Abstract
The reovirus fusion-associated small transmembrane (FAST) proteins are the smallest known viral membrane fusion proteins. With ectodomains of only ∼20–40 residues, it is unclear how such diminutive fusion proteins can mediate cell-cell fusion and syncytium formation. Contained within the 40-residue ectodomain of the p10 FAST protein resides an 11-residue sequence of moderately apolar residues, termed the hydrophobic patch (HP). Previous studies indicate the p10 HP shares operational features with the fusion peptide motifs found within the enveloped virus membrane fusion proteins. Using biotinylation assays, we now report that two highly conserved cysteine residues flanking the p10 HP form an essential intramolecular disulfide bond to create a cystine loop. Mutagenic analyses revealed that both formation of the cystine loop and p10 membrane fusion activity are highly sensitive to changes in the size and spatial arrangement of amino acids within the loop. The p10 cystine loop may therefore function as a cystine noose, where fusion peptide activity is dependent on structural constraints within the noose that force solvent exposure of key hydrophobic residues. Moreover, inhibitors of cell surface thioreductase activity indicate that disruption of the disulfide bridge is important for p10-mediated membrane fusion. This is the first example of a viral fusion peptide composed of a small, spatially constrained cystine loop whose function is dependent on altered loop formation, and it suggests the p10 cystine loop represents a new class of viral fusion peptides.
Keywords: Fusion Protein, Membrane Fusion, Reovirus, Site-directed Mutagenesis, Virus Entry, Fusion Loop, Fusion Peptide
Introduction
Membrane fusion is an essential process involved in an abundance of biological events, including sperm-egg fusion, multinucleated myotube formation, exocytosis, and enveloped virus entry (1). The fusion reaction is catalyzed by specialized membrane fusion proteins designed to alter bilayer structure and bring about membrane merger. The membrane fusion proteins of various enveloped viruses represent some of the best characterized examples of such fusion catalysts (2). Detailed structural and functional analysis of diverse enveloped virus membrane fusion proteins reveals a common pathway of protein-mediated membrane fusion. Triggered exposure of a hydrophobic fusion peptide (FP),3 normally sequestered within the complex pre-fusion ectodomain structure of these proteins, results in FP insertion into the donor and/or target membranes. Subsequent structural remodeling of these large ectodomains generates a trimeric hairpin structure that draws the two membranes together, driving lipid mixing (also referred to as hemifusion) and pore formation and expansion (3, 4). Despite a wealth of information, the precise mechanisms by which FPs and conformational remodeling of enveloped virus fusion proteins mediate lipid bilayer rearrangement and fusion remain unclear.
The reovirus fusion-associated small transmembrane (FAST) proteins are a singular family of viral membrane fusion proteins. Although many nonenveloped viruses encode proteins involved in membrane permeabilization (5–7), some of which can induce low levels of cell-to-cell fusion (8), the FAST proteins are the only known examples of nonenveloped virus proteins that evolved specifically to induce cell-to-cell fusion (9). In contrast to the enveloped virus fusion proteins, which are structural proteins that have evolved primarily to mediate virus-cell fusion, the FAST proteins are nonstructural proteins and therefore are not involved in virus entry (10). Following their expression in virus-infected cells, the FAST proteins traffic to the plasma membrane where they induce cell-cell fusion and multinucleated syncytium formation, promoting virus dissemination (11). Although the FAST proteins (95–198 residues) approximate the size of the soluble NSF attachment protein receptor proteins involved in intracellular vesicle fusion (12), the FAST proteins need only be present in one of the two membranes undergoing fusion (13). The FAST proteins are also both necessary and sufficient to mediate the actual merger of lipid bilayers (14), although they exploit or recruit cellular co-factors to enhance the pre-fusion (i.e. membrane attachment and apposition) and post-fusion (i.e. pore expansion) stages of biological cell-cell fusion (15, 16). Determining how these diminutive fusogens function from only the donor membrane to mediate the lipid rearrangements required to fuse two membranes together is an area of active investigation.
The homologous avian reovirus (ARV) and Nelson Bay reovirus (NBV) p10 proteins are two representatives of the FAST protein family (10). As with all FAST proteins, the p10 proteins assume a bitopic Nexoplasmic/Ccytoplasmic topology in the plasma membrane, using their single transmembrane domain as a reverse signal-anchor to direct membrane insertion (10, 17–19). In contrast to the enveloped virus fusion proteins that position the majority of their mass external to the membrane in which they reside, the p10 FAST proteins have very small, approximately equally sized ecto- and endodomains (40 and 36 residues, respectively, for ARV p10). Important p10 functional motifs include a tri-glycine motif within the transmembrane domain and a polybasic region and palmitoylated di-cysteine motif, both of which lie in the membrane-proximal region of the endodomain (20). There are also two adjacent motifs in the ectodomain that are involved in p10 membrane fusion activity as follows: a 9-residue conserved region present in all p10 isolates from both avian and bat species, and a stretch of 11 moderately apolar residues termed the hydrophobic patch (HP) (Fig. 1A). Mutations in all of the above motifs decrease or eliminate p10-induced syncytium formation, although the role of these motifs in the membrane fusion reaction has not been determined.
FIGURE 1.
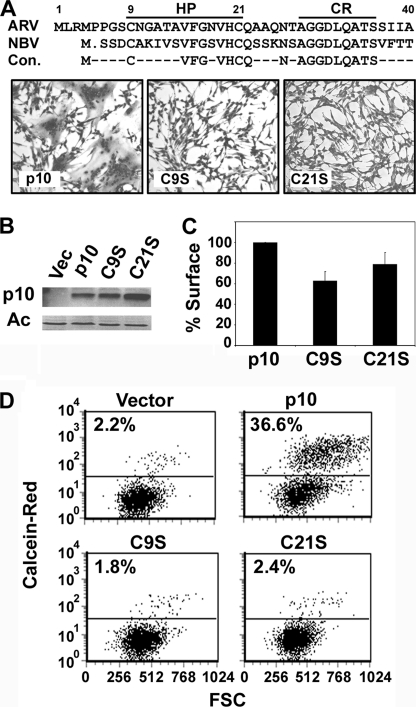
ARVp10 ectodomain contains two essential cysteine residues. A, aligned sequences of the ARV and NBV p10 ectodomains and the consensus sequence (Con.) indicating identical residues (top panel). The locations of the cysteine-flanked 11-residue HP and adjacent 9-residue conserved region (CR) are indicated. QM5 cells were transfected with either authentic ARV p10 or with the C9S or C21S substitution constructs. Cells were fixed and stained with Wright-Giemsa at 32 h post-transfection to detect syncytium formation by bright field microscopy at ×100 (bottom panel). B, cells were transfected as in A, and p10 protein expression was analyzed by Western blotting at 24 h post-transfection with α p10 antiserum or α actin (Ac) as a loading control. Vec, vector. C, surface expression was determined by live cell-labeling transfected QM5 cells with α p10 antibodies and fluorescently conjugated secondary antibodies at 24 h post-transfection, followed by flow cytometry analysis. Results are presented as the mean ± S.D. of the percent expression relative to authentic p10 from one of two experiments conducted in triplicate. D, pore formation activity was analyzed by the transfer of fluorescent calcein red-orange from target Vero cells to QM5 donor cells co-transfected with the indicated constructs and pEGFP as a marker for transfection. EGFP-positive QM5 cells were gated, and the acquisition of the calcein red-orange dye is shown versus forward scatter (FSC). Representational dot plots are from one of two experiments conducted in triplicate.
We previously noted similarities between the p10 HPs and the FPs present in all enveloped virus fusion proteins (13). FPs are small (∼20–30 residues) regions of mostly apolar residues, frequently enriched in glycine, and usually highly conserved within the fusion proteins of different strains of the same virus (21). The enveloped virus FPs are critical for fusion activity and highly sensitive to mutation, and synthetic versions of these peptides insert into lipid bilayers to promote lipid mixing (21, 22). The FPs of most class I viral fusion proteins are located at or near the N terminus of the fusion subunit, whereas the FPs of some class I and all of the class II and III fusion proteins exist as internal fusion loops (23). In almost all instances, the FP is hidden within the pre-fusion structure of the fusion protein complex and becomes exposed in the activated and triggered conformation to allow membrane insertion.
Although the p10 FAST protein HPs share several of these FP features, including hydrophobic partitioning and lipid mixing activity (13), they also have several unexpected features. For example, the p10 HP is relatively small (∼11 residues) and less hydrophobic than typical FPs, and the sequence is not highly conserved in the homologous ARV and NBV p10 proteins (Fig. 1A). It is also extremely unlikely that the limited size of the p10 ectodomain could sequester the HP within a complex, pre-fusion tertiary structure. Furthermore, although the p10 HP is N-proximal, CD analysis suggests it lacks the helical propensity typical of the N-terminal fusion peptides (13, 24). Similar to the fusion loops contained within the fusion proteins of the class II and class III enveloped viruses (23), the presence of two conserved cysteine residues flanking the p10 HP suggests the p10 HP might exist as a disulfide-stabilized loop. However, this loop would be considerably smaller than typical disulfide-stabilized fusion loops; there is no direct evidence this loop is formed, and the influence of loop formation (if it occurs) on p10 fusion activity has not been determined.
The exceptional features of the FAST proteins in general, and of their ectodomains in particular, imply that the mechanism by which the FAST proteins mediate membrane fusion is unlikely to adhere to the concept of dramatic ectodomain conformational changes that force FP exposure and membrane insertion, followed by hairpin formation to drive close membrane apposition and merger. To provide additional mechanistic insights into this unusual family of viral membrane fusion proteins, we sought to more clearly define the features of the p10 HP, the presumed FP of this FAST protein. We now directly demonstrate that the p10 HP exists as a small, disulfide-stabilized fusion loop and that loop formation is essential for p10-mediated pore formation. Results further indicate that loop formation is remarkably sensitive to the amino acid content of the loop and adjacent flanking residues and that dynamic alteration of the loop is required for this motif to function as an FP. These results impact on models of how the small p10 ectodomain might promote the merger of biological membranes.
EXPERIMENTAL PROCEDURES
Cells and Reagents
QM5 and Vero cells were grown and maintained as described previously (17). Rabbit antisera generated against full-length ARV p10 and reptilian reovirus (RRV) p14, as well as the ARV p10 endodomain and RRV p14 ectodomain, were described previously (16, 17, 25). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch), HRP-conjugated neutravidin (Pierce), Alexa 647-conjugated goat anti-rabbit IgG (Invitrogen), calcein red-orange (Invitrogen), neutravidin-immobilized beads (Pierce), maleimide-PEG2-biotin (Pierce), 5′,5-dithiobis(2-nitrobenzoic acid) (DTNB) (Sigma), and Bacitracin (Sigma) were purchased from the indicated commercial sources.
Molecular Cloning
ARV p10, NBV p10, and RRV p14 clones in a pcDNA3 vector were described previously (16). The QuikChange site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions to generate point mutations and insertions for all cysteine shift and alanine insertion constructs. Oligonucleotide primers were purchased from either Operon or IDT, and all constructs were confirmed by sequencing.
Transfections and Syncytial Indexing
QM5 cell transfections were carried out using Lipofectamine (Invitrogen) as described previously (16). For syncytial indexing of ARV p10 and mutant constructs, 50% confluent QM5 monolayers in 12-well dishes were transfected with 1 μg of plasmid DNA and incubated for 6 h before replacing the transfection mixture with Earle's 199 growth media (Invitrogen) supplemented with 10% fetal bovine serum (Sigma). Transfected cells were incubated at 37 °C, methanol-fixed, and stained with Wright-Giemsa at the indicated times, and syncytial nuclei from five random fields were counted from photographs taken at ×200 magnification on a Zeiss Axiovert 200 microscope.
SDS-PAGE and Western Blots
Cell lysate preparation in radioimmunoprecipitation assay buffer (Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate) and analysis by SDS-PAGE and Western blotting were carried out as described previously (26). Protein samples were transferred from 15% polyacrylamide gels to polyvinylidene difluoride membranes using the iBlot dry transfer system (Invitrogen). For Western blotting, a 1:10,000 or 1:30,000 dilution of anti-ARV p10 rabbit antiserum was used for biotinylation experiments or to assess overall protein expression levels, respectively, followed by a 1:10,000 dilution of HRP-conjugated goat anti-rabbit secondary antibody. As a biotinylation control, cell lysates were also probed with HRP-conjugated neutravidin at a 1:10,000 dilution. Western blots were developed with ECL+ reagent (GE Healthcare) and visualized either on a Typhoon 9410 Variable Imager (Amersham Biosciences) or a Kodak 4000-mm Pro CCD imager.
Surface Expression by Flow Cytometry
Transfected QM5 cells were incubated at 37 °C for 24 h in Earle's 199 media (Invitrogen) supplemented with 10% fetal bovine serum and a 1:25 dilution of rabbit antisera directed against ARV p10 (to inhibit syncytium formation). Live cells were then labeled with anti-ARV p10 primary antibody followed by Alexa 647-conjugated goat anti-rabbit antibody, as described previously (26). Cells were resuspended with EDTA and fixed in 3.7% formaldehyde, and 20,000 cells were quantified on a FACSCalibur (BD Biosciences). The fluorescence was analyzed using FCS Express 2.0 (De Novo Software).
Biotinylation Assay
Free thiol groups from cysteine residues were identified by labeling them with maleimide-PEG2-biotin (Pierce). At 24 h post-transfection, QM5 cells were washed twice with Hanks' buffered salt solution (HBSS) and incubated in HBSS in the absence or presence of 0.1 mm dithiothreitol (DTT) for 5 min at room temperature. Cells were washed three times in HBSS and then incubated at 4 °C for 25 min in the presence of 1 μg/ml maleimide-PEG2-biotin. Cells were washed five times in HBSS, the last of which was supplemented with 1% bovine serum albumin, twice in PBS, resuspended in 50 mm EDTA, and then lysed in radioimmunoprecipitation assay buffer with protease inhibitors. Biotinylated cell lysates were incubated at 4 °C overnight with neutravidin-immobilized beads, and then the beads were washed three times with cold radioimmunoprecipitation assay buffer supplemented with protease inhibitors. The biotinylated p10 proteins were removed from the beads by boiling samples for 10 min in 2× Laemmli protein sample buffer supplemented with 100 mm DTT, resolved by SDS-PAGE, and detected by Western blotting with ARV p10 antiserum, as described above.
FACS-based Pore Formation Assay
Pore formation was detected by following the transfer of the small aqueous calcein red-orange dye from target Vero cells to QM5 donor cells, as described previously (16, 26). Briefly, subconfluent QM5 cells in 6-well plates were co-transfected with 0.2 μg of plasmid encoding the enhanced green fluorescent protein (pEGFP) and 1.8 μg of pcDNA3 constructs encoding the indicated FAST proteins and at 6 h post-transfection were overseeded with Vero cells labeled with 10 μm calcein red-orange. Cells were co-cultured for 7 more h at 37 °C to allow fusion to proceed, and the cells were then resuspended, fixed, and analyzed by FACS, as described above. 10,000 EGFP-positive cells were gated on a FACSCalibur and analyzed for acquisition of calcein red-orange using FCS Express 2.0.
Inhibition of Cell Surface Thiol:Disulfide Oxidoreductases
The membrane-impermeant inhibitors of thiol:disulfide oxidoreductases, Bacitracin (5, 7.5, or 15 mm), or DTNB (2.5 or 7.5 mm) were added to cells 3 h post-transfection with plasmids encoding RRV p14, NBV p10, or ARV p10. Cells were incubated an additional 4 h at 37 °C for RRV p14 and NBV p10-transfected cells and 27 h more for ARVp10-transfected cells. Cell-cell fusion was then quantified by syncytial indexing, as described above.
RESULTS
Conserved Cysteine Residues in the ARV p10 Ectodomain Are Essential for Membrane Fusion Activity
Previous studies demonstrated an important role for the ARV and NBV p10 HPs and the two cysteines that flank these motifs (Cys-9 and Cys-21 in ARV p10) in syncytium formation (13, 27). However, the ARV p10 constructs all contained an N-terminal double hemagglutinin epitope tag that significantly reduced p10 fusion activity, and the influence of the cysteines on the actual membrane fusion reaction (i.e. pore formation) was also not determined. It was therefore unclear whether the cysteine residues are actually essential for p10-induced membrane fusion. To address this issue, we created serine substitutions of either cysteine (C9S and C21S) in an authentic ARV p10 background and assessed the effects on fusion pore formation and syncytiogenesis. QM5 cell monolayers transfected with authentic ARV p10 induced extensive syncytium formation, commencing at 12–16 h post-transfection and progressing through to 32–36 h post-transfection (Fig. 1A). No such cell-cell fusion was observed in cells transfected with the C9S or C21S constructs, even as late as 48–72 h post-transfection. The loss of cell-cell fusion activity was not due to decreased expression levels, as observed by Western blotting of whole-cell lysates (Fig. 1B). Cell surface trafficking of p10 was also largely unaffected by the cysteine substitutions, as determined by flow cytometry (Fig. 1C). The modest 20–40% decrease in cell surface expression of C9S and C21S had no effect on cell-cell fusion, as determined by titration of the plasmid dose used for transfection to generate equivalent cell surface expression levels (data not shown).
FAST protein-mediated cell-cell membrane fusion is refractory to standard assays used to detect hemifusion, the earliest event in membrane fusion (28). However, a quantitative fluorescent cell-cell pore formation assay was recently developed for use with the FAST proteins (29). This pore formation assay was used to directly examine the influence of the cysteines on the p10 membrane fusion reaction. Subconfluent QM5 cells were co-transfected with the p10 constructs and pEGFP (as a marker for transfection), and 6 h post-transfection cells were overseeded with target Vero cells labeled with the small fluorescent aqueous dye calcein red-orange. The cells were co-cultured for 7 h to allow fusion to proceed, and the trypsinized cells were then fixed in suspension, and EGFP-positive cells were gated by flow cytometry and analyzed for acquisition of the red aqueous dye. Although authentic p10 induced extensive pore formation and content mixing, the C9S and C21S constructs failed to induce any dye transfer above background levels observed in vector-transfected cells (Fig. 1D). The ectodomain cysteine residues therefore play an essential role in p10-mediated pore formation and membrane fusion.
Cysteine Residues Flanking the Fusion Peptide of ARV p10 Form an Essential Intramolecular Disulfide Bond
Previous results indicated p10 does not form disulfide-stabilized multimers (13), leading to speculation that the ectodomain cysteines may participate in an intramolecular disulfide bond. To directly test this hypothesis, we utilized surface biotinylation to assess the status of the thiol groups of the two cysteine residues in the p10 ectodomain. At 24 h post-transfection, live cells were either left untreated or subjected to a 5-min treatment with 0.1 mm DTT at room temperature to reduce disulfide bonds. Free thiol groups on the surface of transfected cells were then labeled by incubating cells with the thiol-reactive, membrane-impermeable biotinylation reagent maleimide-PEG2-biotin. Biotinylated proteins were then isolated with neutravidin beads and fractionated by SDS-PAGE, and Western blots were probed with antiserum specific for the ARV p10 endodomain.
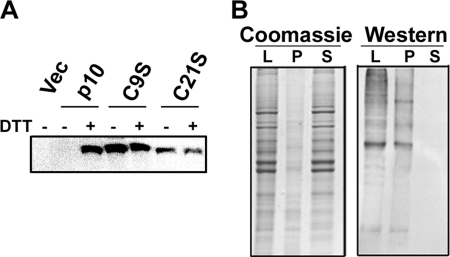
In the absence of DTT treatment, no p10 protein was detected by biotinylation and Western blotting (Fig. 2A). Conversely, p10 was clearly detected following DTT treatment prior to the biotinylation reaction. These results were consistent with the absence of free thiols in the surface-localized ectodomain of p10, suggesting the cysteines were engaged in disulfide bond formation and only susceptible to biotinylation following reduction of the disulfide bond. No p10-specific bands migrating above authentic p10 were detected on nonreducing gels (13), indicating p10 does not form intermolecular disulfide linkages with heterologous cell proteins. Similar analysis was performed on C9S- and C21S-transfected cells. With only one cysteine present in the ectodomain of these constructs, the free thiol group should be available for biotinylation without prior DTT treatment. Such was the case; both C9S and C21S were readily biotinylated to the same degree in the absence or presence of prior DTT treatment (Fig. 2A). To confirm that the biotinylation reagent was membrane-impermeable and therefore labeling the p10 ectodomain and not the palmitoylated endodomain di-cysteine motif, cells were treated with DTT and reacted with the biotin reagent, and cell lysates were fractionated into the membrane pellet and soluble cytosolic fractions. Probing Western blots of these fractions with HRP-conjugated neutravidin detected biotinylated proteins in the membrane fraction but not the cytosolic fraction (Fig. 2B), indicating that the maleimide reagent was indeed unable to traverse the plasma membrane to any significant degree. The two conserved cysteine residues flanking the p10 HP therefore form an intramolecular disulfide bond, creating an 11-residue cystine loop. Based on these results, and considering the amino acid content and lipid mixing abilities of the p10 HP (13), we conclude that p10 membrane fusion activity is dependent on the ability of the HP to assume a disulfide-stabilized fusion loop structure.
FIGURE 2.
Cysteine residues in the p10 ectodomain form an intramolecular disulfide bond. A, at 24 h post-transfection with the indicated p10 constructs, QM5 cells were incubated for 5 min in HBSS with or without 0.1 mm DTT. Free thiols were then labeled with maleimide-PEG2-biotin, and biotinylated proteins were isolated from cell lysates using immobilized neutravidin beads and resolved by SDS-PAGE, and Western blots were probed with ARV p10-specific antiserum. Vec, vector. B, QM5 cells were treated with DTT followed by maleimide-PEG2-biotin as in A, and cell lysates (L) were fractionated into membrane pellets (P) and soluble (S) fractions. Gels were then either Coomassie-stained or processed for Western blotting using HRP-conjugated neutravidin to detect biotinylated proteins.
Formation and Function of the Cystine Loop Is Highly Sensitive to Location and Context of Cysteine Residues
Previous mutational analyses indicate the ARV and NBV p10 HPs are adversely affected by substitutions at numerous sites (13, 27). At the time of the mutational studies, the presence of a cystine loop had not been demonstrated. It is therefore unclear whether the effects of substitutions reflected a direct role for a specific residue within the HP or indirect effects on formation of the cystine loop. To examine topological factors influencing formation of the p10 cystine loop, a series of cysteine shift constructs were created by amino acid substitutions to effectively manipulate the size and position of the loop within the ectodomain (Fig. 3A). For example, Cys-9 was substituted with a serine, and the serine at position 8 (Ser-8) was replaced with a cysteine (S8C/C9S), effectively extending the loop one residue toward the N terminus and altering the geometry of the loop (e.g. changing the residues at the apex of the loop). Eight such shift constructs were generated (S1–S8; Fig. 3A). Some shift constructs only moved a single cysteine one residue position at a time to increase or decrease the size of the loop by one amino acid (S1, S2, S4, and S5). Other shift constructs maintained the position of the loop in the p10 ectodomain and the hydrophobic residues at the apex of the loop (i.e. Val-15 and Phe-16) while increasing or decreasing the loop size by two amino acids (S3 and S6, respectively). Other constructs maintained the size of the loop yet shifted the location and position of amino acids within the loop by shifting both cysteines in the same direction simultaneously (S7, S8; Fig. 3A). The effects of these substitutions on surface expression, membrane fusion activity (syncytium formation and pore formation), and loop formation were examined in transfected cells.
FIGURE 3.
p10 fusion activity and intramolecular disulfide bond formation are highly sensitive to the location and context of the cysteine residues. A, point substitutions were introduced in the p10 ectodomain to create cysteine shift constructs (S1–S8). Substituted residues are underlined, and arrows depict the relative shift of the Cys-9 and Cys-21 residues. B, surface expression of each shift construct (S1–S8) was determined by live cell staining transfected cells followed by flow cytometric analysis at 24 h post-transfection. C, syncytium-inducing capacity of the p10 shift constructs at 32 h post-transfection was determined using Wright-Giemsa-stained monolayers to quantify the average numbers of syncytial nuclei in five random microscopic fields at ×200, and results are presented relative to authentic p10 set at 100%. Results in B and C are presented as the mean ± S.D. relative to authentic p10 from one of two experiments conducted in triplicate. D, pore formation was quantified based on the transfer of the calcein red-orange dye from target Vero cells to donor QM5 cells co-transfected with the indicated constructs and pEGFP as a marker for transfection. Results shown are representational dot plots of calcein fluorescence versus forward scatter (FSC) of EGFP-gated cells from one of two experiments conducted in triplicate. E, at 24 h post-transfection with the indicated p10 plasmid constructs, cells were incubated in HBSS with or without 0.1 mm DTT; free thiol groups were labeled with maleimide-PEG2-biotin; biotinylated proteins were isolated with immobilized neutravidin beads and fractionated by SDS-PAGE, and Western blots were probed with p10-specific antiserum. V, vector.
Although all of the shift constructs were expressed on the surface of transfected cells at levels approximately equivalent to authentic p10 (Fig. 3B), none of these constructs was capable of inducing either syncytium formation (Fig. 3C) or pore formation (Fig. 3D). More surprising, when the formation of the cystine loop was investigated using the biotinylation assay, none of the shift constructs, with the exception of S5, retained the ability to form an intramolecular disulfide bond, indicated by the susceptibility of their cysteine residues to efficient biotinylation in the absence of DTT treatment (Fig. 3E). Formation of the p10 cystine loop is therefore highly sensitive to the amino acid content around the cysteine residues and/or changes in the size/location of the loop in the p10 ectodomain. The function of the p10 cystine loop was also sensitive to changes in the geometry of the loop. The S5 construct (H20C/C21S), which shortened the loop by one residue from the C terminus, could not be biotinylated in the absence of prior treatment with DTT (Fig. 3E), confirming formation of the cystine loop, yet this construct was devoid of membrane fusion activity (Fig. 3, C and D). Therefore, not only is the formation of the cysteine loop essential for p10 fusion activity, but the sequence and/or arrangement of amino acids within the fusion loop is also critical to p10-mediated membrane fusion activity.
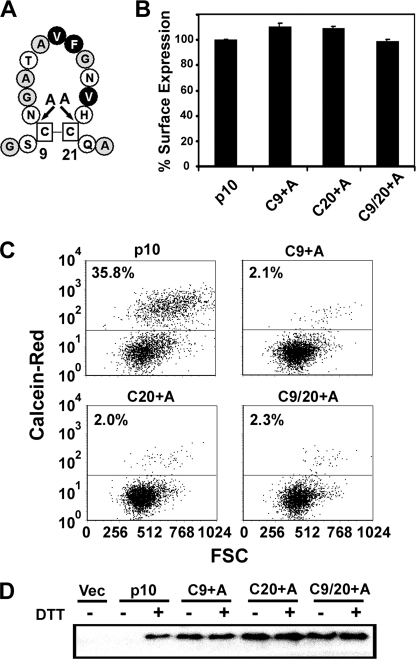
A minimum of two, and as many as four, amino acid substitutions were used to shift the cystine loop in the above constructs. It was therefore conceivable that the negative effects of these substitutions reflected changes to the primary sequence within the loop rather than just changes to the architecture of the cystine loop. For instance, creation of the S4 construct (C9S/N10C) simultaneously decreased the size of the loop by one residue from the N-terminal side and removed the asparagine within the loop (Fig. 3A). Three additional constructs were therefore created to manipulate the size and geometry of the loop without replacing residues normally present in the loop. Alanine residues were inserted within the loop, immediately adjacent to either cysteine residue, either one at a time or concurrently (Fig. 4A). These insertions effectively increased the size of the loop by one amino acid in either direction (Cys-9 + Ala and Cys-20 + Ala), or conserved the relative position of residues within the loop by increasing its size by two amino acids (Cys-9/20 + Ala). Once again, despite normal levels of protein trafficking and expression in the plasma membrane of transfected cells (Fig. 4B), the syncytium formation (data not shown) and pore formation (Fig. 4C) activities of these insertion mutants were abolished. In addition to membrane fusion activity, intramolecular disulfide bond formation was also disrupted in these alanine insertion constructs, as evidenced by the presence of free thiols susceptible to biotinylation in non-DTT-treated cells (Fig. 4D). The dramatic effects on formation of the p10 cystine loop due to diverse, conservative changes in the amino acid context of the cysteines highlights the complex factors that govern disulfide bond formation and stability (30).
FIGURE 4.
Fusion activity and intramolecular disulfide bond formation are dependent on the size, location, and residue context of the cysteine residues. A, alanine residues were inserted in the ARV p10 ectodomain adjacent to Cys-9 and Cys-21, either individually or in combination (Cys-9 + Ala, Cys-20 + Ala, and Cys-9/20 + Ala, respectively) to change the size and geometry of the cystine loop. Hydrophobic, apolar, and polar residues are indicated on black, gray, or white backgrounds, respectively. B, surface expression of cells transfected with the indicated p10 constructs was determined by FACS analysis following live cell labeling with p10-specific antiserum and fluorescently conjugated secondary antibodies. Results represent the mean ± S.D. relative to authentic p10 surface expression from one of two experiments conducted in triplicate. C, calcein red-orange-labeled Vero cells were incubated with donor QM5 cells co-transfected with the indicated p10 constructs and pEGFP as a transfection marker. Pore formation was detected using FACS analysis to quantify transfer of the calcein dye from target Vero to donor QM5 cells. Results are presented as representational dot plots of EGFP-gated QM5 cells versus forward scatter (FSC) from one of two experiments conducted in triplicate. D, QM5 cells transfected with the indicated p10 plasmids were treated with or without DTT; free thiols were labeled with maleimide-PEG2-biotin, and p10-biotinylation was detected by precipitating biotinylated proteins with immobilized neutravidin beads, followed by Western blotting with p10-specific antiserum. Vec, vector.
Efficient p10-mediated Membrane Fusion Requires Reduction of the Intramolecular Disulfide Bond by Cell Surface Thioredoxins
Enveloped virus FPs frequently undergo structural transitions as they interact with membranes, and such structural plasticity appears to be essential for their membrane fusion activity (21, 31). This observation seemed at odds with our demonstration that the p10 FP is likely constrained within the essential cystine loop. However, the cell surface reduction or isomerization of disulfide bonds has been reported to be essential for the function of several enveloped virus fusion proteins (32, 33). It therefore seemed possible that cell surface-localized members of the thioredoxin superfamily of oxidoreductases could disrupt the p10 cystine loop. Two membrane-impermeable cell surface inhibitors of thiol:disulfide oxidoreductases were employed to test the importance of disulfide bond reduction in p10 protein function as follows: the nonspecific free thiol-binding reagent DTNB (34), and the thiol-disulfide isomerase CXXC catalytic domain-binding reagent Bacitracin (35).
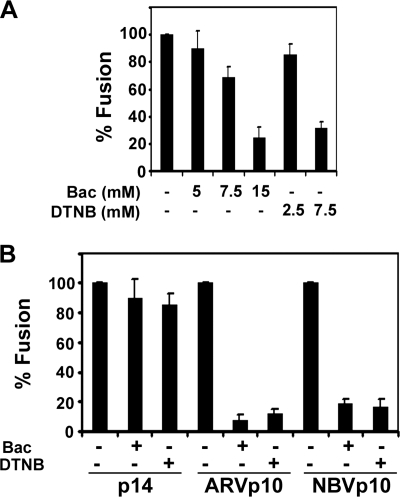
Following a 3-h transfection, QM5 cells were further incubated with regular growth media supplemented with either Bacitracin or DTNB at 37 °C. At various times post-transfection, cells were fixed and stained with Wright-Giemsa, and syncytial nuclei were quantified. Preliminary experiments titrated the Bacitracin and DTNB concentrations to determine appropriate levels to avoid off-target effects, using the reptilian reovirus p14 FAST protein as a control (p14 does not contain any cysteine residues in its ectodomain and therefore cannot form disulfide bonds). Bacitracin concentrations of 5–7.5 mm had no significant effect on p14-induced syncytium formation, whereas concentrations of 15 mm had a pronounced inhibitory effect (Fig. 5A). Similarly, p14-induced cell-cell fusion was unaffected by 2.5 mm DTNB, although 7.5 mm DTNB reduced syncytium formation by 70% (Fig. 5A). Unlike the p14 negative control, low doses of either DTNB (2.5 mm) or Bacitracin (5 mm) inhibited ARV p10-induced fusion activity by >90% (Fig. 5B). Similar results were obtained in cells transfected with the homologous NBV p10 FAST protein, which was inhibited by >80% by these inhibitors of the thioreductase superfamily (Fig. 5B). These results suggest that at some point in the fusion reaction the cystine loop present in both NBV and ARV plasma membrane-localized p10 must be reduced by cell surface thioredoxins in order for fusion to proceed.
FIGURE 5.
Reduction of the intramolecular disulfide bond by cell surface oxidoreductases is essential for efficient p10-mediated membrane fusion. A, at 3 h post-transfection with RRV p14, QM5 cells were incubated with the indicated concentrations of the cell surface thiol:disulfide oxidoreductase inhibitors Bacitracin (Bac) or DTNB. Cells were fixed 4 h later and Wright-Giemsa-stained, and syncytium formation was quantified by counting syncytial nuclei from five random microscopic fields at ×200 magnification. B, QM5 cells were transfected with either p14, ARV p10, or NBV p10, and 3 h later were incubated with 5 mm Bacitracin or 2.5 mm DTNB. At 7 h post-transfection with p14 or NBV p10, or 30 h post-transfection with ARV p10, cells were fixed and Wright-Giemsa-stained, and fusion was quantified by syncytial indexing as described in A. All results are presented as the mean ± S.D. relative to authentic p10 fusion activity from one of two experiments conducted in triplicate.
DISCUSSION
Membrane fusion mediated by viral fusogens is dependent upon the action of FPs. The structural features of the FPs vary between the different classes of viral fusogens, and the precise role of these motifs in the fusion reaction remains a matter of speculation. The present results indicate the following: 1) the p10 HP exists as a disulfide-stabilized loop, and loop formation is highly sensitive to the amino acid context of the cysteines that form the loop; 2) formation of this loop is essential for p10-induced membrane fusion; 3) the amino acid content and/or geometry of residues within the loop affects fusion activity independent of loop formation; and 4) cell surface-localized thiol:disulfide oxidoreductase activity is required for p10 membrane fusion activity. The present demonstration that the p10 HP forms a cystine loop coupled with previous results demonstrating the HP has membrane destabilizing properties (13) indicate the p10 HP functions as a disulfide-stabilized fusion loop. The p10 FP is somewhat reminiscent of the larger fusion loops present in some class I, and all class II and III enveloped virus fusogens. However, several fundamental differences in the physical and functional properties of these fusion loops define the p10 HP as a distinct class of viral fusion loops, the features of which impact on models of how the p10 HP might function as an FP.
Although previous mutagenic analysis indicated the importance of the two cysteine residues conserved in the ectodomains of the ARV and NBV p10 FAST proteins (13, 27), there was no direct evidence to support speculation that these cysteines might participate in disulfide bond formation. Using a cell surface biotinylation assay in combination with cysteine substitutions and reducing agent, we demonstrate that the p10 ectodomain cysteines form an intramolecular disulfide bond creating an 11-residue cystine loop structure (Fig. 2). Substitution analysis further indicated that the residues flanking the two cysteines have a profound effect on loop formation. Formation of the p10 cystine loop in the lumen of the endoplasmic reticulum is presumably catalyzed by a member of the thioredoxin superfamily of enzymes (36). Enzyme-substrate recognition and binding by thioredoxins is believed to be primarily hydrophobic in nature and may involve only a very limited region of the substrate (37). The p10 cysteines involved in disulfide bond formation are flanked by polar residues (Ser-Cys-Asn and His-Cys-Gln). Cysteine shift mutants that either slightly increase or slightly decrease the hydrophobicity of these residues on either side of the two cysteines all eliminated disulfide bond formation (Fig. 3). It therefore seems unlikely that all of the various p10 substitutions would impede formation of the cystine loop by altering p10 recognition by the thio-disulfide oxidoreductase responsible for disulfide bond formation. Previous studies have shown a correlation between changes in side chain volume and disulfide stability, suggesting that the geometry of the disulfide or strains in adjacent bonds influence disulfide formation or stability (30). We therefore favor the hypothesis that steric factors, either within the p10 loop itself or flanking the two cysteine residues, alter the formation or stability of the disulfide bond.
The correlation between formation of the cystine loop and membrane fusion activity, coupled with previous results indicating the p10 HP has membrane destabilizing properties (13), indicates the p10 HP functions as an FP and is a member of the fusion loop class of viral FPs. The specific features of the p10 cystine loop, however, are distinct from the fusion loops of the enveloped viruses. In the case of Ebola virus, the FP is contained within a 46-residue disulfide-stabilized structure of anti-parallel β-strands that serves as a scaffold to present the partly helical 16-residue FP (38, 39). The 16-residue FP of avian sarcoma/leukosis virus also appears to be helical and may be presented within a 35-residue disulfide-stabilized order-turn-order structure (40, 41). The internal FPs of all of the class II enveloped virus fusion proteins are also presented at the tips of elongated, disulfide-stabilized structures composed of anti-parallel β-strands (42–44), whereas the FPs of the class III enveloped virus fusion proteins, such as the vesicular stomatitis virus G protein, are composed of two 10–11-residue flexible loops contained at the tips of three elongated β-strands that are stabilized by disulfide bonds (45, 46). All of the viral FPs that function as fusion loops are therefore components of larger extended structures, with the disulfide bonds serving to stabilize the higher order structure of the overall fusion domain and not the FP per se (23).
The features of the enveloped virus fusion loops contrast markedly with those of the p10 cystine loop. Most notably, rather than stabilizing an extended structure that contains the FP, the cysteines in the p10 ectodomain stabilize an 11-residue loop that appears to constitute the actual FP. Unlike the fusion loops of the α-retroviruses and Ebola virus, where proline residues may be important for formation of the turn in the loop (47, 48), the p10 cystine loop lacks prolines, and loop formation directly relies on formation of the intramolecular disulfide bond. The p10 cystine loop resembles the tips of the fusion loops of the class II and class III enveloped viruses, which exist as open, compact, or flexible loops (49–51). However, unlike the class III enveloped virus fusion loops, the p10 cystine loop is not bipartite. The small size of the p10 ectodomain also makes it unlikely that the fusion loop is shielded from the aqueous environment within a complex tertiary structure, as is the case with the class II enveloped virus fusion loops. In addition, the p10 disulfide bond is required to directly maintain the geometry of the actual FP, as opposed to stabilizing a more complex structure that serves to expose the FP in the fusion loops of the class II and III enveloped virus fusogens. The p10 HP therefore represents a new class of viral FPs that function as a small, spatially constrained cystine loop.
In addition to the formation of the cystine loop, the specific geometry or amino acid content of residues within the loop also determine the role of this structure as an FP. The S5 cysteine shift mutant was the only substitution that formed the cystine loop but was unable to support membrane fusion (Fig. 3). This construct moved the C-terminal cysteine one residue to the left, which had two consequences. First, the conserved His-20 residue adjacent to Cys-21 was deleted from the loop. How this histidine might contribute to the function of the p10 HP as an FP is unclear. Second, the noose was shortened by one amino acid, effectively shifting the arrangement of residues within the loop. Although there is considerable sequence diversity in the ARV and NBV HPs (Fig. 1A), a valine and phenylalanine are conserved near the apex of the cystine loop, and an additional valine is conserved on one side of the loop (Fig. 4A). Mutational analysis indicates that very conservative changes in any of these three residues eliminates p10 membrane fusion activity (13, 27). Similar residues at the apex of the fusion loops in the class II and class III enveloped viruses are implicated in membrane insertion (52, 53), and the importance of positioning of residues within the fusion loops of enveloped viral fusion proteins has also been demonstrated (54). It is therefore conceivable that the loss of fusion activity displayed by the S5 shift mutant reflects repositioning of the conserved hydrophobic residues within the p10 loop. An emerging hypothesis for the function of FPs in membrane fusion suggests that shallow insertion of amphipathic structures into the outer leaflet of bilayers induces high curvature stresses that are resolved by bilayer fusion (1, 3, 55). We note that the p10 HP is more amphiphilic than hydrophobic in nature, similar to the situation with the fusion loop of vesicular stomatitis virus G, which contains aromatic residues for membrane interaction but is much less hydrophobic than most FPs (50). Displacement of the hydrophobic residues from the apex of the p10 cystine loop may therefore reduce membrane insertion of the amphiphilic p10 HP and prevent induction of the membrane curvature needed to promote membrane fusion.
The features of the p10 cystine loop suggest it may function as a cystine noose. Cystine nooses are an unusual structural motif that typically contain ∼4–10 residues constrained by an intramolecular disulfide bond (56). The ϕ and ψ angles of noosed residues fall outside the most favored regions of the Ramachandran plot, suggesting the disruption of most secondary structures. The steric effects of such extreme ϕ and ψ angles produce a three-dimensional structure that provides significant surface accessibility for the side chains contained within these small loops (57, 58). In the case of the G protein of respiratory syncytial virus, the cystine noose generates a global fold with some limited internal mobility of the side chains (59), whereas for measles virus hemagglutinin, the cystine noose assumes a rigid, amphipathic structure with hydrophilic side chains clustering on one surface and hydrophobic side chains on the other surface (60). Translating these observations to p10, we predict that the function of the cystine loop is to generate a noose structure that forces solvent exposure of the hydrophobic side chains for membrane interaction. The observation that p10 is rapidly degraded by the endoplasmic reticulum-associated degradation pathway that recognizes exposed hydrophobic residues (61), and that substitutions of the conserved hydrophobic residues or the cysteines (which we have now shown would eliminate noose formation) also prevented p10 degradation, supports the concept that the p10 loop functions to promote solvent exposure of hydrophobic residues. Although cystine nooses are known to promote membrane interaction and membrane disruption, as with small antimicrobial peptides such as the defensins (62), p10 would be the first example of a cystine noose involved in membrane fusion. However, the p10 fusion loop is slightly larger than most cystine nooses, many of which are flanked by more than one disulfide bond, and the preponderance of glycine and alanine residues in the p10 HP is also not a common feature of cystine nooses. Determining whether the p10 fusion loop functions as a cystine noose clearly requires determination of a high resolution structure. Given the demonstrated importance of flanking residues and the unknown role of the adjacent conserved region, such structural studies are probably best performed in the context of the entire 40-residue p10 ectodomain.
Although formation of a spatially constrained cystine loop is essential for p10 membrane fusion activity, the negative effects of two membrane-impermeant inhibitors of cell surface thiol oxidoreductase activity on p10 membrane fusion activity suggest disruption of the loop may be required for membrane fusion (Fig. 5). Cell surface-localized thiol:disulfide oxidoreductases are involved in a range of biological functions, including thrombus formation, gamete fusion, and activation of enveloped virus fusion proteins (32, 63, 64). In the case of the enveloped virus fusogens, it is unclear whether interfering with disulfide bond disruption or rearrangement affects the structure of the disulfide-stabilized fusion loops or the dramatic structural remodeling of their complex tertiary structures that is required for fusion activity. This is clearly not the case with p10, where the single disulfide bond in the small ectodomain is the only target for thiol-disulfide exchange, implying changes in the cystine loop FP are directly required for p10 membrane fusion activity. Structural plasticity is a hallmark feature of many enveloped viral FPs (31), presumably a reflection of how these peptides interact both with polar lipid headgroups and the hydrophobic interior of the membrane. If a cystine noose forces solvent exposure of the hydrophobic residues in the p10 HP, these residues may interact with the membrane in which p10 resides, inducing positive curvature of the donor membrane toward the target membrane. Such is not the case for the enveloped virus FPs that are usually sequestered within the complex pre-fusion tertiary structure of the fusion protein. Reduction or reduction and re-oxidation of the p10 cysteine residues by cell surface thiol:disulfide oxidoreductases may be required to allow structural transitions in the p10 HP that facilitate dynamic interactions of this FP motif with the donor and/or target membranes.
Unlike the paradigmatic enveloped virus fusion proteins, the rudimentary ectodomain of the FAST proteins precludes massive conformational changes in a complex tertiary structure as a means to regulate FP exposure, close membrane apposition, and membrane merger. As we have now shown, the p10 FAST protein uses FP that relies on dynamic structural changes to a small, spatially constrained cystine loop as a means to drive membrane fusion. Further detailed structural and functional analysis of this novel FP, and of the role of the remainder of the small p10 ectodomain, including the conserved region adjacent to the HP, should provide additional insights into the mechanism of action of this singular family of viral membrane fusion proteins.
This research was supported in part by a grant from the Canadian Institutes of Health Research (to R. D.).
- FP
- fusion peptide
- FAST
- fusion-associated small transmembrane
- HP
- hydrophobic patch
- ARV
- avian reovirus
- NBV
- Nelson Bay reovirus
- RRV
- reptilian reovirus
- HRP
- horseradish peroxidase
- HBSS
- Hanks' buffered salt solution
- DTT
- dithiothreitol
- EGFP
- enhanced green fluorescent protein
- DTNB
- 5′5-dithiobis(2-nitrobenzoic acid)
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.Martens S., McMahon H. T. (2008) Nat. Rev. Mol. Cell Biol. 9, 543–556 [DOI] [PubMed] [Google Scholar]
- 2.Earp L. J., Delos S. E., Park H. E., White J. M. (2005) Curr. Top. Microbiol. Immunol. 285, 25–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernomordik L. V., Kozlov M. M. (2008) Nat. Struct. Mol. Biol. 15, 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melikyan G. B. (2008) Retrovirology 5, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanovic T., Agosto M. A., Zhang L., Chandran K., Harrison S. C., Nibert M. L. (2008) EMBO J. 27, 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowling W., Denisova E., LaMonica R., Mackow E. R. (2000) J. Virol. 74, 6368–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan S. H., Wirblich C., Forzan M., Roy P. (2001) J. Virol. 75, 8356–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falconer M. M., Gilbert J. M., Roper A. M., Greenberg H. B., Gavora J. S. (1995) J. Virol. 69, 5582–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan R., Corcoran J., Shou J., Stoltz D. (2004) Virology 319, 131–140 [DOI] [PubMed] [Google Scholar]
- 10.Shmulevitz M., Duncan R. (2000) EMBO J. 19, 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salsman J., Top D., Boutilier J., Duncan R. (2005) J. Virol. 79, 8090–8100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizo J., Rosenmund C. (2008) Nat. Struct. Mol. Biol. 15, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmulevitz M., Epand R. F., Epand R. M., Duncan R. (2004) J. Virol. 78, 2808–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Top D., de Antueno R., Salsman J., Corcoran J., Mader J., Hoskin D., Touhami A., Jericho M. H., Duncan R. (2005) EMBO J. 24, 2980–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salsman J., Top D., Barry C., Duncan R. (2008) PLoS Pathog 4, e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Top D., Barry C., Racine T., Ellis C. L., Duncan R. (2009) PLoS Pathog. 5, e1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corcoran J. A., Duncan R. (2004) J. Virol. 78, 4342–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawe S., Corcoran J. A., Clancy E. K., Salsman J., Duncan R. (2005) J. Virol. 79, 6216–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racine T., Hurst T., Barry C., Shou J., Kibenge F., Duncan R. (2009) J. Virol. 83, 5951–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shmulevitz M., Salsman J., Duncan R. (2003) J. Virol. 77, 9769–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamm L. K., Han X., Li Y., Lai A. L. (2002) Biopolymers 66, 249–260 [DOI] [PubMed] [Google Scholar]
- 22.Pécheur E. I., Sainte-Marie J., Bienven E. A., Hoekstra D. (1999) J. Membr. Biol. 167, 1–17 [DOI] [PubMed] [Google Scholar]
- 23.White J. M., Delos S. E., Brecher M., Schornberg K. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han X., Bushweller J. H., Cafiso D. S., Tamm L. K. (2001) Nat. Struct. Biol. 8, 715–720 [DOI] [PubMed] [Google Scholar]
- 25.Racine T., Barry C., Roy K., Dawe S. J., Shmulevitz M., Duncan R. (2007) J. Biol. Chem. 282, 25613–25622 [DOI] [PubMed] [Google Scholar]
- 26.Clancy E. K., Duncan R. (2009) J. Virol. 83, 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng L. T., Plemper R. K., Compans R. W. (2005) J. Virol. 79, 1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clancy E. K., Barry C., Ciechonska M., Duncan R. (2010) Virology 397, 119–129 [DOI] [PubMed] [Google Scholar]
- 29.Barry C., Duncan R. (2009) J. Virol. 83, 12185–12195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldenberg D. P., Bekeart L. S., Laheru D. A., Zhou J. D. (1993) Biochemistry 32, 2835–2844 [DOI] [PubMed] [Google Scholar]
- 31.Reichert J., Grasnick D., Afonin S., Buerck J., Wadhwani P., Ulrich A. S. (2007) Eur. Biophys. J. 36, 405–413 [DOI] [PubMed] [Google Scholar]
- 32.Fenouillet E., Barbouche R., Jones I. M. (2007) Antioxid. Redox. Signal. 9, 1009–1034 [DOI] [PubMed] [Google Scholar]
- 33.Jain S., McGinnes L. W., Morrison T. G. (2007) J. Virol. 81, 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feener E. P., Shen W. C., Ryser H. J. (1990) J. Biol. Chem. 265, 18780–18785 [PubMed] [Google Scholar]
- 35.Mou Y., Ni H., Wilkins J. A. (1998) J. Immunol. 161, 6323–6329 [PubMed] [Google Scholar]
- 36.Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 37.Hatahet F., Ruddock L. W. (2007) FEBS J. 274, 5223–5234 [DOI] [PubMed] [Google Scholar]
- 38.Lee J. E., Fusco M. L., Hessell A. J., Oswald W. B., Burton D. R., Saphire E. O. (2008) Nature 454, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freitas M. S., Gaspar L. P., Lorenzoni M., Almeida F. C., Tinoco L. W., Almeida M. S., Maia L. F., Degrève L., Valente A. P., Silva J. L. (2007) J. Biol. Chem. 282, 27306–27314 [DOI] [PubMed] [Google Scholar]
- 40.Delos S. E., White J. M. (2000) J. Virol. 74, 9738–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng S. F., Wu C. W., Kantchev E. A., Chang D. K. (2004) Eur. J. Biochem. 271, 4725–4736 [DOI] [PubMed] [Google Scholar]
- 42.Gibbons D. L., Vaney M. C., Roussel A., Vigouroux A., Reilly B., Lepault J., Kielian M., Rey F. A. (2004) Nature 427, 320–325 [DOI] [PubMed] [Google Scholar]
- 43.Modis Y., Ogata S., Clements D., Harrison S. C. (2004) Nature 427, 313–319 [DOI] [PubMed] [Google Scholar]
- 44.Rey F. A., Heinz F. X., Mandl C., Kunz C., Harrison S. C. (1995) Nature 375, 291–298 [DOI] [PubMed] [Google Scholar]
- 45.Heldwein E. E., Lou H., Bender F. C., Cohen G. H., Eisenberg R. J., Harrison S. C. (2006) Science 313, 217–220 [DOI] [PubMed] [Google Scholar]
- 46.Roche S., Bressanelli S., Rey F. A., Gaudin Y. (2006) Science 313, 187–191 [DOI] [PubMed] [Google Scholar]
- 47.Delos S. E., Gilbert J. M., White J. M. (2000) J. Virol. 74, 1686–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito H., Watanabe S., Sanchez A., Whitt M. A., Kawaoka Y. (1999) J. Virol. 73, 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kielian M., Rey F. A. (2006) Nat. Rev. Microbiol. 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roche S., Albertini A. A., Lepault J., Bressanelli S., Gaudin Y. (2008) Cell. Mol. Life Sci. 65, 1716–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiasny K., Heinz F. X. (2006) J. Gen. Virol. 87, 2755–2766 [DOI] [PubMed] [Google Scholar]
- 52.Allison S. L., Schalich J., Stiasny K., Mandl C. W., Heinz F. X. (2001) J. Virol. 75, 4268–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannah B. P., Cairns T. M., Bender F. C., Whitbeck J. C., Lou H., Eisenberg R. J., Cohen G. H. (2009) J. Virol. 83, 6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balliet J. W., Gendron K., Bates P. (2000) J. Virol. 74, 3731–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martens S., Kozlov M. M., McMahon H. T. (2007) Science 316, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 56.Lapthorn A. J., Janes R. W., Isaacs N. W., Wallace B. A. (1995) Nat. Struct. Biol. 2, 266–268 [DOI] [PubMed] [Google Scholar]
- 57.Janes R. W., Peapus D. H., Wallace B. A. (1994) Nat. Struct. Biol. 1, 311–319 [DOI] [PubMed] [Google Scholar]
- 58.Lapthorn A. J., Harris D. C., Littlejohn A., Lustbader J. W., Canfield R. E., Machin K. J., Morgan F. J., Isaacs N. W. (1994) Nature 369, 455–461 [DOI] [PubMed] [Google Scholar]
- 59.Sugawara M., Czaplicki J., Ferrage J., Haeuw J. F., Power U. F., Corvaïa N., Nguyen T., Beck A., Milton A. (2002) J. Pept. Res. 60, 271–282 [DOI] [PubMed] [Google Scholar]
- 60.Pütz M. M., Hoebeke J., Ammerlaan W., Schneider S., Muller C. P. (2003) Eur. J. Biochem. 270, 1515–1527 [DOI] [PubMed] [Google Scholar]
- 61.Shmulevitz M., Corcoran J., Salsman J., Duncan R. (2004) J. Virol. 78, 5996–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider J. J., Unholzer A., Schaller M., Schäfer-Korting M., Korting H. C. (2005) J. Mol. Med. 83, 587–595 [DOI] [PubMed] [Google Scholar]
- 63.Cho J., Furie B. C., Coughlin S. R., Furie B. (2008) J. Clin. Invest. 118, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellerman D. A., Myles D. G., Primakoff P. (2006) Dev. Cell 10, 831–837 [DOI] [PubMed] [Google Scholar]