Abstract
The polyene macrolide antibiotic filipin is widely used as a probe for cholesterol and a diagnostic tool for type C Niemann-Pick disease. Two position-specific P450 enzymes are involved in the post-polyketide modification of filipin during its biosynthesis, thereby providing molecular diversity to the “filipin complex.” CYP105P1 and CYP105D6 from Streptomyces avermitilis, despite their high sequence similarities, catalyze filipin hydroxylation at different positions, C26 and C1′, respectively. Here, we determined the crystal structure of the CYP105P1-filipin I complex. The distal pocket of CYP105P1 has the second largest size among P450 hydroxylases that act on macrolide substrates. Compared with previously determined substrate-free structures, the FG helices showed significant closing motion on substrate binding. The long BC loop region adopts a unique extended conformation without a B′ helix. The binding site is essentially hydrophobic, but numerous water molecules are involved in recognizing the polyol side of the substrate. Therefore, the distal pocket of CYP105P1 provides a specific environment for the large filipin substrate to bind with its pro-S side of position C26 directed toward the heme iron. The ligand-free CYP105D6 structure was also determined. A small sub-pocket accommodating the long alkyl side chain of filipin I was observed in the CYP105P1 structure but was absent in the CYP105D6 structure, indicating that filipin cannot bind to CYP105D6 with a similar orientation due to steric hindrance. This observation can explain the strict regiospecificity of these enzymes.
Keywords: Antibiotics, Cytochrome P450, Enzyme Structure, Spectroscopy, X-ray Crystallography, Filipin, Macrolide, Streptomyces avermitilis
Introduction
Macrolide compounds have toxic effects on a wide variety of organisms including pathogens, and therefore, their clinical use as antibiotics has been widely developed (1). Post-polyketide modifications of macrolides by cytochrome P450 (P450 or CYP)2 hydroxylases provide molecular diversity to these macrolides during their biosynthesis (2, 3). P450s are hemoproteins whose fifth axial heme iron ligand is a thiolate group found in a variety of organisms (4, 5). A majority of P450s catalyze monooxygenation (hydroxylation or epoxidation) of hydrophobic substrates (6) using a dioxygen bound as the sixth iron ligand as well as various redox systems responsible for the cleavage of the O-O bond (7–9). Understanding the molecular mechanisms of P450 enzymes during the biosynthesis of natural products would facilitate their potential uses in producing new drugs (10). The crystal structures of macrolide monooxygenases complexed with their substrates or analogues have been determined for P450eryF (CYP107A1; erythromycin biosynthesis) (11, 12), P450 EryK (CYP113A1; erythromycin biosynthesis) (13), P450 PikC (CYP107L1; narbomycin and pikromycin biosynthesis) (14), and P450epoK (CYP167A1; epothilone biosynthesis) (12) (see supplemental Fig. S1).
The 28-membered polyene macrolide antibiotic filipin is widely used as a probe for cholesterol in biological membranes (15, 16) and a prominent diagnostic tool for type C Niemann-Pick disease (17, 18). Filipin, originally isolated from Streptomyces filipinensis as a filipin complex (19), is composed of four components (see Fig. 1) (20). The major component (53%) is filipin III, and its stereochemical configuration has been determined (21, 22). Filipin I (4%) lacks two hydroxyl groups of filipin III located at positions C1′ and C26 (23). Filipin II (25%) is 1′-deoxyfilipin III (24). Filipin IV (18%) is isomeric to filipin III and is probably epimeric at C1′ or C3 (25). A solution NMR study has shown that the large 28-membered ring is rigid, stabilized by both intramolecular hydrogen bonds of syn 1,3-polyols and a conjugated pentaene moiety, whereas the lateral aliphatic chain is highly flexible (26).
FIGURE 1.
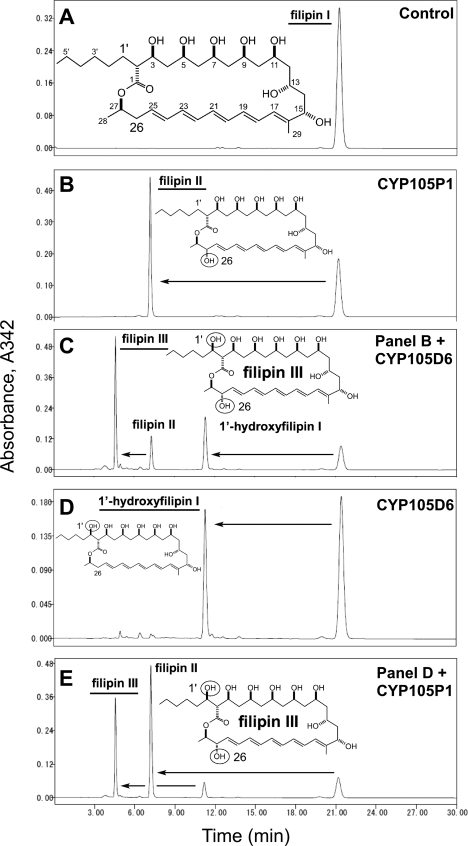
Analytical HPLC of the reaction products from filipin I by CYP105P1 and CYP105D6. A, control reaction without P450 enzymes is shown. B, reaction with CYP105P1 is shown. C, shown is the reaction with CYP105D6 on the sample in panel B. D, reaction with CYP105D6 is shown. E, reaction with CYP105P1 on the sample in panel D is shown. Structures of filipin I, filipin II (1′-deoxyfilipin III), and filipin III are also shown. The configurations of the stereogenic centers in filipin III are 1′R, 2R, 3S, 5S, 7S, 9R, 11R, 13R, 15S, 26S, and 27R.
A gene cluster for filipin biosynthesis was recently identified in the genome of Streptomyces avermitilis (27). The gene cluster contains two P450 genes, CYP105P1 (PteC, SAV413) and CYP105D6 (PteD, SAV412) as well as genes encoding modular polyketide synthases (pteA1-pteA5), ferredoxin (fdxI, pteE), and putative zinc binding dehydrogenase (pteB). The filipin biosynthetic gene cluster is regulated by the negative regulator aveI along with the biosynthetic genes for other antibiotics including avermectin and oligomycin (28). Analysis of P450 gene deletion mutants revealed that CYP105P1 and CYP105D6 catalyze hydroxylations at positions C26 and C1′, respectively.3 The amino acid sequence identity between CYP105P1 and CYP105D6 is 36.7% when the whole lengths of both sequences are used for alignment.
We previously reported the crystal structures of CYP105P1 in three different states (29). The ligand-free wild-type structure provides a unique state in which the His-72 residue in the BC loop is ligated to the heme iron atom. When compared with the 4-phenylimidazole-bound wild-type and ligand-free H72A mutant structures, it is suggested that the high flexibility of the BC loop of this enzyme is a key feature for incorporating the large hydrophobic filipin substrate. In this report we present two crystal structures of the filipin hydroxylases: the structures of CYP105P1-filipin I complex and ligand-free CYP105D6. Our present study provides a concrete structural basis for filipin hydroxylation at position C26 and an insight into the different substrate specificities of these similar P450 enzymes, both belonging to the CYP105 family.
EXPERIMENTAL PROCEDURES
Protein Preparation and Spectroscopy
CYP105P1 protein was expressed and purified as described previously (29). The primers used to amplify the CYP105D6 gene were 5′-CCC ATA TGA CTG AGA CCG AAA TCC GCC TC-3′ and 5′-GGA CTA GTT CAG TGG TGG TGG TGC CAG ACG ACG GGG AGC TCG ATC-3′ (bold type and underlined sequences represent the restriction endonuclease sites and His4 tag, respectively). The expression plasmid was constructed using pET-17b (Novagen, Madison, WI). CYP105D6 protein was expressed in Escherichia coli C43 (DE3) cultured in Terrific Broth medium containing 12 g/liter Bacto-tryptone, 24 g/liter yeast extract, 8 g/liter glycerol, 17 mm KH2PO4, 72 mm K2HPO4, and 100 mg/l ampicillin at 25 °C for 24 h. After the correction of the cells by centrifugation, cells were suspended in 20 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 10 mm imidazole, 0.1 mm dithiothreitol, and 10% (v/v) glycerol. Cell extracts were obtained by sonication and followed by centrifugation to remove cell debris. A fraction containing the protein was purified on a HiTrap Chelating HP 5-ml column (GE Healthcare) with a linear gradient of 10–500 mm imidazole. After dialysis, the protein was further purified on a Resource Q column (GE Healthcare) with a linear gradient of 0–0.5 m NaCl. The final step of purification was on a Superdex 200 column (GE Healthcare) separated with 10 mm Tris-HCl (pH 7.5), 0.15 m NaCl, 0.1 mm dithiothreitol, and 10% (v/v) glycerol. The purified enzyme appeared as a single band corresponding to a molecular mass of 44 kDa on SDS-PAGE (data not shown). The absorbance ratio of proteins purified in this manner was greater than 2.0 at 420 nm as compared with that at 280 nm. The P450 content measured by CO difference spectroscopy was also checked to verify purification quality.
Purification of Filipin I
Spores of S. avermitilis strain ΔpteC/pteD were inoculated into a medium containing 5 g/liter glucose, 15 g/liter soy flour, and 5 g/liter yeast extract (pH 7.0) and cultured with agitation at 30 °C for 48 h. A portion of the culture (1% seed) was inoculated into filipin production medium containing 20 g/liter dextrin, 2 g/liter glucose, 15 g/liter soy flour, 3 g/liter yeast extract, and 3 g/liter CaCO3 (pH 7.0) and cultured at 30 °C for 5 days on a rotary shaker. Mycelia from the culture medium (3 liters) were collected using a Büchner funnel and extracted twice with acetone. The sample was concentrated using a rotary evaporator, transferred to a separating funnel, and extracted twice with ethyl acetate. Solid anhydrous sodium sulfate was added for dehydration and then concentrated to dryness. The sample was dissolved with chloroform and then placed into a silica gel column (60 × 400 mm) eluted with chloroform/methanol (1:0, 10:1, and 4:1). The eluate was collected in 15-ml fractions. The retention time of a yellow band containing filipin I was about 30 min in chloroform/methanol (4:1). The sample was concentrated to dryness, dissolved with methanol, and filtered. The compound was finally purified by preparative HPLC (Pegasil-ODS 20 × 250 mm, Senshu Scientific Co., Tokyo, Japan) eluted with acetonitrile/methanol/water 55:20:25 at 340 nm of UV detection at a 9 ml/min flow rate. The separated compound was concentrated by evaporation and extracted twice with ethyl acetate. The extract was concentrated to dryness using an evaporator and dried in a vacuum desiccator.
Spectroscopy
UV-visible absorption spectra measurements and titration experiments were performed essentially using the same methods as described previously (29). For the titrations of filipin I to CYP105P1, 1 ml of assay buffer containing 50 mm potassium phosphate (pH 7.5), 0.1 mm dithiothreitol, 0.1 mm EDTA, and 10% (v/v) glycerol was used. The protein concentration was 5.1 μm, and 2 mm filipin I stock solution was added. A nonlinear fitting with a quadratic equation was applied to determine the Kd using Kaleidagraph (Synergy, Reading, PA): ΔA = (Bmax/2E){(Kd + E + L) − {(Kd + E + L)2 − 4E/L}1/2}, where Bmax is the maximum absorbance difference extrapolated to infinite ligand enzyme concentration, L is the ligand concentration, and E is the total enzyme concentration.
Measurement of Filipin Hydroxylase Activity
The reaction mixture (200 μl) contained 50 mm potassium phosphate (pH 7.5), 0.1 mm EDTA, 0.1 mm dithiothreitol, 10% (v/v) glycerol, 1 mm NADPH, 0.05 units of spinach ferredoxin:NADP+ reductase (Sigma), 0.015 mg of spinach ferredoxin (Sigma), 0.2 mm filipin I, and 1 μm P450 enzyme. The reaction was started by adding NADPH, and the mixture was incubated at 30 °C for 90 min. The reaction was terminated by mixing with 1.5-fold volume of ethyl acetate. The mixture was centrifuged at 6000 × g for 1 min to separate phases, and a portion of the ethyl acetate layer was concentrated to dryness using a centrifugal evaporator. The sample was dissolved with methanol and subjected to analytical HPLC (Pegasil-ODS 4.6 × 250 mm, Senshu Scientific Co.) at a 0.8 ml/min flow rate. The peak position of each compound was determined according to a previous study; the structure of each compound was analyzed by fast atom bombardment mass spectrometry, 1H NMR and 13C NMR.3 Filipin III, filipin II, 1′-hydroxyfilipin I, and filipin I eluted at 4.6, 7.2, 11.3, and 21.2 min, respectively.
Crystallography
For crystallization, protein was concentrated to >20 mg/ml in 10 mm Tris-HCl (pH 7.5), 0.5 m NaCl, and 0.1 mm EDTA (protein solution buffer). Before crystallization, filipin I was dissolved in dimethyl sulfoxide, mixed with a CYP105P1 sample, and concentrated using an Ultrafree Centrifugal Filter Device (Millipore, Billerica, MA). After three mixing cycles with filipin I and concentration by the centrifugal filter device, three more mixing cycles with the protein solution buffer and concentration were performed to remove unbound filipin I from the solution. Crystallization was performed using the sitting drop vapor diffusion method. CYP105P1 crystals complexed with filipin I were grown at 25 °C by mixing 1 μl of the protein solution (10 mg/ml protein) and 1 μl of the reservoir solution containing 2.0 m (NH4)2SO4, 0.2 m Li2SO4, and 0.1 m N-cyclohexyl-3-aminopropanesulfonic acid (pH 10.5). CYP105D6 crystals were grown at 25 °C by mixing 1 μl of the protein solution (8 mg/ml protein) and 1 μl of the reservoir solution containing 4.0 m sodium formate (pH 8.0). X-ray diffraction data were collected at the BL-5A and NW12A stations at the Photon Factory, High Energy Accelerator Research Organization (KEK), Tsukuba, Japan. After cryoprotection with 20% (v/v) glycerol, crystals were flash-cooled in a nitrogen stream at 100 K. Diffraction images were processed using the HKL2000 program suite (30). The initial phases were determined by molecular replacement using MOLREP (31). The ligand-free CYP105P1 structure was used as a search model. Manual model rebuilding, introduction of water molecules, and refinement were performed using Coot (32) and Refmac5 (33). The topology and parameter file for filipin I was generated based on the solution NMR structure of filipin III (26) using the PRODRG server (34). In the final refinement stage, bulk solvent correction and TLS (parameterization of the translation, libration, and screw rotation displacements of pseudorigid bodies) refinement with the groups defined by the TLSMD server (35) was applied. Data collection and refinement statistics are shown in Table 1. Figures were prepared using PyMol (DeLano Scientific LLC, Palo Alto, CA).
TABLE 1.
Data collection and refinement statistics
| Data set | CYP105P1-Filipin I complex | CYP105D6 Ligand-free |
|---|---|---|
| Data collection statistics | ||
| Beam line | PF-BL5A | PF-AR NW12A |
| Wavelength (Å) | 1.000 | 1.000 |
| Space group | P41212 | P3121 |
| Unit cell (Å) | a = b = 91.368 | a = b = 67.533 |
| c = 151.239 | c = 182.089 | |
| Resolution (Å)a | 50.00-1.80 (1.86-1.80) | 50.00-2.30 (2.38-2.30) |
| Total reflections | 829,838 | 238,132 |
| Unique reflections | 59,975 | 22,279 |
| Completeness (%)a | 99.9 (100.0) | 100.0 (100.0) |
| Redundancya | 13.8 (13.9) | 10.7 (9.8) |
| Mean I/σ(I)a | 38.7 (3.2) | 25.3 (4.1) |
| Rmerge (%)a | 8.8 (46.5) | 9.8 (46.0) |
| Refinement statistics | ||
| Protein Data Bank code | 3ABA | 3ABB |
| Resolution range (Å) | 39.74–1.80 | 33.77–2.30 |
| No. of reflections | 55,867 | 21,082 |
| R-factor/ Rfree (%) | 18.8/23.8 | 16.0/22.1 |
| No. of atoms | 3775 | 3274 |
| TLS groups (residue no.) | 7–82, 83–146, 147–323, 324–403 | 11–92, 93–192, 193–408 |
| Average B-factor (Å2) | ||
| Protein | 18.3 | 23.2 |
| Heme | 21.4 | 16.6 |
| Filipin I | 25.9 | |
| Water | 30.8 | 30.6 |
| SO42− | 46.1 | |
| r.m.s.d. from ideal values | ||
| Bond lengths (Å) | 0.028 | 0.022 |
| Bond angles (degrees) | 2.138 | 2.016 |
| Ramachandran Plot (%)b | ||
| Favored | 98.7 | 96.8 |
| Allowed | 1.0 | 2.9 |
| Outlier | 0.3 | 0.3 |
a Values in parentheses are for the highest resolution shell.
b Determined by RAMPAGE server (49).
RESULTS
Spectral Characterizations and Measurements of Filipin Hydroxylase Activities
Recombinant proteins of CYP105P1 and CYP105D6 with His4 tag at the C termini were expressed in E. coli cells and purified to homogeneity. Spectral characterization of purified CYP105P1 has been described previously (29). UV-visible absorption spectra of purified CYP105D6 in the ferric (resting), dithionite-reduced, and dithionite-reduced plus CO states are shown in supplemental Fig. S2. These spectra show that the protein was folded properly.
To examine the substrate specificities of CYP105P1 and CYP105D6 in vitro, the purified enzymes and filipin I were incubated with the electron transport system of spinach ferredoxin and reductase (Fig. 1). Filipin I was produced and purified from a mutant S. avermitilis strain in which both CYP105P1 and CYP105D6 genes were deleted (ΔpteC/pteD).3 After incubation with CYP105P1, 50.2% of filipin I was converted to filipin II (Fig. 1B). The reaction product was extracted by ethyl acetate and then incubated with CYP105D6 (Fig. 1C). The second reaction resulted in 43.0% conversion to filipin III, and 15.8, 24.4, and 16.8% of filipin II, 1′-hydroxyfilipin, and filipin I were detected. Therefore, in the second reaction catalyzed by CYP105D6, 73.1% of filipin II and 69.2% of filipin I were converted to filipin III and 1′-hydroxyfilipin I, respectively. After incubation with CYP105D6, 34.6% of filipin I was converted to 1′-hydroxyfilipin I (Fig. 1D). Subsequent reaction with CYP105P1 resulted in 27.5% conversion to filipin III and 47.5, 7.6, and 17.4% of filipin II, 1′-hydroxyfilipin, and filipin I were detected. Therefore, in the second reaction catalyzed by CYP105P1, 57.9% of 1′-hydroxyfilipin I and 73.2% of filipin I were converted to filipin III and filipin II, respectively. These results indicated that these enzymes hydroxylate filipin I at different positions. The activity against filipin I was higher for CYP105P1 than CYP105D6. In addition to filipin I, CYP105P1 and CYP105D6 can hydroxylate 1′-hydroxyfilipin I and filipin II, respectively. CYP105D6 showed preference to filipin II over filipin I, and CYP105P1 showed preference to filipin I over 1′-hydroxyfilipin I.
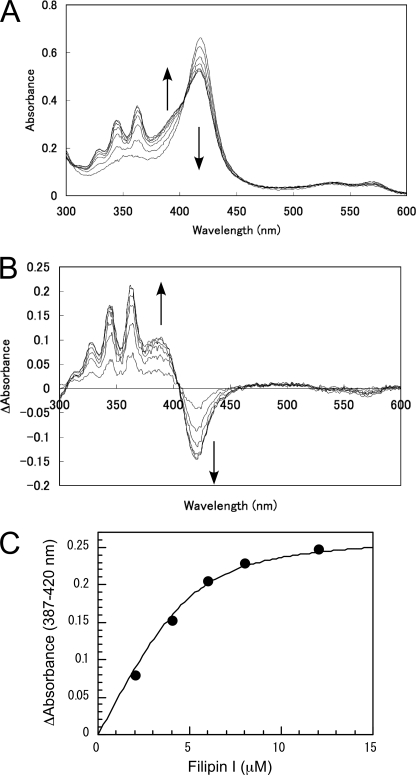
Figs. 2 and 3 show spectral titration results of CYP105P1 and CYP105D6 with filipin I, respectively. The spectra of CYP105P1 illustrate a typical type I spectral shift of the Soret peak (419 nm) to 391 nm. The spectral change of CYP105D6 was relatively smaller, but the difference spectrum clearly shows a type I shift. Filipin I has three absorption maxima around the 320–360-nm region and a shoulder at 305 nm due to vibrational progression of a polyene (19). These peaks exhibited perturbations on binding to CYP105P1 and CYP105D6, and positive peaks were observed in the difference spectra at 329–330, 346–347, and 362–368 nm. The titration curve of CYP105P1 indicated strong binding to the ligand with a stoichiometry of about 1:1 ∼ 2:1. Due to the high affinity, the Kd value of CYP105P1 was difficult to determine. The Kd value was estimated to be 0.66 ± 0.18 μm for CYP105D6. Therefore, CYP105P1 showed higher binding affinity to filipin I than CYP105D6, exhibiting a good correlation with the catalytic activity.
FIGURE 2.
Spectral changes of CYP105P1 (ferric resting state) upon the addition of increasing concentrations of filipin I (A), its difference spectra (B), and the titration curve calculated using the values of absorption differences at 387 and 422 nm (C).
FIGURE 3.
Spectral changes of CYP105D6 (ferric resting state) upon the addition of increasing concentrations of filipin I (A), its difference spectra (B), and the titration curve calculated using the values of absorption differences at 387 and 420 nm (C). A nonlinear fitting with a quadratic equation was applied to the titration curve.
Structure of CYP105P1-Filipin I Complex
The crystal structure of CYP105P1-filipin I complex was determined at 1.8 Å resolution and refined to an R factor of 18.8% (Rfree = 23.8%). The crystal contains one molecule in the asymmetric unit and exhibits a high Matthews coefficient (3.51 Å3/Da) and solvent content (65.0%). The final model contains residues from Asp-7 to His-403, including all four residues of the His tag: one heme, one filipin I molecule, 592 waters, and three sulfate ions. Fig. 4A shows the overall structure of CYP105P1.
FIGURE 4.
Overall structures of CYP105P1-filipin I complex (A) and unliganded CYP105D6 (B), illustrated by ribbon representation. Heme and ligands are shown as stick models. The BC and FG loop regions are shown in dark gray. a.a., amino acids.
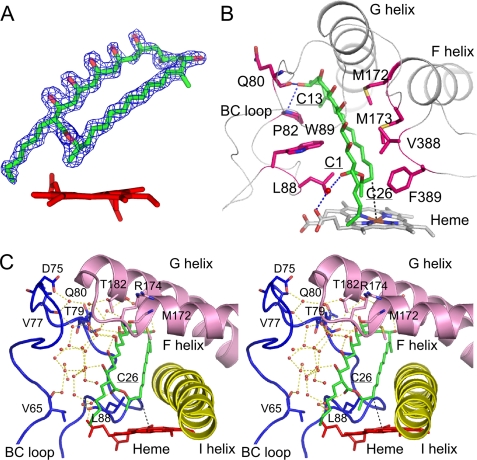
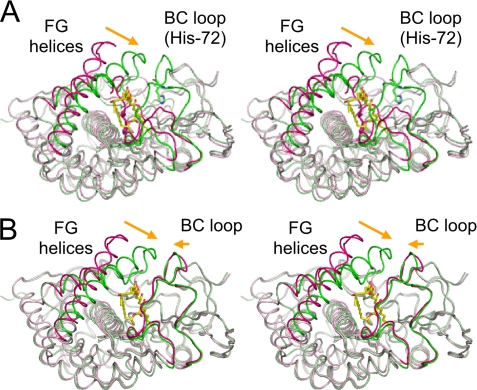
The electron density map for bound filipin I was clearly observed in the distal pocket as shown in Fig. 5A. Superimpositions with the ligand-free wild-type and H72A mutant structures are shown in Fig. 6. The FG helices in the ligand-free structures adopt an open conformation and close on substrate binding. This region often adopts closing motion on ligand binding (36–39). The BC loop region consists of 33 amino acid residues and does not contain a helix, whereas most P450 structures have a B′ helix in this region (4). The BC loop region in the ligand-free wild-type structure has a unique conformation due to ligation of His-72 to the heme iron and completely covers the distal pocket (Fig. 6A), but this histidine ligation state is not detectable in solution (29). The BC loop adopts an open conformation in the H72A mutant structure (29) and is only slightly closed in the complex structure (Fig. 6B). Compared with the H72A mutant structure, the Cα atoms of Asp-176 in the FG loop and Asp-75 in the BC loop shift by 8.7 and 2.6 Å, respectively. Root mean square deviations (r.m.s.d.) with previously determined structures are shown in supplemental Table S1. The long axis of the 28-membered filipin I ring was inclined at about 60 degrees from the vertical axis of the heme plane, and the pro-S hydrogen side of position C26 is directed toward the heme (Fig. 5). The distance between the C26 atom and heme iron is 5.0 Å, which appears to be appropriate for a monooxygenase reaction (discussed later). The filipin I molecule is surrounded by the heme, BC loop, FG loop, G helix, I helix, and C-terminal loop regions. The amino acid residues forming the pocket are as follows: Thr-79—Pro-82 and Ser-86—Trp-89 in the BC loop; Met-172—Met-173 in the FG loop; Thr-182—Glu-183, Gly-186—Met-187, and Leu-189—Gly-190 in the G helix; Met-228—Asn-229, Gly-232—Thr-233, and Ile-236—Ala-238 in the I helix; Val-388—Phe-389 in the C-terminal loop (Fig. 5B). Among these, Gln-80 and Pro-82 are the only two residues to form direct hydrogen bonds with filipin I. The flat 28-membered filipin I ring is sandwiched between two hydrophobic faces. The β-face of the ring (see Rose et al. (40) for the definition) is recognized by Pro-82, Leu-88, and Trp-89, and the α-face is recognized by Met-172, Met-173, Val-388, and Phe-389. The C1 hydroxyl group of filipin I interacts with a carboxylate moiety of heme through water-mediated hydrogen bonds. The pocket at the polyol side of filipin I is filled with numerous water molecules (Fig. 5C), and they mediate interactions with the FG loop, G helix, and BC loop regions. About 30 water molecules are involved in this hydrogen-bonding network. The main chain atoms of Val-65, Val-77, Met-172, and Arg-174 and the side chain atoms of Asp-75, Thr-79, and Thr182 are involved in the water-mediated recognition of the polyol side of filipin I. In contrast, the pentaene side of filipin I forms hydrophobic interactions with the I helix. Another important aspect for substrate recognition is the K helix and subsequent loop region (Fig. 7A). A pocket is formed between this region and the heme, and the alkyl chain moiety of filipin I is bound at this pocket. Three Gly residues, Gly-284, Gly-287, and Gly-288, are clearly important to form this pocket. Moreover, in this region there are several water-mediated hydrogen bonds with the C3 hydroxyl group of filipin I to stabilize substrate binding. The environment around the C1′ atom appears not to hinder the binding of 1′-hydroxy filipin I.
FIGURE 5.
Interactions between CYP105P1 and filipin I. A, shown is an Fobs − Fcalc omit electron density map of the filipin I molecule contoured at 4.0 σ. B, hydrophobic interactions are observed at both sides of the 28-membered ring. Labels for atoms of filipin I are underlined. C, a stereographic figure shows interactions with the BC loop, FG helices, and I helix. The water molecules mediate hydrophilic interactions with the polyol group of filipin I. The extensive hydrogen-bonding network and residues involved in it are shown as dotted lines and stick models, respectively. The distance between the C26 atom of filipin I and the heme iron is 5.0 Å, and the pro-S hydrogen side is directed toward the heme.
FIGURE 6.
Closing motion of CYP105P1. Shown is a stereographic superimposition of the filipin I complex structure with the ligand-free wild-type (A) and H72A mutant (B) structures. BC loop and FG helices are colored magenta and green in ligand-free and complex structures, respectively. The filipin I molecule is shown as yellow sticks. In the ligand-free wild-type structure (A), the side chain of His-72 is ligated to the heme iron as the sixth ligand, and the BC loop sinks into the heme to completely cover the distal pocket.
FIGURE 7.
Interactions between filipin I and a region from the K helix to β1–5 strand. A, a small pocket of CYP105P1 to accommodate the alkyl side chain is shown. This pocket is formed by a kink in a loop after the K helix that contains three glycine residues. Filipin I and water molecules are shown as green sticks and red spheres, respectively. B, superimposition of CYP105P1 (gray) and CYP106D6 (green) structures is shown. The side chains of Ser-290 and Ile-293 of CYP105D6 are shown by the dot surface of van der Waals radii. C, amino acid sequence alignment at a region from the K helix to subsequent strands is shown. Secondary structures of CYP105P1 are indicated above the sequence. Completely and relatively conserved regions are highlighted by black and white inverse characters and boxes, respectively. Residues labeled in panel B are underlined.
Comparison of the Substrate Binding Pocket with Other P450 Enzymes
The volumes of substrate binding distal pockets were calculated using the complex structures of CYP105P1 and several P450s (Table 2). Supplemental Fig. S3 illustrates the ligand binding pockets of CYP105P1, P450nor (CYP55A1), P450eryF, and P450cam (CYP101A). Among the P450s acting upon macrolide substrates, P450 EryK has the largest pocket size, and CYP105P1 is the second largest. The pocket sizes basically correlate with substrate sizes. The substrate of P450 EryK (erythromycin D) is far larger than that of P450eryF (6-deoxyerythronolide B) due to insertion of two deoxysugar units (supplemental Fig. S1). The crystal structures of two CYP105 family enzymes, P450 MoxA and P450 SU-1 (CYP105A1), have been reported (41, 42). In contrast to the two position-specific filipin hydroxylases described in this study, both these enzymes can hydroxylate a wide variety of compounds. Their pocket sizes are completely different due to conformational differences. The P450 SU-1 structure complexed with one of its substrates, 1α,25-dihydroxyvitamin D3, adopts a closed conformation. In contrast, the crystal structure of P450 MoxA adopts an open conformation, although it binds a 2-morpholinoethanesulfonic acid (MES) molecule that is derived from a crystallization buffer. P450nor is closely related to the CYP105 family, but it catalyzes the reduction of nitric oxide to nitrous oxide using NADH as the direct electron donor (43). P450nor has an unusually large distal pocket even in the closed conformation in complex with an NADH analogue, nicotinic acid adenine dinucleotide (38). The distal pocket of P450nor is filled with numerous water molecules that form a proton channel, whereas most P450 enzymes have relatively tight hydrophobic pockets for their substrates.
TABLE 2.
Distal pocket volumes of P450 enzymes
| P450 | Source organism | PDB code | Substrate/Liganda | Distal pocket volumeb |
|---|---|---|---|---|
| Å3 | ||||
| CYP105P1 | S. avermitilis | 3ABA | Filipin I (622.4) | 2166 |
| P450 EryK (CYP113A1) | Saccharopolyspora erythraea | 2JJO | Erythromycin D (703.9) | 2483 |
| P450eryF (CYP107A1) | S. erythraea | 1JIO | 6-Deoxyerythronolide B (386.5) | 1247 |
| P450epoK (CYP167A1) | Sorangium cellulosum | 1Q5D | Epothilone B (507.7) | 1316 |
| P450 PikC (CYP107L1) | Streptomyces venezuelae | 2C7X | Narbomycin (509.7) | 1860 |
| P450 SU-1 (CYP105A1) | Streptomyces griseolus | 2ZBZ | 1α,25-Dihydroxyvitamin D3 (416.6) | 1537 |
| P450 MoxA (CYP105) | Nonomuraea recticatena | 2Z36 | MES (195.2) | 3285 |
| P450nor (CYP55A1) | Fusarium oxysporum | 1XQD | Nicotinic acid adenine dinucleotide (665.4) | 3470 |
| CYP2B4 | Rabbit | 1SUO | 4-(4-Chlorophenyl)imidazole (178.6) | 790 |
| P450cam (CYP101A1) | Pseudomonas putida | 1DZ4 | Camphor (152.2) | 374 |
a Values in parentheses are the molecular weights of the substrate or ligand. See supplemental Fig. S1 for the structures of substrates and ligands.
b Calculated by CASTp server with probe radius of 1.4 Å (50).
Structure of CYP105D6
The substrate-free crystal structure of CYP105D6 was determined at 2.3 Å resolution and refined to an R factor of 16.0% (Rfree = 22.1%). The crystal contains one molecule in the asymmetric unit and exhibits a normal Matthews coefficient (2.66 Å3/Da) and solvent content (53.9%). The final model contains residues from Ser-11 to His-408, including all four residues of the C-terminal His tag, one heme, and 277 waters. However, nine residues in the BC loop ranging from Arg-82 to Leu-90 and six residues in the FG loop ranging from Gly-181 to Ala-186 were not included due to a disorder (Fig. 4B).
In ligand-free P450 structures, the BC loop region is relatively flexible and sometimes disordered. For example, the ligand-free open structures of P450 PikC (14), P450 StaP (44), and CYP231A2 (45) have disordered BC loops. It is a notable feature of CYP105D6 that a total of 15 residues are disordered in both the BC and FG loops, whereas the ligand-free CYP105P1 structure has only four disordered residues in the BC loop (29). The overall structure of CYP105D6 is similar to those of ligand-free CYP105P1 structures. r.m.s.d. for 361 Cα atoms was 2.4 Å with the ligand-free wild-type CYP105P1 structure, and r.m.s.d. for 361 Cα atoms was 2.1 Å with the H72A structure. The CYP105D6 structure shows relatively low structural similarity to the structure of CYP105P1-filipin I complex (r.m.s.d. for 363 Cα atoms = 2.5 Å), as the ligand-free CYP105D6 structure is in an open state. Fig. 7B shows a superimposition of CYP105D6 and CYP105P1-filipin I complex in the region from the K helix to the β1–5 strand. It is clearly visible that CYP105D6 lacks a pocket for the alkyl side chain of filipin. The three glycine residues in CYP105P1 are replaced by bulky residues in CYP105D6, and a deletion of one residue takes place in CYP105D6 (Fig. 7C). The side chains of Ser-290 and Ile-293 in CYP105D6 appear to hinder the binding of the alkyl side chain, and thus, the C26 atom of filipin I cannot approach the heme iron.
DISCUSSION
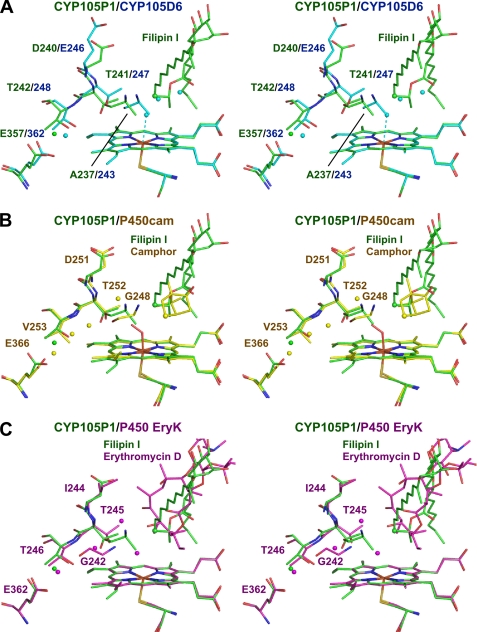
Fig. 8A shows superimposition of the active site structures of CYP105P1-filipin I and CYP105D6. The distances of C25, C26, C27, and C28 of filipin I from the heme iron of CYP105P1 is 6.7, 5.0, 5.3, and 4.5 Å, respectively. Superimposition with the ferrous dioxygen complex of P450cam suggests that the C26 atom is most appropriately positioned for a monooxygenase reaction, as C26 is more closely located to the C5 of camphor than C28 (Fig. 8B). Moreover, C26 of filipin I is located close to the O2 atoms of P450cam oxy-complex on the superimposition (about 3.1 Å to both oxygen atoms), but C28 is not (>3.4 Å). Thr-252 residue of P450cam is proposed to play an important role in protonation required for oxygen activation (46). The preceding acidic residue, Asp-251, is suggested to help proper positioning of Thr-252 and catalytic waters. The Asp/Glu-Thr pair is conserved in CYP105P1 (Asp-240—Thr-241) and CYP105D6 (Glu-246—Thr-247). In the case of P450eryF and CYP158A2, the Thr residue is replaced with Ala, but a hydroxyl group of their substrates substitutes for the Thr and helps to deliver the protons (47, 48). In the active site of CYP105P1, no hydroxyl group of the filipin I or 1′-hydroxyfilipin I substrate is positioned near the heme iron. Fig. 8C shows superposition with P450 EryK-erythromycin D. The target of hydroxylation site of erythromycin D (C12) is positioned 5.3 Å from the heme iron. A water molecule chain, which is thought to deliver proton from the bulk solvent to the active site, is present in P450cam and P450 EryK (Fig. 8, B and C) (13, 46). A conserved Glu residue (Glu-366 in P450cam and Glu-362 in P450 EryK) is involved in holding this water molecule chain. This Glu residue is also present in CYP105P1 (Glu-357) and CYP105D6 (Glu-362) and holds a water molecule in both structures (Fig. 8A). However, there are no water molecules near the active site of CYP105P1, as in the cases of P450eryF and P450 PikC (supplemental Fig. S4) (14, 47). In the substrate-free structure of CYP105D6, several water molecules are present near the heme iron (Fig. 8A). A water molecule is positioned 2.8 Å from the heme iron. These waters may be displaced on substrate binding, as type I spectral change is observed when filipin I is titrated (Fig. 3). In conclusion, a detailed catalytic mechanism of the filipin hydroxylases remains to be elucidated, but the general mechanism proposed for bacterial macrolide monooxygenases seems to be conserved.
FIGURE 8.
Active site structures of CYP105P1-filipin I (green) superimposed with CYP105D6 (A, cyan), P450cam-camphor-O2 (B, yellow), and P450 EryK-erythromycin D (C, magenta). Water molecules and the hydroxylation target positions of substrates (C26 of filipin I, C5 of camphor, and C12 of erythromycin D) are shown as spheres. A water molecule is positioned 2.8 Å from the heme iron in the CYP105D6 structure (A).
Compared with the previously reported structures (29), the CYP105P1-filipin I complex determined in this study provides clear structural insights into the mechanisms of substrate recognition. Filipin I is bound in a large pocket observed in the ligand-free H72A structure (29). The environment of the binding pocket specific for the shape and chemical nature of the substrate explains the strict regio- and stereospecificity as well as the efficient catalysis of 26S-hydroxylation by CYP105P1. The FG helices region adopts an open-close motion on substrate binding as similar to many other P450s, and this movement appears to be sufficient for providing an entrance for the large substrate (see the supplemental movie). However, it is also possible that the filipin molecule enters through the region around the BC loop. This loop is thought to be highly flexible because it adopts distinct conformations among the three previously reported structures (29). Moreover, spectroscopic analysis indicates that the His-ligated conformation of CYP105P1 in which the BC loop blocks substrate binding is not predominant in solution. A combination of crystallographic and kinetic analyses recently revealed that substrate binding by P450 EryK involved at least two steps because there was a pre-existing equilibrium between the open and closed subpopulations (13). There may also be a similar open-close equilibrium in the BC loop of CYP105P1.
Two similar P450s catalyzing hydroxylations at different positions on the same substrate is an interesting feature. Filipin I is expected to bind to CYP105D6 in a “flipped” orientation relative to its binding with CYP105P1. However, the detailed mechanisms for CYP105D6 substrate recognition remain to be elucidated because we could not obtain a complex structure with the substrate. It is difficult to speculate on the possible binding mode of filipin I to this protein because the disordered regions are too long at the distal pocket. However, structural comparisons with the CYP105P1-filipin I complex revealed that filipin I cannot bind to CYP105D6 with a similar orientation due to steric hindrance. This observation explains the strict regiospecificity of CYP105D6, which cannot catalyze hydroxylation of filipin I at position C26.
The measurements of the catalytic activities against filipin I indicated that the 1′-hydroxylating activity of CYP105D6 was relatively less productive than the C26-hydroxylating activity of CYP105P1. Moreover, spectral titration analysis indicated that the filipin I binding to CYP105D6 was weaker than CYP105P1. Although the production mechanism of filipin complex by S. filipinensis remains uncharacterized, our results probably explain why a natural filipin complex contains filipin II (1′-deoxyfilipin III), whereas 1′-hydroxyfilipin I is absent. When CYP105P1 and CYP105D6 were simultaneously incubated with filipin I, 51.8, 20.9, 2.3, and 25.0% of filipin III, filipin II, 1′-hydroxyfilipin I, and filipin I were detected (data not shown). Filipin IV present in the filipin complex has been suggested to be a epimer of filipin III at C1′ or C3 (25). If filipin IV is the 1′-epimer of filipin III, a possible CYP105D6 counterpart in S. filipinensis likely to have ambiguity in its stereospecificity. The intrinsic flexibility of the alkyl side chain (26) may reduce the stereospecificity of its hydroxylation reaction.
Supplementary Material
Acknowledgments
We thank the staff of the Photon Factory for X-ray data collection and Dr. Jean-Marc Lancelin for providing the atomic coordinates of filipin III.
This work was supported by Grants-in-aid for Scientific Research 20248009 (to H. S.) and 20310122 (to H. I.) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S4, and a movie.
The atomic coordinates and structure factors (codes 3ABAM and 3ABBM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
H. Ikeda, M. Doi, A. Arisawa, Y. Fujii, and S. Ōmura, unpublished data.
- P450 or CYP
- cytochrome P450
- HPLC
- high performance liquid chromatography
- r.m.s.d.
- root-mean square deviations
- MES
- 2-morpholinoethanesulfonic acid.
REFERENCES
- 1.Omura S. (ed) (2002) Macrolide Antibiotics: Chemistry, Biology, and Practice, 2nd Ed., Academic Press, San Diego, CA [Google Scholar]
- 2.Xue Y., Sherman D. H. (2001) Metab. Eng. 3, 15–26 [DOI] [PubMed] [Google Scholar]
- 3.Fjaervik E., Zotchev S. B. (2005) Appl. Microbiol. Biotechnol. 67, 436–443 [DOI] [PubMed] [Google Scholar]
- 4.Ortiz de Montellano P. R. (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd Ed., Kluwer Academic/Prenum Publishers, New York [Google Scholar]
- 5.Lamb D. C., Waterman M. R., Kelly S. L., Guengerich F. P. (2007) Curr. Opin. Biotechnol. 18, 504–512 [DOI] [PubMed] [Google Scholar]
- 6.Isin E. M., Guengerich F. P. (2007) Biochim. Biophys. Acta 1770, 314–329 [DOI] [PubMed] [Google Scholar]
- 7.McLean K. J., Sabri M., Marshall K. R., Lawson R. J., Lewis D. G., Clift D., Balding P. R., Dunford A. J., Warman A. J., McVey J. P., Quinn A. M., Sutcliffe M. J., Scrutton N. S., Munro A. W. (2005) Biochem. Soc. Trans. 33, 796–801 [DOI] [PubMed] [Google Scholar]
- 8.Munro A. W., Girvan H. M., McLean K. J. (2007) Nat. Prod. Rep. 24, 585–609 [DOI] [PubMed] [Google Scholar]
- 9.Poulos T. L. (2007) Drug Metab. Rev. 39, 557–566 [DOI] [PubMed] [Google Scholar]
- 10.Guengerich F. P. (2002) Nat. Rev. Drug Discov. 1, 359–366 [DOI] [PubMed] [Google Scholar]
- 11.Cupp-Vickery J. R., Poulos T. L. (1995) Nat. Struct. Biol. 2, 144–153 [DOI] [PubMed] [Google Scholar]
- 12.Nagano S., Li H., Shimizu H., Nishida C., Ogura H., Ortiz de Montellano P. R., Poulos T. L. (2003) J. Biol. Chem. 278, 44886–44893 [DOI] [PubMed] [Google Scholar]
- 13.Savino C., Montemiglio L. C., Sciara G., Miele A. E., Kendrew S. G., Jemth P., Gianni S., Vallone B. (2009) J. Biol. Chem. 284, 29170–29179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman D. H., Li S., Yermalitskaya L. V., Kim Y., Smith J. A., Waterman M. R., Podust L. M. (2006) J. Biol. Chem. 281, 26289–26297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wachtler V., Balasubramanian M. K. (2006) Trends Cell Biol. 16, 1–4 [DOI] [PubMed] [Google Scholar]
- 16.Gimpl G., Gehrig-Burger K. (2007) Biosci. Rep. 27, 335–358 [DOI] [PubMed] [Google Scholar]
- 17.Butler J. D., Comly M. E., Kruth H. S., Vanier M., Filling-Katz M., Fink J., Barton N., Weintroub H., Quirk J. M., Tokoro T. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 556–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler J. D., Blanchette-Mackie J., Goldin E., O'Neill R. R., Carstea G., Roff C. F., Patterson M. C., Patel S., Comly M. E., Cooney A. (1992) J. Biol. Chem. 267, 23797–23805 [PubMed] [Google Scholar]
- 19.Whitfield G. B., Brock T. D., Ammann A., Gottlieb D., Carter H. E. (1955) J. Am. Chem. Soc. 77, 4799–4801 [Google Scholar]
- 20.Bergy M. E., Eble T. E. (1968) Biochemistry 7, 653–659 [DOI] [PubMed] [Google Scholar]
- 21.Rychnovsky S. D., Richardson T. I. (1995) Angew. Chem. Int. Ed. Engl. 34, 1227–1230 [Google Scholar]
- 22.Richardson T. I., Rychnovsky S. D. (1996) J. Org. Chem. 61, 4219–4231 [DOI] [PubMed] [Google Scholar]
- 23.Pandey R. C., Rinehart K. L., Jr. (1970) J. Antibiot. 23, 414–417 [DOI] [PubMed] [Google Scholar]
- 24.Edwards D. M. F. (1989) J. Antibiot. 42, 322–324 [DOI] [PubMed] [Google Scholar]
- 25.Pandey R. C., Narasimhachari N., Rinehart K. L., Jr., Millington D. S. (1972) J. Am. Chem. Soc. 94, 4306–4310 [DOI] [PubMed] [Google Scholar]
- 26.Volpon L., Lancelin J. (2000) FEBS Lett. 478, 137–140 [DOI] [PubMed] [Google Scholar]
- 27.Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H., Shiba T., Sakaki Y., Hattori M., Omura S. (2003) Nat. Biotechnol. 21, 526–531 [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Chen J., Jiang Y., Zhang W., Jiang W., Lu Y. (2009) FEMS Microbiol Lett. 298, 199–207 [DOI] [PubMed] [Google Scholar]
- 29.Xu L. H., Fushinobu S., Ikeda H., Wakagi T., Shoun H. (2009) J. Bacteriol. 191, 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Vagin A., Teplyakov A. (1997) J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 35.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 36.Park S. Y., Yamane K., Adachi S., Shiro Y., Weiss K. E., Maves S. A., Sligar S. G. (2002) J. Inorg. Biochem. 91, 491–501 [DOI] [PubMed] [Google Scholar]
- 37.Poulos T. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13121–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima R., Fushinobu S., Su F., Zhang L., Takaya N., Shoun H. (2004) J. Mol. Biol. 342, 207–217 [DOI] [PubMed] [Google Scholar]
- 39.Zhao B., Guengerich F. P., Bellamine A., Lamb D. C., Izumikawa M., Lei L., Podust L. M., Sundaramoorthy M., Kalaitzis J. A., Reddy L. M., Kelly S. L., Moore B. S., Stec D., Voehler M., Falck J. R., Shimada T., Waterman M. R. (2005) J. Biol. Chem. 280, 11599–11607 [DOI] [PubMed] [Google Scholar]
- 40.Rose I. A., Hanson K. R., Wilkinson K. D., Wimmer M. J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 2439–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasutake Y., Imoto N., Fujii Y., Fujii T., Arisawa A., Tamura T. (2007) Biochem. Biophys. Res. Commun. 361, 876–882 [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto H., Shinkyo R., Hayashi K., Yoneda S., Yamada M., Kamakura M., Ikushiro S., Shiro Y., Sakaki T. (2008) Biochemistry 47, 4017–4027 [DOI] [PubMed] [Google Scholar]
- 43.Nakahara K., Tanimoto T., Hatano K., Usuda K., Shoun H. (1993) J. Biol. Chem. 268, 8350–8355 [PubMed] [Google Scholar]
- 44.Makino M., Sugimoto H., Shiro Y., Asamizu S., Onaka H., Nagano S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11591–11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho W. W., Li H., Nishida C. R., de Montellano P. R., Poulos T. L. (2008) Biochemistry 47, 2071–2079 [DOI] [PubMed] [Google Scholar]
- 46.Nagano S., Poulos T. L. (2005) J. Biol. Chem. 280, 31659–31663 [DOI] [PubMed] [Google Scholar]
- 47.Nagano S., Cupp-Vickery J. R., Poulos T. L. (2005) J. Biol. Chem. 280, 22102–22107 [DOI] [PubMed] [Google Scholar]
- 48.Zhao B., Guengerich F. P., Voehler M., Waterman M. R. (2005) J. Biol. Chem. 280, 42188–42197 [DOI] [PubMed] [Google Scholar]
- 49.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 50.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.