Abstract
ACSL3 is a member of the long chain acyl-CoA synthetase (ACSL) family that plays key roles in fatty acid metabolism in various tissues in an isozyme-specific manner. Our previous studies showed that ACSL3 was transcriptionally up-regulated by the cytokine oncostatin M (OSM) in HepG2 cells, accompanied by reduced cellular triglyceride content and enhanced β-oxidation. In this study, we investigated the molecular mechanism underlying the OSM-induced activation of ACSL3 gene transcription in HepG2 cells. We showed that OSM treatment resulted in a coordinated elevation of mRNA levels of ACSL3 and peroxisome proliferator-activated receptor δ (PPARδ). The effect of OSM on ACSL3 mRNA expression was inhibited by cellular depletion of PPARδ. By utilizing a PPARδ agonist, L165041, we demonstrated that activation of PPARδ led to increases in ACSL3 promoter activity, mRNA level, and protein level in HepG2 cells. Analysis of the ACSL3 promoter sequence identified two imperfect PPAR-responsive elements (PPRE) located in the ACSL3 promoter region −944 to −915, relative to the transcription start site. The up-regulation of ACSL3 promoter activity by PPARδ was abolished by deletion of this PPRE-containing region or mutation to disrupt the binding sites. Direct interactions of PPARδ with ACSL3-PPRE sequences were demonstrated by gel mobility shift and chromatin immunoprecipitation assays. Finally, we provided in vivo evidence showing that activation of PPARδ by L165041 in hamsters increased ACSL3 mRNA and protein levels in the liver. These new findings define ACSL3 as a novel molecular target of PPARδ in HepG2 cells and provide a regulatory mechanism for ACSL3 transcription in liver tissue.
Keywords: Cytokine, Fatty Acid Metabolism, Gene Transcription, Liver, PPAR, ACSL3, PPARδ, Oncostatin M
Introduction
Perturbed fatty acid (FA)2 metabolism is one underlying contributor to the development of obesity, diabetes, and cardiovascular diseases. One family of enzymes that plays critical roles in FA metabolism is the long chain acyl-CoA synthetases (ACSL) (1, 2). ACSLs catalyze the formation of fatty acyl-CoAs from ATP, CoA, and long chain fatty acids. This reaction is the first step in lipid metabolism after FA entry into the cell and the activation process is necessary for FA cellular utilization in different metabolic pathways including the catabolic pathway for the degradation of FA via the β-oxidation system and the anabolic pathway for the synthesis of phospholipids, cholesterol esters, and triglycerides (TG).
Since the initial report on ACSL1 in 1990 (3), major progress has been made in the identification and characterization of other members of the acyl-CoA synthetase family. A total of 5 isoforms of ACSL have been described in humans and rodents (3–7). Although all isozymes of this family carry out the same enzymatic reaction, they differ greatly in substrate preferences, enzyme kinetics, tissue, and subcellular compartment distribution, all of which contribute to their different cellular functions and metabolic outcomes (2, 8).
The isozyme ACSL3 has been characterized in human, rat, mouse, and recently hamster (2, 5, 8, 9). It consists of 720 amino acids with a molecular mass of ∼79–80 kDa and its sequence is highly homologous among these species (10). ACSL3 preferentially utilizes laurate and myristate among C8-C22 saturated FAs and arachidonate and eicosapentaenoate among C16-C20 unsaturated FAs. This substrate specificity is distinct from that of ACSL1, which uses C10-C18 saturated FAs and C16-C20 unsaturated fatty acids with equivalent activities (5). Although there is no report on membrane association of ACSL3, this isozyme has been found in the subcellular fraction of lipid droplets in the human hepatocyte HuH7 cells (11) as well as in lipid droplets from lipolytically stimulated 3T3-L1 adipocytes (12).
In rats, ACSL3 mRNA was shown to be highly expressed in brain and testis, and to a lesser extent in other organs such as adipose tissue, lung, kidney, heart, muscle, or liver (8). This tissue-specific expression pattern of ACSL3 mRNA was recently confirmed at the protein level in hamsters (9). By utilizing a highly specific antibody developed against the C-terminal region of hamster ACSL3, we have shown that the ACSL3 protein is abundant in brain and testis, detectable in liver and adipose tissue, barely seen in muscle, and untraceable in the heart (9).
In the initial study of rat ACSL3, it was proposed that the expression of ACSL3 in brain was developmentally regulated because its mRNA level in adult brain was only ∼10% of the maximum of 15 days after birth (5); however, the molecular and cellular mechanisms underlying this regulation in the brain have remained elusive. Although it is unclear what functional role ACSL3 plays during brain development, this enzyme has been shown to promote β-oxidation of FAs in HepG2 hepatoma cells. Our previous studies showed that the transcription of ACSL3 along with ACSL5 was activated by the cytokine oncostatin M (OSM) in HepG2 cells (13). Using a specific inhibitor to the upstream kinase of the extracellular signal-regulated kinase (ERK), we further demonstrated that the ERK signaling pathway was activated by OSM in HepG2 cells and in OSM-treated livers of hamsters. This activation was critically required for OSM to induce ACSL3 transcription. The increased expression of ACSL3 was associated with a decreased cellular TG content and an enhanced FA β-oxidation in OSM-treated cells. Moreover, depletion of ACSL3 by specific small interference RNA (siRNA) transfection abolished the OSM effects on FA metabolism. That cell culture data, coupled with the hypolipidemic effects of OSM observed in hamster experiments (13), led us to speculate that the increased ACSL3 expression in liver might contribute to the reductions of plasma and hepatic-free FAs and TG in OSM-treated animals. Recent studies in our laboratory further demonstrated that expressions of ACSL3 mRNA and protein in liver were specifically increased after feeding hamsters with a fat- and cholesterol-enriched diet, providing the first in vivo evidence for the regulated expression of ACSL3 in liver tissue (9). Altogether, these recent new findings suggest that despite the low expression level, ACSL3 may play an important regulatory role in FA and TG metabolism in liver, a major organ in lipid metabolism of mammals.
The peroxisome proliferator-activated receptors (PPARs) are nutritional sensors and have profound roles in FA metabolism (14–16). Three subtypes, PPARα, PPARγ, and PPARδ/β, have been identified with distinct tissue distribution and biological activities. PPARs contain a signature type II zinc finger DNA binding motif and a hydrophobic ligand binding domain. When PPARs are activated by ligands, they heterodimerize with retinoid X receptor α (RXRα) and then bind to specific sequences on the DNA referred to as PPAR response elements (PPREs) to activate the transcription of target genes. PPARα is expressed in liver, heart, muscle, and kidney where it stimulates FA catabolism. PPARγ is highly enriched in adipocyte and macrophage. It is the primary member of this family involved in adipocyte differentiation, lipid storage, and glucose homeostasis. In contrast to the other two family members, PPARδ is ubiquitously expressed in many tissues including liver and its function in lipid metabolism has recently begun to gain intensive interest (14).
The facts that dietary fatty acids are natural activators of PPARs and that fatty acyl-CoAs, the products of ACSL family enzymes, are substrates for most downstream metabolic pathways led us to hypothesize the existence of an intrinsic molecular link between ACSL and PPARs. Consistent with this hypothesis, PPARα has been identified as the molecular mediator for fenofibrate-stimulated expression of ACSL1 mRNA in rat hepatocytes, Fa-32 hepatoma cells, and in liver and adipose tissue through a PPRE motif embedded in the ACSL1 promoter region (17).
To gain insight into the regulatory mechanism for ACSL3 expression in liver tissue, we asked whether PPAR was involved in the OSM-induced activation of ACSL3 gene transcription. Here, we show that OSM induced coordinated increases in ACSL3 and PPARδ mRNA expression in HepG2 cells, whereas the mRNA expression of PPARα and PPARγ was not affected by OSM treatment. Through a battery of molecular and cellular assays we demonstrate, for the first time, that ACSL3 is a molecular target of PPARδ in liver cells and PPARδ is involved in the OSM-mediated transcriptional up-regulation of ACSL3. PPARδ regulates ACSL3 gene transcription through its binding to two imperfect PPRE motifs that are located in close proximity in the region −944 to −915 relative to the transcription start site (TSS). Finally, we provide in vivo evidence demonstrating that the hepatic expression of ACSL3 mRNA and protein were increased by ligand-mediated activation of PPARδ in hamsters.
EXPERIMENTAL PROCEDURES
Cells and Reagents
The human hepatoma cell line HepG2 was obtained from American Type Culture Collection and cultured in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (Summit Biotechnology, Fort Collins, CO), 1 mm streptomycin, and 1 mm penicillin. Rabbit antibody against hamster ACSL3 was generated as described previously (9). Anti-PPARδ (sc-7197) and anti-histone deacetylase (sc-7872) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and monoclonal anti-β-actin and anti-γ-tubulin antibody were obtained from Sigma. PPARα agonist WY-14643 was obtained from Sigma and PPARγ agonist 15d-PGJ2 and PPARδ agonist L165041 were obtained from AXXORA (La Jolla, CA). The plasmid (PPRE)3-tk-luc was generously provided by Dr. Jun-ichi Abe, University of Rochester School of Medicine.
Human ACSL3 Promoter Luciferase Reporters
A series of unidirectional deletion constructs of human ACSL3 gene promoter luciferase reporters were made using the phACSL3Luc plasmid as the template, which contains a DNA fragment 3600 bp upstream of the TSS of the ACSL3 gene (18). The desired promoter region was amplified by PCR. The gel-purified PCR product was cloned into pCR2.1-TOPO TA vector (Invitrogen) initially and subcloned into pGL3-basic vector (Promega, Madison, WI) at the KpnI and XhoI sites. The PPRE mutant reporter (PPREmu) was generated using the pACSL3-1116 plasmid as template, the QuikChange site-directed mutagenesis kit (Stratagene), and the respective oligonucleotides as described below. The mutated nucleotides are in bold and underlined: PPREMU1F, 5′-GGCTGGCTCAGGAAGGCAGGGTTTCACCTACTTAAGGAAAAG-3′; PPREMU1R, 5′-CTTTTCCTTAAGTAGGTGAAACCCTGCCTTCCTGAGCCAGCC-3′; PPREMU2F, 5′-GGGTTTCACCTACTTAAGGAAACCAACACAGGCAAACTAAAC-3′; PPREMU2R, 5′-GTTTAGTTTGCCTGTGTTGGTTTCCTTAAGTAGGTGAAACCC-3′. The nucleotide sequence and correct orientation of each deletion or mutation reporter was verified by DNA sequencing.
Transient Transfections of Reporter Constructs
ACSL3 promoter reporters of wild-type, deletion, and mutation were cotransfected with a control Renilla luciferase expression vector (pRL-SV40) to normalize for transfection efficiency in a DNA ratio of 200:1. To measure OSM or agonist effects, 1 day post-transfection, cells were cultured in medium containing 10% fetal bovine serum and treated with OSM (100 ng/ml) or PPARδ agonist L165041 (25 μm) for 16 h prior to cell lysis for dual luciferase activity measurements. The firefly luciferase activity was normalized to the Renilla luciferase activity in each sample. Three to five separate transfections were conducted for each reporter in which 3–6 wells were used for each condition.
RNA Isolation and Real Time RT-PCR
Total RNA was extracted from HepG2 cells or hamster livers using UltraspecTM total RNA isolation reagent (Biotecx Laboratories). Two μg of total RNA was reverse transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) using random primers according to the manufacturer's instructions. Real time PCR was performed on the cDNA using an ABI Prism 7900-HT Sequence Detection System. Human ACSL3, PPARα, PPARγ, PPARδ, and glyceraldehyde-3-phosphate dehydrogenase pre-developed TaqMan Assay Reagents (Applied Biosystems) were used to assess mRNA expression in HepG2 cells with or without treatment. The relative ACSL3 and CPT1A mRNA levels in hamster livers were measured with SYBR PCR master mix (Applied Biosystems) using hamster-specific primers (9). All values are reported as mean ± S.D. of triple measurements of each cDNA sample.
siRNA Transfection
Pre-designed siRNAs targeted to human PPARδ mRNA (catalog number AM51331; ID 5465), PPARα mRNA (catalog number AM51331; ID 5348), and Silencer Negative control siRNA with a scrambled sequence (catalog number 4618G) were obtained from Ambion. Another PPARδ siRNA was obtained from Dharmacon (catalog number L-003435-00-005; ID 5467). For promoter activity analyses, HepG2 cells in suspension were mixed with siRNA in the SilencerTM siRNA transfection reagent for 10 min according to the vendor's instructions and plated in 96-well plates at a density of 1 × 104 cells/well. Twenty-four h later, cells were transfected with phACSL3Luc and pRL-SV40. Transfectants were cultured in minimum essential medium containing 10% fetal bovine serum for 24 h prior to the addition of L165041 at a concentration of 10 μm. Cells were harvested 24 h later for measuring dual luciferase activities. For real time PCR, 1 × 105 cells were mixed with siRNA in the Silencer siRNA transfection reagent for 10 min and plated in 12-well plates. After 2 days, transfected cells were cultured in minimum essential medium containing 0.5% fetal bovine serum overnight and then treated with OSM overnight prior to cell lysis.
Construction of PPARδ Expression Plasmid
The coding region of human PPARδ cDNA was amplified by PCR from a full-length cDNA clone (ID 3630487) obtained from Open Biosystems, Huntsville, AL. The amplified cDNA product was gel-purified using the QiaQuick PCR purification kit (Qiagen, Inc., Valencia, CA) and cloned into pcDNA4/HisMax topo TA vector (Invitrogen) to yield plasmid pHis-PPARδ. The sequence and orientation of the insert were verified by DNA sequencing. The expression of His-tagged PPARδ in HepG2 cells after transient transfection was confirmed by Western blot analysis with anti-His antibody.
Western Blot Analysis
ACSL3 protein expression in HepG2 cells and hamster livers was assessed by Western blot analysis using a rabbit antibody recognizing the C-terminal region of the hamster ACSL3. This anti-ACSL3 antibody also specifically recognized ACSL3 of human, mouse, and rat origins (9). PPARδ protein expression in nuclear extracts of HepG2 cells was detected by rabbit anti-PPARδ antibody (sc-7197) from Santa Cruz. The specific immunoreactive bands were visualized using an ECL plus kit (GE Healthcare) and quantified with the Kodak Molecular Imaging Software (Kodak).
ACSL Activity Assay
HepG2 cells were untreated or treated with 25 μm L165041 for 24 h. The cell lysate preparation and ACSL3 activity assay were conducted as previously described (13).
Electrophoretic Mobility Shift Assays
HepG2 nuclear extracts were prepared as described (19). A double-stranded oligonucleotide, designated as ACSL3-PPRE, was end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Human recombinant proteins RXRα (catalog number 31133) was obtained from Active Motif (Carlsbad, CA) and PPARδ (catalog number 10007451) were purchased from Cayman Chemical (Ann Arbor, MI). The identities of PPARδ and RXRα were confirmed by Western blots using rabbit anti-PPARδ polyclonal antibody (Santa Cruz, sc-7197) and rabbit anti-RXRα polyclonal antibody (Santa Cruz, sc-553), respectively (data not shown). For visualizing band shift, 150 ng of PPARδ and 50 ng of RXRα recombinant proteins were incubated together with labeled ACSL3 probe in a binding buffer containing 0.75 mm EDTA, 18 mm HEPES (pH 7.9), 0.5 mm dithiothreitol, 5% glycerol in a final volume of 20 μl. For supershift assays, 1 μg of anti-PPARδ antibody (Santa Cruz, sc-7197x) was used. For electrophoretic mobility shift assay using nuclear extracts, each binding reaction was composed of 25 mm Tris (pH 7.5), 1 mm MgCl2, 60 mm KCl, 5% glycerol, 1 μg of poly(dI-dC), and 30 μg of HepG2 nuclear extract in a final volume of 20 μl. The nuclear extracts were incubated with 0.4 to 0.5 ng of 32P-labeled ACSL3-PPRE (1 × 105 cpm) for 30 min at room temperature. The reaction mixtures were loaded onto a 6% polyacrylamide gel and run in 0.5× TBE buffer at 30 mA for 2 h at 4 °C. Gels were dried and visualized on a PhosphorImager. For competition assays, 100× unlabeled ACSL3-PPRE, ACSL3-PPREmu, or PPRE (a double-stranded nucleotide containing a consensus PPRE sequence motif), or a nonspecific oligonucleotide containing a Sp1 binding site, was added to the reaction mixture. The sense sequences of electrophoretic mobility shift assay probe and competitors are as follows: ACSL3-PPREwt, 5′-GAAGGCAACCTTTCACCTACTTAAGGAAAAGTTCACAGGC-3′; ACSL3-PPREmu, 5′-GAAGGCAGGGTTTCACCTACTTAAGGAAACCAACACAGGC-3′; PPRE, 5′-GGGCGAGGTCAGAGGTCAGAAGG-3′; and nonspecific, 5′-TTTGAAAATCACCCCACTGCAAACTCCTCCCCCTGCT-3′. The mutated nucleotides are bold and underlined.
Chromatin Immunoprecipitation (ChIP)
The ChIP assays were conducted according to the described protocol using aliquots of lysate obtained from 2 × 107 HepG2 cells (20). Briefly, cells were fixed in 1.42% formaldehyde for 15 min at room temperature. Cells were lysed and the chromatin was sheared to an average length of 0.2–1 kb by the sonication method for 15 min in an ultrasonic water bath. Samples were immunoprecipitated at 4 °C with 2 μg of rabbit anti-PPARδ antibody (sc-7197X) or rabbit IgG as negative control. Immunocomplexes were isolated by binding to protein A-agarose beads after extensive washing with the immunoprecipitation buffer. Chelex 100 slurry was added to the washed beads, boiled for 10 min, and centrifuged. The supernatant contained the sheared chromatin. An aliquot of the sheared chromatin was used in the PCR as a control for the amount of input DNA used in precipitations and diluted 10-fold prior to PCR. The bound and the input DNA were analyzed by PCR with primers that amplify a 164-bp fragment of the human ACSL3 promoter region −1020 to −857, relative to the TSS. The sequences of ChIP primers are: ACSL3-ChIP primer forward, 5′-CAAGTTCTGGCGGCTTCCTG-3′; ACSL3 ChIP primer reverse, 5′-CCCAGTACTAGTAGGATTGGTCTC-3′.
In the ChIP assay, a 238-bp promoter fragment corresponding to the −3600 to −3363 region of the ACSL3 promoter upstream sequence lacking a PPRE motif was also PCR amplified as a negative control. Sequences of the control primers are: ACSL3-ChIP primer 1 forward, 5′-GATCTAGCTGGGATTGTTGATGAAGG-3′; and ACSL3 ChIP primer 1 reverse, 5′-CTATTCTGAAAATCCTAGGGCCC-3′.
The PCR conditions included a denaturation step at 94 °C for 3 min, then 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and extended at 72 °C for 7 min. The PCR products were visualized on a 2% agarose gel stained with ethidium bromide. Intensities of the PCR products were scanned and quantified with the Kodak Image Station 1000 System. Additionally, real time quantitative PCR was performed using the two primer sets and chromatin DNA contained in the immunoprecipitates.
Regulation of ACSL3 Expression in Vivo
Eighteen male Golden Syrian hamsters with body weights of 100–120 g were purchased from Harlan Sprague-Dawley. Hamsters were housed (3 animals/cage) under controlled temperature (72 °F) and lighting (12 h light/dark cycle). Animals had free access to autoclaved water and normal rodent chow diet. L165041 was dissolved in a vehicle containing 0.25% carboxyl methylcellulose sodium and 5% Triton X-100 at a concentration of 0.5 mg/ml (21). After an acclimatization period of 7 days, hamsters were randomly divided into 2 groups. One group was given a daily dose of 10 mg/kg of L165041 intraperitoneally, and the other group was given an equal volume of vehicle intraperitoneally. The treatment lasted for 7 days. Overnight fasting serum was collected before and after the drug treatment. Four h after the last drug treatment, all animals were sacrificed. Livers were immediately removed, cut into small pieces, and stored at −80 °C for RNA and protein isolations.
To measure serum total cholesterol (TC) and TG levels, blood samples (0.2 ml) were collected from the retro-orbital plexus using heparinized capillary tubes under anesthesia (2–3% isoflurane and 1–2 liters/min of oxygen) after a 16-h fast (5 p.m. to 9 a.m.) before and after drug treatments. Serum was isolated at room temperature and stored at −80 °C. Standard enzymatic methods were used to measure TC and TG by using commercially available kits purchased from Stanbio Laboratory (TX). Each sample was assayed in duplicate. Animal use and experimental procedures were approved by the Institutional Animal Care and Use Committee of the Veterans Affairs Palo Alto Health Care System.
Statistical Analysis
Significant differences between control and treatment groups or between wild-type and mutated vectors were assessed by two-tailed Student's t test or one-way analysis of variance with Dunnett's post test where applicable. Values of p < 0.05 were considered statistically significant.
RESULTS
Involvement of PPARδ in OSM-mediated Up-regulation of ACSL3 Transcription
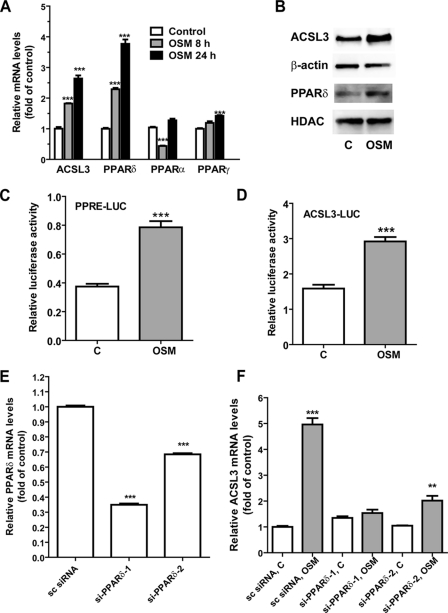
To explore a possible role of PPARs in OSM-induced activation of ACSL3 gene transcription, we first examined the mRNA levels of PPARα, PPARγ, and PPARδ along with ACSL3 in HepG2 cells without and with OSM treatments of 8 and 24 h by real time RT-PCR assay (Fig. 1A). OSM at a concentration of 50 ng/ml increased ACSL3 mRNA 1.8-fold and PPARδ mRNA 2.2-fold at 8 h. At 24 h, OSM increased ACSL3 mRNA 2.6-fold and PPARδ mRNA 3.7-fold. By contrast, the changes in PPARα mRNA fluctuated during the OSM treatment with a reduction at 8 h and returned to the baseline at 24 h; and the mRNA expression of PPARγ was only slightly elevated by OSM treatment to 1.4-fold of control after 24 h. These results demonstrated that among the three subtypes of PPARs, PPARδ mRNA expression appeared to be coordinately induced with ACSL3 mRNA expression by OSM in HepG2 cells. Western blot analysis to detect ACSL3 and PPARδ protein expression in control and OSM-treated cells showed that both ACSL3 and PPARδ protein levels were increased by the OSM treatment (Fig. 2B), thereby confirming the results of mRNA analysis. Actin was used as a protein loading control for total cell lysate and histone deacetylase 1 (HDAC1) was a control for nuclear extracts, both of which were not affected by the OSM treatment.
FIGURE 1.
Involvement of PPARδ in OSM-mediated up-regulation of ACSL3 gene transcription. A, HepG2 cells were treated with 50 ng/ml of OSM for 8 and 24 h. Total RNA was isolated and mRNA levels of PPARα, PPARγ, PPARδ, and ACSL3 were quantified by real time quantitative PCR. The figure shown is representative of 3 independent experiments. In B, HepG2 cells were treated with 100 ng/ml of OSM for 24 h. Total cell lysates and nuclear extracts were isolated. Total cell lysates were used for detecting ACSL3 and nuclear extracts were used for PPARδ detection by Western blotting. β-Actin and histone deacetylase were used as loading control for whole cell lysate and nucler extracts, respectively. In C and D, suspended HepG2 cells were transfected with plasmids (PPRE)3-tk-luc or phACSL3Luc along with a Renilla luciferase expression vector (pRL-SV40) and then seeded in a 96-well culture plate for 24 h. Transfectants were treated with OSM or its vehicle (phosphate-buffere saline) for 16 h prior to cell lysis for dual luciferase assays. Firefly luciferase activity was normalized to Renilla luciferase activity in each sample. The figure shown is representative of 3 independent experiments. In E, cells were transfected with siRNAs targeted to PPARδ, or a control nonspecific siRNA for 2 days prior to cell lysis to analyze PPARδ and glyceraldehyde-3-phosphate dehydrogenase mRNA levels by real time PCR. In F, cells were transfected with siRNAs for 24 h before the addition of OSM or vehicle for 16 h. ***, p < 0.001 compared with untreated control in A–D and F or compared with nonspecific siRNA in E. **, p < 0.01 compared with control. Each value represents the mean ± S.D. of triplicate assays per condition.
FIGURE 2.
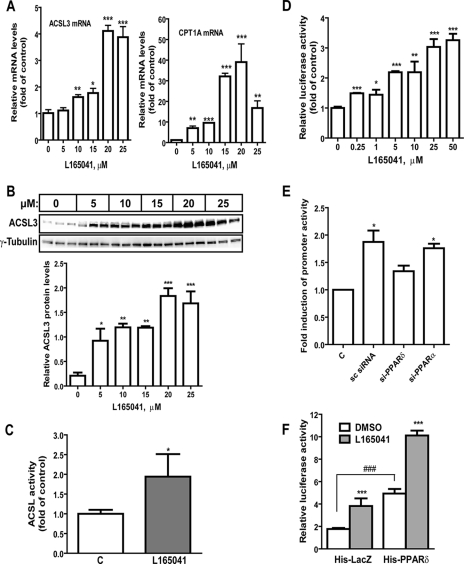
Activation of PPARδ leads to increased ACSL3 expression. A, HepG2 cells were treated with different doses of L165041 for 16 h. Total RNA was isolated and mRNA levels of ACSL3 and CPT1A were determined by quantitative real time PCR. The figure shown is representative of 3 separate assays. B, HepG2 cells were treated with the indicated doses of L165041 for 24 h with triplicate dishes for each condition. Total cell lysates were probed with anti-ACSL3 antibody, followed by anti-γ-tubulin antibody. The signal intensity of ACSL3 was normalized to that of tubulin and presented in the lower panel. The figure shown is representative of 2 separate assays. C, HepG2 cells were treated with 25 μm L165041 for 24 h. Cells were lysed and ACSL activity in 30 μg of cytosolic protein was measured. The figure shown is representative of 2 separate assays. D, HepG2 cells were co-transfected with ACSL3 promoter luciferase reporter (pACSL3-2700) and pRL-SV40 for 24 h prior to treatment of L165041 at the indicated doses for 16 h. Triplicate wells were used in each condition. The normalized luciferase activities in untreated samples were set as 1 and luciferase activities in treated samples were plotted relative to control. The figure shown is representative of 2 independent experiments. E, HepG2 cells were transfected with si-PPARδ-1, si-PPARα, or scrambled control siRNA for 24 h, followed by cotransfection of pACSL3-1116 and pRL-SV40 plasmids. After 24 h of reporter transfection, cells were treated with 10 μm L165041 or DMSO for 16 h before cell lysis for dual luciferase activity assay. The figure shown is representative of 3 independent experiments in which 4 wells were used in each transfection condition. F, HepG2 cells were cotransfected with pACSL3-2700, pHis-PPARδ, and pRL-SV40 in a DNA ratio of 1:1:0.05 or with the same amount of DNA containing pACSL3-2700, pHis-LacZ, and pRL-SV40 for 24 h prior to treatment of L165041 for 16 h. The figure shown is representative of 2 separate transfection experiments in which 8 wells were used in each transfection condition. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 compared with untreated control; ###, p < 0.001 compared with untreated His-LacZ control. In A–D, each value represents the mean ± S.D. of triplicate assays per condition (A–D); in E, each value represents the mean ± S.D. of 4 wells per condition; in F, each value represents the mean ± S.D. of 8 wells per condition.
To determine whether OSM treatment increased the trans-activating activity of PPARδ, HepG2 cells were transfected with a PPRE-driven luciferase reporter construct or the ACSL3 promoter luciferase reporter (phACSL3Luc) along with the plasmid pRL-SV40, a Renilla luciferase expression vector used for the normalization of transfection efficiency. One day post-transfection, OSM was added to the culture medium and cells were lysed 16 h later to measure both luciferase activities. OSM treatment led to a 2.3-fold increase (p < 0.001) in PPRE reporter activity (Fig. 1C) and a 1.8-fold increase (p < 0.001) in ACSL3 promoter activity (Fig. 1D).
Next, we examined the role of PPARδ in OSM-induced up-regulation of ACSL3 mRNA expression by transfecting HepG2 cells with two siRNA targeting to different coding regions of PPARδ. The mRNA levels of PPARδ were reduced to 35% in si-PPARδ-1 transfectants and 68% in si-PPARδ-2-transfected cells as compared with the cells transfected with the scrambled control siRNA (Fig. 1E). This siRNA-mediated down-regulation of PPARδ resulted in a complete counteraction (si-PPARδ-1) and a partial inhibition (si-PPARδ-2) of the increase in ACSL3 mRNA expression induced by OSM (Fig. 1F). Altogether, these results demonstrate that PPARδ mRNA expression, protein expression, and its transactivting activity are up-regulated by OSM in HepG2 cells, and are required for OSM stimulation of ACSL3 gene transcription.
Ligand-induced Activation of PPARδ Elevates ACSL3 mRNA and Protein Expression in HepG2 Cells
To examine a direct functional role of PPARδ in ACSL3 expression, HepG2 cells were treated with different concentrations of L165041, a specific PPARδ agonist for 24 h. Real time PCR and Western blot analysis were performed to examine the expression of ACSL3 mRNA and protein in untreated and treated cells. Ligand activation of PPARδ strongly increased ACSL3 mRNA (Fig. 2A, left panel), carnitine palmitoyltransferase 1A (CPT1A) mRNA, a known target gene of PPARs (Fig. 2A, right panel), and ACSL3 protein (Fig. 2B) expression in an agonist concentration-dependent manner. We also detected a 1.9-fold increase in total ACSL enzymatic activity in HepG2 cells treated with L165041 at a concentration of 25 μm (Fig. 2C). The inducing effects of L165041 on ACSL3 were exerted at the transcriptional level as evidenced by a similar increase in ACSL3 promoter activity by L165041 treatment (Fig. 2D). To further demonstrate that the effect of L165041 on ACSL3 transcription is mediated through PPARδ, HepG2 cells were first transfected with si-PPARδ-1, si-PPARα, or the scrambled control siRNA. The next day, cells were transfected with the ACSL3 promoter for 24 h, followed by a 24-h treatment of L165041. The results showed that the L165041-induced increase in ACSL3 promoter activity was not affected by PPARα siRNA but was significantly attenuated by PPARδ siRNA transfection (Fig. 2E). Real time PCR analysis confirmed the effective depletion of endogenous PPARδ and PPARα mRNA expression in the siRNA-transfected cells (data not shown).
To firmly demonstrate the function of PPARδ in the activation of ACSL3 transcription, the ACSL3 promoter reporter pGL3-ACSL3Luc was cotransfected with a PPARδ expression plasmid (pHis-PPARδ) or the control plasmid (pHis-LacZ). Transfectants were treated with L165041 or vehicle (DMSO). A 2.8-fold increase (p < 0.001) in ACSL3 promoter activity was observed following PPARδ transfection, and the activity was further enhanced by the treatment of L165041 (Fig. 2F). The increased promoter activity in cells transfected with pHis-LacZ resulted from the activation of endogenous PPARδ by L165041.
Next, we examined the combined effects of OSM and L165041 on ACSL3 promoter activity (Fig. 3A), mRNA expression (Fig. 3B), and protein expression (Fig. 3C). The combination of OSM, an inducer of PPARδ expression and L165041, a ligand that binds and stimulates the activity of PPARδ, increased ACSL3 promoter activity, mRNA, and protein expression to levels significantly higher than that of each individual treatment, thereby providing additional evidence for an activating role of PPARβ in ACSL3 gene expression.
FIGURE 3.
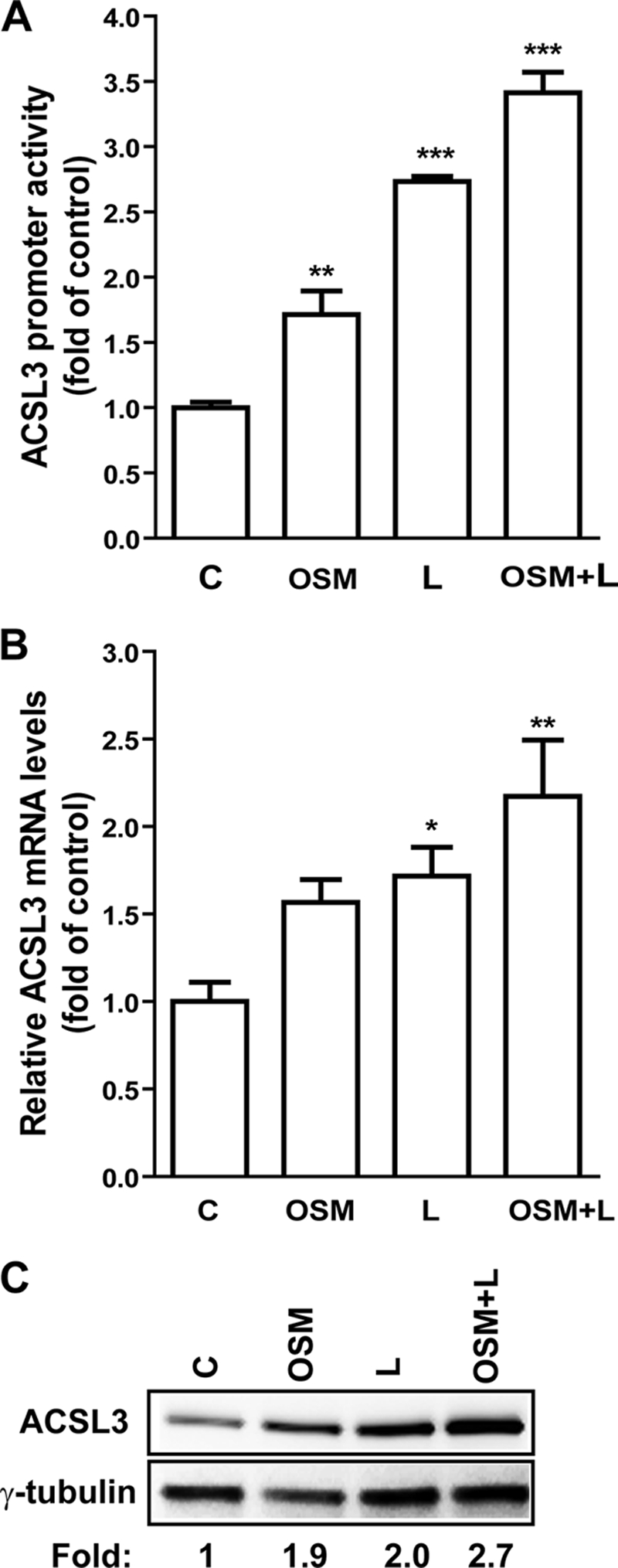
Induction of ACSL3 gene transcription by OSM, L165041, and the combined treatment. A, HepG2 cells were cotransfected with pACSL3-1116 or pRL-SV40 for 24 h. Transfectants were treated with OSM, L165041 (L), or OSM + L165041 (OSM + L) for 16 h before harvesting cells for dual luciferase assays. B, cells were treated with OSM, L165041, or the combination for 16 h prior to cell lysis to harvest RNA for real time PCR analysis. C, cells were treated with OM, L165041, or the combination for 24 h prior to harvesting protein for Western blotting. Each figure shown is representative of three separate experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 as compared with control. Each value represents the mean ± S.D. of triplicate assays per condition.
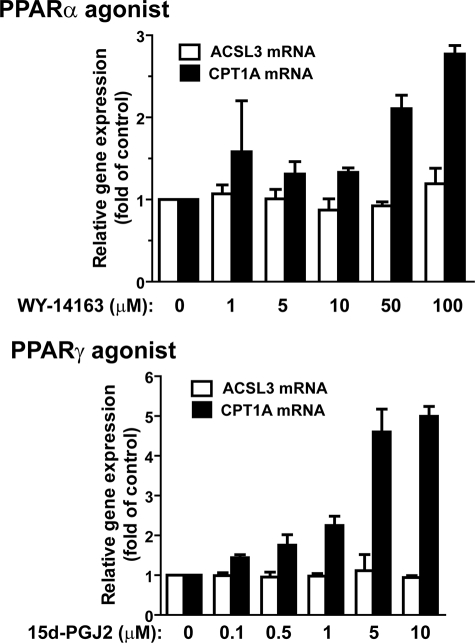
We were also interested in knowing whether ACSL3 gene transcription could be activated by PPARα and PPARγ. Treating HepG2 cells with PPARα agonist WY-14643 or PPARγ agonist 15d-PGJ2 over broad concentration ranges had no effect on ACSL3 mRNA expression. In contrast, the mRNA level of CPT1A was significantly elevated by these two agonists dose-dependently in HepG2 cells (Fig. 4). These data, combined with the results in Fig. 1 showing that OSM did not induce PPARα or PPARγ mRNAs, suggest that PPARδ is the only member of the PPAR family involved in the OSM-regulated ACSL3 expression in the HepG2 cell culture system.
FIGURE 4.
The expression of ACSL3 mRNA was not affected by activation of PPARα or PPARγ. HepG2 cells were treated with PPARα agonist WY-14643 or PPARγ agonist 15d-PGJ2 at the indicated concentrations for 16 h. Total RNA was isolated to assess the mRNA levels of ACSL3, CPT1A, and glyceraldehyde-3-phosphate dehydrogenase. The figures shown are representative of 2 independent experiments. Each value represents the mean ± S.D. of triplicate assays per condition.
Mapping ACSL3 Promoter to Identify PPARδ Responsive cis-Acting Elements
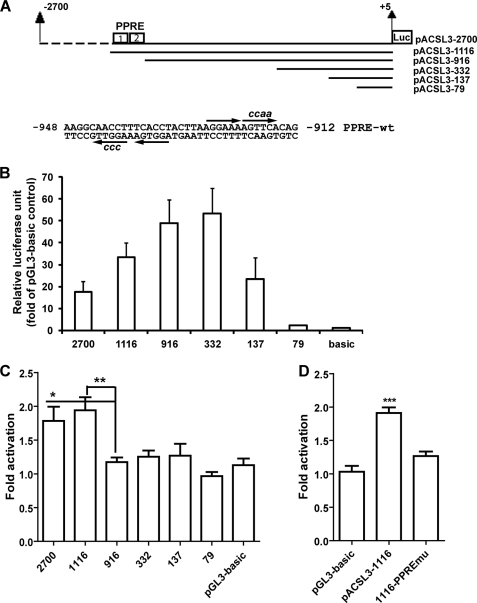
To define the promoter region responsible for PPARδ-induced up-regulation of ACSL3, a series of unidirectional deletion constructs (Fig. 5A) were made in the backbone of pGL3-basic vector and tested in reporter assays (Fig. 5B). Deletion of the 5′ promoter region up to −333 bp upstream of the TSS did not reduce the basal promoter activity. Further deletion to −80 reduced the basal promoter activity to the level of the promoter-less pGL3-basic vector, indicating that the proximal promoter functional region resides within the ∼252-bp region from −332 to −79, relative to the TSS. This result was in line with a previous report of ACSL3 promoter activity (18). Importantly, the summarized results of 5 separate transfection experiments clearly indicate that the response of the ACSL3 promoter to PPARδ activation was abolished by deleting the sequences located between −1116 and −916 (Fig. 5C). Analysis of the nucleotide sequence within this 200-bp segment of the ACSL3 promoter by MatInspector software revealed the presence of two putative PPRE motifs in close proximity (Fig. 5A) and thus these sites were designated as PPRE1 and PPRE2. PPRE1 is located on the antisense strand of the promoter spanning the region −944 to −932 and is composed of two half-sites separated by a single nucleotide. PPRE2 resides in the sense strand of the ACSL3 promoter spanning the region −926 to −915. The PPRE2 site comprises two half-sites next to each other. We mutated both PPRE sites on the reporter pACSL3-1116 and tested the response of the mutated reporter (1116-PPREmu) to L165041. The induction of ACSL3 promoter activity by this agonist was totally abolished by disruption of the PPRE sites (Fig. 5D), thereby confirming the role of these nucleotide sequences in mediating the PPARδ-induced activation of ACSL3 gene transcription.
FIGURE 5.
Analysis of basal and L165041-induced luciferase activities in HepG2 cells transfected with ACSL3 promoter constructs. A, schematic presentation of the deletion constructs of the ACSL3 promoter luciferase reporters and the nucleotide sequences of the putative PPRE sequences. Each arrow indicates the half-site and the lower case letters are mutated nucleotides. Nucleotide position was defined relative to the TSS. B and C, ACSL3 promoter luciferase reporters were transiently cotransfected with pRL-SV40 into HepG2 cells for 24 h, followed with a 16-h treatment of L165041. After normalization, the relative basal luciferase activity of each reporter was expressed as the fold of pGL3-basic (B) and the fold-activation by L165041 was calculated by dividing the normalized luciferase activity in treated cells with the value in untreated cells of each construct (C). Results shown are mean ± S.D. of 5 separate transfections in which triplicate samples were measured for each condition. *, p < 0.05; **, p < 0.01 as compared with the construct without PPRE sites. D, the ACSL3 promoter wild-type (pACSL-1116) and PPRE-mutated site construct vector (1116-PPREmu) were cotransfected with pRL-SV40 into HepG2 cells. The plasmid pGL3-basic was included in the transfection as a negative control for L165041 treatment. ***, p < 0.001 as compared with pGL3-basic. Each value represents the mean ± S.D. of triplicate assays per condition.
Interaction of PPARδ with ACSL3 Promoter in Vitro and in Vivo
To detect a direct interaction of PPARδ with ACSL3-PPRE promoter sequences, we first performed gel mobility shift assays using a radiolabeled double-stranded oligonucleotide probe containing the ACSL3 promoter sequence −947 to −910 and human recombinant PPARδ and RXRα proteins (Fig. 6A). One major complex was detected (lane 2). The formation of this complex was inhibited by a 100-fold molar excess of unlabeled wild-type oligonucleotide (lane 3), but it was not inhibited by a 100-fold molar excess of oligonucleotide containing the mutated PPRE sites (lane 4). Furthermore, this complex was completely supershifted in the presence of anti-PPARδ antibody. These data demonstrate that the PPARδ-RXRα heterodimer can bind to the PPRE sites located at positions −948 to −912 of the ACSL3 promoter. In addition to the recombinant proteins, we also examined the interaction of endogenous PPARδ with the ACSL3-PPRE sequences by the gel shift assay using the labeled probe and nuclear extracts of HepG2 cells (Fig. 6B). Again, one major complex was detected (lane 2). The formation of this complex was inhibited by a 100-fold molar excess of unlabeled wild-type oligonucleotide (lane 3) and by a 100-fold molar excess of unlabeled oligonucleotide containing a canonical PPRE site (lane 4), but it was not inhibited by a 100-fold molar excess of oligonucleotide containing the mutated PPRE sites (lane 5) nor by a 100-fold molar excess of a nonspecific oligonucleotide lacking a PPRE sequence (lane 6). We further compared the relative abundance of this complex formed with nuclear extracts prepared from untreated (lanes 7-9) and OSM-treated (lanes 10-12) HepG2 cells. The results showed that the signal intensity of the complex formed with 32P-labeled ACSL3-PPRE probe was clearly increased by OSM treatment at all three different doses, which was consistent with the observed increase in the expression of PPARδ mRNA and protein in OSM-treated cells (Fig. 1, A and B).
FIGURE 6.
Electrophoretic mobility shift assay and ChIP analyses of PPARδ association with PPRE sites of ACSL3 promoter in vitro and in vivo. A, labeled ACSL3-PPRE probe was incubated with 150 ng of PPARδ and 50 ng of RXRα recombinant proteins in the absence (lane 2) or presence of 100-fold molar excess of unlabeled wild-type probe (lane 3) or mutated probe (lane 4). The binding reaction was also carried out in the presence of 1 μg of anti-PPARδ antibody (lane 5). B, labeled ACSL3-PPRE probe was incubated with 30 μg of nuclear extracts (lanes 2–6) with and without the indicated competitors. In lanes 7–12, different amounts of nuclear extracts of control or OSM-treated cells were examined in the binding assays. C, anti-PPARδ antibody was used in a ChIP analysis followed by PCR to amplify a 164-bp fragment of the human ACSL3 promoter region −1020 to −857 and a 238-bp fragment of the ACSL3 promoter region (−3600 to −3363) from genomic DNA isolated from HepG2 cells untreated or treated with OSM or L165041. Normal rabbit IgG was included in the assay as negative controls for nonspecific binding. The PCR product was separated on a 2% agarose gel and stained with ethidium bromide. Input represents the starting material before immunoprecipitation. The data shown are representative of 2 separate ChIP assays with similar results. D, real time PCR analysis was conducted to amplify the ACSL3-PPRE promoter region (−1020 to −857). Each immunoprecipitated DNA sample was analyzed in triplicates. The data are expressed as relative increases for the ratio of signal from PPARδ antibody-enriched chromatin relative to a control IgG. The ratio of untreated cells was set as 1, and ratios in treated samples were plotted relative to the control. *, p < 0.05; **, p < 0.01, as compared with control. Each value represents the mean ± S.D. of triplicate assays per condition.
To examine the in vivo interaction of PPARδ with the ACSL3 promoter, ChIP assays were conducted to detect the binding of PPARδ to the region containing PPRE of the ACSL3 promoter in intact HepG2 cells. Cells were untreated or treated with OSM or L165041 for 24 h. Chromatin fragments were immunoprecipitated with a rabbit anti-PPARδ antibody or rabbit IgG as a negative control. The bound and input DNA were analyzed by PCR with one primer set that amplified a 164-bp fragment of the human ACSL3 promoter region from −1020 to −857 encompassing the PPRE sites. Another primer set amplified a 238-bp promoter fragment corresponding to the −3600 to −3363 region of the ACSL3 promoter without a putative PPRE site. This PCR was conducted in parallel as a negative control for nonspecific amplification. Fig. 6C shows that the amount of PPARδ bound to the ACSL3-PPRE sequence was increased by both OSM and L165041 treatment, whereas the faint nonspecific binding signal of the IgG immunoprecipitates was not enriched by the treatments. In addition, only a scant background signal was detected from PCR amplification of the same chromatin preparations using primers for the sequence upstream of the PPRE sites and again this nonspecific amplification signal was not enhanced by the treatment. We further performed real time PCR to amplify the promoter region −1020 to −857 to determine the relative amounts of chromatin DNA in various precipitates. Fig. 6D shows that compared with untreated cells, the amount of PPARδ cross-linked to the PPRE-ACSL3 sequence was increased 2.1- and 2.6-fold by OSM and L165041, respectively. Thus, the results of ChIP assays provide further support to the in vitro binding assay and demonstrate a direct interaction of PPARδ with ACSL3-PPRE sequences in vivo, and this interaction was further enhanced by treating cells with OSM or the agonist.
Activation of PPARδ Increases the Hepatic ACSL3 Expression in Hamsters
The results described above established the important role of PPARδ as the trans-activator for ACSL3 gene transcription in the HepG2 cell system. To examine whether this regulatory mechanism also operates in the liver tissue in vivo, L165041 was administered to hamsters intraperitoneally for 1 week at a daily dose of 10 mg/kg, whereas control hamsters received an equal volume of the vehicle by the same route. Measurement of ACSL3 mRNA levels in control and ligand-treated livers by real time RT-PCR demonstrated a 1.8-fold increase (p < 0.05) by L165041 treatment (Fig. 7A), whereas the mRNA level of CPT1A was increased as well up to 1.5-fold (p < 0.05). Examination of ACSL3 protein abundance in hamster liver samples by Western blotting corroborated the results of mRNA and showed higher ACSL3 protein levels in ligand-treated livers as compared with control (Fig. 7B). Measurement of serum lipid levels showed that L165041 treatment reduced serum TG levels by 28% (p < 0.05) and TC levels by 21% (p < 0.05) as compared with vehicle control animals. Together, these data demonstrate that activation of PPARδ up-regulates the liver expression of ACSL3 mRNA and protein in hamsters, thereby confirming ACSL3 as a direct target gene of PPARδ in liver tissue.
FIGURE 7.
Effects of activation of PPARδ on liver ACSL3 expression and plasma TG and TC levels. Hamsters (n = 9) were treated with 10 mg/kg of L165041 or vehicle (n = 9) for 7 days. At the end of treatment, animals were sacrificed and serum and liver tissues were isolated. In A, total RNA was isolated from each liver sample and the relative mRNA abundance of ACSL3 and CPT1A were determined by conducting real time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase. *, p < 0.05 as compared with control group. Data shown are mean ± S.E. of 7–9 samples per group. In B, total protein extracts were individually prepared from 6 randomly chosen liver samples of the vehicle or treatment groups. Equal amounts of homogenate proteins (50 μg) were resolved by SDS-PAGE and ACSL3 protein was detected by immunoblotting using a rabbit anti-ACSL3 antibody. The membrane was reprobed with an anti-β-actin antibody. In C, levels of TG and TC in serum samples of treated and vehicle control groups were individually measured. Data shown are mean ± S.E. of 8 to 9 samples per group. *, p < 0.05.
DISCUSSION
Hepatic lipid metabolism is coordinately regulated by a complex interplay between hormones, transcription factors, and energy substrates to meet the metabolic needs of the body. The ACSL family of enzymes is an important and integral part of this network that converts inactive and unusable FAs to CoA-conjugated FAs as substrates for most downstream pathways that metabolize FA (2). From this perspective, one could postulate that the expression level of ACSL enzymes is likely to be governed by transcription factors within this metabolic network to adapt to the rapid changes in metabolic demands for FAs.
The PPAR nuclear receptor subfamily of transcription factors are master players in FA metabolism (14, 22). Depending on the tissues, individual members of the family may exert dominant roles over the other subtypes in certain cellular functions by regulating different sets of target genes. PPARα has been well recognized as the major regulator of FA catabolism in liver tissue by activation of many genes involved in β-oxidation (16, 23), whereas PPARγ has a predominant role in the activation of genes involved in lipid synthesis and glucose homeostasis in adipose tissue (24, 25). The first example that connected the ACSL family with PPAR nuclear receptors was the report in 1995 that showed regulation of ACSL1 expression by PPARα (17). In this current study we have identified PPARδ as the predominant transactivator for ACSL3 gene transcription in liver-derived HepG2 cells and in the liver tissue of hamsters in a pharmacologically responsive manner. Thus, ACSL3 is the second enzyme of the ACSL family as a direct target gene for PPARs, activated by PPARδ, whereas ASCL1 is activated by PPARα.
Our investigation initially set out to elucidate the mechanism underlying the OSM-induced transcription of ACSL3 in HepG2 cells as an entry point to unravel the cellular mechanisms that regulate the transcription of this enzyme in liver tissue under different metabolic or pharmacologic conditions to gain a better understanding to our previous intriguing observation that ACSL3 expression in liver is most responsive to changes in nutritional status (9). Our first observation of this current study was that OSM induced a significant elevation of PPARδ mRNA without affecting PPARα or PPARγ gene expression. To our knowledge, this is the first report on the regulation of PPARδ expression by OSM. This new finding provided an important clue for a possible role of PPARδ in ACSL3 transcription. The concomitant induction of a PPRE luciferase reporter activity and ACSL3 promoter activity by OSM added more circumstantial evidence to link PPARδ with ACSL3 gene transcription, which was further strengthened by the loss of the OSM inducing effect on ACSL3 mRNA expression in PPARδ-depleted cells through siRNA-mediated knockdown. This set of data established the involvement of PPARδ in OSM-mediated up-regulation of ACSL3. By utilizing the well characterized PPARδ agonist L165041, we demonstrated a direct impact of PPARδ activation, separate from the context of OSM stimulation, on ACSL3 expression at the promoter, mRNA, and protein levels. Although some studies have reported receptor-independent effects of PPAR agonist (26, 27), the up-regulation of ACSL3 transcription by this agonist requires the presence of PPARδ because its effect on ACSL3 promoter activity was largely abolished in PPARδ-depleted cells. Additional evidence is provided by the data showing that overexpression of PPARδ alone increased ACSL3 promoter activity, which was further stimulated by the addition of L165041.
PPARs regulate gene transcription through direct binding to the PPRE motifs imbedded in the promoter region of target genes (28). In this report, we have mapped the PPARδ response DNA elements to two PPRE-like sequences. These two PPRE sites are separated by 5 nucleotides and located in the ACSL3 promoter region −944 to −915, relative to the TSS. It is interesting that the promoter basal activity is localized to the 252-bp proximal region −332 to −79 of ACSL3 promoter, whereas the PPRE sites are approximately 900 bp upstream of the TSS. This characteristic has been reported in other nuclear receptor target genes where PPRE binding sites are not in proximity of the annotated TSS of a gene but are often located distally (29, 30). We confirmed the functional importance of these two PPRE sites through deletion and mutation analysis. Deletion of the PPRE-containing segment eliminated the agonist-mediated up-regulation of ACSL3 promoter activity. Mutation of both PPRE sites together abolished the inducing effect of L165041, whereas a single site mutation at either PPRE1 or PPRE2 could not completely prevent the induction (data not shown). This suggests that both PPRE motifs are functional binding sites of PPARδ.
The direct binding of PPARδ to these motifs on the ACSL3 promoter was first suggested by conducting gel shift and supershift assays using recombinant PPARδ-RXRα. We found one DNA-protein complex formed. The specificity of this interaction was demonstrated by the loss of binding in the presence of 100-fold molar excess of specific competitors, whereas retaining binding in the presence of 100-fold molar excess of oligonucleotides that did not have a PPRE sequence or a mutated PPRE sequence from the ACSL3 promoter. By using nuclear extracts of HepG2 cells we further demonstrated that endogenous binding of the PPARδ complex to the ACSL3-PPRE sequences was enhanced by OSM treatment, corroborating the results of mRNA, protein, and enzyme activity analyses.
Additional evidence for a direct binding of PPARδ to the PPRE sites of the ACSL3 promoter was obtained by ChIP assays. The amount of PPARδ bound to the ACSL3 promoter containing chromatin in the immunoprecipitates of anti-PPARδ was substantially higher than that of the control IgG, and it was enriched further in OSM- or L165041-treated cells, thereby demonstrating a PPARδ-mediated and ligand-induced binding event. Altogether, these results provide strong support for a functional role of PPARδ in regulating ACSL3 gene transcription through these PPRE sequences.
With the previous observed association of ACSL3 induction and the reduced cellular TG content in OSM-treated HepG2 cells (11), we were very interested in knowing whether activation of PPARδ could stimulate ACSL3 expression in vivo, particularly in liver tissue, thus linking a metabolic transcriptional activator to our phenotypic observations and providing novel mechanistic insights. The results obtained from hamsters confirmed our findings in HepG2 cells. We detected a clear increase in both mRNA and protein levels of ACSL3 in L165041-treated livers as compared with control liver samples. Indeed, the TG plasma level was reduced by 28% in the ligand-treated hamsters. Thus, the increase in ACSL3 expression in liver is likely a contributing factor for the hypolipidemic effect of OSM and the PPARδ agonist. Because PPARδ agonists have been previously reported to lower plasma TG in different animal models (31), future studies of liver-specific knockdown or overexpression of ACSL3 will be required to assess to what extent the increased expression of ACSL3 in liver contributes to the overall TG lowering effect of PPARδ activation.
In conclusion, we have demonstrated that OSM increases the transcription of PPARδ, which in turn engages in OSM-induced ACSL3 transcription through its direct binding to the PPRE sequences imbedded in the ACSL3 promoter to activate gene transcription in HepG2 cells. We further demonstrated that this PPARδ-governed regulatory mechanism of ACSL3 transcription operates in liver tissue in vivo and has an impact on the plasma level of TG. This work adds new understanding to how the ACSL family of enzymes is transcriptionally regulated.
Acknowledgment
We thank Dr. Michael R. Briggs for critical review of this manuscript.
The work was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service) and National Center for Complementary and Alterative Medicine Grants 1RO1 AT002543-01A1 and 1R21AT003195-01A2.
- FA
- fatty acid
- ACSL3
- long chain acyl-CoA synthetase-3
- ChIP
- chromatin immunoprecipitation
- CPT1A
- carnitine palmitoyltransferase 1A
- OSM
- oncostatin M
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- PPAR-responsive element
- RXR
- retinoid X receptor
- siRNA
- small interference RNA
- TC
- total cholesterol
- TG
- triglyceride
- TSS
- transcription start site
- ERK
- extracellular signal-regulated kinase
- RT
- reverse transcription.
REFERENCES
- 1.Mashek D. G., Coleman R. A. (2006) Curr. Opin. Lipidol. 17, 274–278 [DOI] [PubMed] [Google Scholar]
- 2.Soupene E., Kuypers F. A. (2008) Exp. Biol. Med. 233, 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki H., Kawarabayasi Y., Kondo J., Abe T., Nishikawa K., Kimura S., Hashimoto T., Yamamoto T. (1990) J. Biol. Chem. 265, 8681–8685 [PubMed] [Google Scholar]
- 4.Oikawa E., Iijima H., Suzuki T., Sasano H., Sato H., Kamataki A., Nagura H., Kang M. J., Fujino T., Suzuki H., Yamamoto T. T. (1998) J. Biochem. 124, 679–685 [DOI] [PubMed] [Google Scholar]
- 5.Fujino T., Kang M. J., Suzuki H., Iijima H., Yamamoto T. (1996) J. Biol. Chem. 271, 16748–16752 [DOI] [PubMed] [Google Scholar]
- 6.Kang M. J., Fujino T., Sasano H., Minekura H., Yabuki N., Nagura H., Iijima H., Yamamoto T. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2880–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujino T., Yamamoto T. (1992) J. Biochem. 111, 197–203 [DOI] [PubMed] [Google Scholar]
- 8.Mashek D. G., Li L. O., Coleman R. A. (2006) J. Lipid Res. 47, 2004–2010 [DOI] [PubMed] [Google Scholar]
- 9.Wu M., Liu H., Chen W., Fujimoto Y., Liu J. (2009) Lipids 44, 989–998 [DOI] [PubMed] [Google Scholar]
- 10.Fujino T., Man-Jong K., Minekura H., Suzuki H., Yamamoto T. T. (1997) J. Biochem. 122, 212–216 [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., Takano T. (2007) J. Lipid Res. 48, 1280–1292 [DOI] [PubMed] [Google Scholar]
- 12.Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y., Abidi P., Kim A., Chen W., Huang T. T., Kraemer F. B., Liu J. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 2198–2205 [DOI] [PubMed] [Google Scholar]
- 14.Lee C. H., Olson P., Evans R. M. (2003) Endocrinology 144, 2201–2207 [DOI] [PubMed] [Google Scholar]
- 15.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. (2001) Science 294, 1866–1870 [DOI] [PubMed] [Google Scholar]
- 16.Latruffe N., Vamecq J. (1997) Biochimie 79, 81–94 [DOI] [PubMed] [Google Scholar]
- 17.Schoonjans K., Watanabe M., Suzuki H., Mahfoudi A., Krey G., Wahli W., Grimaldi P., Staels B., Yamamoto T., Auwerx J. (1995) J. Biol. Chem. 270, 19269–19276 [DOI] [PubMed] [Google Scholar]
- 18.Minekura H., Kang M. J., Inagaki Y., Suzuki H., Sato H., Fujino T., Yamamoto T. T. (2001) Gene 278, 185–192 [DOI] [PubMed] [Google Scholar]
- 19.Li H., Chen W., Zhou Y., Abidi P., Sharpe O., Robinson W. H., Kraemer F. B., Liu J. (2009) J. Lipid Res. 50, 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson J. D., Denisenko O., Bomsztyk K. (2006) Nat. Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 21.Choi K. C., Lee S. Y., Yoo H. J., Ryu O. H., Lee K. W., Kim S. M., Baik S. H., Choi K. M. (2007) Biochem. Biophys. Res. Commun. 357, 62–67 [DOI] [PubMed] [Google Scholar]
- 22.Sprecher D. L. (2007) Am. J. Cardiol. 100, n20–n24 [DOI] [PubMed] [Google Scholar]
- 23.Peters J. M., Hennuyer N., Staels B., Fruchart J. C., Fievet C., Gonzalez F. J., Auwerx J. (1997) J. Biol. Chem. 272, 27307–27312 [DOI] [PubMed] [Google Scholar]
- 24.Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., Koder A., Evans R. M. (1999) Mol. Cell 4, 585–595 [DOI] [PubMed] [Google Scholar]
- 25.Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Eto K., Tsubamoto Y., Okuno A., Murakami K., Sekihara H., Hasegawa G., Naito M., Toyoshima Y., Tanaka S., Shiota K., Kitamura T., Fujita T., Ezaki O., Aizawa S., Kadowaki T. (1999) Mol. Cell 4, 597–609 [DOI] [PubMed] [Google Scholar]
- 26.Crosby M. B., Svenson J. L., Zhang J., Nicol C. J., Gonzalez F. J., Gilkeson G. S. (2005) J. Pharmacol. Exp. Ther. 312, 69–76 [DOI] [PubMed] [Google Scholar]
- 27.Scatena R., Bottoni P., Giardina B. (2008) PPAR Res. 2008, 256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degenhardt T., Saramäki A., Malinen M., Rieck M., Väisänen S., Huotari A., Herzig K. H., Müller R., Carlberg C. (2007) J. Mol. Biol. 372, 341–355 [DOI] [PubMed] [Google Scholar]
- 30.Sanderson L. M., Degenhardt T., Koppen A., Kalkhoven E., Desvergne B., Müller M., Kersten S. (2009) Mol. Cell Biol. 29, 6257–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. (2003) Cell 113, 159–170 [DOI] [PubMed] [Google Scholar]