Abstract
Regulator of calcineurin 1 (RCAN1) inhibits the protein phosphatase calcineurin and is required for appropriate immune responses, synaptic plasticity, vascular tone, angiogenesis, and cardiac remodeling. Expression of the RCAN1–4 isoform is under the control of the calcineurin-responsive transcription factor NFAT. Typically, NFATs act in cooperation with other transcription factors to achieve maximal activation of gene expression. In this study, we identify the CCAAT/enhancer binding protein β (C/EBPβ) as an NFAT binding partner that cooperates with NFAT to regulate RCAN1–4 expression. Numerous C/EBPβ binding sites are conserved in the RCAN1–4 proximal promoter. Overexpression of C/EBPβ increased activity of both the endogenous mouse Rcan1–4 gene and a human RCAN1–4 luciferase reporter. Binding of C/EBPβ to multiple sites in the promoter was verified using electrophoretic mobility shift assays and chromatin immunoprecipitation. A direct interaction between C/EBPβ and NFAT was demonstrated by co-immunoprecipitation of proteins and complex formation at NFAT-C/EBPβ composite sites. Depletion of endogenous C/EBPβ decreased maximal activation of RCAN1–4 expression by calcineurin, whereas inhibition of calcineurin did not alter the ability of C/EBPβ to activate RCAN1–4 expression. Together, these findings suggest that calcineurin/NFAT activation of RCAN1–4 expression is in part dependent upon C/EBPβ, whereas activation by C/EBPβ is not dependent on calcineurin and may provide a calcineurin-independent pathway for regulating RCAN1–4 expression. Importantly, nuclear localization, C/EBPβ DNA binding activity and occupancy of the Rcan1–4 promoter increased in mouse models of heart failure demonstrating in vivo activation of this pathway to regulate Rcan1–4 expression and ultimately shape the dynamics of calcineurin-dependent signaling.
Keywords: C/EBP Transcription Factor, Calcineurin, Cardiac Hypertrophy, Signal Transduction, Transcription Regulation, RCAN1
Introduction
Regulator of calcineurin 1 (RCAN1/MCIP1/DSCR1) influences intracellular signaling cascades primarily inhibiting the calcium-activated protein phosphatase calcineurin (1). In mice, Rcan1 is required for appropriate immune responses (2), synaptic plasticity (3), vascular tone (4), angiogenesis (5, 6), and cardiac remodeling (7). Increased expression of RCAN1 in cardiomyocytes reduces pathological remodeling of the myocardium in response to pressure overload and myocardial infarction (8–10). In transgenic mice, forced expression of RCAN1 can inhibit both vascularization and metastasis of tumors (5). In humans, RCAN1 protein levels are elevated in the brains of patients with Alzheimer disease (11), and trisomy of RCAN1 in humans is thought to contribute to diverse phenotypes occurring in individuals with Down syndrome (12). Thus, controlling RCAN1 levels may be beneficial in a variety of human pathologies.
To this end, we have sought to determine the molecular mechanisms that control RCAN1 levels. The RCAN1 gene encodes two major protein products, RCAN1–1 and RCAN1–4, that differ only at their N termini (1). Expression of each is initiated from a promoter upstream of an N-terminal exon that is unique to that isoform. Both RCAN1–1 and RCAN1–4 can bind to and inhibit the catalytic activity of calcineurin. Expression of the RCAN1–4 isoform is under the control of calcineurin through a cluster of bindings sites for the calcineurin-responsive transcription factor nuclear factor of activated T cells (NFATs)2 located in an independent promoter upstream of exon 4 (13). Through this mechanism, RCAN1–4 forms a negative feedback loop that helps shape the dynamics of calcineurin signaling and may protect cells from unrestrained calcineurin activity. The family of NFAT transcription factors show a characteristic ability to cooperate with other transcription factors, such as AP1, MEF2, or GATA, in DNA binding and transactivation (14, 15). This property allows integration of calcium signaling with diverse signaling pathways. We therefore hypothesized that NFAT does not act alone at the RCAN1–4 promoter but partners with other transcription factors.
The CCAAT/enhancer binding proteins (C/EBP) are a family of basic leucine zipper transcription factors (16). The bZIP domain in the C terminus mediates dimerization and DNA binding. The N terminus contains both an activation domain and negative regulatory elements that can either repress DNA binding activity or mask the activation domain. C/EBP family members can homo- and heterodimerize with other C/EBP proteins or additional transcription factors such as p300/CBP, CREB, NF-κB, or AP1. C/EBPβ was initially identified as a gene involved in the acute response of the liver but also participates in adipocyte and lymphocyte differentiation, mammary gland development, and osteoblast proliferation (16). Changes in phosphorylation can regulate C/EBPβ-DNA binding activity, including activation by the mitogen-activated protein kinase ERK1/2 (17). C/EBPβ responds to hypoxia during pulmonary remodeling. Hypoxia and hypoxia reperfusion increase its DNA binding activity in heart, lung, and liver (18). Composite NFAT-C/EBPβ enhancer complexes have been characterized in the promoters of several genes including secretory phospholipase A2 (19), the peroxisome proliferator-activated receptor γ2 (PPARγ2) (20), and insulin (21). These diverse examples of NFAT-C/EBPβ interactions make C/EBPβ a potential candidate for cooperating with NFAT in regulation of the RCAN1–4 promoter.
In this study, we used a cross-species comparison of a 500 bp sequence in the proximal promoter of RCAN1–4 to identify the location of numerous conserved C/EBPβ DNA-binding sites that are adjacent to or overlap with conserved NFAT-binding sites. We then demonstrated that C/EBPβ can bind to several sites within this genomic region and cooperate with NFAT to regulate expression of RCAN1–4.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Adenoviruses
C2C12 myoblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum and antibiotics under standard conditions. Cells were transfected using Lipofectamine PLUSTM (Invitrogen). The constitutively active calcineurin (Cn), RCAN1–4, and NFAT expression vectors were described previously (22). The FLAG-tagged C/EBPβ expression plasmids LAP, LAP*, and LIP were a generous gift from Dr. Sheng-Chung Lee (23) along with the short hairpin RNA construct si-C/EBPβ from Dr. Jessica Schwartz (24). The RCAN1.4-Luc reporter plasmid was described previously (13). Luciferase assays were performed using the luciferase assay system (Promega) and normalized to β-galactosidase activity from a cotransfected pCMV-lacZ reporter. Data were derived from a minimum of three independent experiments carried out in duplicate. Error bars indicate S.D. Neonatal rat ventricular myocytes were isolated as described previously (25) and cultured for 2 days in Dulbecco's modified Eagle's medium:M199 (3:1) supplemented with 10% fetal bovine serum then transferred to serum-free medium prior to adenoviral infection. Ad-RCAN1.4-Luc, Ad-CMV-βGal, Ad-Cn, and Ad-MEK1CA were described previously (25, 26) and used at a multiplicity of infection of 100.
Animal Studies
Eight-week-old, male C57BL/6 mice were subjected to pressure overload by severe transverse aortic constriction (sTAC) as described previously (27). Control animals underwent sham operations. The cardiac-specific constitutively active calcineurin transgenic mice (αMHC-CnA*) were described previously (28). The University of Texas Southwestern Animal Care and Use Committee approved all procedures.
Cell Fractionation and Western Blot Analysis
Left ventricles were flash frozen, ground in liquid nitrogen, and then homogenized in lysis buffer containing 10 mm Hepes, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 1 mm dithiothreitol, 0.02% Nonidet P-40, 0.5 mm sodium bisulfite, 4 mm sodium orthovanadate, 100 mm sodium fluoride, and a protease inhibitor mixture (Roche Applied Science) and incubated on ice for 15 min. Lysates were centrifuged for 5 min at 2,000 rpm to obtain nuclear and cytoplasmic fractions. The pellet was resuspended in lysis buffer adjusted to 1% Nonidet P-40 and then centrifuged again to obtain nuclei. The nuclear pellets were resuspended in nuclei lysis buffer containing 20 mm Hepes, pH 7.9, 1.5 mm MgCl2, 420 mm KCl, 0.5 mm dithiothreitol, 25% glycerol, 2 mm EDTA, and protease inhibitors and then vortexed. The nuclear lysate was incubated 4 °C for 45 min on a rotating platform and centrifuged for 10 min at 12,000 rpm, and the supernatant used for EMSA. For Western blot analysis, cells were lysed in a buffer containing 10 mm Tris-HCl, pH 7.5, 4% glycerol, 0.1% Triton X-100, 0.5 mm sodium bisulfite, 4 mm sodium orthovanadate, 100 mm sodium fluoride, and protease inhibitors. The solubilized lysates were run on SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore). Polyclonal RCAN1 antibody was described previously (29). C/EBPβ (sc-150) and glyceraldehyde-3-phosphate dehydrogenase (sc-25778) antibodies were purchased from Santa Cruz Biotechnology. Anti-lamin A/C (catalog no. 2032) was from Cell Signaling.
Immunoprecipitation
C2C12 myoblasts were washed with cold phosphate-buffered saline twice and lysed in ice-cold modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 0.5 mm sodium bisulfite, 1 mm sodium orthovanadate, 1 mm sodium fluoride, and protease inhibitors) then precleared by centrifugation. Soluble extracts were incubated with anti-NFATc1 (Thermo Scientific, MA3-024) overnight at 4 °C. Reaction mixtures were incubated with protein A-agarose (Roche Applied Sciences) at 4 °C for 3 h and then washed three times with modified radioimmune precipitation assay buffer. Immunocomplexes were separated on SDS-PAGE.
Electrophoretic Mobility Shift Assays
EMSA was performed as described previously (24) using 6 μg of nuclear extract (prepared as described above) and 1 μg of poly(dI-dC) (Pierce). The oligonucleotides used to make probes for EMSA are listed in supplemental Table S1. Forward and reverse oligonucleotides were annealed directly or used for amplification of the indicated promoter region by PCR to make double-stranded templates, which were then labeled with [γ-32P]ATP. Extracts and labeled templates were combined and incubated for 20 min at room temperature, and then the protein-DNA complexes were separated on 4% nondenaturing polyacrylamide gels and autoradiographed. In template competition experiments, a 50-fold molar excess of unlabeled template was added to 10 min prior to addition of the labeled template.
Chromatin Immunoprecipitation (ChIP)
Transfected or control C2C12 myoblasts were cross-linked with 1% formaldehyde for 10 min at 37 °C, washed twice with phosphate-buffered saline, harvested in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1), and then sonicated to shear chromatin to an average length of 500 bp (40% output, 15-s continuous pulse for four times). Sonicated samples were centrifuged at 15,000 rpm for 10 min at 4 °C. ChIP assays were performed on the supernatants using an assay kit (Upstate Biotechnology) according to the manufacturer's instructions. DNA-protein complexes were immunoprecipitated with anti-C/EBPβ (Santa Cruz Biotechnology, sc-150) antibody or control IgG. Immunoprecipitated DNA was amplified for semiquantitative PCR with the primers 5′-GCA AGA CAC ATC AGT TGA GG-3′ and 5′-CCT TAG TCA TTT TCC CTA TGC-3′ to detect the mouse Rcan1–4 promoter region. Primers for quantitative real-time PCR were 5′-TGG GAA CTA TGC CGC AAG AG-3′ and 5′-GGT GGA AAA GGC GCT AAG GT-3′ for the Rcan1–4 promoter with 5′-CTG AGC AGG TCA CAA CAG GC-3′ and 5′-GCT ATG AGG AAT GGC TGC AT-3′ for a genomic control from a noncoding region of mouse chromosome 13.
For in vivo experiments, tissue from the left ventricle was finely chopped and transferred into a tube containing 1× phosphate-buffered saline and protease inhibitors. The tissue was fixed with 1% formaldehyde at room temperature for 15 min. Cross-linking was terminated by the addition of glycine to a final concentration of 0.125 m. The cross-linked tissue was washed with phosphate-buffered saline twice, resuspended in 6× volume of cell lysis buffer (5 mm PIPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40, plus protease inhibitors) and incubated on ice for 15 min. The tissue was homogenized in a Dounce homogenizer then centrifuged at 2,000 rpm for 5 min. The pellet was resuspended in nuclei lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS, plus protease inhibitors) and incubated on ice for 20 min. DNA was sheared, immunoprecipitated, and analyzed as above.
Quantitative Real-time PCR Quantification of mRNA
Total RNA was isolated using Tripure reagent (Roche Applied Science) and treated with DNase (Roche Applied Science). Random hexamers (Roche Applied Science) and SuperScript II (Invitrogen) were used for reverse transcription reactions following the manufacturer's protocols. Real-time PCR was performed using SYBR green on an ABI7000 cycler (Applied Biosystems). Data for each transcript were normalized to cyclophilin 1 transcripts as an internal control using the ΔΔCT method.
RESULTS
The RCAN1–4 Promoter Contains Numerous Conserved C/EBPβ-binding Sites
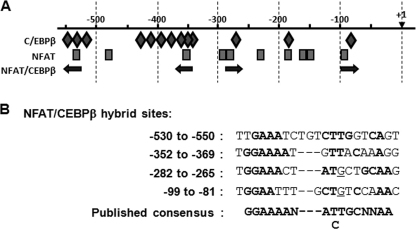
We postulated that other transcription factors act in conjunction with NFAT at the RCAN1–4 promoter. Using MatInspectorTM, we previously identified fifteen potential NFAT-binding sites in the human genomic sequence 1,000 bp upstream of the RCAN1–4 start of transcription (13). NFAT binding to several of these sites has been verified in both the human and mouse promoter (30, 31). The MatInspectorTM program also indicated the presence of 16 putative C/EBPβ-binding sites (data not shown). No other DNA-binding sites were as abundant as these two in the MatInspectorTM analysis. A cross-species genomic sequence alignment was carried out using the Evolutionary Conserved Regions browser (32) to determine which of the NFAT and C/EBPβ sites are conserved. Comparison of the human sequence to that of dog, mouse, rat, and frog indicated high conservation within the 550 bp immediately upstream of the start of transcription. An rVISTA search for DNA-binding sites conserved between the mouse and human sequence indicated 10 potential NFAT DNA-binding sites and 13 potential C/EBPβ DNA-binding sites conserved between the two species (Fig. 1A and supplemental Fig 1). Four of the C/EBPβ sites were located adjacent to NFAT sites, with configurations similar to composite NFAT-C/EBPβ DNA-binding sites previously identified in the promoters of other genes (Fig. 1B) (20). Three of the C/EBPβ sites overlapped directly with NFAT sites, and several of the C/EBPβ sites were not in the immediate vicinity of any of the conserved NFAT sites. Interestingly, none of the NFAT binding sites had a configuration typical of a composite NFAT/AP1 site (GGAAAaxxxTGAxTCA) (33) suggesting that AP1 may not participate in controlling expression of this calcineurin/NFAT-regulated gene.
FIGURE 1.
The RCAN1–4 promoter region contains several potential NFAT-C/EBPβ hybrid binding sites. A, schematic of C/EBPβ- and NFAT-binding sites conserved between human and mouse. C/EBPβ sites are indicated by diamonds, and NFAT sites are rectangles. Potential NFAT-C/EBPβ hybrid sites are indicated by arrows. B, comparison of the nucleotide sequences of the four putative NFAT-C/EBPβ hybrid-binding sites to the published consensus sequence. The start of transcription is indicated as +1.
C/EBPβ Can Activate Expression of RCAN1–4
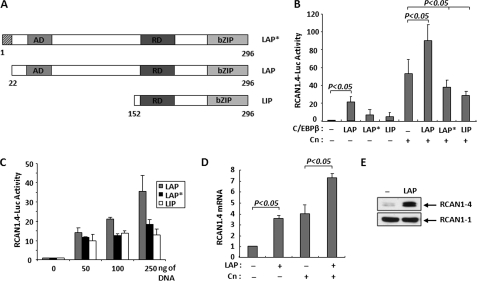
There are three different variants of C/EBPβ, generated by alternative translation initiation of the same mRNA transcript (34) (Fig. 2A). The 35-kDa LAP isoform is primarily a transcriptional activator. The 21-kDa LIP isoform inhibits transcription, whereas the 38-kDa LAP* isoform can either activate or repress transcription depending upon context (23). To test whether C/EBPβ can regulate RCAN1–4 expression, C2C12 myoblasts were co-transfected with a reporter plasmid carrying the human RCAN1–4 promoter driving luciferase expression (RCAN1.4-Luc), a control CMV-βGal reporter, and vectors expressing each of the individual C/EBPβ isoforms. The LAP isoform produced a more than a 20-fold increase in reporter activity, whereas the LIP and LAP* isoforms resulted in more modest increases (Fig. 2B). Co-transfection with a vector expressing constitutively active calcineurin (Cn) increased activity of the RCAN1.4-Luc reporter as published previously (13). Activation by calcineurin and LAP together was additive. Conversely, co-transfection with either LAP* or LIP expression vectors decreased calcineurin activation of the reporter (Fig. 2B). Activation by LAP was dose-dependent (Fig. 2C), whereas activation by LIP and LAP* was not.
FIGURE 2.
C/EBPβ activates RCAN1–4 expression. A, structure of LAP*, LAP, and LIP, indicating DNA-binding (bZIP), regulatory (RD), and activation (AD) domains. B, C2C12 myoblasts were cotransfected with the RCAN1.4-Luc reporter, and expression plasmids encoding LAP, LAP*, or LIP and constitutively activated Cn. Activity is presented as fold activation over the RCAN1.4-Luc reporter alone. C, co-transfection with increasing concentrations of LAP, LAP*, or LIP expression plasmids was used to assess the dose response of the RCAN1.4-Luc reporter. D, C2C12 myoblasts were transfected with LAP and Cn as indicated. Endogenous Rcan1–4 mRNA levels were quantified and normalized to 18 S RNA transcript levels 24 h later. E, Western blot analysis of endogenous RCAN1–4 protein in C2C12 myoblasts 24 h after transfection with a control vector (−) or one expressing LAP. The RCAN1–1 isoform is provided as a loading control. All transfections were carried out in duplicate in a minimum of three independent experiments. Error bars indicate S.D.
To test whether C/EBPβ could also regulate expression of the endogenous mouse Rcan1–4 gene within the context of native chromatin, C2C12 myoblasts were transfected with LAP and calcineurin expression vectors. Changes in Rcan1–4 transcript levels were quantified by real-time PCR. Transfection with either LAP or Cn alone increased the level of endogenous Rcan1.4 transcripts roughly 4-fold over control transfected cultures (Fig. 2D). Similar to our results with the RCAN1.4-Luc reporter, the effect of LAP and Cn was additive. There was a corresponding increase in endogenous RCAN1–4 protein levels in LAP transfected cells (Fig. 2E), consistent with the observed changes in transcript levels. These studies demonstrate that C/EBPβ can promote expression from both an ectopic RCAN1.4-Luc reporter and the endogenous gene in its native chromatin configuration.
Depletion of Endogenous C/EBPβ Reduces Calcineurin-dependent Activation of RCAN1.4 Expression
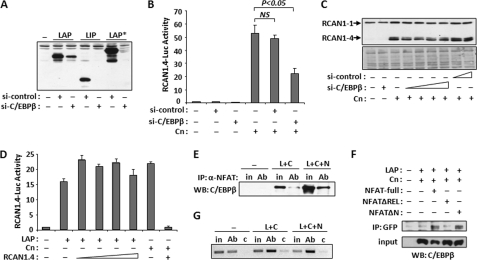
To test whether C/EBPβ and calcineurin/NFAT activation of RCAN1–4 transcription are interdependent, we used a short hairpin siRNA directed against C/EBPβ (si-C/EBPβ) to deplete endogenous C/EBPβ protein from C2C12 cells. Si-C/EBP is capable of depleting all three isoforms as demonstrated in Fig. 3A. Neither si-C/EBPβ nor a scrambled control siRNA (si-control) altered the baseline activity of the RCAN1.4-Luc reporter in C2C12 myoblasts (Fig. 3B). The control siRNA also did not alter activation of the RCAN1.4-Luc reporter by calcineurin, however, depletion of endogenous C/EBPβ by cotransfection with si-C/EBPβ reduced calcineurin activation of the RCAN1.4-Luc reporter (Fig. 3B) suggesting that activation of RCAN1–4 expression by the calcineurin/NFAT pathway is in part dependent upon the presence of endogenous C/EBPβ in C2C12 myoblasts under these conditions. Consistent with these results, si-C/EBPβ also reduced calcineurin-dependent increases in RCAN1–4 protein levels (Fig. 3C). Importantly, si-C/EBPβ reduced, but did not abolish, calcineurin-dependent increases in RCAN1–4 expression and protein levels. These results suggest that there may be both C/EBPβ-dependent and C/EBPβ-independent aspects to calcineurin/NFAT control of RCAN1–4 expression.
FIGURE 3.
C/EBPβ and NFATc1 form a complex and can bind the Rcan1–4 promoter. A, Western blot analysis showing depletion of LAP, LIP, and LAP* when cotransfected with a control (si-control) or C/EBPβ-specific (si-C/EBPβ) siRNA. B, luciferase activity was measured 24 h after co-transfection with RCAN1.4-Luc, si-control, si-C/EBPβ, and Cn. C, Western blot analysis for RCAN1 24 h after co-transfection with Cn and increasing amounts of si-control or si-C/EBPβ. Lower panel, Ponceau S staining of total protein. D, luciferase activity was assayed 24 h after transfection of C2C12 myoblasts with RCAN1.4-Luc, LAP, Cn, and increasing amounts of RCAN1.4 expression plasmid. E, C2C12 myoblasts were co-transfected with control vector (−), LAP and Cn (L+C), or LAP and Cn and NFATc1 (L+C+N) expression vectors. NFATc1-specific antibodies (Ab) were used for immunoprecipitation (IP), and the complexes were probed for the presence of C/EBPβ by Western blot (WB). Input samples (in) were run alongside for comparison (F). C2C12 myoblasts were transfected with plasmids encoding full-length NFATc1 fused to GFP (NFAT-full), amino acids 219–716 fused to GFP (NFATΔN) or 1–460 fused to GFP (NFATΔREL) along with C/EBPβ. Anti-GFP was used for co-immunoprecipitation and then probed for the presence of C/EBPβ. G, C2C12 myoblasts were transfected as in E. Protein-DNA complexes were precipitated with anti-C/EBPβ (Ab) or control IgG (c) and then assayed for the presence of the Rcan1–4 promoter by PCR. An input sample was used as a control. All experiments were carried out in triplicate. Error bars indicate S.D. NS, not significant.
Activation of RCAN1–4 Expression by C/EBPβ Is Not Dependent on Calcineurin Activity
To test whether activation of RCAN1–4 expression by C/EBPβ requires activation of calcineurin/NFAT, we carried out the reciprocal experiment. C2C12 myoblasts were co-transfected with the RCAN1.4-Luc reporter plasmid and a LAP expression vector with and without increasing levels of a plasmid expressing the RCAN1–4 protein to inhibit calcineurin activity. Activation of the RCAN1.4-Luc vector by LAP was not reduced by co-transfection with an RCAN1–4 expression vector (Fig. 3D), whereas the RCAN1–4 expression plasmid completely inhibited activation of the RCAN1.4-Luc reporter by calcineurin, verifying that the protein produced by the RCAN1–4 expression construct was capable of inhibiting calcineurin in these experiments (Fig. 3D). Taken together, these results suggest that, under these conditions, activation of the RCAN1–4 promoter by C/EBPβ is independent of the calcineurin/NFAT, whereas activation by calcineurin is at least in part dependent upon C/EBPβ.
C/EBPβ Binds Both NFATc1 and the RCAN1–4 Promoter
Co-immunoprecipitation was used to test for a direct interaction between C/EBPβ and NFAT proteins. C2C12 myoblasts were transfected with vectors encoding the LAP isoform of C/EBPβ and the NFATc1 isoform of NFAT. Co-transfection with Cn was included to ensure translocation of NFATc1 to the nucleus. Anti-NFATc1 antibodies were used for immunoprecipitation and the precipitated complexes and then probed for the presence of C/EBPβ protein by Western blot. A small amount of C/EBPβ protein co-immunoprecipitated with endogenous NFATc1 (Fig. 3E, lane 4). Both input and co-immunoprecipitated C/EBPβ protein increased in the presence of transfected NFATc1 (Fig. 3E, lane 6). To map the region of NFATc1 that interacts with C/EBPβ, we used NFATc1-GFP fusion constructs with N and C-terminal deletions of regions of NFATc1. C/EBPβ co-immunoprecipitated with full-length NFATc1-GFP and an N-terminal deletion (NFATΔN) containing amino acids 219–716 fused to GFP (Fig. 3F). C/EBPβ did not co-immunoprecipitate with NFATc1 when the C terminus, containing the REL DNA-binding/protein dimerization domain, was deleted (NFATΔREL). These findings are consistent with earlier reports demonstrating the importance of the REL domain of NFATc4 for interaction with C/EBP proteins (20).
ChIP was used to verify C/EBPβ binding to the endogenous mouse Rcan1–4 promoter. C2C12 myoblasts were transfected with empty control vector or vectors encoding LAP, NFATc1, and Cn. Anti-C/EBPβ or control IgG was used to immunoprecipitate protein-DNA complexes. The endogenous Rcan1–4 promoter was present in the ChIP complexes selected by immunoprecipitation of endogenous C/EBPβ (Fig. 3G, lane 2) but not when control IgG was used in the assay (Fig. 3G, lane 3). C/EBPβ binding to the Rcan1–4 promoter increased when cells were transfected with vectors to express excess LAP and Cn (Fig. 3G, lane 5).
C/EBPβ Binds Multiple Sites in the RCAN1–4 Promoter
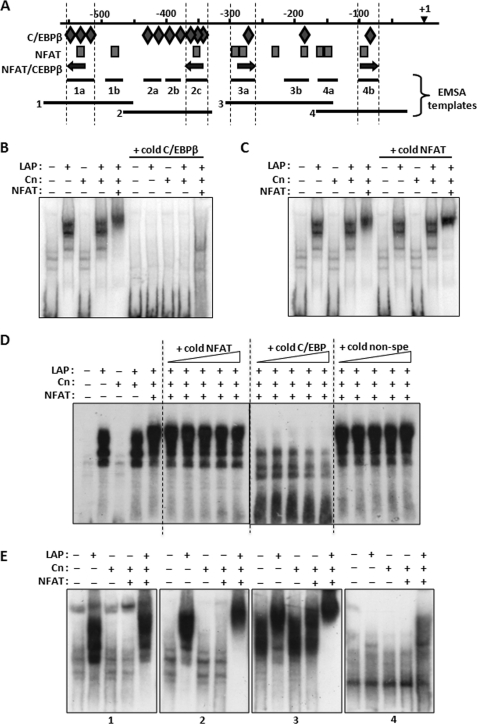
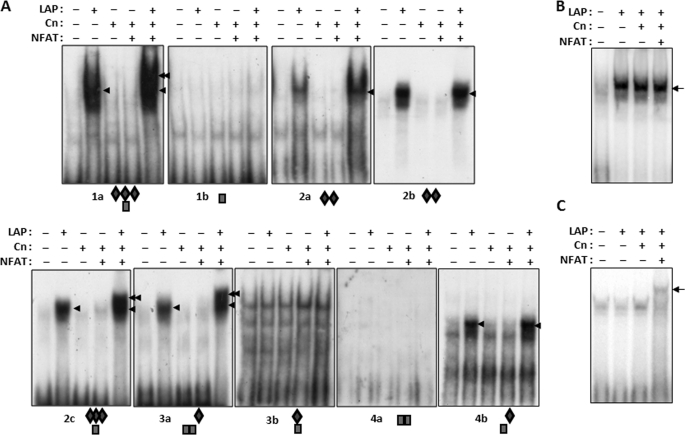
Although the ChIP assay demonstrates that C/EBPβ binds to the mouse Rcan1–4 promoter somewhere within a 500-bp region flanking the PCR probes used to detect the target, it does not identify the specific location of binding or determine the number of binding sites. Therefore, EMSAs were used to determine which of the 13 conserved C/EBPβ DNA-binding sites in the RCAN1-4 promoter were capable of binding C/EBPβ. Initially, we tested four different templates (1, 2, 3, and 4) that subdivided the RCAN1–4 promoter region from −26 to −580 (Fig. 4A). Each of these four templates contained a variety of putative C/EBPβ and NFAT sites including one each of the potential hybrid NFAT/C/EBPβ-binding sites. Template 2, spanning −465 to −329, contained the highest density of C/EBPβ sites. EMSAs using nuclear extracts from C2C12 cells transfected with a LAP expression vector indicated LAP-dependent binding to this template (Fig. 4B). Extracts from cells transfected with Cn alone did not shift the template and the combination of Cn and LAP produced a pattern similar to that of LAP alone suggesting that calcineurin activity is not required for LAP to bind free DNA template. Co-transfection with LAP, NFATc1, and Cn together supershifted the LAP EMSA pattern suggesting that NFATc1 participates in the LAP-DNA complex on template 2. Binding was competed away with a cold double-stranded oligomer containing a C/EBPβ consensus template verifying the presence of C/EBPβ in the EMSA complex (Fig. 4B). A cold NFAT consensus template did not compete away the EMSA complex (Fig. 4, C and D) suggesting that either NFATc1 does not need to bind DNA to participate in the complex or the C/EBPβ-NFATc1 complex binds to template 2 with a higher affinity than to the NFAT consensus template.
FIGURE 4.
C/EBPβ binds multiple sites on the RCAN1–4 promoter. A, a schematic of the location of the various templates used for EMSA experiments is shown. B and C, EMSAs were carried out using template 2, and nuclear extracts from C2C12 myoblasts were transfected with LAP, Cn, and NFATc1 expression vectors. 40-fold excess unlabeled template containing a consensus binding site specific for C/EBPβ (B) or for NFAT (C) was used to compete for binding. D, gradients of excess cold C/EBPβ, NFAT, and nonspecific (non-spe) template (ranging from 20 to 500-fold) were used to compete for binding to template 2. E, EMSA analysis was carried out using fragments 1, 2, 3, and 4 as templates.
Each of the four RCAN1–4 templates were tested in turn and found to form C/EBPβ-dependent EMSA complexes. Transfection with LAP, Cn, and NFATc1 combined supershifted the complex relative to LAP alone on templates 1, 2, and 3 but not on template 4 (Fig. 4E). Each of these binding activities could be competed away with excess unlabeled C/EBPβ consensus template but not by a cold NFAT consensus template (data not shown). Fine mapping of the RCAN1–4 promoter was carried out using smaller templates containing various combinations of C/EBPβ- and NFAT-binding sites as indicated by the diamonds and rectangles below each gel in Fig. 5. All of the templates containing putative C/EBPβ binding sites showed positive binding for LAP, except for 3b (shown on Fig. 5), which shifted under all conditions. The EMSA data suggest that there are a minimum of six sites capable of binding C/EBPβ within 600 bp of the RCAN1–4 start of transcription. Neither of the fragments containing NFAT sites alone (Fig. 5, 1b and 4a) formed complexes, even when cells were transfected with excess NFATc1. It may be that these are not true NFAT-binding sites or that NFAT requires the presence of some other transcription factor binding partner to act at these sites. Three of the four putative hybrid NFAT-C/EBPβ sites (Fig. 5, 1a, 2c, and 3a) were supershifted when the cells were co-transfected with NFATc1 in addition to LAP and Cn (indicated by the double arrows in Fig. 5), suggesting formation of an NFAT-C/EBPβ-DNA complex at these sites.
FIGURE 5.
C/EBPβ can bind to the RCAN1–4 promoter alone and at NFAT-C/EBPβ hybrid sites. A, the RCAN1–4 promoter was further subdivided and assayed by EMSA using nuclear extracts prepared from transfected C2C12 myoblasts. The single arrowhead indicates the location of the C/EBPβ complex, whereas the double arrowhead indicates the location of a potential NFAT-C/EBPβ complex. Conserved C/EBPβ (B) or NFAT (C) consensus binding sites were used as templates in EMSA using nuclear extracts from transfected C2C12 myoblasts.
The pattern of EMSA complex formation suggests that a hybrid NFAT-C/EBPβ-binding site is required for formation of the NFAT-C/EBPβ-DNA supershifted complex. To verify that protein-protein interactions between NFAT and C/EBPβ are not sufficient to form a complex at an isolated NFAT- or C/EBPβ-binding site, EMSAs were carried out using consensus binding templates for C/EBPβ (Fig. 5B) or NFAT (Fig. 5C) alone. A LAP-dependent complex was formed on the C/EBPβ and an NFAT-dependent complex formed on the NFAT template; however, an NFAT-C/EBPβ complex did not form on either template suggesting that a hybrid-binding site is required for this regulatory interaction.
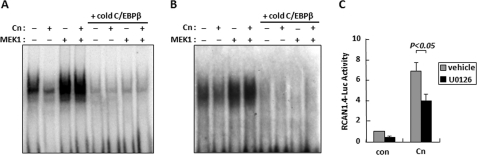
Activation of MEK1 Promotes Binding of C/EBPβ to the RCAN1–4 Promoter in Cardiomyocytes
C/EBPβ DNA binding activity increases in response to activation of the MEK1/ERK1/2 MAPK signaling cascade (17). We tested the ability of MEK1 to promote C/EBPβ DNA binding and activate the RCAN1–4 promoter in cultured NRCMs. Nuclear extracts for EMSA were prepared from NRCMs infected with either a control Ad-CMV-βGal adenovirus or ones expressing constitutively active forms of calcineurin (Ad-Cn) and MEK1 (Ad-MEK1CA). Infection with Ad-MEK1CA increased DNA binding activity toward a C/EBPβ consensus template verifying the ability of the MEK1/ERK1/2 pathway to activate C/EBPβ in NRCMs (Fig. 6A). Infection with Ad-MEK1CA also increased DNA binding activity toward template number 2 from the human RCAN1–4 promoter (Fig. 6B). This activity was competed away with an excess of cold C/EBPβ consensus template, suggesting that the positive EMSA signal in the Ad-MEK1CA-infected samples was due to MEK1 activation of C/EBPβ rather than some other MEK1-reponsive transcription factor.
FIGURE 6.
MEK1 activity promotes C/EBPβ binding to the RCAN1–4 promoter. EMSA was performed with nuclear extracts from neonatal rat ventricular myocytes infected with a Cn and/or constitutively active MEK1 adenovirus as indicated. A C/EBPβ consensus site (A) or template 2 (B) were used for EMSA. Excess cold C/EBPβ consensus binding template was used as a competitor in both experiments. C, neonatal rat ventricular myocytes were infected with Ad-RCAN1.4-Luc along with control adenoviruses or Ad-Cn as indicated. Two days after infection, cultures were treated with dimethyl sulfoxide (vehicle) or the MEK1 inhibitor U0126 (10 μm) for 4 h and then assayed for luciferase activity. Transfections were carried out in triplicate. Error bars indicate S.D. con, control.
To test the ability of MEK1 to activate transcription from the RCAN1.4 promoter, NRCMs were infected with the Ad-RCAN1.4-Luc reporter along with Ad-CVM-βGal as a control for normalization. As expected, co-infection with constitutively active calcineurin activated the RCAN1.4-Luc reporter (Fig. 6C). A 4-h incubation with the ERK1/2 inhibitor U0126 reduced calcineurin-dependent activation, suggesting that maximal activation of the RCAN1.4-Luc by calcineurin/NFAT requires parallel activation of the MEK1/ERK1/2 pathway. These results suggest a potential mechanism for cross-talk between calcineurin and MAPK signaling cascades.
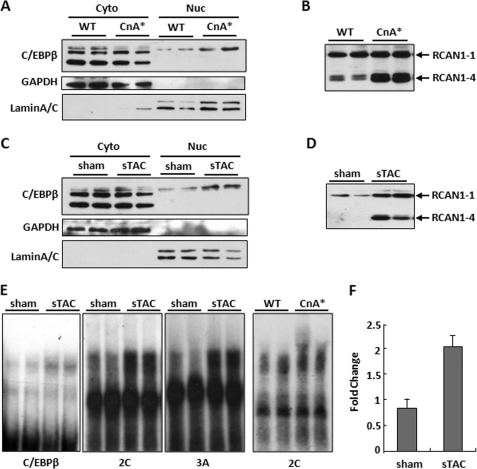
C/EBPβ DNA Binding Activity Increases in Pressure Overload-induced Heart Failure
Two different mouse models of heart failure were used to look for changes in nuclear localization and/or DNA binding activity of C/EBPβ. There was an increase in nuclear localized C/EBPβ protein in mice carrying a cardiac-specific calcineurin transgene (αMHC-CnA*) (Fig. 7A). These mice develop cardiac hypertrophy that progresses to failure (28). There was a similar increase in nuclear C/EBPβ protein levels in the hearts of mice subjected to sTAC (Fig. 7C). Mice subjected to sTAC develop decompensated hypertrophic heart failure (27). We verified that RCAN1.4 protein levels were elevated in both models as reported previously (Fig. 7, B and D). To test for activation of C/EBPβ DNA binding activity in the setting of pressure overload-induced heart failure, nuclear extracts were made from the left ventricles of sham-operated controls and sTAC hearts. DNA binding activity was increased toward both a consensus C/EBPβ-binding site and templates 2C and 3A from the RCAN1–4 promoter (Fig. 7E). Likewise, DNA binding activity also increased in hearts from αMHC-CnA* transgenic mice compared with DNA binding activity in hearts of wild type siblings (Fig. 7E). Furthermore, chromatin immunoprecipitation demonstrated increased binding of C/EBPβ to the endogenous mouse Rcan1–4 promoter in sTAC hearts compared with control sham-operated hearts (Fig. 7F). Taken as a whole, these findings indicate that C/EBPβ is activated in pressure overload-induced heart failure in mice and participates in increasing expression of Rcan1–4.
FIGURE 7.
C/EBPβ DNA binding activity increases in hypertrophic failing hearts. Nuclear (Nuc) and cytoplasmic (Cyto) protein fractions were isolated from the hearts of transgenic mice with cardiac-specific expression of a constitutively active calcineurin transgene (CnA*) and wild type (WT) littermates (A) or from sham operated and sTAC mice 1 week after banding (C). Western blot analysis for C/EBPβ indicated an increase in nuclear C/EBPβ in both models of heart failure. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lamin A/C were used to verify enrichment of cytoplasmic and nuclear fractions, respectively. B and D, Western blot analysis for RCAN1 shows increased levels of the RCAN1–4 isoform in both models. E, nuclear extracts from sham sTAC and CnA* hearts were used for EMSA using the C/EBPβ consensus template, 2C, or 3A from the human RCAN1–4 promoter. F, ChIP assays using C/EBPβ-specific antibodies indicate increased occupancy of the Rcan1–4 promoter in the hearts of mice subjected to sTAC compared with sham controls. Error bars indicate S.D.
DISCUSSION
Feedback inhibition of calcineurin by RCAN1–4 is activated during diverse adaptive processes, helping to shape immune responses, synaptic plasticity, angiogenesis, and cardiac remodeling. Therefore, mechanisms that act in conjunction with NFAT to control RCAN1–4 expression have the potential to influence a broad range of biological systems. This study provides evidence that the transcription factor C/EBPβ cooperates with NFAT to regulate expression of the calcineurin regulatory protein RCAN1–4. EMSA assays demonstrate that C/EBPβ is capable of binding to several conserved sites in the RCAN1–4 promoter. Several of these sites are composite NFAT-C/EBPβ-binding sites, whereas others are not, suggesting that C/EBPβ may cooperate with NFAT at some sites and act independently at others. Immunoprecipitation experiments verify binding of C/EBPβ both to NFATc1 and to the endogenous mouse Rcan1–4 promoter. Depletion of C/EBPβ indicates that maximal activation of RCAN1–4 expression by calcineurin/NFAT is at least in part dependent on the presence of C/EBPβ, whereas C/EBPβ activation of an RCAN1–4-Luc reporter plasmid is independent of calcineurin activity. Thus, it appears that C/EBPβ can both cooperate with NFAT to facilitate feedback inhibition of calcineurin activity and may also serve as an independent pathway by which RCAN1–4 expression can be controlled and calcineurin signaling inhibited.
Our findings identify RCAN1–4 as a point of interaction between MAPK- and calcineurin-dependent intracellular signaling cascades. MEK1, an activator of C/EBPβ DNA binding activity, increases RCAN1–4 expression and binding of C/EBPβ to the promoter, whereas an MEK1 inhibitor suppresses calcineurin activation of RCAN1–4 expression. In mouse models of heart failure, nuclear translocation of C/EBPβ increases, as does its occupancy of the Rcan1–4 promoter, demonstrating that C/EBPβ participates in control of Rcan1–4 expression in vivo during cardiac stress. In response to increased workload, the heart undergoes compensatory hypertrophic growth. We postulate that MEK1 activity may help protect the heart via C/EBPβ-dependent activation of RCAN1–4 expression, thus limiting calcineurin-dependent hypertrophic growth of cardiomyocytes.
Typically, promoters are bound by a complex array of proteins encompassing basal transcription factors along with regulated factors, both positive and negative, which together confer control over transcriptional activity. Therefore, it is likely that many other transcription factors, in addition to NFAT and C/EBPβ, influence RCAN1–4 expression either directly or indirectly. To date, there is evidence of RCAN1–4 regulation by GATA-binding proteins (30), activating transcription factor 6 (ATF6) (35), and early growth response protein 1 (EGR-1) (36). Interestingly, there is precedence for each of these factors to cooperate with C/EBPβ. There is evidence for a protein kinase A-dependent interaction between GATA and C/EBPβ (37). ATF6 and C/EBPβ are activated in response to endoplasmic reticulum stress and can cooperate in controlling expression of a number of genes (38). In the case of EGR-1, MEK1-dependent interaction between Egr-1 and C/EBPβ leads to activation of the low density lipoprotein receptor gene (39). Although the proposed Egr-1 binding site on the human RCAN1–4 promoter is not conserved in rodents, it is possible that Egr-1 acts indirectly on RCAN1.4 expression via a protein-protein interaction with C/EBPβ as it does on the low density lipoprotein receptor promoter (39). Thus, GATA, ATF6, Egr-1, and C/EBPβ may act in concert or individually with NFAT proteins to shape the magnitude and duration of a calcineurin-mediated signal.
The major immunosuppressive drugs cyclosporine A and FK506 act by targeting calcineurin. Because of their toxic side effects, there has been substantial interest in pursuing the RCAN family of proteins as alternative pathways for controlling calcineurin activity. Unfortunately, although current small molecule screens have yielded compounds that activate RCAN1–4 expression, these compounds act via the calcineurin/NFAT pathway (29), with the net result likely being increased calcineurin activity rather than suppression. Based on the findings presented here, the involvement of C/EBPβ may provide a calcineurin-independent mechanism through which endogenous RCAN1–4 expression can be enhanced or suppressed to control calcineurin-dependent processes that are involved in a broad range of developmental and adaptive responses.
Supplementary Material
This work was supported, in whole or in part, by National Institute of Health Grants HL072016 and HL097768 (to B. A. R.) and HL075173, HL080144, and HL090842 (to J. A. H.). This work was also supported by American Heart Association Grants 0655202Y (to B. A. R.), 0970518N, and 0640084N (to J. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. 1.
- NFAT
- nuclear factor of activated T cells
- ERK
- extracellular signal-regulated kinase
- si
- small interfering
- Luc
- luciferase
- Ad
- adenovirus
- sTAC
- severe transverse aortic constriction
- EMSA
- electrophoretic mobility shift assay
- ChIP
- chromatin immunoprecipitation
- PIPES
- 1,4-piperazinediethanesulfonic acid
- GFP
- green fluorescent protein
- MAPK
- mitogen-activated protein kinase
- ATF6
- activating transcription factor 6
- EGR-1
- early growth response protein 1.
REFERENCES
- 1.Rothermel B. A., Vega R. B., Williams R. S. (2003) Trends Cardiovasc. Med. 13, 15–21 [DOI] [PubMed] [Google Scholar]
- 2.Ryeom S., Greenwald R. J., Sharpe A. H., McKeon F. (2003) Nat. Immunol. 4, 874–881 [DOI] [PubMed] [Google Scholar]
- 3.Hoeffer C. A., Dey A., Sachan N., Wong H., Patterson R. J., Shelton J. M., Richardson J. A., Klann E., Rothermel B. A. (2007) J. Neurosci. 27, 13161–13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riper D. V., Jayakumar L., Latchana N., Bhoiwala D., Mitchell A. N., Valenti J. W., Crawford D. R. (2008) Arch. Biochem. Biophys. 472, 43–50 [DOI] [PubMed] [Google Scholar]
- 5.Baek K. H., Zaslavsky A., Lynch R. C., Britt C., Okada Y., Siarey R. J., Lensch M. W., Park I. H., Yoon S. S., Minami T., Korenberg J. R., Folkman J., Daley G. Q., Aird W. C., Galdzicki Z., Ryeom S. (2009) Nature 459, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryeom S., Baek K. H., Rioth M. J., Lynch R. C., Zaslavsky A., Birsner A., Yoon S. S., McKeon F. (2008) Cancer Cell 13, 420–431 [DOI] [PubMed] [Google Scholar]
- 7.Vega R. B., Rothermel B. A., Weinheimer C. J., Kovacs A., Naseem R. H., Bassel-Duby R., Williams R. S., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 669–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill J. A., Rothermel B., Yoo K. D., Cabuay B., Demetroulis E., Weiss R. M., Kutschke W., Bassel-Duby R., Williams R. S. (2002) J. Biol. Chem. 277, 10251–10255 [DOI] [PubMed] [Google Scholar]
- 9.Rothermel B. A., McKinsey T. A., Vega R. B., Nicol R. L., Mammen P., Yang J., Antos C. L., Shelton J. M., Bassel-Duby R., Olson E. N., Williams R. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3328–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E., Doevendans P. A., Crijns H. J., Heeneman S., Lips D. J., van Bilsen M., Williams R. S., Olson E. N., Bassel-Duby R., Rothermel B. A., De Windt L. J. (2004) Circ. Res. 94, e18–26 [DOI] [PubMed] [Google Scholar]
- 11.Harris C. D., Ermak G., Davies K. J. (2007) FEBS J 274, 1715–1724 [DOI] [PubMed] [Google Scholar]
- 12.Park J., Oh Y., Chung K. C. (2009) BMB Rep. 42, 6–15 [DOI] [PubMed] [Google Scholar]
- 13.Yang J., Rothermel B., Vega R. B., Frey N., McKinsey T. A., Olson E. N., Bassel-Duby R., Williams R. S. (2000) Circ. Res. 87, E61–68 [DOI] [PubMed] [Google Scholar]
- 14.Macian F. (2005) Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 15.Rao A., Luo C., Hogan P. G. (1997) Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 16.Nerlov C. (2007) Trends Cell Biol. 17, 318–324 [DOI] [PubMed] [Google Scholar]
- 17.Zhu S., Yoon K., Sterneck E., Johnson P. F., Smart R. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S. F., Zou Y. S., Mendelsohn M., Gao Y., Naka Y., Du Yan S., Pinsky D., Stern D. (1997) J. Biol. Chem. 272, 4287–4294 [DOI] [PubMed] [Google Scholar]
- 19.Yang T. T., Ung P. M., Rincón M., Chow C. W. (2006) J. Biol. Chem. 281, 11541–11552 [DOI] [PubMed] [Google Scholar]
- 20.Yang T. T., Chow C. W. (2003) J. Biol. Chem. 278, 15874–15885 [DOI] [PubMed] [Google Scholar]
- 21.Lawrence M. C., McGlynn K., Park B. H., Cobb M. H. (2005) J. Biol. Chem. 280, 26751–26759 [DOI] [PubMed] [Google Scholar]
- 22.Rothermel B., Vega R. B., Yang J., Wu H., Bassel-Duby R., Williams R. S. (2000) J. Biol. Chem. 275, 8719–8725 [DOI] [PubMed] [Google Scholar]
- 23.Su W. C., Chou H. Y., Chang C. J., Lee Y. M., Chen W. H., Huang K. H., Lee M. Y., Lee S. C. (2003) J. Biol. Chem. 278, 51150–51158 [DOI] [PubMed] [Google Scholar]
- 24.Cui T. X., Piwien-Pilipuk G., Huo J. S., Kaplani J., Kwok R., Schwartz J. (2005) Mol. Endocrinol. 19, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 25.Ni Y. G., Berenji K., Wang N., Oh M., Sachan N., Dey A., Cheng J., Lu G., Morris D. J., Castrillon D. H., Gerard R. D., Rothermel B. A., Hill J. A. (2006) Circulation 114, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicol R. L., Frey N., Pearson G., Cobb M., Richardson J., Olson E. N. (2001) EMBO J. 20, 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothermel B. A., Berenji K., Tannous P., Kutschke W., Dey A., Nolan B., Yoo K. D., Demetroulis E., Gimbel M., Cabuay B., Karimi M., Hill J. A. (2005) Physiol. Genomics 23, 18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. (1998) Cell 93, 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush E., Fielitz J., Melvin L., Martinez-Arnold M., McKinsey T. A., Plichta R., Olson E. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2870–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minami T., Horiuchi K., Miura M., Abid M. R., Takabe W., Noguchi N., Kohro T., Ge X., Aburatani H., Hamakubo T., Kodama T., Aird W. C. (2004) J. Biol. Chem. 279, 50537–50554 [DOI] [PubMed] [Google Scholar]
- 31.Wu H., Kao S. C., Barrientos T., Baldwin S. H., Olson E. N., Crabtree G. R., Zhou B., Chang C. P. (2007) J. Biol. Chem. 282, 30673–30679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M. (2002) Genome Res. 12, 832–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kel A., Kel-Margoulis O., Babenko V., Wingender E. (1999) J. Mol. Biol. 288, 353–376 [DOI] [PubMed] [Google Scholar]
- 34.Descombes P., Schibler U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 35.Belmont P. J., Tadimalla A., Chen W. J., Martindale J. J., Thuerauf D. J., Marcinko M., Gude N., Sussman M. A., Glembotski C. C. (2008) J. Biol. Chem. 283, 14012–14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schabbauer G., Schweighofer B., Mechtcheriakova D., Lucerna M., Binder B. R., Hofer E. (2007) Thromb. Haemost. 97, 988–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tremblay J. J., Hamel F., Viger R. S. (2002) Endocrinology 143, 3935–3945 [DOI] [PubMed] [Google Scholar]
- 38.Donati G., Imbriano C., Mantovani R. (2006) Nucleic Acids Res. 34, 3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F., Lin M., Abidi P., Thiel G., Liu J. (2003) J. Biol. Chem. 278, 44246–44254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.