 14, 123–133 [DOI] [PubMed] [Google Scholar]
- 4.Radosavljevic M., Cuillerier B., Wilson M. J., Clément O., Wicker S., Gilfillan S., Beck S., Trowsdale J., Bahram S. (2002) Genomics 79, 114–123 [DOI] [PubMed] [Google Scholar]
- 5.Eagle R. A., Trowsdale J. (2007) Nat. Rev. Immunol. 7, 737–744 [DOI] [PubMed] [Google Scholar]
- 6.Takada A., Yoshida S., Kajikawa M., Miyatake Y., Tomaru U., Sakai M., Chiba H., Maenaka K., Kohda D., Fugo K., Kasahara M. (2008) J. Immunol. 180, 1678–1685 [DOI] [PubMed] [Google Scholar]
- 7.Eagle R. A., Traherne J. A., Hair J. R., Jafferji I., Trowsdale J. (2009) Eur. J. Immunol. 39, 3207–3216 [DOI] [PubMed] [Google Scholar]
- 8.Bacon L., Eagle R. A., Meyer M., Easom N., Young N. T., Trowsdale J. (2004) J. Immunol. 173, 1078–1084 [DOI] [PubMed] [Google Scholar]
- 9.Ebihara T., Masuda H., Akazawa T., Shingai M., Kikuta H., Ariga T., Matsumoto M., Seya T. (2007) Int. Immunol. 19, 1145–1155 [DOI] [PubMed] [Google Scholar]
- 10.Wittenbrink M., Spreu J., Steinle A. (2009) Eur. J. Immunol. 39, 1642–1651 [DOI] [PubMed] [Google Scholar]
- 11.Eagle R. A., Traherne J. A., Ashiru O., Wills M. R., Trowsdale J. (2006) Hum. Immunol. 67, 159–169 [DOI] [PubMed] [Google Scholar]
- 12.Hamerman J. A., Ogasawara K., Lanier L. L. (2004) J. Immunol. 172, 2001–2005 [DOI] [PubMed] [Google Scholar]
- 13.Gasser S., Orsulic S., Brown E. J., Raulet D. H. (2005) Nature 436, 1186–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern-Ginossar N., Gur C., Biton M., Horwitz E., Elboim M., Stanietsky N., Mandelboim M., Mandelboim O. (2008) Nat. Immunol. 9, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 15.Nice T. J., Coscoy L., Raulet D. H. (2009) J. Exp. Med. 206, 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohishi K., Nagamune K., Maeda Y., Kinoshita T. (2003) J. Biol. Chem. 278, 13959–13967 [DOI] [PubMed] [Google Scholar]
- 17.Thomas M., Boname J. M., Field S., Nejentsev S., Salio M., Cerundolo V., Wills M., Lehner P. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eagle R. A., Flack G., Warford A., Martínez-Borra J., Jafferji I., Traherne J. A., Ohashi M., Boyle L. H., Barrow A. D., Caillat-Zucman S., Young N. T., Trowsdale J. (2009) PLoS One 4, e4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böhme U., Cross G. A. (2002) J. Cell Sci. 115, 805–816 [DOI] [PubMed] [Google Scholar]
- 20.Kodukula K., Gerber L. D., Amthauer R., Brink L., Udenfriend S. (1993) J. Cell Biol. 120, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chesla S. E., Li P., Nagarajan S., Selvaraj P., Zhu C. (2000) J. Biol. Chem. 275, 10235–10246 [DOI] [PubMed] [Google Scholar]
- 22.Groh V., Bahram S., Bauer S., Herman A., Beauchamp M., Spies T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12445–12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pende D., Rivera P., Marcenaro S., Chang C. C., Biassoni R., Conte R., Kubin M., Cosman D., Ferrone S., Moretta L., Moretta A. (2002) Cancer Res. 62, 6178–6186 [PubMed] [Google Scholar]
- 24.Nedvetzki S., Sowinski S., Eagle R. A., Harris J., Vély F., Pende D., Trowsdale J., Vivier E., Gordon S., Davis D. M. (2007) Blood 109, 3776–3785 [DOI] [PubMed] [Google Scholar]
- 25.Boissel N., Rea D., Tieng V., Dulphy N., Brun M., Cayuela J. M., Rousselot P., Tamouza R., Le Bouteiller P., Mahon F. X., Steinle A., Charron D., Dombret H., Toubert A. (2006) J. Immunol. 176, 5108–5116 [DOI] [PubMed] [Google Scholar]
- 26.Schrambach S., Ardizzone M., Leymarie V., Sibilia J., Bahram S. (2007) PLoS One 2, e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eleme K., Taner S. B., Onfelt B., Collinson L. M., McCann F. E., Chalupny N. J., Cosman D., Hopkins C., Magee A. I., Davis D. M. (2004) J. Exp. Med. 199, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao W., Xi X., Hao Z., Li W., Kong Y., Cui L., Ma C., Ba D., He W. (2007) J. Biol. Chem. 282, 18922–18928 [DOI] [PubMed] [Google Scholar]
- 29.Cao W., Xi X., Wang Z., Dong L., Hao Z., Cui L., Ma C., He W. (2008) Int. Immunol. 20, 981–991 [DOI] [PubMed] [Google Scholar]
- 30.Waldhauer I., Steinle A. (2006) Cancer Res. 66, 2520–2526 [DOI] [PubMed] [Google Scholar]
- 31.Suemizu H., Radosavljevic M., Kimura M., Sadahiro S., Yoshimura S., Bahram S., Inoko H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2971–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.