Abstract
Our study of the mouse Ate1 arginyltransferase, a component of the N-end rule pathway, has shown that Ate1 pre-mRNA is produced from a bidirectional promoter that also expresses, in the opposite direction, a previously uncharacterized gene (Hu, R. G., Brower, C. S., Wang, H., Davydov, I. V., Sheng, J., Zhou, J., Kwon, Y. T., and Varshavsky, A. (2006) J. Biol. Chem. 281, 32559–32573). In this work, we began analyzing this gene, termed Dfa (divergent from Ate1). Mouse Dfa was found to be transcribed from both the bidirectional PAte1/Dfa promoter and other nearby promoters. The resulting transcripts are alternatively spliced, yielding a complex set of Dfa mRNAs that are present largely, although not exclusively, in the testis. A specific Dfa mRNA encodes, via its 3′-terminal exon, a 217-residue protein termed DfaA. Other Dfa mRNAs also contain this exon. DfaA is sequelogous (similar in sequence) to a region of the human/mouse HTEX4 protein, whose physiological function is unknown. We produced an affinity-purified antibody to recombinant mouse DfaA that detected a 35-kDa protein in the mouse testis and in several cell lines. Experiments in which RNA interference was used to down-regulate Dfa indicated that the 35-kDa protein was indeed DfaA. Furthermore, DfaA was present in the interchromatin granule clusters and was also found to bind to the Ggnbp1 gametogenetin-binding protein-1 and to the Abt1 activator of basal transcription that interacts with the TATA-binding protein. Given these results, RNA interference was used to probe the influence of Dfa levels in luciferase reporter assays. We found that DfaA acts as a repressor of TATA-box transcriptional promoters.
Keywords: DNA transcription, Promoters, Repressor Protein, Transcription Promoter, Ubiquitin, Ate1, Fanconi Anemia, N-end Rule, Arginylation, Ubiquitin
Introduction
This study stems from a serendipitous finding (described below) in our 2006 investigation of the mouse Arg-tRNA-protein transferase (R-transferase), encoded by the Ate1 gene. The Ate1 R-transferase is a component of the N-end rule pathway of protein degradation. In eukaryotes, this pathway is a part of the ubiquitin-proteasome system. The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue (1–13). N-terminal degradation signals of the N-end rule pathway are called N-degrons. The main determinant of an N-degron is a destabilizing N-terminal residue of a substrate protein. Among such residues, some are primary destabilizing N-terminal residues, which are recognized directly by ubiquitin ligases of the N-end rule pathway, called N-recognins. Other destabilizing N-terminal residues, called secondary or tertiary destabilizing residues, must be modified through deamidation and/or arginylation (6, 7, 14) or, alternatively, through N-terminal acetylation (13) before the corresponding proteins can be targeted by cognate N-recognins. In particular, the destabilizing activity of N-terminal Asp and Glu requires their conjugation, by the Ate1 R-transferase (6, 7, 12, 14–16), to Arg, one of the primary destabilizing residues. In eukaryotes that produce nitric oxide, R-transferase arginylates not only N-terminal Asp and Glu but also Cys, after its conversion to Cys-sulfinate or Cys-sulfonate, in reactions that require nitric oxide and oxygen (6, 17). Alternative splicing of the mouse Ate1 pre-mRNA produces at least six isoforms of R-transferase, a metabolically unstable protein whose enzymatic activity and the in vivo half-life are down-regulated by heme (7, 12, 14–16).
We found that the transcriptional promoter of mouse Ate1 is bidirectional, driving the expression of both Ate1 and an oppositely oriented, previously uncharacterized gene (14), which was termed Dfa (divergent from Ate1). We wished to explore Dfa because the proximity and head-to-head arrangement of Ate1 and Dfa (Fig. 1A) suggested that the corresponding gene products, R-transferase and Dfa, might be functionally linked, either directly or through belonging to a specific regulatory circuit. This work is the beginning of Dfa studies. We show that Dfa is transcribed from both the bidirectional PAte1/Dfa promoter and other nearby promoters. The resulting transcripts are alternatively spliced, yielding a complex set of Dfa mRNAs that are present largely, although not exclusively, in the testis. A specific Dfa mRNA encodes, via its 3′-terminal exon, a 217-residue protein, termed DfaA. Other Dfa mRNAs also contain this exon (Fig. 1, D–F). DfaA is sequelogous (similar in sequence) (18) to a region of the previously described human/mouse HTEX4 protein, whose physiological function is unknown. We produced an affinity-purified antibody to the 26-kDa recombinant mouse DfaA, whose apparent molecular mass, upon SDS-PAGE, is 35 kDa. This antibody detected a 35-kDa protein in the mouse testis, in mouse NIH-3T3 cells, and in human HeLa and HEK-293T cells. Experiments in which RNAi4 was used to down-regulate Dfa in NIH-3T3 cells indicated that the 35-kDa protein (recognized by anti-DfaA antibody) was indeed DfaA. This protein is both nuclear and cytoplasmic. Biochemical fractionations suggested an association of DfaA with membranes or other rapidly sedimenting structures. Transient expression of a GFP-DfaA fusion protein indicated that DfaA was present preferentially in the interchromatin granule clusters (IGCs). In addition, DfaA was found to interact with specific proteins, including the Abt1 transcriptional activator. Given these results, RNAi was used to down-regulate Dfa in assays with NIH-3T3 cells and a luciferase reporter expressed from a TATA-box transcriptional promoter. We found that DfaA acts as a repressor of this promoter but does not influence the bidirectional PAte1/Dfa promoter, which contains a CpG island and lacks the TATA-box. Contrary to our expectation, no functional or mechanistic connections between the Dfa protein(s) and isoforms of the Ate1 R-transferase were detected so far, apart from the proximity of their head-to-head oriented genes and the antisense orientation of some among the Dfa and Ate1 transcripts (Fig. 1, A and D).
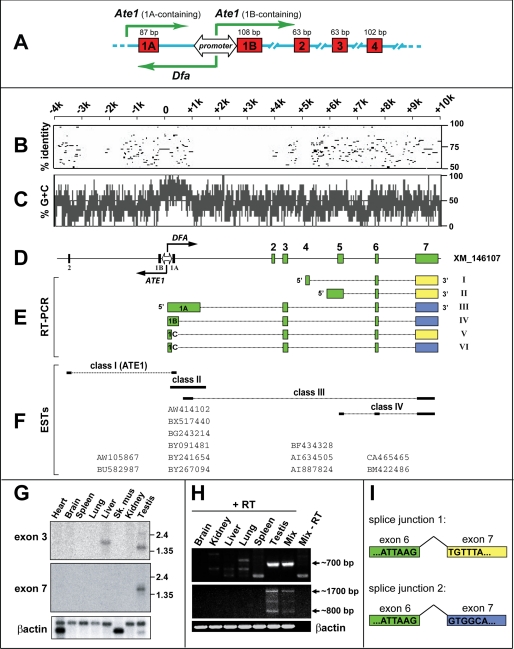
FIGURE 1.
Genomic characterization of the mouse Dfa gene. A, bidirectional promoter upstream of the mouse Ate1 exon 1B (see Introduction and Refs. 12, 14). Green arrows indicate transcriptional units oriented in both directions from the PAte1/Dfa promoter and also from an unmapped “upstream” promoter that mediates the expression of Ate1 transcripts containing exon 1A (14). The locations and sizes of some Ate1 exons are shown as well. B, to make expression patterns of Dfa easier to follow, the orientation of the Dfa and Ate1 transcriptional units was flipped 180° in this and other panels, in comparison with A. Percent identity (from 50 to 100%) of each gap-free segment between ∼14 kb of the mouse and human genomic DNA segments that encompass the Ate1 and Dfa genes. Position of identities are shown with respect to mouse DNA. Note short regions of significant conservation, including an ∼200-bp segment that contains the bidirectional PAte1/Dfa promoter. C, (G + C) content (%) over ∼14 kb of genomic DNA that encompasses the 5′-regions of Dfa and Ate1 reveals a CpG island at the center of the bidirectional PAte1/Dfa promoter (see the main text). D, shown are the relative positions of Ate1 exons 1A, 1B, and 2, the bidirectional PAte1/Dfa promoter, and Dfa exons 1–7. The two oppositely oriented arrows at the PAte1/Dfa promoter indicate the directions of Ate1 and Dfa transcription, respectively. E, different species of RT-PCR DNA fragments that were amplified from mouse testis total RNA in this work and their positions vis à vis specific Dfa exons that are shown in D. A yellow box exon 7 (Dfa cDNAs I, II, and V) signifies the use of splice junction 1 to produce a Dfa transcript (see panel I). A blue box exon 7 (Dfa cDNAs III, IV, and VI) signifies the use of splice junction 2 (see panel I) to produce a Dfa transcript. F, comparison and classification of ESTs in the NCBI data base that encompassed the Ate1/Dfa locus. These ESTs are arranged according to their positions vis à vis specific Ate1 or Dfa exons (see D) and specific RT-PCR DNA fragments isolated in this study (see E). G, Northern analyses of Dfa expression in mouse tissues. Upper panel, Dfa exon 3-specific probe. Middle panel, Dfa exon 7-specific probe. Lower panel, β-actin mRNA probe was used to verify the uniformity of total RNA inputs. H, RT-PCR DNA fragments amplified from total RNA isolated from indicated mouse tissues. Upper panel, ∼0.7-kb Dfa-specific RT-PCR DNA fragment amplified from testis RNA using forward and reverse primers annealing to the class IV EST-CA465465 (see F) (forward 5′-CCAGACCACAGAGCCAGCAC-3′; reverse 5′-TTTGCCCAGGCCATTTTCGGC-3′). DNA sequence analyses of RT-PCR-amplified DNA fragments from tissues other than the testis indicated that they were nonspecific (unrelated to the Dfa/Ate1 locus) (data not shown). Middle panel, ∼1.7 and ∼0.8 kb Dfa-specific RT-PCR DNA fragments amplified from testis RNA using a forward primer annealing genomic DNA in the vicinity of the bidirectional PAte1/Dfa promoter (5′-GCCCTTGTATTCCACCACCG-3′) and a reverse primer annealing to the class IV EST-CA465465 (reverse 5′-TTTGCCCAGGCCATTTTCGGC-3′). Bottom panel, β-actin-specific RT-PCR DNA fragments derived from all tissues (mix, equimolar mixture of cDNA isolated from brain, kidney, liver, lung, spleen, and testis; ±RT, indicates the presence or absence of reverse transcriptase used in generating 1st strand cDNA). I, diagram of the Dfa pre-mRNA splicing that involved depicting the alternative splice site selection between Dfa exons 6 and 7. See E and the main text for additional details.
EXPERIMENTAL PROCEDURES
RT-PCR
Total RNA from the brain, kidney, liver, lung, spleen, and testis (C57 mice) was isolated using the RNeasy kit (Qiagen, Valencia, CA), according to manufacturer's instructions. First-strand cDNA synthesis was carried out using the Superscript kit and oligo(dT) primer (Invitrogen). These samples were then used to carry out either standard PCR or 5′-RACE, using Invitrogen GeneRacer kit (catalog no. L1500-01, version J). The following oligonucleotide primers were employed to detect, by RT-PCR, the Dfa cDNA-I (Fig. 1E): forward 5′-CGACTGGAGCACGAGGACACTGA-3′ (GeneRacer 5′ Primer); reverse 5′-TTTGCCCAGGCCATTTTCGGC-3′ (CA465465-Rev). The primers for the nested reaction were as follows: forward 5′-GGACACTGACATGGACTGAAGGAGTA-3′ (GeneRacer 5′ Nested Primer); reverse 5′-CCTGTTGGAACTTTTGGACTAACAG-3′ (CA465465-Rev2). The primers that yielded the Dfa cDNA-II were as follows: forward 5′-GGACACTGACATGGACTGAAGGAGTA-3′ (GeneRacer 5′ Nested Primer); reverse 5′-CCTGTTGGAACTTTTGGACTAACAG-3′ (CA465465-Rev2). The primers that yielded the Dfa cDNA-III were as follows: forward 5′-GCCCTTGTATTCCACCACCG-3′; reverse 5′-TTTGCCCAGGCCATTTTCGGC-3′ (CA465465-Rev). The primers that yielded the Dfa cDNA-IV were as follows: forward 5′-GCCCTTGTATTCCACCACCG-3′; reverse 5′-TTTGCCCAGGCCATTTTCGGC-3 (CA465465-Rev). The primers that yielded the Dfa cDNA-V and cDNA-VI were as follows: forward 5′-CCTCCTCTGCCTGCCAGG-3′; reverse 5′-TTTGCCCAGGCCATTTTCGGC-3′ (CA465465-Rev). 3′-RACE reactions were performed using nested PCRs and Invitrogen GeneRacer kit (version J) according to the manufacturer's protocol and a forward primer specific for the Dfa exon 7, which is shared by Dfa cDNAs characterized so far (Fig. 1E) as follows: 5′-CCGAAAATGGCCTGGGCAAAGG-3′; GeneRacer 3′ Primer 5′-GCTGTCAACGATACGCTACGTAACG-3′, and GeneRacer 3′ Nested Primer 5′-CGCTACGTAACGGCATGACAGTG-3′.
Northern Hybridization
Northern blots of mRNAs from mouse tissues, containing 2 μg of mouse poly(A)+ RNA per lane (Clontech), were first probed with a 180-bp DNA fragment specific for the Dfa exon 3 (Fig. 1G). This fragment was produced by PCR from the RT-PCR-derived Dfa cDNA III, using primers CB120 (5′-GATGAAGCTGCTGATGCTGC-3′) and CB121 (5′-CTTAGGTTCTCTCACAGAATC-3′). After hybridization with the exon 3-specific probe, the membrane was stripped and rehybridized with a 556-bp DNA probe specific for Dfa exon 7, which is shared by Dfa cDNAs characterized so far (Fig. 1G). This probe was produced by PCR from the RT-PCR-derived Dfa cDNA III, using primers 5′-GAGAGAACCTAAGATTGGCCTGGGC-3′ and 5′-TTTGCCCAGGCCATTTTCGGC-3′. Northern DNA probes were 32P-labeled using the RediprimeII random prime labeling system (Amersham Biosciences). Hybridization with a DNA probe specific for exon 3 was carried out for 2 h at 68 °C in ExpressHyb solution (Clontech). The blot was then washed once for 30 min at room temperature in 2× SSC, 0.1% SDS, once for 30 min at 55 °C in 0.5× SSC, 0.1% SDS, and once for 30 min at 52 °C in 0.1× SSC, 0.1% SDS, followed by autoradiography. Hybridization with a probe specific for exon 7 was carried out for 12 h at 65 °C in ExpressHyb solution. The blot was then washed once for 10 min at room temperature in 2× SSC, 0.1% SDS, once for 30 min at 55 °C in 2× SSC, 0.1% SDS, and once for 30 min at 55 °C in 1× SSC, 0.1% SDS followed by autoradiography.
Antibody to Mouse Dfa
His10-DfaA, an N-terminally His10-tagged isoform encoded by Dfa cDNA I (Fig. 1E), was expressed in Escherichia coli using the pET-16b vector (Novagen, EMD Chemicals, Gibbstown, NJ). Briefly, a 1-liter culture of E. coli BL21(DE3) was grown in LB medium at 37 °C to an A600 of ∼0.6, then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for additional 3 h at 37 °C. Cells were harvested by centrifugation at 2000 × g for 10 min at 4 °C. The cell pellet was resuspended in 50 ml of 10 mm imidazole, 20 mm Tris-HCl (pH 8.0), 1 mg/ml lysozyme, plus EDTA-free protease inhibitors (Roche Applied Science), followed by a 30-min incubation on ice, and one cycle of freeze-thaw. The resulting suspension was centrifuged at 100,000 × g for 35 min at 4 °C. Inclusion bodies containing His10-DfaA were solubilized by resuspension in 30 ml of ice-cold 6 m guanidine HCl, 10 mm imidazole, 0.5 m KCl, 40 mm Tris-HCl (pH 8.0), also containing 0.5 mm phenylmethylsulfonyl fluoride (PMSF). The resulting solubilized His10-DfaA was clarified by centrifugation at 100,000 × g for 35 min at 4 °C and was thereafter purified by Ni2+-agarose affinity chromatography as described previously (11), except for the presence of 6 m guanidine HCl. Imidazole-eluted, purified His10-DfaA was dialyzed overnight at 4 °C against 10% glycerol, 0.15 m KCl, 0.5 mm PMSF, 40 mm HEPES (pH 7.6) and was thereafter further purified by SDS-12% PAGE. ∼0.9 mg of His10-DfaA that was excised from a 12% preparative-scale polyacrylamide gel was used to inject two rabbits (six times over the course of 3 months) to produce antisera (Covance, Berkeley, CA).
Total IgG was isolated by affinity chromatography, using GammaBind G-Sepharose (GE Healthcare). Pooled IgG fractions were dialyzed against phosphate-buffered saline (PBS: 0.15 m NaCl, 50 mm potassium phosphate (pH 7.5)) containing 10% glycerol and then passed through a column of Affi-Gel-10 (Bio-Rad) with the immobilized (purified) His6-Aat, an unrelated E. coli protein described previously (19). The flow-through fraction was incubated with Affi-Gel-10 beads conjugated to purified mouse His10-DfaA. After 2 h at 4 °C, with gentle rocking, the sample was loaded into 5-ml disposable columns; the flow-through was collected, and the column was washed with 20 bed volumes of PBS. Anti-DfaA IgG was eluted with 0.2 m glycine (pH 2.8) into tubes containing one-third of elution volume of 1 m K2HPO4 (pH 8.5). The resulting fractions were pooled, dialyzed against PBS (0.15 m NaCl, 50 mm potassium phosphate (pH 7.5)) containing 10% glycerol, and stored at −20 °C.
Construction of shRNAi Plasmids
Of the four different Dfa-specific target sequences tested, the most potent microRNA-like short hairpin (sh) RNA that down-regulated Dfa expression targeted the sequence GCCACCCTCACTTGAAATCAAA in exon 7 of Dfa (Figs. 1D and 2A). To construct a corresponding shRNAi-expressing plasmid, termed pEN-DFAsh4, 0.1 ml of the DFAsh4 oligonucleotide (5′-AGCGACCACCCTCACTTGAAATCAAATAGTGAAGCCACAGATGTATTTGATTTCAAGTGAGGGTGGC-3′), at 50 μm, was mixed with 0.1 ml of 50 μm DFAsh4-rev (5′-GGCAGCCACCCTCACTTGAAATCAAATACATCTGTGGCTTCACTATTTGATTTCAAGTGAGGGTGGT-3′) and incubated for at 95 °C for 10 min. This mixture of single-stranded (unannealed) oligonucleotides was incubated in a water bath at 70 °C for 10 min. The bath was then turned off, and the temperature was allowed to decrease to room temperature overnight. The resulting double-stranded oligo was phosphorylated by T4 polynucleotide kinase and ligated to BfuAI-cut pEN-hU6miR2c (a gift from J. Zavzavadjian, California Institute of Technology, Pasadena, CA) (20), yielding pEN-DFAsh4 (the pEN_hU6miR2c plasmid (20) containing Dfa-sh4). For use in stable transfections, an NheI-digested fragment containing a floxed PGK-hygromycin cassette (12) was inserted into the single NheI site of pEN-DFAsh4.
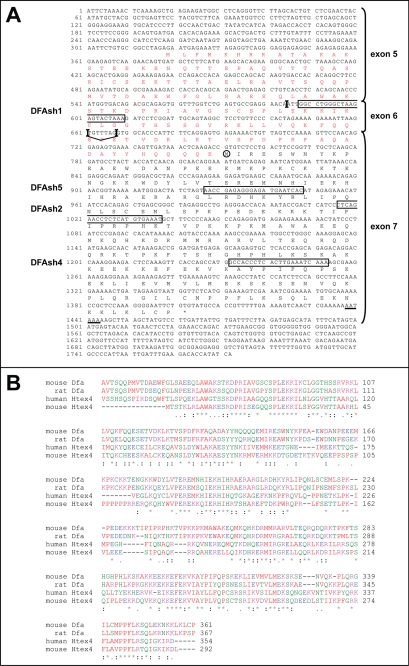
FIGURE 2.
Amino acid sequence of Dfa. A, deduced mouse Dfa amino acid sequence encoded by Dfa cDNA-II (see Fig. 1E). This sequence includes the 217-residue sequence (in black letters) that is encoded by exon 7 and includes the DfaA isoform (see the main text). DfaB, a larger isoform shown here, differs from DfaA in containing an N-terminal extension (in red letters) that is encoded by exons 5 and 6 and is produced by selection of the splice junction 1 in Dfa cDNA-II (Fig. 1, E and I). Exon junctions, including alternative junctions at nucleotide positions 661 and 668, are indicated by vertical bars. The corresponding Dfa exons (see Fig. 1, D and E) are indicated on the right. The deduced N-terminal Met residue of the DfaA isoform is circled. Dfa-coding sequences that were targeted by RNAi (shRNA) in the present study are boxed and are denoted on the left. Underlined nucleotides denote the deduced poly(A) signal. Nucleotide 1 is the 5′-end of Dfa cDNA-II mapped by RACE (see under “Experimental Procedures”). B, ClustalW2-based amino acid sequence alignments of mouse and rat Dfa versus human and mouse Htex4 (see the main text). Note that the sequences encoded by Dfa exons 6 and 7 are sequelogous (18) to Htex4. Residues of similar physicochemical properties are similarly colored. Specifically, A, V, F, P, M, I, L, and W are shown in red; D and E are shown in blue; R, H, and K are shown in magenta, and S, T, Y, H, C, N, G, and Q are shown in green. *, identical residues in all of the aligned sequences; :, highly conserved; ., weakly conserved.
Gel Filtration of Dfa from Mouse Testis
Whole-testis extract was prepared by homogenizing pooled testes of 60 C57BL/6 mice, using a tissue homogenizer (Biospec Products, Bartlesville, OK) in TMSD Buffer (0.25 m sucrose, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 10 mm Tris-HCl (pH 7.6)) also containing 0.5 mm PMSF, added from a freshly prepared stock solution in isopropyl alcohol. Nuclear (pellet) and cytosolic (supernatant) fractions were prepared by centrifugation at 800 × g for 10 min at 4 °C. The supernatant was frozen with liquid N2 and stored at −80 °C. The nuclear fraction was washed three times in TMSD, then resuspended in Nuclear Buffer (25% glycerol, 0.42 m NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20 mm HEPES (pH 7.9), containing 0.5 mm PMSF), frozen with liquid N2, and stored at −80 °C. In the next step, the nuclear fraction was thawed on ice and centrifuged at 16,000 × g for 90 min at 4 °C, yielding the post-nuclear supernatant. The cytosolic fraction was thawed and centrifuged at 28,000 × g for 90 min at 4 °C, yielding post-lysosomal supernatant. Before gel filtration, samples of nuclear and cytosolic extracts were dialyzed against Buffer G (10% glycerol, 0.15 m NaCl, 1 mm dithiothreitol, 40 mm HEPES (pH 7.9) containing 0.1 mm PMSF) overnight at 4 °C. The resulting samples were clarified by centrifugation in the SWTi41 rotor (Beckman Instruments, Fullerton, CA) at 28,000 rpm for 2 h at 4 °C. The samples obtained were concentrated using Amicon Ultracentrifugal filter devices (Millipore, Billerica, MA). The resulting samples (5–20 mg of total protein in 1.5 ml) were subjected to gel filtration, using fast protein liquid chromatography apparatus and the HiLoad 16/60 Superdex 200 (GE Healthcare) pre-equilibrated with Buffer G, at 0.5 ml/min. 2-ml fractions were collected and analyzed, in particular, by immunoblotting with anti-DfaA antibody.
GFP-Dfa and Immunofluorescence
The plasmid pEGFP-DfaA was constructed by ligating the DfaA ORF from cDNA I into KpnI/BamHI-cut pEGFP-C1 (Clontech). Poly-d-lysine (1 mg/ml)-coated coverslips were placed in 35-mm tissue culture dishes. 1.5 × 105 NIH-3T3 cells were then seeded overnight and transfected with the pEGFP-C1 (control) or pEGFP-DfaA plasmids. The cells were then grown to ∼70% confluence. For GFP localization studies, coverslips were fixed in 4% formaldehyde for 10 min at room temperature, then washed three times in PBS, and directly mounted using Vectashield H-1500 mounting medium (Vector Laboratories, Burlingame, CA). For analyses that also involved immunofluorescence staining, cells were fixed in 0.2% Triton X-100, 2% formaldehyde, in PBS for 10 min at room temperature followed by acetone at −20 °C for 5 min. After fixation, coverslips were washed three times in PBS, blocked by incubation with 1% goat serum for 10 min at room temperature, followed by incubation with an anti-SC35 antibody (Abcam, Cambridge, MA) for 2 h at room temperature. For indirect immunofluorescence, the coverslips were then washed in PBS at incubated with a Cy3-conjugated goat anti-mouse IgG (Abcam) for 1 h at room temperature in the dark. Coverslips were again washed with PBS, mounted using Vectashield H-1500 mounting medium, and examined by fluorescence microscopy, using Zeiss Axiophot.
Expression of DfaA in Saccharomyces cerevisiae
A colony of SC295 S. cerevisiae that had been transformed with pCB172 (expressing FLAG-tagged mouse DfaA) was inoculated into 2 liters of the plasmid-retaining SD (−Leu) medium, followed by growth at 30 °C to A600 of ∼1.0. An equal volume of YPD medium was then added, and the culture was grown to A600 of ∼4.0. Cells were harvested by centrifugation, washed once with cold PBS, and frozen in liquid N2. The frozen pellet (∼10 g wet weight) was then ground to a fine powder in liquid N2 using a mortar and pestle and resuspended (6 ml of buffer per 1 g of pellet) in yeast Lysis Buffer (10% glycerol 0.05% Nonidet P-40, 0.2 m KCl, 50 mm HEPES (pH 7.5)) containing leupeptin, antipain, pepstatin A, and aprotinin, each at 5 μg/ml. The suspension was centrifuged at 11,200 × g for 30 min, and the supernatant was mixed, with slow rocking, with 1 ml of anti-FLAG M2 affinity gel (Sigma) at 4 °C for 2 h. The affinity beads were collected by a brief spin in a microcentrifuge, then washed once with 20 bed volumes of yeast Lysis Buffer containing 0.8 m KCl, twice with 20 bed volumes of yeast Lysis Buffer, then again with 20 bed volumes with Lysis Buffer lacking Nonidet P-40. The antibody-bound FLAG-DfaA was eluted with yeast Lysis Buffer lacking Nonidet P-40 and containing FLAG peptide (Sigma) at 0.2 mg/ml.
Yeast two-hybrid assay was carried out using the Matchmaker kit (BD Biosciences). Briefly, S. cerevisiae AH109 (MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80, LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) was transformed with pAS2-DfaA, which expressed Gal4DBD-DfaA. The resulting strain (AH109-Dfa2) was mated with S. cerevisiae Y187 (MATα ura3-52 his3-200 ade2-101 trp1-90 leu2-3, 112, gal4Δ gal80Δ met− URA3::GAL1UAS-GAL1TATA-lacZ MEL1) that had been pre-transformed with a mouse testis cDNA pACT library (Clontech). The resulting diploids were incubated on SC plates containing 5 mm 3-aminotriazole (Sigma) and lacking His, Trp, and Leu. The resulting ∼2.5 × 106 Trp+, Leu+, His+ transformants were further assayed for their growth on His-lacking, Ade-lacking media and also for their β-galactosidase activity. Colonies that were Trp+, Leu+, His+, and Ade+ and exhibited β-galactosidase activity were then grown in Leu-lacking, Trp-containing media to facilitate the loss of the pAS2-DfaA plasmid. pACT plasmids were recovered from the resulting cell clones, followed by sequencing of cDNA inserts to identify potential Dfa-binding proteins. Vector “swapping” experiments to verify detected interactions were carried out as follows. The DfaA ORF was excised from pAS2-DfaA, using digestion with BamHI and SalI. In addition, the Abt1 ORF as well as the GgnBP1 ORF were excised from the corresponding pACT2-based plasmids, using digestion with BamHI and XhoI. The excised DfaA ORF was subcloned into pACT2, and the excised Abt1 ORF and (separately) the GgnBP1 ORF were subcloned into pAS2, followed by two-hybrid assays with the resulting plasmids.
Expression of Recombinant Proteins in BL21(DE3) E. coli
The mouse DfaA cDNA was amplified from pACT-DfaA using PCR and primers flanked by the BamHI sites. The resulting DNA fragment was subcloned at position 1 of the BamHI-cut pET-Duet1 vector (Novagen, EMD Chemicals, Gibbstown, NJ), yielding pCB157. A triple HA-tagged wild-type mouse Ggnbp1 cDNA was amplified from pcDNA3.1-Ggnbp1 using primers flanked by the NdeI and XhoI sites. The resulting DNA fragment was subcloned at position 2 of the NdeI/XhoI-cut pET-Duet, yielding pCB182. A third pET-Duet-based plasmid, pCB183, encoding His6-DfaA at position 1 and Ggnbp1 in position 2, was also constructed to coexpress these proteins in E. coli. 50-ml cultures of E. coli BL21(DE3) were grown at 37 °C to an A600 of ∼0.6 in LB containing ampicillin (50 μg/ml). The cultures were placed on ice for 15 min, followed by the addition of isopropyl 1-thio-β-d-galactopyranoside to the final concentration of 0.25 mm and incubation for 4 h at room temperature. Cells were harvested by centrifugation at 2,000 × g for 10 min at 4 °C. The pellets were resuspended in 5 ml of 0.15 m NaCl, 10 mm imidazole, 20 mm Tris-HCl (pH 8.0), containing lysozyme at 1 mg/ml, followed by incubation on ice for 30 min and one freeze-thaw cycle. The resulting suspensions were centrifuged at 100,000 × g for 30 min at 4 °C. Inclusion bodies were solubilized by resuspension in 5 ml of ice-cold 6 m guanidine hydrochloride, 0.5 m KCl, 10 mm imidazole, 40 mm Tris-HCl (pH 8.0), also containing 0.5 mm phenylmethylsulfonyl fluoride. The resulting suspensions were clarified by centrifugation at 100,000 × g for 30 min at 4 °C. The N-terminally His10-tagged DfaA was detected in the soluble and insoluble fractions by SDS-12% PAGE and immunoblotting using anti-DfaA antibody (see above). The N-terminally HA-tagged GgnBP1 was also detected by immunoblotting, using a monoclonal anti-HA antibody (Sigma).
Expression of Recombinant Proteins in Mammalian Cells
A cDNA encoding the N-terminally triple FLAG-tagged FLAG3-DfaA (f3DfaA) was amplified using a three-PCR strategy in which the DfaA moiety was amplified from the pACT-DfaA plasmid, and a DNA segment encoding an overlapping N-terminal triple-FLAG sequence was amplified by PCR from a separate plasmid (a gift from Dr. K. Piatkov). The final ORF encoding f3DfaA was amplified using primers flanked by the NheI and BamHI sites. The resulting DNA fragment was subcloned into NheI/BamHI-cut pcDNA3.1 (Invitrogen), yielding pCB180. Mouse NIH-3T3 cells were grown in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) containing 10% fetal bovine serum. Cells were transfected with specific plasmids using Lipofectamine-Plus (Invitrogen). After 48 h, cells were trypsinized, washed in PBS, and lysed by incubation for 10 min on ice, with frequent mixing, in Lysis Buffer (0.5% Triton X-100, 10% glycerol, 0.5 m NaCl, 1 mm dithiothreitol, 40 mm HEPES (pH 7.9)) also containing leupeptin, antipain, pepstatin A, and aprotinin (each at 5 μg/ml). The extracts was clarified at 10,000 × g for 20 min at 4 °C.
Immunoprecipitation and Immunoblotting
These experiments used anti-FLAGM2 beads (Sigma), anti-HA monoclonal antibody (12CA5; Roche Applied Science), and the affinity-purified anti-DfaA antibody (see above). Except for experiments that employed anti-FLAGM2 beads, immunoprecipitations were performed by incubating clarified lysates from transfected NIH-3T3 cells with the desired primary antibodies (at dilutions indicated under “Results”) for 2 h at 4 °C, with gentle rotation. Agarose beads with immobilized protein A (Repligen, Waltham, MA) were then added, and the lysates were incubated for 1 more h at 4 °C, with gentle rotation. The beads were then washed three times in 10% glycerol, 0.15 m NaCl, 1 mm dithiothreitol, 40 mm HEPES (pH 7.9), followed by elution of the bound proteins with SDS-sample buffer, SDS-12.5% PAGE, a transfer of fractionated proteins to Immobilon-P polyvinylidene difluoride membranes (Millipore), and immunoblotting with antibodies indicated under “Results,” using SuperSignal West Pico or SuperSignal West Dura chemiluminescent reagents (Thermo Scientific, Rockford, IL).
Transient Transfection and Luciferase Assay
NIH-3T3 cells were grown in 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and supplemented with penicillin/streptomycin/glutamine (Mediatech). The medium was changed every 2–3 days, and cultures were re-seeded at ∼50% confluency before allowing them to reach ∼100% confluency. About 20 h before transfection, the cultures were seeded at 1.5 × 105 cells per in 3.5-cm wells. Cultures at 60–70% confluency were transfected according to the manufacturer's protocol, using 0.25 μg of pRL-CMV plasmid (Promega, Madison, WI) expressing Renilla luciferase from the TATA-box-containing PCMV promoter, as well as varying amounts of the pCB180 and pEN-DFAsh4 plasmids (or a plasmid expressing a nonspecific shRNAi), as described under “Results.” Total DNA among various transfection mixtures was normalized by the addition of the pcDNA3.1 vector DNA. 5 μl of Lipofectamine and 5 μl of Plus reagent (Invitrogen) were used per well. About 48 h post-transfection, cells in each well were lysed by incubation in 0.5 ml of Passive Lysis Buffer (Promega) on an orbital shaker at room temperature for 15 min. The extracts were clarified by centrifugation at 16,000 × g for 10 min at 4 °C. For luciferase assays, 50 μl of extract was mixed with 0.2 ml of the LARII reagent (Promega), followed by the addition of 0.2 ml of Stop & Glo reagent (Promega). The activity of Renilla luciferase was then measured over 10-s intervals using a luminometer.
RESULTS AND DISCUSSION
Bidirectional PAte1/Dfa Transcriptional Promoter
As described in our previous study of the mouse Ate1 R-transferase (14), the mean G + C content of ∼30 kb of the mouse genomic DNA, from ∼10 kb upstream to ∼20 kb downstream of the Ate1 exon 1B, is ∼40%. In contrast, an ∼800-bp region containing both exons 1A and 1B of Ate1, from ∼680 bp upstream of exon 1B to ∼120 bp downstream of exon 1B, has a mean G + C content of 75% (Fig. 1C). Closer inspection identified 85 CpG dinucleotide repeats in this short region (14). About half of these CpGs resided in a segment directly upstream of the Ate1 exon 1B that includes the highly conserved 192-bp region 1, which was demonstrated to function as the core bidirectional promoter element, located between the alternative Ate1 exons 1A and 1B (Fig. 1, A and D) (14). In particular, we showed, using in vivo transcription assays, that the above CpG-rich 192-bp mouse DNA segment (Fig. 1, A and D), which is highly conserved at least among mammals, can drive transcription in both the direction of mapped Ate1 transcripts and in the opposite direction (14). This evidence prompted our interest in exploring a previously undescribed mouse gene, termed Dfa, that was expressed in the direction opposite that of Ate1 from the above bidirectional promoter, termed PAte1/Dfa (Fig. 1, A and D) (see Introduction). To make expression patterns of Dfa easier to follow, the orientation of the Dfa and Ate1 transcriptional units was flipped 180° in Fig. 1 after its A, in which the orientations of Dfa and Ate1 are identical to those in previously published diagrams (14).
High density of CpG repeats is a characteristic feature of the previously identified bidirectional promoters in mammalian genomes (21–30). Bidirectional promoters are abundant in multicellular eukaryotes; >20% of mammalian genes are located within 1 kb of each other and oriented head-to-head. In addition, some bidirectionally transcribed chromosomal segments appear to be coregulated and functionally linked (31–35). Bidirectional transcription involves more than 50% of promoters in the yeast S. cerevisiae (36). In the parasitic eukaryote Giardia lamblia, most promoters are bidirectional (37).
Expressed Sequence Tags (ESTs) That Encompass Dfa
Initially, we carried out BLAST-based analyses of EST databases in the region of mouse genomic DNA (chromosome 7) directly upstream of the Ate1 gene. Our analyses of several of the resulting ESTs (including their additional sequencing, beyond the regions present in databases) defined four classes of such sequences. Class 1 included ESTs (AW105867 and BU582987) that contained genomic sequences in the immediate vicinity of the PAte1/Dfa promoter spliced to genomic sequences encoding exon 2 of Ate1. The class 1 ESTs corresponded to mRNAs encoding isoforms of the Ate1 R-transferase containing exon 1A, specifically Ate11A7A, Ate11A7B, and Ate11A7AB (14). The ESTs of class 2 included AW414102, BX517440, BG243214, BY091481, BY241654, and BY267094, which were derived from the immediate vicinity of the PAte1/Dfa promoter but also contained sequences located as far away as 1.6 kb upstream of Ate1. The ESTs of class 3 (BF434328, AI634505, and AI887824) were derived from transcripts that were initiated in the immediate vicinity of PAte1/Dfa, extended in the direction opposite to that of Ate1, and were clearly spliced, terminating ∼10 kb upstream of Ate1. The class 4 of ESTs (CA465465 and BM422486) shared 3′-exons with ESTs of class 3 but were unlike other ESTs in that the 5′-ends of the corresponding transcripts were ∼6 kb away from the bidirectional PAte1/Dfa promoter, in the direction opposite that of the Ate1 gene (Fig. 1F).
The ESTs derived from transcripts initiated in the immediate vicinity of the PAte1/Dfa promoter (class 1–3 ESTs) were obtained from various cellular sources, including tumor cells. In contrast, all ESTs that corresponded to transcripts initiated (in the direction opposite the orientation of Ate1) between 6 and 10 kb away from PAte1/Dfa (class 4 ESTs) were detected only in the testis. At the time of our initial analyses of the mouse Dfa gene, GenBankTM contained an entry for a putative locus, XM_146107 (now called Dfa), that had been predicted by automated analysis of the annotated mouse genomic sequence (NT_081265), using the GNOMON gene prediction method (www.ncbi.nlm.nih.gov). That entry assisted our analyses and assembly of the intron/exon structure of the Dfa gene, as most of the exons proposed by XM_146107 were similar to those in relevant ESTs derived from databases and from our own RT-PCR analyses as well (Fig. 1, E and H). A more recent GenBankTM accession number XM_001479657 encodes a hypothetical mouse protein (LOC100043163) whose sequence is identical to the sequence encoded by the exons 5 and 6 of the Dfa cDNA II (Fig. 1, D and E). In addition, the GenBankTM accession number XM_001479095 encodes a hypothetical mouse protein (LOC100047902) whose sequence is identical to the sequence encoded by the exon 7 of Dfa (Fig. 1, D and E).
Class 3 ESTs (Fig. 1F) suggested that transcripts specified by genomic sequences in the vicinity of the PAte1/Dfa promoter could be spliced to regions between 6 and 10 kb from PAte1/Dfa, in the direction opposite the orientation of Ate1. To verify this, we isolated total RNA from C57BL6 mouse brain, kidney, liver, lung, spleen, and testis, followed by RT-PCR analyses (Fig. 1H). Forward primers used in these reactions were specific for genomic DNA in the vicinity of the PAte1/Dfa promoter, although reverse primers were specific for the sequence of a class 4 EST called EST-CA465465 (GenBankTM). No specific RT-PCR products were amplified using these primers (as well as additional forward primers) from the mouse brain, kidney, liver, lung, or spleen RNA, whereas at least six different Dfa cDNAs were amplified from the testis RNA (Fig. 1, E and H).
Dfa cDNAs III–VI, the three largest cDNAs that had been isolated by RT-PCR, correspond to class 3 ESTs in that the transcripts that gave rise to those cDNAs were initiated in the vicinity of the bidirectional PAte1/Dfa promoter, were extended in the direction opposite that of the Ate1 gene, and contained exons encoded by genomic DNA between ∼4 and ∼10 kb away from PAte1/Dfa. The 5′-rapid amplification of cDNA ends (5′-RACE) technique (38) revealed that a transcript that corresponded to the Dfa cDNA III was initiated just 161 nucleotides from the exon 1B of Ate1, between exons 1A and 1B. Specifically, the Dfa cDNAs III and IV were antisense to Ate1-encoding transcripts that contained the exon 1A of Ate1, its most upstream alternative exon (Fig. 1E). These Dfa transcripts extended for up to ∼1.2 kb before splicing to the Dfa (XM_146107) exons 3, 6, and 7. The Dfa cDNAs V and VI are derived from transcripts that were spliced to Dfa exon 3 at a location 805 nucleotides upstream of the splice site that produced the Dfa cDNA III (Fig. 1E).
In addition to Dfa transcripts initiated at or close to the bidirectional PAte1/Dfa promoter, we also isolated two RT-PCR products, Dfa cDNAs I and II, that were similar to class 4 ESTs in that the corresponding transcripts were initiated between 5 and 6 kb away from PAte1/Dfa, in the direction opposite that of the Ate1 gene (Fig. 1H). The Dfa cDNA II encompasses the class 3 EST-CA465465 (as well as the putative genomic locus XM_001479657 and XM_001479095) and contains the exons 5–7 that are present in XM_146107 (Fig. 1E). 5′-RACE analysis of the Dfa cDNA II mapped its 5′-end to a region ∼5.8 kb away of the bidirectional PAte1/Dfa promoter. Dfa cDNA I is distinct in that it contains an alternative, much smaller 5′-exon (exon 4), located ∼1 kb upstream of genomic DNA that contains the 5′-exon specific for Dfa cDNA II (Fig. 1E). As described above, the 5′-ends of the Dfa cDNAs I and II are located ∼5 kb away from the 5′-ends of class 1–3 Dfa ESTs (Fig. 1, E and F). Thus, transcripts that give rise to the Dfa cDNAs I and II and to cDNAs represented by class 4 ESTs are expressed, most likely, not from the bidirectional PAte1/Dfa promoter but from other currently unknown promoters. In addition to the above Dfa isoforms, we have also isolated (but not yet further characterized) an RT-PCR cDNA from testis that contains the putative exon 2 of XM_146107 (data not shown).
Together, specific cDNA clones from EST databases and our own RT-PCR results (Fig. 1) confirmed the existence of the Dfa gene and its head-to-head arrangement vis à vis Ate1 (Fig. 1, A and D). These results revealed a complex set of Dfa-encoded mRNAs that are produced by alternative splicing (39) of primary Dfa transcripts that are expressed from several promoters, including PAte1/Dfa (Fig. 1, D–F).
To explore evolutionarily conserved Dfa-relevant genomic segments that contain transcriptional promoters, other regulatory elements, or exons that might have been overlooked by RT-PCR analyses, we produced a percent identity plot (40, 41) of gap-free segments that are conserved between ∼14 kb of the mouse genomic DNA in the vicinity of the bidirectional PAte1/Dfa promoter on the mouse chromosome 7F3 and the corresponding human genomic sequence on chromosome 10q26.13. This analysis (Fig. 1B) showed that, similarly to the Ate1 exons 1A, 1B, and 2 and the bidirectional PAte1/Dfa promoter, the genomic sequences of specific mouse Dfa exons that had been identified by RT-PCR contained more gap-free DNA segments with greater than 50% identity to the corresponding human genomic DNA than nearby less sequelogous (less similar in sequence) (18) DNA regions. A low DNA sequelogy between mouse and human DNA in the region from +1 to +4 kb relative to the PAte1/Dfa promoter (within Dfa) strongly suggests the absence of additional Dfa exons or regulatory elements in this region.
At the same time, our percent identity plot analyses revealed a high density of nearly identical, gap-free mouse-versus-human DNA segments between the Dfa exons 3 and 5 (Fig. 1B), indicating that most of this genomic region was under a drift-reducing selection after the divergence of rodent and monkey lineages. Transcripts that give rise to the Dfa cDNAs I and II, as well as to class 4 Dfa ESTs, have been found to initiate in this region (see above), strongly suggesting that the evolutionary stability of specific nucleotide sequences in this area stems, at least in part, from the likely presence of additional Dfa transcriptional promoters (other than PAte1/Dfa). We did not include this genomic segment in our reporter assays (see below), but an examination of genomic DNA in the interval of ∼4–6 kb from the PAte1/Dfa promoter (in the direction opposite to that of Ate1) by PromoterInspector (Genomatix) (42) did suggest the presence of a putative promoter in that region (data not shown).
Our Northern analyses in the earlier study of mouse Ate1 (14) used a DNA probe in the vicinity of the PAte1/Dfa promoter (upstream of the Ate1 exon 1B) that detected transcripts from the DNA strand complementary to the Ate1-specific strand. Those preliminary investigations of what we now call Dfa revealed mRNA species of 1.5–1.7 kb in the heart, brain, spleen, lung, liver, and kidney with a much higher level of this mRNA in the testis where additional, less intense Dfa-specific RNA bands were present as well, at >4.5, ∼3, ∼2.3, and <1.35 kb (see Fig. 5C in Ref. 14). In this work, we carried out Northern analyses with total RNA from various mouse tissues using a Dfa exon 3-specific 181-bp DNA probe, and also an exon 7-specific 556-bp probe. The exon 3-specific probe detected a single ∼1.7-kb RNA in the liver, as well as a major ∼1.5-kb and a minor ∼2.5-kb RNA in the testis (Fig. 1G). Stripping and reprobing the same Northern blot with a Dfa exon 7-specific DNA probe revealed a major ∼1.5-kb RNA in the testis, in addition to a few minor RNAs close to ∼1.5 kb (Fig. 1G). Given a relatively low sensitivity of the second (reprobed) Northern blot and the much higher expression of Dfa in the testis, in comparison with other tissues, these results are consistent with the previously observed presence of low abundance, Dfa-specific mRNAs in tissues other than testis (see Fig. 5C in Ref. 14).
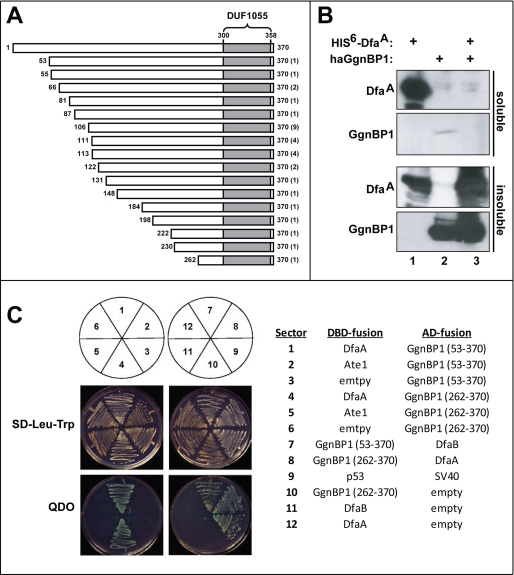
FIGURE 5.
Two-hybrid assay detects interaction of DfaA with Ggnbp1. A, diagram of different segments of the mouse GgnBP1 protein that were isolated in 16 independent “two-hybrid-positive” clones from 32 separate S. cerevisiae colonies. The number of times each segment GgnBP1 was isolated in two-hybrid assays is shown in parentheses to the right. The DUF1055 domain of GgnBP1 (between residues 300 and 358) is shaded. B, soluble (upper two panels) and insoluble (solubilized by guanidine-HCl) proteins from BL21 (DE3) E. coli cells that had been transformed with pET-Duet1 plasmids (see “Experimental Procedures”). Lane 1, E. coli expressing mouse His6-DfaA. Lane 2, E. coli expressing N-terminally HA-tagged mouse haGgnBP1. Lane 3, E. coli expressing both His6-DfaA and haGgnBP1. Note a decrease in the fraction of soluble His6-DfaA in the presence of coexpressed (and virtually entirely insoluble) haGgnBP1 (cf. lanes 1 and 3). C, indicated DBD fusion and activation domain (AD) fusion proteins were expressed in S. cerevisiae YH109 and assayed for their ability to grow on SD medium lacking either Leu and Trp or lacking Leu, Trp, His, and Ade (QDO (Quadruple DropOut) media; see the main text and see under “Experimental Procedures”).
Sequelogs of Mouse Dfa
To address the presence of proteins similar to Dfa, in the mouse or other species, we focused on the ORF encoded entirely by the Dfa exon 7 that is present in all of examined Dfa-specific RT-PCR products. With the exception of the 5′-exon of the Dfa cDNA III, the 3′-terminal exon 7 is the largest Dfa exon (Fig. 1, D and E). This ORF encodes a 217-residue protein, termed DfaA, with a calculated molecular mass of 26 kDa and a deduced pI of 9.84 (Fig. 2A). BlastP analyses showed that this amino acid sequence exhibits 37% identity and 52% sequelogy (sequence similarity) (18) to human HTEX4, a protein of unknown function encoded by the major histocompatibility complex class I gene cluster on human chromosome 6 (Fig. 2B) (43, 44). In addition, the amino acid sequence encoded by the mouse Dfa exons 5 and 6 in Dfa cDNA II (upstream of the exon 7-encoded 217-residue sequence) is 48% identical and 73% sequelogous to human HTEX4, indicating that the sequelogy of HTEX4 and Dfa involves more than exon 7 of Dfa (Fig. 2B). Similarly to mouse Dfa, the human HTEX4 pre-mRNA (and presumably its mouse counterpart as well) undergoes extensive alternative splicing in the testis (44).
Analyses by tBLASTn that employed the 217-residue C-terminal region of DfaA as a query (see Fig. 2A) identified two HTEX4-like genes on mouse chromosome 17 that encoded proteins (of unknown function) that are sequelogous to mouse DfaA. In addition, although no sequelogs of Dfa were found to be encoded by yeast, nematode, fly, or fish genomes, all examined mammalian genomes contained HTEX4-like genes, including putative Dfa genes (data not shown). Moreover, sequence alignments also showed that those HTEX4-like genes, in different vertebrates, that mapped close to the corresponding ATE1 genes (encoding R-transferase) (Fig. 1, A and D) were more sequelogous to mouse Dfa than to other HTEX4-like genes of the same species (data not shown). Thus, Dfa is a member of a distinct gene family at least in mammals.
Close examination of Dfa-specific genomic DNA sequences shows that a splice site selection between the common Dfa exons 6 and 7 (Fig. 1I) determines whether or not the DfaA ORF can be extended in the 5′ direction. Although the AG-GT splice junction-2 was found between exons 6 and 7 in Dfa cDNAs III, IV, and VI, an AG-TG splice junction-1 was found between these exons in Dfa cDNAs I, II, and V (Fig. 1, E and I). In the case of Dfa cDNA II, the selection of splice junction-1 makes possible a contiguous ORF formed by the exons 5–7 in Dfa cDNA II. This putative ORF encodes a 361-residue protein, termed DfaB. Its calculated molecular mass is 42 kDa, and its deduced pI is 9.65. DfaA and DfaB share the 217-residue C-terminal region (encoded by exon 7) and differ by the presence of the 144-residue N-terminal extension in DfaA (encoded by exons 5 and 6 of cDNA II) (Fig. 2A). Although a more detailed study of Dfa isoforms will be required for their comprehensive characterization, our results strongly suggest that most, if not all, Dfa isoforms share the 217-residue C-terminal sequence, encoded by exon 7.
Expression, Intracellular Localization, and Possible Complexes Containing Dfa
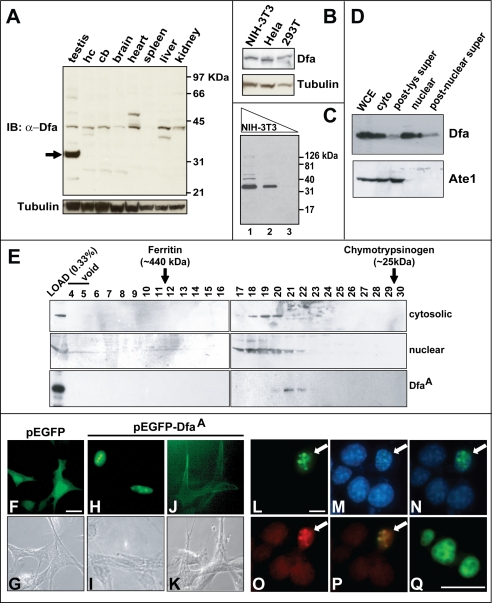
To detect endogenous Dfa, we produced an affinity-purified polyclonal rabbit antibody, termed anti-Dfaex7, to the E. coli-expressed 217-residue mouse DfaA, which is encoded by the Dfa exon 7 (Fig. 1, D and E). (As mentioned above, the 26-kDa recombinant DfaA migrated, upon SDS-PAGE, as an ∼35-kDa protein.) Immunoblotting analyses with this antibody, affinity-purified against immobilized DfaA (see “Experimental Procedures”), detected a putative ∼42-kDa Dfa protein that was present in whole-cell extracts from the mouse testis, hippocampus, cerebellum, total brain, heart, spleen, liver, and kidney (Fig. 3A). Additional, tissue-specific putative Dfa species were also observed in most of the tissues examined. For example, a putative ∼47-kDa Dfa protein was present in heart extracts, and a putative ∼38-kDa Dfa was present in the hippocampus and the liver (Fig. 3A). Testis extracts contained, in addition to the putative ∼42-kDa Dfa isoform and several other minor bands, a major putative Dfa of ∼35 kDa (i.e. of the same apparent molecular mass as the recombinant DfaA protein) (Fig. 3A). An apparently identical Dfa isoform was also detected in extracts from mouse NIH-3T3 cells, as well as human HeLa and 293T cells (Fig. 3B).
FIGURE 3.
Characterization of mouse Dfa. A, endogenous Dfa detected by SDS-PAGE and immunoblotting (IB), with affinity-purified anti-Dfaex7 antibody (see “Experimental Procedures”) of extracts from mouse testis, hippocampus (hc), cerebellum (cb), total brain (brain), heart, spleen, liver, and kidney. Lower panel, immunoblotting of the same extracts using an anti-tubulin antibody. Arrow on the left indicates a major ∼35-kDa Dfa species in the testis. B, endogenous 35-kDa Dfa detected using anti-Dfaex7 and immunoblotting of extracts from mouse NIH-3T3 cells and human HeLa and HEK-293T cells. C, 35-kDa Dfa, detected by immunoblotting with anti-Dfaex7, in serially diluted extracts from NIH-3T3 cells. Lanes 1–3, 20, 4, and 0.4 μg of total protein, respectively. D, relative levels of the 35-kDa Dfa (detected with anti-Dfaex7, upper panel) and Ate1 (detected with anti-Ate1; lower panel) in specific fractions of a mouse testis extract. WCE, whole-cell extract; cyto, cytosolic fraction; post-lys super, post-lysosomal supernatant (28,000 × g for 90 min); post-nuclear super, post-nuclear supernatant (16,000 × g for 90 min). E, immunoblotting, using anti-Dfaex7, of fractions collected from a Superdex-200 column fractions 4–30. Upper panel, cytosolic subfraction of mouse testis extract. Middle panel, same but a nuclear extract. Lower panel, gel filtration pattern, on the same column, of the recombinant mouse DfaA that had been purified from S. cerevisiae (see “Experimental Procedures”). F–K, Subcellular localization of eGFP-DfaA transiently expressed in NIH-3T3 cells. F and G, fluorescent and phase contrast images, respectively, of cells that had been transiently transfected with the control plasmid pEGFP-C1, expressing eGFP. H and I, same but cells were transfected with pEGFP-DfaA, which expressed eGFP-DfaA. Note the predominantly nuclear localization of eGFP-DfaA. J and K, same but with cells (also transfected with pEGFP-DfaA) in which eGFP-DfaA was apparently associated plasma membranes (<10% of transfected cells). L–P, subnuclear localization of eGFP-DfaA transiently expressed in NIH-3T3 cells. L, fluorescent image showing large and “condensed” nuclear speckles in cells expressing eGFP-DfaA (white arrow). M, 4′,6-diamidino-2-phenylindole staining of the same field as in L reveals four additional non-transfected cells. N, merged image of L and M (4′,6-diamidino-2-phenylindole and EGFP). O, immunofluorescent image, produced using an anti-SC35 antibody (see the main text) and showing condensed nuclear speckles in the nucleus of a cell that expressed eGFP-DfaA (see L). P, merged image of L and O, suggesting colocalization of SC-35 and eGFP-DfaA in the nuclear IGC (see the main text). Q, higher magnification of three large, condensed EGFP-DfaA-containing nuclear speckles, one of them with visible “cavities,” in a single nucleus (see also the main text). Bars in F, L, and Q indicate 10 μm.
These findings suggest that different cell types, if they produce Dfa at all, tend to produce different isoforms of Dfa. The 35-kDa protein, a major species in the testis and in NIH-3T3 cells, is actually Dfa, because it can be specifically down-regulated, in 3T3 cells, using RNAi specific for Dfa (see below). The other less abundant Dfa species, for example the ones detected in heart and liver extracts (Fig. 3A), will remain tentative until Dfa−/− (Dfa-lacking) mouse mutants are constructed and employed as null-Dfa controls for antibody specificity. Under the immunoblotting conditions used, the anti-Dfaex7 antibody could detect ∼0.1 ng of recombinant DfaA (data not shown) and could also detect the band of endogenous DfaA in ∼400 ng of total protein extracted from mouse 3T3 cells (Fig. 3C). Thus, DfaA comprised ∼0.025% of soluble proteins in these cells, i.e. ∼8 × 104 DfaA molecules per cell.
To address the intracellular distribution of Dfa, we used immunoblotting with the affinity-purified anti-Dfaex7 antibody to analyze fractions of extract from mouse testis (Fig. 3D). A previously characterized, affinity-purified antibody to the mouse Ate1 R-transferase (Fig. 1A) (6, 7, 14) was also employed, in parallel, to compare the levels of Ate1 in the same fractions. (Ate1 and Dfa are expressed from the bidirectional PAte1/Dfa promoter (Fig. 1, A and D).) We found that the bulk of Ate1 was present in the cytosol fraction of extracts from the testis (Fig. 3D). In contrast, the endogenous 35-kDa DfaA (the only Dfa isoform for which the specificity of anti-Dfaex7 antibody was confirmed using RNAi (see below)), was present in both nuclear and cytoplasmic fractions. Moreover, the bulk, although not all of DfaA, could be pelleted by centrifugation of cytoplasmic and nuclear fractions of a detergent-free testis extract for 90 min at 28,000 and 16,000 × g, respectively (Fig. 3D), suggesting an association of DfaA with membranes and/or cytoskeleton.
We also asked whether DfaA was present in the testis as a part of an endogenous complex stable enough to be detected by gel filtration of testis extracts, using immunoblotting of fractions with anti-Dfaex7 antibody. Purified recombinant mouse DfaA (expressed in S. cerevisiae; see under “Experimental Procedures”) eluted from a Superdex-200 column in a single peak centered at fraction 21 (Fig. 3E, bottom panels) and corresponding to the molecular mass of 30–50 kDa for a globular protein, in agreement with the apparent molecular mass of monomeric DfaA fractionated by SDS-PAGE (the actual molecular mass of the recombinant DfaA was 26 kDa; see above). A small proportion of DfaA in either the cytosolic or nuclear testis extract also eluted at the position of monomer, but the bulk of DfaA migrated in the region corresponding to the molecular mass of 100–150 kDa (Fig. 3E, upper and middle panels). The composition and function of this endogenous DfaA complex(es) remain to be determined.
In addition, we transiently expressed, in mouse NIH-3T3 cells, an EGFP-DfaA fusion. Although the EGFP protein lacks known nuclear localization signals, EGFP (or GFP) alone is known to be present, in diffuse patterns, in both the cytoplasm and the nucleus, because, at least in part, of the low size of EGFP (27 kDa) that allows its nuclear localization signal-independent transport to the nucleus (Fig. 3F) (45). Despite this drawback of EGFP as a location marker, the results with EGFP-DfaA were meaningfully interpretable, because in ∼95% of EGFP-DfaA-expressing cells the bulk of this fusion was predominantly nuclear, in contrast to EGFP alone (Fig. 3, H and cf. F). A predominantly nuclear localization of DfaA was consistent with it containing the sequence KKKPK, a putative nuclear localization signal (Fig. 2A). Moreover, in contrast to EGFP alone, the nuclear EGFP-DfaA protein often exhibited a speckled pattern (Fig. 3, H and L) that under higher magnification appeared as cavity-containing structures (Fig. 3Q). In a minority (∼5%) of cells that expressed EGFP-DfaA, it was associated, in particular, with plasma membrane regions (Fig. 3J), in agreement with biochemical fractionation data that suggested an association of DfaA with rapidly sedimenting structures (Fig. 3D).
IGCs are interconnected, dynamic nuclear structures that are enriched in pre-mRNA splicing factors, including the non-small nuclear ribonucleoprotein splicing factor SC-35, which is commonly employed as an IGC marker (46). During transcriptional repression, IGCs undergo an actin-dependent reorganization from diffuse interconnected speckles to larger, condensed (and apparently unconnected) speckles associated with RNA polymerase II (47, 48). When stained for SC-35, IGCs from transcriptionally inactive cells contain a cavity that sequesters, in particular, the hyperphosphorylated large subunit of RNA polymerase II (49). At higher magnifications, most of DfaA-containing nuclear speckles, observed in the bulk of cells that expressed EGFP-DfaA, were large, condensed, and contained characteristic cavities (Fig. 3Q). Confirming their identity as IGCs, we found that the IGC marker SC-35 colocalized with EGFP-DfaA-containing nuclear speckles (Fig. 3, L–Q). Interestingly, although IGCs (stained with anti-SC-35 antibody) were small and scattered throughout the nucleus in nontransfected cells (Fig. 3O), the IGCs in cells that expressed EGFP-DfaA (cells marked with white arrows in Fig. 3, L–P) were larger and more condensed. Thus expression of EGFP-DfaA induces IGC reorganization to an architecture characteristic of transcriptionally repressed cells (Fig. 3O).
Dfa-Specific RNAi Confirms the Identity of a Putative Dfa Isoform Detected by Anti-Dfaex7 Antibody
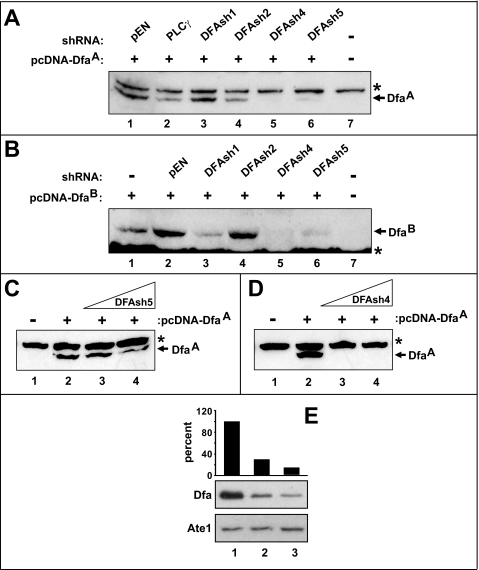
To down-regulate the expression of Dfa in cultured cells and to verify specificity of the anti-Dfaex7 antibody, we employed microRNA-like shRNAs (DFAsh1, DFAsh2, DFAsh4, and DFAsh5) specific to four different regions mRNA encoding Dfa (Fig. 2A), using transient coexpression of these shRNAs with DfaA or DfaB in NIH-3T3 cells. Because the DFAsh1 sequence is complementary to a sequence in exon 6 (Fig. 2A), the ability of DFAsh1 to inhibit the expression of a transfected Dfa cDNA was confined to the DfaB isoform (Fig. 4, A and B) whose cDNA contains exons 5–7 (Figs. 1E and 2A). Whereas DFAsh2 was relatively ineffective at reducing the expression of transiently transfected DfaA or DfaB cDNAs (Fig. 4, A and B), DFAsh4 and DFAsh5 reduced the expression of both Dfa isoforms to undetectable and barely detectable levels, respectively (Fig. 4, A and B). To determine the relative efficacies of DFAsh4 and DFAsh5, we examined the effects of varying their levels on expression of the transiently cotransfected DfaA cDNA. Although the expression of DfaA was inversely proportional to input amounts of the DFAsh5-expressing plasmid in a cotransfection mixture (Fig. 4C), even the lowest examined levels of the DFAsh4-expressing plasmid reduced DfaA expression to levels undetectable by immunoblotting (Fig. 4D).
FIGURE 4.
RNAi of Dfa. A, relative efficacies of different shRNAs in down-regulating the expression of exogenous (triple-FLAG-tagged) f3DfaA in NIH-3T3 cells, measured using immunoblotting with anti-FLAG antibody. Lane 1, pEN, the parental pEN_hU6miR2c vector for shRNAs. Lane 2, shRNA specific for phospholipase Cγ (PLCγ). Lanes 3–6, DFAsh1, DFAsh2, DFAsh4, and DFAsh5, specific for different regions of Dfa mRNAs (see Fig. 2A). Lane 7, control 3T3 cells, mock-transfected. B, same as in A but with f3DfaB. Equal numbers of cells were transfected with equal amounts of a Dfa-expressing plasmid, an shRNA-expressing plasmid, or control vectors. C, effect of increasing amounts of transfected DFAsh5 on the levels of exogenous f3DfaA. Lane 1, transfected with a vector alone. Lane 2, transfected with 1 μg of pcDNA-FLAG-DfaA (pCB180), expressing f3DfaA. Lane 3, same as lane 2 but cells were also cotransfected also with 1 μg of pEN-DFAsh5, expressing DFAsh5 shRNA. Lane 4, same as lane 3 but 2 μg of pEN-DFAsh5. Total amounts of DNA in each transfection were equalized using an empty DNA vector. D, same as in C but with DFAsh4 shRNA. Asterisks in A– D denote a protein cross-reacting with anti-FLAG antibody. E, comparing, using anti-Dfaex7 immunoblots, the levels of endogenous DfaA in independently produced NIH-3T3 cell clones, either a control cell line or stably transfected with pEN-DFAsh4 DfaA (two representative examples, out of a number of cell clones thus produced, are shown). Upper panel, quantitation of the levels of endogenous DfaA that was detected by immunoblotting with anti-DfaA (middle panel). Lower panel, relative levels of the Ate1 R-transferase in the same cell clones, determined by immunoblotting with anti-Ate1 antibody. Lane 1, NIH-3T3 cells stably transfected with a vector alone. Lanes 2 and 3, same but with a plasmid expressing DFAsh4 shRNA (30 and 13% of control DfaA levels, respectively, in different 3T3 clones).
To determine whether the expression of endogenous DfaA could be strongly inhibited with DFAsh4, we isolated individual NIH-3T3 colonies stably transfected with the constitutively active pEN-DFAsh4 (Fig. 4E). Immunoblotting of whole-cell extracts using the anti-Dfaex7 antibody showed that expression of the endogenous 35-kDa DfaA was reduced by up 87% in different clonally derived DFAsh4-expressing 3T3 cell cultures (Fig. 4E and data not shown). In addition to establishing a method for knocking down the expression of endogenous DfaA in a cell line, these experiments validated the specificity of our anti-Dfaex7 antibody. Interestingly, while isolating and propagating the above cell clones, we noticed fewer viable pEN-DFAsh4-containing 3T3 cell colonies than those containing the control (non-Dfa-specific) pEN-shGFP plasmid, under conditions where similar initial amounts of cells were plated (data not shown). In addition, individual colonies containing DFAsh4 grew at slightly but significantly lower rates than those expressing GFP-specific shRNAi (∼1.1 doublings/day ± 0.012; n = 35 versus 1.3 doublings/day ± 0.019; n = 5, respectively (p < 0.03)). We also noticed that the early growth of DFAsh4-expressing cell colonies was particularly delayed, in comparison with colonies expressing GFP-specific shRNAi. Together, these findings validated the use of anti-Dfaex7 antibody (by confirming that the antibody-recognized 35-kDa protein was indeed encoded by Dfa in 3T3 cells) and also suggested that DfaA may be required for viability of mammalian cells.
Dfa Interactions with GgnBP1 and Abt1
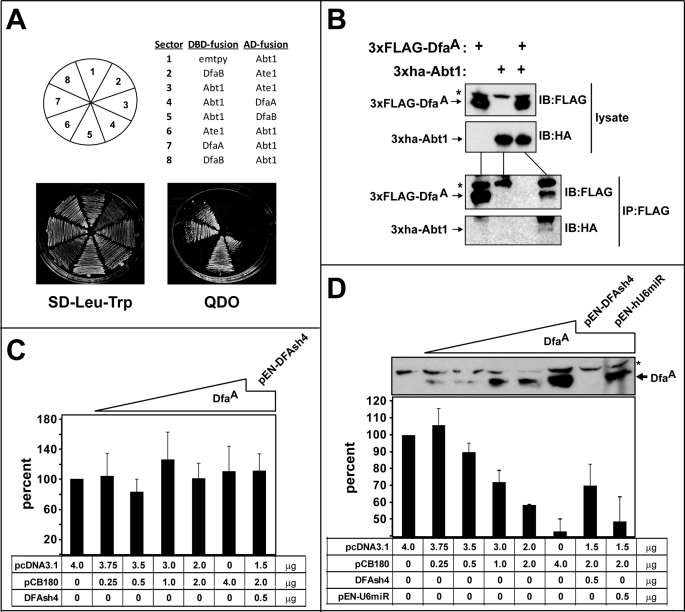
To search for DfaA-binding proteins, we employed the yeast two-hybrid assay, with a Gal4-DNA-binding-DfaA fusion as a bait and a mouse testis cDNA library based on the Gal4-activation domain. We isolated 16 independent DNA clones that encoded different (overlapping) segments of the mouse gametogenesis binding protein-1 (50, 51) (GgnBP1; GenBankTM accession number Q6K1E7) from 32 yeast colonies (Fig. 5A). Sequence analyses of the 16 pACT2-based encoding segments of GgnBP1 mapped the DfaA-binding region of GgnBP1 to its C-terminal residues 262–370 that encompass the C-terminal DUF1055 domain (domain of unknown function) that is present in many mammalian deubiquitylating enzymes (Fig. 5A). To further verify the GgnBP1-DfaA interaction and to examine the ability of GgnBP1 to interact with DfaB, we coexpressed Gal4-DNA-binding domain (DBD) fusions (DfaA, DfaB, Ate1, GgnBP(53–370), GgnBP1(262–370), and p53 (the latter a control)) with Gal4 activation domain fusions (DfaA, DfaB, GgnBP(53–370), and GgnBP1(262–370)), and the SV40 large T antigen (the latter as a control) in appropriately marked S. cerevisiae strains (Fig. 5C). These strains were assayed for their ability grow on SD media lacking Leu, Trp, His, and Ade (QDO, Quadrupole DropOut) as the evidence for interaction of the corresponding fusions (Fig. 5C). These assays confirmed, in the framework of two-hybrid assays, that either DfaA or DfaB interacted with GgnBP1(53–370) (residues 53–370, the largest GgnBP1 fragment isolated in this screen) or GgnBP1(262–370) (residues 262–370 of GgnBP1; the smallest “positive” GgnBP1 fragment) (Fig. 5C). “Vector swapping” two-hybrid assays further validated these Dfa-GgnBP1 interactions (Fig. 5C). In sum, the C-terminal DUF1055 domain of GgnBP1, which included GgnBP1(262–370), was sufficient for an interaction of GgnBP1 with either DfaA or DfaB (Fig. 5C).
GgnBP1 was originally identified in two-hybrid analyses as a protein that interacted with Ggn1 (gametogenetin 1) (50, 51). Ggn1 is a germ cell-specific protein that binds to FANCL (Fanconi anemia complementation group L), the protein product of the gene mutated in gcd (germ cell-deficient) mice (52, 53). GgnBP1 is associated with the Golgi and the plasma membrane, is testis-specific, and is expressed primarily in meiotic spermatocytes (51). More recently, mouse GgnBP1 was found, using a two-hybrid assay, to interact with the mitochondrial fission factor FIS1 and to participate, through the DUF1055 domain of GgnBP1, in the mitochondrial morphogenesis during spermatogenesis (54).
Although all of the above GgnBP1-interacting partners have been robustly identified through yeast two-hybrid assays, none of these interactions could be verified, so far, by other means, e.g. through coimmunoprecipitation of proteins expressed in mammalian cells (50–54). For reasons unknown (possibly because Dfa exists in multiple, differently located intracellular pools), our attempts to coimmunoprecipitate GgnBP1 with Dfa in mammalian cells were also unsuccessful (data not shown). Given specific membrane locations of GgnBP1 (see above), and the association of DfaA with both nuclear and membrane fractions (Fig. 3, D, G, and J), a subset of DfaJ that interacts with GgnBP1 is likely to be a small one. Recombinant mouse DfaA, when expressed alone in E. coli, was detected in both soluble and insoluble fractions after cell lysis, whereas all DfaA became insoluble when it was coexpressed with recombinant mouse Ggnbp1 (Fig. 5B), a finding in agreement with extensive two-hybrid data for a DfaA-Ggnbp1 interaction (Fig. 5).
Two-hybrid assays with DfaA as a bait also yielded three independent cDNA clones encoding overlapping segments of the mouse Abt1 (activator of basal transcription) protein. In additional two-hybrid screens, we found that, similarly to the GgnBP1 protein, Abt1 could interact with both the DfaA and DfaB isoforms (Fig. 6A). To verify and extend these findings, NIH-3T3 cells were transiently cotransfected with plasmids that expressed triple FLAG-tagged f3DfaA and triple HA-tagged h3Abt1 (Fig. 6B). In agreement with the results of two-hybrid assays (Fig. 6A), h3Abt1 was specifically coimmunoprecipitated with f3DfaA, using anti-FLAG-M2 beads (Fig. 6B). Thus, DfaA and Abt1 can interact in both S. cerevisiae and mouse fibroblasts. It should also be noted that no interaction between DfaA and Ate1 (R-transferase; see Introduction) was detected using two-hybrid assay (data not shown), in agreement with other evidence in this study that suggested the absence of a significant functional or mechanistic connection between Dfa and Ate1, apart from their proximity and head-to-tail orientation (Fig. 1A).
FIGURE 6.
DfaA interacts with Abt1 and inhibits transcription from the TATA-containing PCMV promoter. A, indicated Abt1-based and Dfa-based DBD fusion and activation domain (AD) fusion proteins were expressed in S. cerevisiae YH109, and two-hybrid assays for their interaction were carried out (see under “Experimental Procedures”). QDO, quadruple dropout. B, mouse NIH-3T3 cells were transiently transfected with pcDNA-based plasmids that expressed the indicated proteins (tagged with the FLAG or HA epitopes), followed by preparation of extracts, and either SDS-PAGE (followed by immunoblotting with anti-FLAG and anti-HA) or immunoprecipitation with anti-FLAG antibody, followed by SDS-PAGE of immunoprecipitates and immunoblotting with anti-HA antibody. IB, immunoblotting; IP, immunoprecipitation. C, levels of luciferase (expressed from the PAte1/Dfa promoter in the plasmid pCB44 (14)) in extracts from mouse NIH-3T3 cells that had been cotransfected with pCB44 and the indicated plasmids (the empty vector pCDNA; the f3DfaA-expressing pCB180, and the DFAsh4 shRNA-expressing pEN-DFAsh4). Luciferase levels are plotted as percentages of the level that was observed with pcDNA3.1 vector alone (no exogenous DfaA; no DFAsh4 shRNA). Error bars indicate standard deviations among three independent assays. D, same as in C but with luciferase expressed from the TATA-box-containing PCMV promoter. The immunoblot in D shows relative levels of f3DfaA in the corresponding cotransfected cells. An asterisk denotes a protein cross-reacting with anti-FLAG antibody.
Dfa Acts As a Transcriptional Repressor of a TATA-box Promoter
Previous work has shown that Abt1 is associated with the TATA-box-binding protein and enhances transcriptional activity of TATA-containing promoters (55). As described above, Dfa was shown to interact with Abt1 both in yeast-based two-hybrid assays and in coimmunoprecipitation assays with mammalian cells (Fig. 6, A and B), and it was also found to be associated in vivo with condensed nuclear IGCs, a hallmark of transcriptional repression (Fig. 3, L–Q). To determine whether DfaA could influence the in vivo transcription of a reporter gene, we transiently cotransfected NIH-3T3 cells with a plasmid that expressed luciferase from the TATA-containing PCMV promoter, and with increasing amounts of the DfaA-expressing pCB180 plasmid (Fig. 6D). (To control for promoter titration effects, transfection samples were normalized by the addition of the pcDNA3.1 vector DNA.) Addition of increasing amounts of the DfaA-expressing plasmid progressively decreased the expression of luciferase (Fig. 6D), suggesting that DfaA acted as a transcriptional repressor, possibly through its interaction with endogenous Abt1. To verify that the transcriptional repression observed under these conditions was caused by DfaA, we repeated these assays in the presence of DFash4 (see above) and the resulting RNAi-mediated down-regulation of DfaA. In the absence of DfaA-specific RNAi, the addition of 2 μg of the DfaA-expressing pCB180 plasmid caused an ∼42% reduction in luciferase levels, in comparison with expression in the presence of the DfaA-lacking pcDNA3.1 vector (Fig. 6D). In contrast, the DfaA-specific RNAi (verified, using immunoblotting, by a loss of DfaA expression upon RNAi) and the resulting down-regulation of DfaA increased luciferase expression to ∼70% of the control levels (in the absence of exogenous DfaA) (Fig. 6D). In additional control experiments, no effect on DfaA expression or its reporter-repressive effects were observed with cells that were also transfected with the “control” RNAi plasmid pEN-hU6miR (Fig. 6D).
Given these results, we also asked whether DfaA could also repress transcription from the bidirectional and TATA-less PAte1/Dfa promoter, as such a property would potentially be a functional link between Ate1 and Dfa. We measured the levels of luciferase expressed (in the direction of Ate1 transcription) from the PAte1/Dfa promoter under the conditions described above for the PCMV-luciferase setting. In contrast to down-regulating effects of DfaA on the expression of PCMV-luciferase, increasing levels of DfaA did not alter significantly the expression of the PAte1/Dfa-luciferase reporter (Fig. 6C). Although more detailed studies of Dfa as a transcriptional repressor of TATA-containing promoters are clearly necessary, our findings suggest that the repressor activity of DfaA is confined to promoters of this class.
Concluding Remarks
This study of the previously uncharacterized mouse gene, termed Dfa, was carried out to explore the possibility that both proximity and head-to-head orientation of the Dfa gene and the previously known Ate1 gene (encoding R-transferase of the N-end rule pathway) might signify their functional link, either directly or through belonging to a specific regulatory circuit. As shown in this work, Dfa is expressed largely, although not exclusively, in the testis, where it produces a complex set of spliced Dfa mRNAs from both the bidirectional PAte1/Dfa promoter and other nearby promoters. The 3-terminal exon of Dfa, termed DfaA and shared by other mapped Dfa mRNAs, encodes a 217-residue (26 kDa) protein that was found to migrate, upon SDS-PAGE, as a 35-kDa species. An affinity-purified polyclonal antibody to DfaA was prepared and shown to recognize DfaA in cell extracts. Specifically, the antibody detected a 35-kDa protein that was down-regulated upon RNAi-mediated repression of Dfa. The DfaA protein was sequelogous (similar in sequence) (18) to the previously described human/mouse HTEX4 protein, whose physiological function is unknown. DfaA was also found to interact with specific proteins, including the Abt1 transcriptional activator. Dfa-Abt1 interactions could be detected either by the yeast-based two-hybrid assay or through coimmunoprecipitation of the two proteins from mammalian cell extracts. This potential link between DfaA and regulation of transcription was consistent with the observed preferential location of DfaA (as an EGFP-DfaA fusion) in compact (condensed) interchromatin granule clusters, a hallmark of transcriptional repression. Experiments with a luciferase-based transcriptional reporter and DfaA-specific RNAi have shown that DfaA acts as a repressor of a TATA-box-containing transcriptional promoter but does not influence the bidirectional PAte1/Dfa promoter, which contains a CpG island and lacks the TATA-box.
Much remains to be learned about Dfa, its elaborate set of primary transcripts, differential splicing, mature mRNAs, the observed transcriptional-repressor activity of Dfa, and ultimately its specific roles in cell physiology. Contrary to expectation that led us to initiate this work, no functional or mechanistic connections between Dfa proteins and the isoforms of the Ate1 R-transferase were detected so far, apart from the proximity of their head-to-head oriented genes and the antisense orientation of some among Dfa and Ate1 transcripts (Fig. 1, A and D). As this is the first exploration of Dfa and is also a far from complete understanding of the PAte1/Dfa promoter, future studies of Dfa and Ate1 may still uncover a physiologically relevant connection between these adjacent, divergently oriented genes, perhaps outside of domains we explored so far.
Acknowledgments
We are grateful to current and former members of the Varshavsky laboratory for advice and help. We thank K. I. Piatkov for helpful discussions, J. Zavzavadjian and E. Wall for advice with shRNA, and E. Udartseva for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31530 and DK39520 (to A. V.). This work was also supported by a grant from the March of Dimes Foundation.
- RNAi
- RNA interference
- PMSF
- phenylmethylsulfonyl fluoride
- HA
- hemagglutinin
- sh
- short hairpin
- RT
- reverse transcription
- EST
- expressed sequence tag
- IGC
- interchromatin granule cluster
- EGFP
- enhanced GFP
- GFP
- green fluorescent protein
- RACE
- rapid amplification of cDNA ends
- ORF
- open reading frame
- DBD
- DNA-binding domain.
REFERENCES
- 1.Varshavsky A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mogk A., Schmidt R., Bukau B. (2007) Trends Cell Biol. 17, 165–172 [DOI] [PubMed] [Google Scholar]
- 3.Tasaki T., Kwon Y. T. (2007) Trends Biochem. Sci. 32, 520–528 [DOI] [PubMed] [Google Scholar]
- 4.Varshavsky A. (2008) Nat. Struct. Mol. Biol. 15, 1238–1240 [DOI] [PubMed] [Google Scholar]
- 5.Varshavsky A. (2008) J. Biol. Chem. 283, 34469–34489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu R. G., Sheng J., Qi X., Xu Z., Takahashi T. T., Varshavsky A. (2005) Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 7.Hu R. G., Wang H., Xia Z., Varshavsky A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Z., Webster A., Du F., Piatkov K., Ghislain M., Varshavsky A. (2008) J. Biol. Chem. 283, 24011–24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang C. S., Varshavsky A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19188–19193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang C. S., Shemorry A., Varshavsky A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2142–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Piatkov K. I., Brower C. S., Varshavsky A. (2009) Mol. Cell 34, 686–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brower C. S., Varshavsky A. (2009) PLoS ONE 4, e7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang C. S., Shemorry A., Varshavsky A. (2010) Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu R. G., Brower C. S., Wang H., Davydov I. V., Sheng J., Zhou J., Kwon Y. T., Varshavsky A. (2006) J. Biol. Chem. 281, 32559–32573 [DOI] [PubMed] [Google Scholar]
- 15.Kwon Y. T., Kashina A. S., Varshavsky A. (1999) Mol. Cell. Biol. 19, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon Y. T., Kashina A. S., Davydov I. V., Hu R. G., An J. Y., Seo J. W., Du F., Varshavsky A. (2002) Science 297, 96–99 [DOI] [PubMed] [Google Scholar]
- 17.Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshavsky A. (2004) Curr. Biol. 14, R181–R183 [DOI] [PubMed] [Google Scholar]
- 19.Graciet E., Hu R. G., Piatkov K., Rhee J. H., Schwarz E. M., Varshavsky A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3078–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin K. J., Wall E. A., Zavzavadjian J. R., Santat L. A., Liu J., Hwang J. I., Rebres R., Roach T., Seaman W., Simon M. I., Fraser I. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13759–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prioleau M. N. (2009) PLoS Genetics 5, e1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patton J., Block S., Coombs C., Martin M. E. (2006) Gene 369, 35–44 [DOI] [PubMed] [Google Scholar]
- 23.Emoto M., Miki M., Sarker A. H., Nakamura T., Seki Y., Seki S., Ikeda S. (2005) Gene 357, 47–54 [DOI] [PubMed] [Google Scholar]
- 24.Travers M. T., Cambot M., Kennedy H. T., Lenoir G. M., Barber M. C., Joulin V. (2005) Genomics 85, 71–84 [DOI] [PubMed] [Google Scholar]
- 25.Saleh A., Davies G. E., Pascal V., Wright P. W., Hodge D. L., Cho E. H., Lockett S. J., Abshari M., Anderson S. K. (2004) Immunity 21, 55–66 [DOI] [PubMed] [Google Scholar]
- 26.Meier-Noorden M., Flindt S., Kalinke U., Hinz T. (2004) Gene 338, 197–207 [DOI] [PubMed] [Google Scholar]
- 27.Whitehouse C., Chambers J., Catteau A., Solomon E. (2004) Gene 326, 87–96 [DOI] [PubMed] [Google Scholar]
- 28.Trinklein N. D., Aldred S. F., Hartman S. J., Schroeder D. I., Otillar R. P., Myers R. M. (2004) Genome Res. 14, 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takai D., Jones P. A. (2004) Mol. Biol. Evol. 21, 463–467 [DOI] [PubMed] [Google Scholar]
- 30.Adachi N., Lieber M. R. (2002) Cell 109, 807–809 [DOI] [PubMed] [Google Scholar]
- 31.West A. B., Lockhart P. J., O'Farell C., Farrer M. J. (2003) J. Mol. Biol. 326, 11–19 [DOI] [PubMed] [Google Scholar]
- 32.Li Y. Y., Yu H., Guo Z. M., Guo T. Q., Tu K., Li Y. X. (2006) PLoS Comp. Biol. 2, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., Flynn R. A., Young R. A., Sharp P. A. (2008) Science 322, 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Vogelstein B., Velculescu V. E., Papadopoulos N., Kinzler K. W. (2008) Science 322, 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piontkivska H., Yang M. Q., Larkin D. M., Lewin H. A., Reecy J., Elnitski L. (2009) BMC Genomics 10, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L. M. (2009) Nature 457, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teodorovic S., Walls C. D., Elmendorf H. G. (2007) Nucleic Acids Res. 35, 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Smith J. A., Seidman J. G., Struhl K. (eds) (2006) Current Protocols in Molecular Biology, Wiley-Interscience, New York [Google Scholar]
- 39.Chen M., Manley J. L. (2009) Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elnitski L., Riemer C., Burhans R., Hardison R., Miller W. (2005) Curr. Protoc. Bioinformatics, Unit 10.14 [DOI] [PubMed] [Google Scholar]
- 41.Elnitski L., Riemer C., Schwartz S., Hardison R., Miller W. (2003) Curr. Protoc. Bioinformatics, Unit 10.12 [DOI] [PubMed] [Google Scholar]
- 42.Scherf M., Klingenhoff A., Werner T. (2000) J. Mol. Biol. 297, 599–606 [DOI] [PubMed] [Google Scholar]
- 43.Coriton O., Lepourcelet M., Hampe A., Galibert F., Mosser J. (2000) Mamm. Genome 11, 1127–1131 [DOI] [PubMed] [Google Scholar]
- 44.Lepourcelet M., Coriton O., Hampe A., Galibert F., Mosser J. (1998) Immunogenetics 47, 491–496 [DOI] [PubMed] [Google Scholar]
- 45.Seibel N. M., Eljouni J., Nalaskowski M. M., Hampe W. (2007) Anal. Biochem. 368, 95–99 [DOI] [PubMed] [Google Scholar]
- 46.Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F., Spector D. L. (2004) Mol. Biol. Cell 15, 3876–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bregman D. B., Du L., van der Zee S., Warren S. L. (1995) J. Cell Biol. 129, 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamond A. I., Spector D. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 49.Wang I. F., Chang H. Y., Shen C. K. (2006) Exp. Cell Res. 312, 3796–3807 [DOI] [PubMed] [Google Scholar]
- 50.Zhang J., Wang Y., Zhou Y., Cao Z., Huang P., Lu B. (2005) FEBS Lett. 579, 559–566 [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y., Zhao Q., Bishop C. E., Huang P., Lu B. (2005) Mol. Reprod. Dev. 70, 301–307 [DOI] [PubMed] [Google Scholar]
- 52.Lu B., Bishop C. E. (2003) J. Biol. Chem. 278, 16289–16296 [DOI] [PubMed] [Google Scholar]
- 53.Agoulnik A. I., Lu B., Zhu Q., Truong C., Ty M. T., Arango N., Chada K. K., Bishop C. E. (2002) Hum. Mol. Genet. 11, 3047–3053 [DOI] [PubMed] [Google Scholar]
- 54.Aihara T., Nakamura N., Honda S., Hirose S. (2009) Biol. Reprod. 80, 762–770 [DOI] [PubMed] [Google Scholar]
- 55.Oda T., Kayukawa K., Hagiwara H., Yudate H. T., Masuho Y., Murakami Y., Tamura T. A., Muramatsu M. A. (2000) Mol. Cell. Biol. 20, 1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]