Abstract
The transcription factor ZAC1 is expressed in a variety of tissues including the developing heart, but its physiological role is unclear. We examined the role of ZAC1 in regulating expression of the insulin-responsive glucose transporter GLUT4 and whether ZAC1 expression is altered in cardiomyocyte hypertrophy. We demonstrated expression of Zac1 mRNA and protein in rat cardiomyocytes by PCR and Western blotting, respectively. Using a combination of chromatin immunoprecipitation and luciferase assays, we showed that ZAC1 regulates Glut4 expression via a specific binding site in the Glut4 promoter. Overexpression of ZAC1 increased Glut4 mRNA and protein expression and resulted in increased glucose uptake in cardiomyocytes as determined by a fluorescent analog uptake assay. Induction of hypertrophy by phenylephrine or isoproterenol resulted in increased Zac1 expression. We identified a novel putative promoter in the Zac1 gene and demonstrated increased binding of MEF2 to this promoter in response to hypertrophic stimulation. MEF2 regulated transactivation of the Zac1 promoter and ZAC1 protein expression. This work identifies ZAC1 as a novel and previously unknown regulator of cardiomyocyte Glut4 expression and glucose uptake. Our results also implicate MEF2 as a regulator of ZAC1 expression in response to induction of hypertrophy.
Keywords: Cardiac Hypertrophy, Gene Regulation, Glucose Transport, Transcription Factors, Transcription Regulation
Introduction
The heart is a metabolic omnivore, using various substrates as fuel. Fatty acids and glucose are the primary substrates of choice, but reliance on one or the other changes depending on age. During development, the heart primarily uses glucose but shifts largely to fatty acids soon after birth (1). Fuel choice can also be altered in disease, e.g. increased reliance on fatty acids in diabetes or on glucose in pathological cardiac hypertrophy (1). The transcriptional mechanisms underlying fuel selection, either in physiological or pathological hypertrophy, are incompletely understood.
To accommodate this metabolic flexibility, cardiac fuel uptake and anabolism are tightly regulated by transcriptional, translational, and post-translational mechanisms. Only recently has a detailed understanding of transcriptional metabolic control begun to emerge. Studies into the roles of transcription factors such as the peroxisome proliferator-activated receptors (PPARs)2 and nuclear respiratory factors or co-regulators such as PGC-1α (PPARγ co-activator 1α) have improved our understanding of how fuel is selected or metabolized in the heart, as well as how disease may arise when the activity of these factors is altered (1–2). New classes of drugs have been developed that specifically regulate some of these transcription factors, e.g. thiazolidinediones, which regulate PPARγ and have shown efficacy in treating diabetes by acting as insulin sensitizers (3). Altering fuel selection in the heart may improve patient outcomes, e.g. increasing cardiac reliance on glucose following infarction may prove beneficial (1). To seriously consider metabolic therapies as viable treatments, it is critical that the mechanisms underlying fuel selection and energy metabolism are fully elucidated.
The transcription factor, transcriptional co-activator, and repressor ZAC1 regulates cell cycling and apoptosis of transformed cells and is highly expressed in the developing heart, but its physiological role in cardiomyocytes is unknown (4–5). The ZAC1 genomic locus in humans is associated with the development of transient neonatal diabetes mellitus and with type II diabetes in African Americans (6–7). ZAC1 also regulates the expression of PACAP1-R, a potent insulin secretagogue (8). These findings indicate that ZAC1 plays an important role in insulin signaling and/or glucose metabolism.
We report here that ZAC1 independently regulates cardiomyocyte gene expression of the insulin-responsive glucose transporter Glut4. Zac1 expression increases during cardiomyocyte hypertrophy, via a mechanism that appears to involve transactivation of a novel putative promoter containing a functional MEF2-binding site. Together, these findings support a role for ZAC1 as a potential regulator of glucose metabolism downstream of MEF2 signaling and may contribute to our understanding of how glucose transport is regulated during physiological forms of hypertrophy.
EXPERIMENTAL PROCEDURES
Cell Culture and Cardiomyocyte Isolation
COS7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine. Neonatal rat ventricular cardiomyocytes (NRC) were cultured from day-old pups by differential plating as described (9). Animals were treated in accordance with the guidelines of the Canadian Council on Animal Care, the Guide for the Care and Use of Laboratory Animals (49) and the University of Manitoba Animal Protocol Management and Review Committee.
Cloning of Zac1 Alternate Promoter
A 5.4-kb sequence immediately upstream of the ATG translational start codon and encompassing the first 162 nucleotides of exon VII of the mouse Zac1 gene (NCBI sequence database sequence NC_000076.5) was amplified from bacterial artificial chromosome RPCI23-82B19 (CHORI-BacPac) by high fidelity Pfx DNA polymerase (Invitrogen) (primer sequences listed in supplemental Table S1). The gel-purified PCR product was sequentially subcloned into pCR2.1-TOPO (Invitrogen) and pGL3 basic (Promega) to generate luciferase reporter mZac1pr-pGL3. Three point mutations (CTAAAAATA to CTAAGGATC) were generated in the mZac1pr-pGL3 putative MEF2-binding site using the QuikChange XL site-directed mutagenesis kit (Stratagene) to generate mutMEF2-mZac1pr-pGL3.
Luciferase Reporter Gene Assays
COS7 cells were seeded on six-well plates 24 h prior to transfection. Cells were transfected (Lipofectamine 2000, Invitrogen) with reporter plasmid (human GLUT4 promoter luciferase reporter hG4-luc (10); T3GN12G4-hG4-luc bearing a mutated ZAC1-binding site in hG4-luc; mZac1pr-pGL3; or mutMEF2-mZac1pr-pGL3), pCMV-lacZ for transfection normalization, and expression vectors as required (pcDNA1-mycMEF2C, pcDNA1-mycMEF2A, pcDNA1-mycMEF2D, pcDNA3.1-FLAG-HDAC5S/A, pSG5-HA-mZac1b (mZAC1b is a functionally identical splice variant of ZAC1 containing 11 additional amino acids (11)), and pSG4-HA-GRIP1). pcDNA3.1 was added as required to normalize total DNA transfected. Luciferase activity was measured on a TD20/20 luminometer (Turner BioSystems). β-Galactosidase activity was measured by fluorometric assay (12).
Generation of Adenovirus
An adenovirus encoding HA-tagged mZac1b under control of a constitutive cytomegalovirus (CMV) promoter was generated by cloning the HA-mZAC1b coding region from pSG5-HA-mZac1b into pCA3, followed by packaging in 293A cells with co-transfected pJM17. Following amplification in 293A cells, adenovirus titer was determined by Adeno-X kit (Clontech).
Treatment of Cardiomyocytes
Isolated NRC were grown for 24–48 h and then infected with adenoviruses encoding CMV-β-galactosidase (Ad-β-gal) or CMV-FLAG-HDAC5S/A (Ad-HDAC5S/A) as described previously (9) or CMV-myc-MEF2C (Ad-MEF2C) using a multiplicity of infection of 100. In other experiments, cardiomyocytes were infected with adenoviruses encoding CMV-eGFP (Ad-eGFP) or CMV-HA-mZac1b (Ad-mZac1b). Total RNA or protein was isolated following a 48-h incubation in serum-free medium after infection. Alternatively, cardiomyocytes were treated with phenylephrine (PE) or isoproterenol (ISO) at indicated doses for 48 or 72 h.
Western Blot Analysis of Proteins
Protein lysates were isolated as described (9) and assayed using a Coomassie stain-based kit (Pierce) and then resolved on 12% SDS-polyacrylamide gels and wet-transferred onto polyvinylidene difluoride (Bio-Rad). Blots were immuno-labeled with anti-ZAC1 (Santa Cruz Biotechnology) or anti-GLUT4 (Abcam) primary antibodies and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories), detected using Western blotting luminol reagent (Santa Cruz Biotechnology) and visualized on a Fluor-S-Max MultiImager (Bio-Rad). Anti-α-tubulin antibodies (Developmental Studies Hybridoma Bank, University of Iowa) were used for gel loading controls.
Analysis of RNA
Total RNA was isolated using a GenElute mammalian total RNA miniprep kit (Sigma). 1 μg total RNA was reverse transcribed using the Superscript III 1st Strand Synthesis System for RT-PCR (Invitrogen). Using appropriate primers (supplemental Table S1), optimal PCR cycle number (22 or 28 for glyceraldehyde-3-phosphate dehydrogenase and Zac1, respectively) was determined by standard curves to ensure that bands were obtained within the linear range of amplification. PCR products were resolved on 2% agarose gels and quantified by GelDoc (Bio-Rad). Alternatively, mRNA abundance was determined by quantitative RT-PCR using the 2−ΔΔCT method on an iQ5 real-time PCR system (Bio-Rad).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed on isolated NRC as described (9). Immunoprecipitation was carried out using anti-MEF2C (Cell Signaling Technology), anti-ZAC1 (M-300; Santa Cruz Biotechnology) or nonspecific IgG antibodies (negative control), with pull down by protein A-agarose beads. PCR was carried out using appropriate primers (supplemental Table S1). NRC genomic DNA was used as a positive control (input).
Fluorescent Imaging
NRC were plated on coverslips 24 h prior to treatment. Cells were fixed in 4% formalin and permeabilized with 0.1% Triton X-100. After blocking (2% milk powder in phosphate-buffered saline plus 0.1% Tween 20), cells were incubated with anti-α-actinin (Sigma) or anti-GLUT4 antibodies (Abcam), followed by Alexa Fluor 488-conjugated goat-anti-mouse or Alexa Fluor 594-conjugated goat-anti-rabbit secondary antibody (Molecular Probes). Cells were co-labeled with 500 μm 4′,6-diamidino-2-phenylindole and mounted with Prolong Gold mounting medium (Molecular Probes) prior to imaging (Zeiss Axiophot 2).
Glucose Uptake
Isolated NRC were plated on gelatin-coated coverslips. Cells were allowed to attach for 24 h and then starved for 24 h. Cells were infected with Ad-β-gal or Ad-mZac1b for 24 h. Cells were washed twice with Krebs buffer without glucose and then incubated with 0.3 mm fluorescent glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-d-glucose (2-NBDG; Molecular Probes) for 0, 15, or 30 min. Cells were then washed in Krebs buffer for 10 min and imaged for glucose analog accumulation.
Statistical Analysis
Statistical significance of results was determined by one-way analysis of variance with Student Newman-Keuls post hoc test, or by Student's t test as appropriate, with p < 0.05 considered significant. Results represent means ± S.E.
RESULTS
Cardiac Zac1 Expression
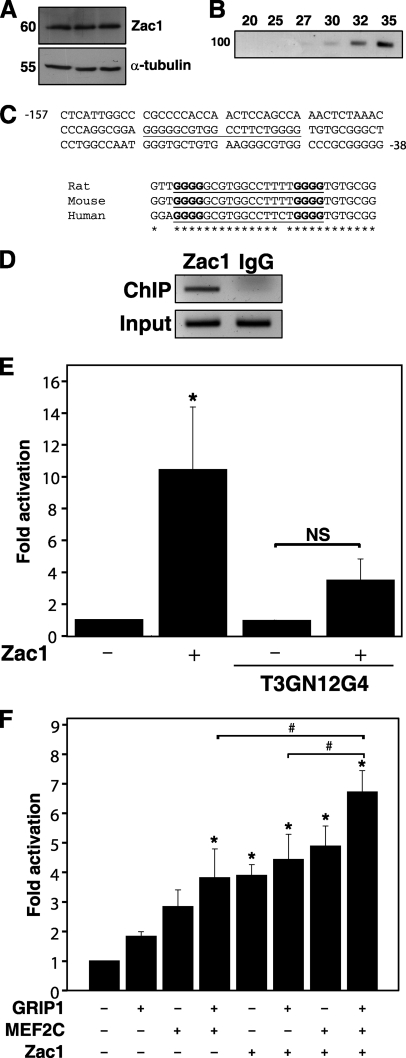
High Zac1 expression has been reported in the developing heart and somites, although expression specifically in cardiomyocytes has not been described (13–14). Using a commercially available specific antibody, we detected a single ∼60-kDa ZAC1 band by Western blotting in isolated neonatal rat cardiomyocytes (Fig. 1A). ZAC1 expression was also observed in skeletal muscle (data not shown). Zac1 mRNA was readily detectable in 1-day-old neonatal rat cardiac muscle by RT-PCR (Fig. 1B).
FIGURE 1.
Transactivation of the GLUT4 promoter by ZAC1. A, Western blot of isolated neonatal rat cardiomyocyte protein lysates (in triplicate), using anti-ZAC1 antibody or α-tubulin loading control (molecular mass indicated in kilodaltons). B, PCR amplification of the Zac1 coding region from total NRC cDNA, generating a single 90-bp predicted amplicon (100 bp standard indicated at left); numbers at top indicate amplification cycles. C, upper panel, partial sequence of the human GLUT4 promoter, with the putative ZAC1 binding site underlined. Numbers denote position relative to the transcription start site. Lower panel, sequence alignment of the rat, mouse, and human putative ZAC1-binding site (underlined). Asterisks denote identical nucleotides across all three species; boldface font indicates critical G4 nucleotides. D, ChIP of NRC using anti-ZAC1 antibody and primers specific for the Glut4 promoter ZAC1-binding site, generating a single 201-bp amplicon. Input DNA was used as a positive control, and nonspecific IgG was used for a negative control. E, COS7 cells were transiently transfected with the wild type or mutant (T3GN12G4) human GLUT4 promoter luciferase reporter, with or without a ZAC1 expression vector. Empty pcDNA3.1 was used for negative controls. Data represents five independent experiments. *, p < 0.05 versus control; NS, not significant. F, luciferase reporter assays performed as in F, plus expression vectors for MEF2C, ZAC1, or GRIP1. Results represent fold activation of reporter alone, for five to seven independent experiments. *, p < 0.05 versus reporter alone; #, p < 0.05 versus samples as indicated.
Regulation of Glut4 Promoter by Zac1
Misexpression of the human ZAC1 locus is associated with transient neonatal diabetes mellitus and with type II diabetes in African Americans (6–7, 15). We therefore hypothesized that ZAC1 may regulate genes involved in glucose metabolism such as the insulin-responsive glucose transporter Glut4. We noted a putative ZAC1-binding site (G4N12G4) from −107 to −88 bp relative to the transcription start site in the human GLUT4 gene promoter. This sequence is similar to reported ZAC1-binding sites containing dual G4 repeats separated by even-length spacers (16) and is highly conserved in the rat and mouse Glut4 gene promoters (Fig. 1C). We employed primers spanning the putative ZAC1-binding site of the rat Glut4 promoter to perform ChIP, confirming the presence of endogenous Zac1 bound to this region in cardiomyocytes (Fig. 1D).
ZAC1 transactivation of the GLUT4 promoter was examined by luciferase assay. ZAC1 strongly activated the GLUT4 promoter, and this activation was attenuated by mutation of the G4N12G4 motif (Fig. 1E). We confirmed activation of this promoter by MEF2C, as reported by others (Fig. 1F) (10). Both ZAC1 and MEF2C bind to and are co-activated by the transcriptional co-activator GRIP1 (11, 17); therefore, we tested the effect of GRIP1 on activation of the GLUT4 promoter. GRIP1 alone had a negligible effect but augmented reporter activity induced by MEF2C. The effectiveness of ZAC1 alone in transactivating the reporter was similar to MEF2C and GRIP1 combined. Reporter activity was highest in the presence of all three factors and was significantly greater than GRIP1+MEF2 or GRIP1+ZAC1, demonstrating an additive effect of these transcriptional regulators (Fig. 1F).
Regulation of Glut4 Expression by ZAC1
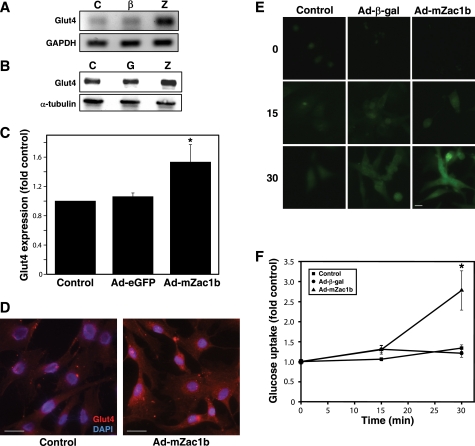
Because ZAC1 was capable of regulating the human GLUT4 promoter, we assessed the effect of ZAC1 on Glut4 expression in intact rat cardiomyocytes. Glut4 mRNA was strongly up-regulated by ZAC1 (Fig. 2A). Protein expression was moderately but significantly up-regulated by ZAC1 (Fig. 2, B and C), a result confirmed by immunofluorescence (Fig. 2D). This increase in ZAC1-mediated Glut4 expression resulted in a significant increase in glucose uptake as measured by fluorogenic assay (Fig. 2, E and F).
FIGURE 2.
Induction of Glut4 expression by ZAC1. A, NRC were untreated (C) or infected for 48 h with adenoviruses encoding β-galactosidase (β) or ZAC1 (Z). Total RNA was isolated for RT-PCR, generating a single 554-bp amplicon using Glut4-specific primers. B, NRC were untreated (C), or infected for 48 h with adenoviruses encoding either eGFP (G) or ZAC1 (Z). Total protein was subjected to Western blotting, and a 50-kDa band corresponding to GLUT4 was obtained. C, densitometric analysis of the Western blot data in B, representing three independent experiments. *, p < 0.01 versus control. D, NRC plated on glass coverslips were untreated or infected with an adenovirus encoding ZAC1 for 48 h and then immunolabeled with anti-GLUT4 (red) and nuclei stained with 4′,6-diamidino-2-phenylindole (blue). Scale bar, 25 μm. E, glucose uptake assay was performed, for the indicated time in minutes, in NRC untreated or previously infected with adenoviruses encoding β-galactosidase or ZAC1. Scale bar, 25 μm. F, quantification of data obtained in E, representing three independent experiments, ∼12 cells per data point per experiment. Results were normalized to control at each time point. *, p < 0.05 versus control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Zac1 Expression in Cardiomyocyte Hypertrophy
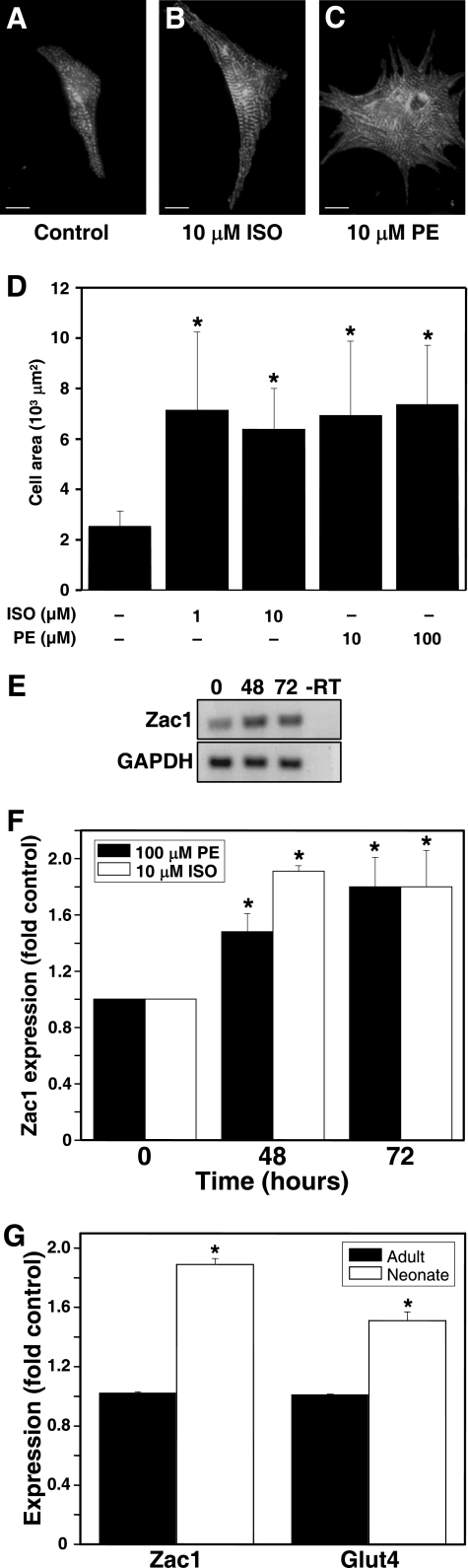
GLUT4 expression is increased in physiological hypertrophy (18–19). We therefore hypothesized that Zac1 expression may be increased in hypertrophic cardiomyocytes. NRC treated with the hypertrophic agents ISO or PE exhibited a significant increase in cardiomyocyte size (Fig. 3, A–D) and Zac1 mRNA expression (Fig. 3, E and F). Shortly after birth, rat cardiomyocytes undergo a transition from hyperplastic to hypertrophic growth (20–21). We found that expression of Zac1 and Glut4 mRNA was significantly higher in NRC compared with adult nonhypertrophic cardiomyocytes (Fig. 3G).
FIGURE 3.
Induction of Zac1 expression during cardiomyocyte hypertrophy. NRC were treated for 48 h with vehicle (A), 10 μm ISO (B), or 10 μm PE (C). Cells were visualized using anti-α-actinin immunostaining. Scale bar, 25 μm. D, increase in cardiomyocyte size following treatment for 48 h with ISO or PE. Results represent three independent experiments (at least 20 cells total per sample). *, p < 0.05 versus control (vehicle-treated) cells. E, Zac1 RT-PCR of NRC treated with PE (0, 48, 72 h; -RT, negative control), generating a single ∼500 bp band. F, quantification of three experiments performed as in E or with 10 μm ISO. *, p < 0.05 versus control cells. G, Zac1 and Glut4 mRNAs are expressed at higher levels in hypertrophying neonatal cardiomyocytes compared with adult cells. Results represent three independent experiments. *, p < 0.05 versus adult. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Identification of a MEF2-binding Site in a Putative Novel Zac1 Promoter
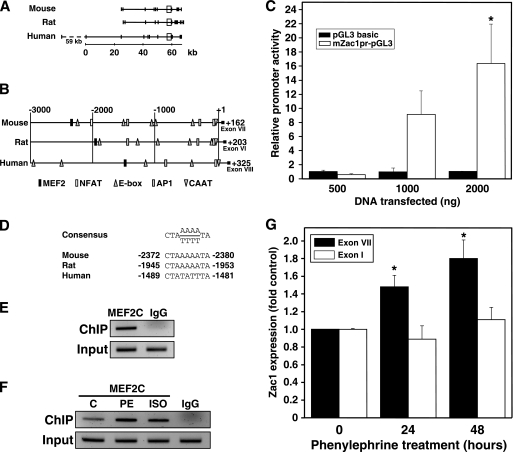
The MEF2 transcription factors are key regulators of hypertrophic gene expression in striated muscle (22–23); thus, MEF2 may regulate Zac1 expression. In silico analysis revealed no MEF2 binding sites within 3000 bp of the canonical transcription start sites upstream of the rat, mouse, or human Zac1 genes. However, we did identify a putative MEF2-binding site between 2400 and −1400 bp relative to the first coding exon in each species (exons VII, VI, and VIII in mouse, rat, and human respectively; Fig. 4, A–B). A CCAAT box was found within 40 bases of the start of each of these exons, and numerous putative binding sites for other transcription factors are also found in this region. A database search revealed a large number of mRNAs and expressed sequence tags initiating in this region (Table 1), suggesting the existence of a proximal alternative transcription start site.
FIGURE 4.
Identification of a putative MEF2-regulated proximal promoter element in the Zac1 gene. A, schematic representation of the mouse, rat, and human genomic loci for Zac1. Scale bar, distance in kilobases. Vertical bars represent exons. Boxes highlight the area immediately upstream of the first coding exon in each species. The dashed line denotes a 59-kb span between alternate first exons in humans. B, representation of a 3-kb sequence upstream from the first coding exon (located at +1 and indicated by roman numerals in the diagram) of mouse, rat, or human Zac1, corresponding to boxed areas in A. Distances in base pairs are indicated at the top. Numbers at right indicate the location of the ATG translation start codon. Symbols indicate the location of putative transcription factor binding sites with strong identity with published consensus sequences. C, luciferase assays were performed in COS7 cells transiently transfected with the mouse Zac1 putative promoter reporter (mZac1pr-pGL3) or with empty vector (pGL3 basic). Results were scaled to empty vector and represent three independent experiments. *, p < 0.01 versus matched control. D, MEF2 consensus binding sequence aligned to sites located in the mouse, rat, and human putative Zac1 promoters. Numbers indicate location in base pairs relative to the start of the first coding exon. E, ChIP was performed on NRC using anti-MEF2C antibody and primers specific for the MEF2-binding site in the Zac1 putative promoter, generating a single 190-bp band. Input DNA was used as a positive control. F, ChIP was performed as in E on cells treated for 24 h with vehicle (C), 10 μm PE, or 1 μm ISO prior to immunoprecipitation. G, NRC were treated with 100 μm PE, and then total RNA was isolated for RT-PCR using primers specific to exon VII (coding region) or exon I (5′ UTR) of the rat Zac1 gene. Data represent three experiments. *, p < 0.05 versus control cells.
TABLE 1.
mRNAs (italicized) and expressed sequence tags originating in the vicinity of the first coding exon of Zac1, reported in NCBI GenbankTM and dbEST databases
| GenBankTM accession no. | Species | Initiation sitea |
|---|---|---|
| AF324471 | Mouse | +4 |
| AA919394 | Mouse | +5 |
| X95504 | Mouse | +75 |
| BX336511 | Human | −75 |
| BP344615 | Human | +20 |
| BP344689 | Human | +20 |
| DA717483 | Human | +31 |
| DA013221 | Human | +45 |
| CN312868 | Human | +66 |
| DR006097 | Human | +68 |
| CN312866 | Human | +109 |
| CN312867 | Human | +129 |
| AW606609 | Human | +175 |
| BC074814 | Human | +276 |
a Numbers are relative to the 5′ boundary of exon VII in mice (ATG at +162) or exon VIII in humans (ATG at +325).
To test this possibility, we cloned a 5.4-kb span of mouse genomic DNA 5′ to the ATG translation start site (+162) in exon VII of the mouse Zac1 gene, which included the exon VII 5′ boundary at +1, the CCAAT box at −42, and a putative MEF2 site at −2372/−2380. Transient transfection of COS7 cells with a luciferase reporter bearing this putative promoter region resulted in a dose-dependent increase in reporter gene expression (Fig. 4C). In contrast, the empty reporter vector showed no change in luciferase activity, suggesting that the cloned region behaves as a promoter.
Mice, rats, and humans possess a putative MEF2-binding site within this region (Fig. 4D). We therefore confirmed that MEF2C is able to bind to a site within this promoter region in rat cardiomyocytes using ChIP (Fig. 4E). Induction of cardiomyocyte hypertrophy by PE or ISO treatment resulted in an increase in MEF2C binding to this site (Fig. 4F).
To examine the mechanism of hypertrophic Zac1 induction, RT-PCR was repeated using primers specific for exon I in the Zac1 5′ untranslated region (Fig. 4G). No induction of this region was observed in hypertrophic cardiomyocytes, in contrast to the significant amplification observed using primers located in the protein coding region, suggesting that induced transcripts excluded upstream sequences and supporting the idea that Zac1 is expressed from the novel putative promoter.
MEF2 Regulates Zac1 Expression
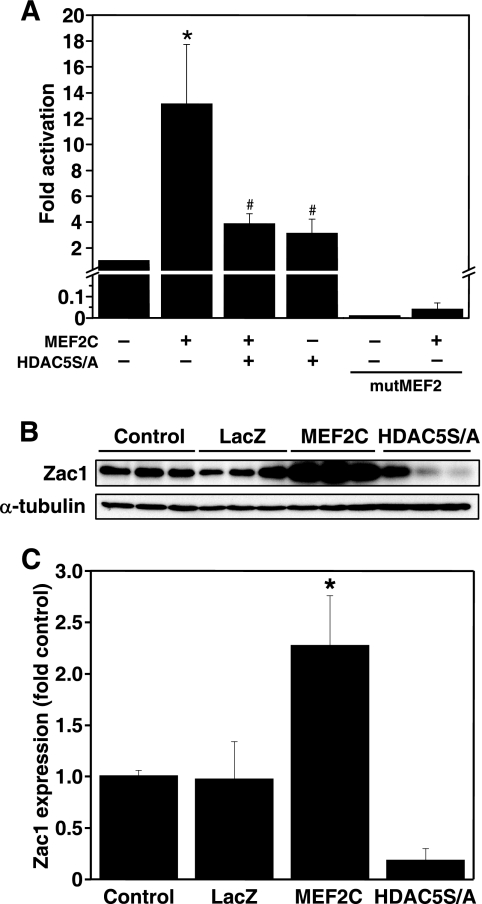
Luciferase assays demonstrated that MEF2C (Fig. 5A), MEF2A, and MEF2D (data not shown) strongly transactivated the putative Zac1 promoter. Transactivation was attenuated by a double serine-to-alanine mutant of histone deacetylase 5 (HDAC5S/A), which constitutively binds to and represses MEF2 (24). Mutation of the MEF2-binding site prevented transactivation by MEF2C. MEF2C overexpression significantly increased ZAC1 expression (Fig. 5, B and C). HDAC5S/A appeared to down-regulate ZAC1 expression; however, this result did not reach statistical significance in the present experiment (Fig. 5, B and C).
FIGURE 5.
MEF2 regulates transactivation of the Zac1 promoter. A, luciferase assays were performed in COS7 cells transfected with wild type mZac1pr-pGL3 or a reporter in which the MEF2-binding site was mutated (mutMEF2), plus expression vectors for MEF2C or HDAC5S/A as indicated. Empty pcDNA3.1 was used for negative controls. All results are reported as fold activation of mZac1pr-pGL3 reporter alone and represent three to eight independent experiments. *, p < 0.05 versus control (column 1); and #, p < 0.05 versus MEF2C-treated (column 2). B, NRC were untreated or infected for 48 h with adenoviruses encoding β-galactosidase (LacZ), MEF2C, or HDAC5S/A. Total protein from each dish was collected for Western blotting, and an ∼65-kDa band corresponding to ZAC1 was obtained. C, densitometry analysis of results from B, representing three experiments normalized to α-tubulin expression. *, p < 0.05 versus control.
DISCUSSION
The transcriptional networks regulating cardiac energy metabolism are gradually being elucidated. In this study, we report that ZAC1 directly regulates expression of the insulin-responsive glucose transporter Glut4 via a highly conserved G4N12G4 motif upstream of the transcription start site. ZAC1 overexpression increases expression of Glut4, resulting in increased glucose uptake by cardiomyocytes. These results are particularly striking in light of the association of ZAC1 defects with diabetes and the recent report that ZAC1 regulates expression of PPARγ, the therapeutic target of the thiazolidinedione class of insulin sensitizers for diabetes treatment (6–7, 25). We also demonstrate that ZAC1 expression is regulated by MEF2, which may explain our observation that Zac1 expression is increased during cardiomyocyte hypertrophy.
Zac1 Is a Novel Regulator of Glut4 Expression
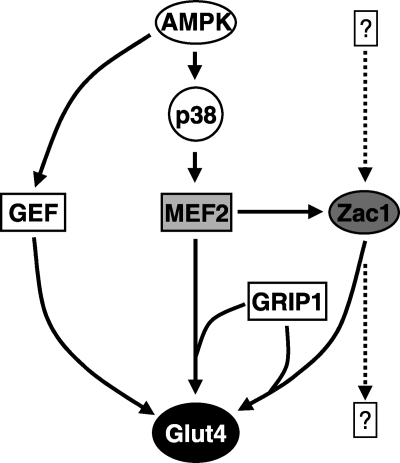
Our results agree with reports that Glut4 expression is regulated by MEF2 (26). We also found that ZAC1 additively regulates GLUT4 transactivation with MEF2 and the steroid receptor co-activator GRIP1, likely via a binding site in the GLUT4 promoter in close proximity to the previously identified MEF2 binding site at −470 bp (26). Because both MEF2C and ZAC1 are co-activated by GRIP1, our data suggest that these three factors work together as part of a common transcriptional complex (Fig. 6) (11, 17). We previously showed that MEF2 regulates expression of the transcriptional co-activator PGC-1α, while others have shown that PGC-1α represses expression of Glut4 in vivo but augments expression of Glut4 in vitro (9, 27–28). This further underscores the complexity of transcriptional pathway interactions that regulate net Glut4 expression.
FIGURE 6.
Regulation of Glut4 expression by MEF2 and ZAC1. Glut4 expression levels result from complex interactions among transcriptional pathways. MEF2 appears to be a central regulator of Glut4 expression, either directly or indirectly via ZAC1, depending on cell context. ZAC1 expression is likely to be regulated by other as yet unidentified signaling pathways. These pathways may link upstream pro-hypertrophic signals with downstream increases in glucose metabolism in situations such as physiological hypertrophy. AMPK, AMP-regulated protein kinase; GEF, Glut4 enhancer factor.
MEF2 Regulates Zac1 Expression
Although MEF2 is insufficient to drive cardiomyocyte hypertrophy unless expressed at very high levels, it nonetheless plays a central role in this process, including regulating hypertrophic gene expression and promoting sarcomere elongation (29–31). MEF2 transactivation activity is increased in cardiac hypertrophy or exercised skeletal muscle, while class II histone deacetylases such as HDAC5, which repress MEF2 activity, are considered antihypertrophic (22–23, 32–34). In the present study, MEF2 transactivated the putative Zac1 promoter and increased ZAC1 protein expression, suggesting a mechanism for our observation that Zac1 expression increases in hypertrophy. Although HDAC5S/A blocked MEF2-mediated transactivation of the Zac1 promoter, inhibition of ZAC1 protein expression did not reach statistical significance; thus, a definitive role for HDAC5 in regulation of ZAC1 expression cannot yet be demonstrated. However, our data is consistent with a previous report that the HDAC inhibitor trichostatin A relieves silencing of LOT1/Zac1 expression (35).
Glut4 Expression in Hypertrophy
Our data demonstrate that MEF2 regulates Zac1 expression and that ZAC1 regulates Glut4 expression. It is thus intriguing to speculate that ZAC1 may provide an accessory regulatory mechanism for Glut4 expression in hypertrophy downstream of or in parallel to MEF2 (Fig. 6). However, we did not observe an increase in Glut4 expression following PE or ISO treatment of cardiomyocytes (data not shown) despite induction of ZAC1 expression. Although this observation might argue against the involvement of ZAC1 in hypertrophy, it must be noted that our results parallel observations made for MEF2. MEF2 is a potent inducer of Glut4 expression and is also induced by PE (22, 26, 36). Yet, in pathological cardiac hypertrophy, MEF2 activity increases while GLUT4 expression decreases (22, 37). In contrast, GLUT4 expression increases in physiological hypertrophy (exercise), which is also associated with increased MEF2 activity (18–19). In agreement with these reports, we demonstrated increased Zac1 and Glut4 mRNA expression in hypertrophic neonatal cardiomyocytes compared with nonhypertrophic adult cells (Fig. 3G). The PE/ISO cardiomyocyte hypertrophy model employed in this study thus likely fails to recapitulate physiological hypertrophy. Further study is required to confirm the role of ZAC1 in mediating Glut4 expression during physiological hypertrophy in vivo, e.g. in response to exercise.
Identification of a Novel Zac1 Promoter
Intriguingly, MEF2-mediated regulation appears to occur via a novel noncanonical promoter element. The genomic region immediately upstream of the first coding exon (exon VII) of the mouse Zac1 gene exhibits several features, suggesting that it contains a novel promoter. First, a putative CCAAT box is conserved at +1 relative to the exon start in humans, or at −42 in mice and rats. Second, multiple mRNAs and expressed sequence tags initiate in this region, lacking the 5′ UTR found in transcripts originating from the canonical Zac1 promoter (Table 1). Third, this region contains a large number of putative transcription factor-binding sites, including sites for myogenic transcription factors such as MyoD and myogenin (E-boxes) or nuclear factor of activated T cells. It is noteworthy that generation of an mRNA transcript from this site would produce an identical open reading frame to transcripts that originate at exon I because only the 5′ UTR would be truncated (38). Furthermore, the NCBI sequence database currently reports eight differentially spliced Zac1 transcripts in humans, two of which utilize an alternative first exon nearly 60 kb upstream of the first exon of the other transcripts, suggesting the presence of alternative promoters for the Zac1 gene.
Our data further support this model. When placed upstream of a promoter-less luciferase reporter, a DNA dose-dependent increase in reporter activity is observed, indicating basal promoter activity. ChIP demonstrates MEF2 binding to this region, complementing luciferase assays demonstrating responsiveness of this putative promoter to MEF2 and HDAC5S/A. Most notably, MEF2 binding increases in response to PE or ISO treatment of cardiomyocytes, suggesting the intriguing possibility that this promoter responds to hypertrophic signaling. We also noted that up-regulation of Zac1 mRNA was detected when primers corresponding to the coding region (but not exon I) were used, further supporting the idea that cardiac hypertrophic expression of Zac1 is not mediated by the canonical Zac1 gene promoter. However, to date, we have been unsuccessful in isolating mRNAs originating from this promoter by 5′ rapid amplification of cDNA ends; thus, we cannot discount the possibility that this region may represent a transcriptional enhancer rather than a promoter.
Physiological Significance
Induction of Zac1 expression during cardiomyocyte growth is particularly striking in light of a recent report (39) that ZAC1 acts via an enhancer element to regulate expression of insulin-like growth factor II, a regulator of cardiomyocyte hypertrophy (40–43). An intriguing model is that activation of MEF2 and/or inhibition of HDACs downstream of hypertrophic growth cues results in increased expression of Zac1, in turn up-regulating insulin-like growth factor II-mediated hypertrophic signaling pathways that reinforce or augment the effect of MEF2 independently.
It was recently reported that MEF2 and Glut4 enhancer factor cooperate to regulate Glut4 expression (44). The cellular fuel sensor AMP-regulated protein kinase directly activates Glut4 enhancer factor and indirectly activates MEF2 via phosphorylation of p38, which in turn phosphorylates and activates MEF2 (44–46). In this way, expression of Glut4 is up-regulated when fuel supply is low, permitting increased glucose uptake and generation of ATP. Our results indicate that ZAC1 interacts with this mechanism to impact Glut4 expression.
It is unknown which MEF2-independent pathways may be involved in up-regulation of Zac1 and which other ZAC1 gene targets may exist. Gain-of-function or loss-of-function studies may provide insight into other genes directly regulated by ZAC1. Although both Zac1-null mice and transgenic mice overexpressing the human ZAC1 locus exist, specific effects on cardiac gene expression or glucose metabolism have not been described (39, 47). Should ZAC1 prove to be an important regulator of glucose uptake, striated muscle-specific overexpression of ZAC1 to increase expression of Glut4 may prove useful in treating diabetes because increased Glut4 expression is beneficial in this disease (48). ZAC1 may therefore represent an intriguing novel target for therapeutic manipulation in the future.
Supplementary Material
Acknowledgments
We thank Drs. Ann Louise Olson (University of Oklahoma Health Science Center) for hG4-luc, Michael Stallcup (University of Southern California) for pSG5-HA-mZac1b and pSG5-HA-GRIP1, and Eric Olson (University of Texas Southwestern Medical Center) for pcDNA1-mycMEF2C, pcDNA1-mycMEF2D, pcDNA1-mycMEF2A, pcDNA3.1-FLAG-HDAC5S/A, and Ad-MEF2C. We also thank Laura Albak for technical assistance.
This work was supported by the University of Manitoba Research Grants program, the Manitoba Medical Services Foundation, and the Canadian Institutes of Health Research (ROP 83898).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- PPAR
- peroxisome proliferator-activated receptor
- Ad
- adenovirus
- eGFP
- enhanced green fluorescent protein
- NRC
- neonatal rat cardiomyocytes
- luc
- luciferase
- CMV
- cytomegalovirus
- HA
- hemagglutinin
- PE
- phenylephrine
- ISO
- isoproterenol
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- HDAC5S/A
- histone deacetylase 5 double serine-to-alanine mutant.
REFERENCES
- 1.Stanley W. C., Recchia F. A., Lopaschuk G. D. (2005) Physiol. Rev. 85, 1093–1129 [DOI] [PubMed] [Google Scholar]
- 2.Huss J. M., Kelly D. P. (2004) Circ. Res. 95, 568–578 [DOI] [PubMed] [Google Scholar]
- 3.Spiegelman B. M. (1998) Diabetes 47, 507–514 [DOI] [PubMed] [Google Scholar]
- 4.Spengler D., Villalba M., Hoffmann A., Pantaloni C., Houssami S., Bockaert J., Journot L. (1997) EMBO J. 16, 2814–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alam S., Zinyk D., Ma L., Schuurmans C. (2005) Dev. Dyn. 234, 772–782 [DOI] [PubMed] [Google Scholar]
- 6.Varrault A., Bilanges B., Mackay D. J., Basyuk E., Ahr B., Fernandez C., Robinson D. O., Bockaert J., Journot L. (2001) J. Biol. Chem. 276, 18653–18656 [DOI] [PubMed] [Google Scholar]
- 7.Sale M. M., Freedman B. I., Langefeld C. D., Williams A. H., Hicks P. J., Colicigno C. J., Beck S. R., Brown W. M., Rich S. S., Bowden D. W. (2004) Diabetes 53, 830–837 [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Henche N., Jamen F., Leroy C., Bockaert J., Brabet P. (2002) Biochim. Biophys. Acta 1576, 157–162 [DOI] [PubMed] [Google Scholar]
- 9.Czubryt M. P., McAnally J., Fishman G. I., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knight J. B., Eyster C. A., Griesel B. A., Olson A. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S. M., Stallcup M. R. (2000) Mol. Cell. Biol. 20, 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waser M., Mesaeli N., Spencer C., Michalak M. (1997) J. Cell Biol. 138, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valente T., Junyent F., Auladell C. (2005) Dev. Dyn 233, 667–679 [DOI] [PubMed] [Google Scholar]
- 14.Varrault A., Ciani E., Apiou F., Bilanges B., Hoffmann A., Pantaloni C., Bockaert J., Spengler D., Journot L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8835–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur E. I., Zlotogora J., Lerer I., Dagan J., Marks K., Abeliovich D. (1997) Eur. J. Hum. Genet. 5, 417–419 [PubMed] [Google Scholar]
- 16.Hoffmann A., Ciani E., Boeckardt J., Holsboer F., Journot L., Spengler D. (2003) Mol. Cell. Biol. 23, 988–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S. L., Dowhan D. H., Hosking B. M., Muscat G. E. (2000) Genes Dev. 14, 1209–1228 [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu L. L., Tsai Y. L., Lee W. C., Cho Y. M., Ho H. Y., Chen S. M., Chen M. T., Kuo C. H. (2005) High Alt. Med. Biol. 6, 256–262 [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto T., Kambara N., Nohara R., Yazawa M., Taguchi S. (2004) J. Appl. Physiol. 97, 843–851 [DOI] [PubMed] [Google Scholar]
- 20.Porrello E. R., Widdop R. E., Delbridge L. M. (2008) Clin. Exp. Pharmacol. Physiol. 35, 1358–1364 [DOI] [PubMed] [Google Scholar]
- 21.Li F., Wang X., Capasso J. M., Gerdes A. M. (1996) J. Mol. Cell Cardiol. 28, 1737–1746 [DOI] [PubMed] [Google Scholar]
- 22.Passier R., Zeng H., Frey N., Naya F. J., Nicol R. L., McKinsey T. A., Overbeek P., Richardson J. A., Grant S. R., Olson E. N. (2000) J. Clin. Invest. 105, 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naya F. J., Wu C., Richardson J. A., Overbeek P., Olson E. N. (1999) Development 126, 2045–2052 [DOI] [PubMed] [Google Scholar]
- 24.McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Nature 408, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barz T., Hoffmann A., Panhuysen M., Spengler D. (2006) Cancer Res. 66, 11975–11982 [DOI] [PubMed] [Google Scholar]
- 26.Mora S., Pessin J. E. (2000) J. Biol. Chem. 275, 16323–16328 [DOI] [PubMed] [Google Scholar]
- 27.Miura S., Kai Y., Ono M., Ezaki O. (2003) J. Biol. Chem. 278, 31385–31390 [DOI] [PubMed] [Google Scholar]
- 28.Michael L. F., Wu Z., Cheatham R. B., Puigserver P., Adelmant G., Lehman J. J., Kelly D. P., Spiegelman B. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Oort R. J., van Rooij E., Bourajjaj M., Schimmel J., Jansen M. A., van der Nagel R., Doevendans P. A., Schneider M. D., van Echteld C. J., De Windt L. J. (2006) Circulation 114, 298–308 [DOI] [PubMed] [Google Scholar]
- 30.Xu J., Gong N. L., Bodi I., Aronow B. J., Backx P. H., Molkentin J. D. (2006) J. Biol. Chem. 281, 9152–9162 [DOI] [PubMed] [Google Scholar]
- 31.Black B. L., Olson E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 32.Wu H., Rothermel B., Kanatous S., Rosenberg P., Naya F. J., Shelton J. M., Hutcheson K. A., DiMaio J. M., Olson E. N., Bassel-Duby R., Williams R. S. (2001) EMBO J. 20, 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song K., Backs J., McAnally J., Qi X., Gerard R. D., Richardson J. A., Hill J. A., Bassel-Duby R., Olson E. N. (2006) Cell 125, 453–466 [DOI] [PubMed] [Google Scholar]
- 35.Abdollahi A., Pisarcik D., Roberts D., Weinstein J., Cairns P., Hamilton T. C. (2003) J. Biol. Chem. 278, 6041–6049 [DOI] [PubMed] [Google Scholar]
- 36.Lu J., McKinsey T. A., Nicol R. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minakawa M., Takeuchi K., Ito K., Tsushima T., Fukui K., Takaya S., Fukuda I. (2003) Eur. J. Cardiothorac Surg 24, 493–501 [DOI] [PubMed] [Google Scholar]
- 38.Bilanges B., Varrault A., Mazumdar A., Pantaloni C., Hoffmann A., Bockaert J., Spengler D., Journot L. (2001) Oncogene 20, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 39.Varrault A., Gueydan C., Delalbre A., Bellmann A., Houssami S., Aknin C., Severac D., Chotard L., Kahli M., Le Digarcher A., Pavlidis P., Journot L. (2006) Dev. Cell 11, 711–722 [DOI] [PubMed] [Google Scholar]
- 40.Adachi S., Ito H., Akimoto H., Tanaka M., Fujisaki H., Marumo F., Hiroe M. (1994) J. Mol. Cell Cardiol. 26, 789–795 [DOI] [PubMed] [Google Scholar]
- 41.Turner J. D., Rotwein P., Novakofski J., Bechtel P. J. (1988) Am. J. Physiol. 255, E513–517 [DOI] [PubMed] [Google Scholar]
- 42.Huang C. Y., Hao L. Y., Buetow D. E. (2002) Cell Biol. Int. 26, 737–739 [DOI] [PubMed] [Google Scholar]
- 43.Armstrong M. T., Lee D. Y., Armstrong P. B. (2000) Dev. Dyn 219, 226–236 [DOI] [PubMed] [Google Scholar]
- 44.Holmes B. F., Sparling D. P., Olson A. L., Winder W. W., Dohm G. L. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E1071–1076 [DOI] [PubMed] [Google Scholar]
- 45.Rauch C., Loughna P. T. (2005) Am. J. Physiol. Cell Physiol. 288, C593–605 [DOI] [PubMed] [Google Scholar]
- 46.Xi X., Han J., Zhang J. Z. (2001) J. Biol. Chem. 276, 41029–41034 [DOI] [PubMed] [Google Scholar]
- 47.Ma D., Shield J. P., Dean W., Leclerc I., Knauf C., Burcelin R., R., Rutter G. A., Kelsey G. (2004) J. Clin. Invest. 114, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galuska D., Ryder J., Kawano Y., Charron M. J., Zierath J. R. (1998) Adv. Exp. Med. Biol. 441, 73–85 [DOI] [PubMed] [Google Scholar]
- 49.National Institutes of Health (1996) Guide for the Care and Use of Laboratory Animals Publication 85-23 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.