Abstract
Insulin and insulin-like growth factor-1 (IGF-1) act on highly homologous receptors, yet in vivo elicit distinct effects on metabolism and growth. To investigate how the insulin and IGF-1 receptors exert specificity in their biological responses, we assessed their role in the regulation of gene expression using three experimental paradigms: 1) preadipocytes before and after differentiation into adipocytes that express both receptors, but at different ratios; 2) insulin receptor (IR) or IGF1R knock-out preadipocytes that only express the complimentary receptor; and 3) IR/IGF1R double knock-out (DKO) cells reconstituted with the IR, IGF1R, or both. In wild-type preadipocytes, which express predominantly IGF1R, microarray analysis revealed ∼500 IGF-1 regulated genes (p < 0.05). The largest of these were confirmed by quantitative PCR, which also revealed that insulin produced a similar effect, but with a smaller magnitude of response. After differentiation, when IR levels increase and IGF1R decrease, insulin became the dominant regulator of each of these genes. Measurement of the 50 most highly regulated genes by quantitative PCR did not reveal a single gene regulated uniquely via the IR or IGF1R using cells expressing exclusively IGF-1 or insulin receptors. Insulin and IGF-1 dose responses from 1 to 100 nm in WT, IRKO, IGFRKO, and DKO cells re-expressing IR, IGF1R, or both showed that insulin and IGF-1 produced effects in proportion to the concentration of ligand and the specific receptor on which they act. Thus, IR and IGF1R act as identical portals to the regulation of gene expression, with differences between insulin and IGF-1 effects due to a modulation of the amplitude of the signal created by the specific ligand-receptor interaction.
Keywords: Gene Expression, Insulin, Insulin-like Growth Factor (IGF), Microarray, Receptors, Transcription Regulation
Introduction
Insulin and insulin-like growth factor-1 (IGF-1)2 are closely related hormones that control different aspects of growth and metabolism in many organisms. They act on specific tyrosine kinase receptors, i.e. the insulin receptor (IR) and the IGF-1 receptor (IGF1R), which, once activated, elicit the activation of a cascade of intracellular proteins leading to the regulation of gene expression, protein synthesis, cell proliferation or death, and glucose and lipid metabolism. Insulin and IGF-1 fully activate their own receptor, but can also bind and activate the other receptor, although with reduced affinity.
In mammals, the conventional view regarding the actions of insulin and IGF-1 is that in vivo insulin mediates mainly a metabolic response, whereas IGF-1 mediates growth promoting effects (1). This is supported by the phenotypes of insulin receptor and IGF-1 receptor knock-out mice. Thus, mice lacking IGF1R display pronounced growth retardation and die shortly after birth due to respiratory insufficiency and failure to thrive (2), whereas mice with knock-out of the IR exhibit only slight growth retardation, but die during the first week of life from severe hyperglycemia and diabetic ketoacidosis (3, 4). In addition, phenotypes of the insulin and IGF-1 knock-out mice are very similar to those of IRKO and IGFRKO, indicating that the ability of the two receptors to compensate for each other is limited (5–7). Combined ablation of IGF-1 and IGF1R results in the same phenotype as lack of IGF1R alone suggesting that IGF-1 signals exclusively through IGF1R (7). These findings suggest distinct patterns of signaling by the IR and IGF1R. However, these differences could also reflect different patterns and timing of IR and IGF1R expression, leading to different responses when genetically inactivating the receptors.
Indeed, evidence exists that insulin and IGF-1 can mediate very similar responses. Thus, in some hyperinsulinemic states in infants, hyperinsulinemia produces early growth, whereas babies with diabetes and hypoinsulinemia are short (8). In states of insulin resistance, IGF-1 can regulate glucose metabolism. For example, administration of IGF-1 to IRKO mice decreases plasma glucose levels by action through the IGF1R on skeletal muscle (9). Similarly, people with type 2 or type 1 diabetes can respond to IGF-1 with a beneficial effect on glucose homeostasis (10–14).
Several studies have tried to elucidate some of the factors controlling the specificity of insulin and IGF-1 effects by focusing on the intracellular signals generated from the activation of the IR or the IGF1R (1, 15). Chimeric receptors consisting of the ligand binding domain of IR and the cytoplasmic domain of IGF1R function more like the IGF1R than the IR regarding their mitogenic capacity (16), whereas chimeric IGF1R containing the carboxyl-terminal β subunit domain of the IR more closely resemble the IR than the IGF1R regarding glycogen synthesis (17). When the extracellular portion of the neurotrophin receptor was fused to the intracellular portions of IR or IGF1R and stably expressed in 3T3-L1 cells (18, 19), activation by nerve growth factor of the TrkC-IR chimeric receptor was more effective in stimulating metabolic responses, whereas the TrkC-IGF1R was more effective in promoting mitogenesis. Structural differences of the β subunit and kinase domains of the IR and the IGF1R leading to differences in substrate interactions have been suggested to be partly responsible for insulin-IGF1 specificity (20).
Thus, whether or not IR and IGF1R have distinct or overlapping functions remains to be elucidated. Studies dependent on endogenous IR and IGF1R may be biased by differences in the level of expression and in affinity of the receptors for their respective ligands, whereas overexpression of the receptors does not circumvent the potential activation of endogenous receptors. In this study, we combined three strategies to assess the contribution of insulin and IGF-1 receptors in the regulation of gene expression induced by their respective ligands: 1) using brown preadipocyte cell lines before and after differentiation, 2) using cells with a knock-out of either IR or IGF1R, or 3) in IR/IGF1R double knock-out cells reconstituted with IR, IGF1R, or both. We found that IR and IGF1R act as identical portals to the regulation of gene expression, with differences between insulin and IGF-1 effects due to a modulation of the amplitude of the signal created by the specific ligand-receptor interaction.
EXPERIMENTAL PROCEDURES
Materials
Antibodies (with catalogue number) to IR (sc-711) and IGFR (sc-713) were purchased from Santa Cruz Biotechnologies. PPARγ antibody (catalogue number 07-466) was purchased from Upstate and β-tubulin antibody (number 2146) was purchased from Cell Signaling. Human insulin was purchased from Sigma and human IGF-1 from Preprotech.
Cell Isolation and Culture
Cells were derived from four strains of C57Bl/6 mice: 1) wild-type mice, 2) mice homozygous for a floxed allele of exon 4 of the insulin receptor (IRlox), 3) mice with a floxed allele of exon 3 of the IGF-I receptor (IGFRlox), and 4) mice with both floxed IR and IGF1R alleles (IRlox/IGFRlox). Preadipocytes were isolated from newborn control wild-type, IRlox, IGFRlox, or IRlox/IGFRlox mice by collagenase digestion of the brown fat pad as described previously (40) and immortalized by infection with a pBABE retrovirus encoding SV40 T-antigen followed by selection with 2 μg/ml of puromycin. For in vitro recombination of the insulin or IGF-I receptor, preadipocytes harboring the floxed allele of the insulin receptor (IRlox), IGF-I receptor (IGFRlox), or both were first plated at a subconfluent density. After 24 h, cells were infected with an adenovirus encoding Cre recombinase at a titer of 500 multiplicity of infection. After 1 h the viral supernatant was replaced with culture medium. Individual colonies were selected, and IR and/or IGF1R recombination was assessed by PCR of genomic DNA. Cells were maintained in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum at 37 °C in a 5% CO2 environment. Experiments were performed in the different KO cells lines using IRlox, IGFRlox, IRlox/IGFRlox, or wild-type cell lines as a control. Because wild-type, IRlox, IGFRlox, or IRlox/IGFRlox cell lines showed similar results, data obtained using the different control cell lines were pooled and referred to as WT in the text.
Retroviral Infection
IR and IGF1R were stably introduced into DKO cells by retroviral infection with a pBABE retrovirus encoding hIR, hIGF1R, or control vectors. Plates (10 cm) of human embryonic kidney 293T cells were transiently transfected with 10 μg of retroviral expression vectors and viral packaging vectors SV-E-MLV-env and SV-E-MLV using TransIT-Express transfection reagent (Mirus Bio Corp.). At 48 h after transfection, virus-containing medium was collected and passed through a 0.45-μm pore size syringe filter. Filter-sterilized Polybrene (hexadimethrine bromide; 12 μg/ml) was added to the virus-loaded medium. This medium was then applied to proliferating (40% confluent) DKO cells. 24 h after infection, cells were treated with trypsin and replated in a medium supplemented with zeocin and hygromycin (Invitrogen) as a selection antibiotic.
Adipocyte Differentiation
Adipocyte differentiation was induced in confluent WT or DKO preadipocytes by treating cells (day 0) with an induction mixture containing 20 nm insulin and 1 nm triiodothyronine, 0.5 mm isobutylmethylxanthine, 1 μm dexamethasone, and 0.125 mm indomethacin for 48 h. After this induction phase (day 2), cells were kept in medium containing insulin and triiodothyronine for the subsequent 6 days, changing the medium every 2 days. Lipid accumulation was visualized at day 8 by oil red O staining. Cells were washed once with phosphate-buffered saline and fixed with 10% buffered formalin for 1 h. Cells were then stained with filtered oil red O solution (5 g/liter in isopropyl alcohol) diluted 2-fold in water for 1 h at room temperature.
Cell Treatment
For all experiments, cells were washed twice and serum starved overnight with Dulbecco's modified Eagle's medium containing 0.1% bovine serum albumin. Cells were then treated with insulin or IGF-1 for 30 min or 6 h and then washed once with cold phosphate-buffered saline and resuspended in RLT lysis buffer (Qiagen).
Sample Preparation for Microarrays and Microarray Analysis
Total RNA from confluent WT preadipocytes treated or not with 10 nm IGF-1 for 30 min or 6 h were extracted using an RNeasy mini kit (Qiagen). Double-stranded cDNA synthesis was reverse transcribed from 15 μg of isolated RNA using the SuperScript Choice system (Invitrogen) using an oligo(dT) primer containing a T7 RNA polymerase promoter site. Double-stranded cDNA was purified with Phase Lock Gel (Eppendorf). Biotin-labeled cRNA was transcribed using a BioArray RNA transcript labeling kit (Enzo). A hybridization mixture containing 15 μg of biotinylated cRNA, adjusted for possible carryover of residual total RNA, was prepared and hybridized to mouse Affymetrix MG-U74A-v2 chips containing 12,488 probesets. The chips were washed, scanned, and analyzed with GENECHIP MAS version 5.0. For each group, 12 chips were used. All chips were subjected to global scaling to a target intensity of 1500 to take into account the inherent differences between the chips and their hybridization efficiencies. To obtain a list of genes differentially regulated by IGF-1, we selected genes that were significantly regulated by p < 0.05 using Student's t tests comparing IGF-1-treated and nontreated cells. We also eliminated probesets with an Affymetrix value lower than 300 for all conditions to reduce the number of false positive.
Analysis of Gene Expression by Quantitative PCR
Total RNA was extracted using an RNeasy mini kit (Qiagen). 1 μg of RNA was reverse transcribed using a high capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. Real time PCR was performed starting with 12.5 ng of cDNA and both sense and antisense oligonucleotides (300 nm each) in a final volume of 10 μl using the SYBR Green PCR master mix (Bio-Rad). Fluorescence was monitored and analyzed in an ABI Prism 7900 HT sequence detection system (Applied Biosystems). Analysis of TBP expression was performed in parallel to normalize gene expression. Amplification of specific transcripts was confirmed by analyzing melting curve profiles at the end of each PCR.
Cell Lysates and Immunoblotting
Cells were washed once with cold phosphate-buffered saline and scraped in radioimmunoprecipitation assay lysis buffer complemented with 1% SDS, 10 mm glycerophosphate, 10 mm NaF, 0.1 mm sodium orthovanadate, and 1% protease inhibitor mixture (Sigma). Protein concentrations were determined using the Bradford protein assay (Bio-Rad). Lysates (20 to 40 μg) were subjected to SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (Amersham Biosciences) and immunoblotted with the appropriate antibodies. Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) from donkey. Proteins on the membranes were visualized using Supersignal West Pico substrate or Supersignal West Dura extended duration substrate (Pierce).
RESULTS
Regulation of Gene Expression by IGF-1 in Wild-type Brown Preadipocytes
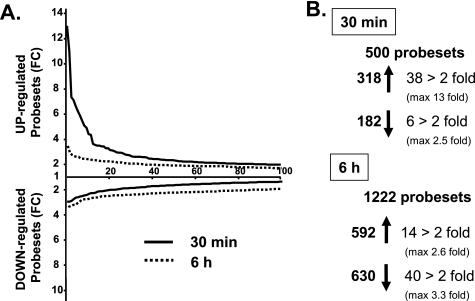
To identify the genes acutely and chronically regulated by insulin/IGF-1 signaling, we stimulated wild-type (WT) brown preadipocytes with 10 nm IGF-1 for 30 min and 6 h. Consistent with the higher expression of IGF1R versus IR in these cells (21), preliminary experiments indicated that IGF-1 was more potent to regulate gene expression than insulin. We then extracted RNA from nonstimulated or IGF-1-stimulated cells and measured gene expression using Affymetrix U74Av2 microarrays containing 12,488 probesets. Of all the probesets, 500 were found to be significantly regulated (p < 0.05) 30 min after IGF-1 treatment, with 318 being up-regulated and 182 being down-regulated (Fig. 1 and supplemental Table S1). Acute IGF-1 stimulation especially led to an up-regulation rather than down-regulation of numerous genes: 38 genes were up-regulated 2-fold or more (with a maximum fold-change of 13 for Egr-2) and only 6 genes were down-regulated 2-fold (50%) or more with the biggest change being a 60% decrease in Cdkn2a (Fig. 1 and Table 1).
FIGURE 1.
Number of probesets significantly regulated by IGF-1. A, fold-change (FC) representation of the 100 probesets most differentially regulated by a 10 nm IGF-1 treatment after 30 min (solid line) or 6 h (dotted line) in brown preadipocytes measured by microarrays. B, total number of probesets significantly up- or down-regulated (p < 0.05) by IGF-1 treatment or significantly regulated more than 2-fold after 30 min or 6 h.
TABLE 1.
Probesets significantly regulated more than 2-fold by a 30-min IGF-1 treatment in WT cells
44 probesets regulated more than 2-fold by 10 nm IGF-1 after 30 min in WT preadipocytes are listed with fold-change, gene symbol, full gene name, and fold-change measured by real time PCR after IGF-1 or insulin treatment (for selected genes).
| Probeset | Fold-change | Symbol | Name | Real time PCR |
|
|---|---|---|---|---|---|
| Fold-change IGF1 | Fold-change insulin | ||||
| 102661_at | 13.03 | Egr2 | Early growth response 2 | 10.88 | 8.04 |
| 160901_at | 11.02 | Fos | FBJ osteosarcoma oncogene | 14.03 | 9.24 |
| 99109_at | 7.33 | Ier2 | Immediate early response 2 | 5.99 | 3.69 |
| 93294_at | 7.15 | Ctgf | Connective tissue growth factor | 2.11 | 1.94 |
| 102362_i_at | 6.62 | Junb | JunB oncogene | 4.17 | 2.97 |
| 98579_at | 6.25 | Egr1 | Early growth response 1 | 7.23 | 5.85 |
| 93943_f_at | 5.49 | Zfp36l2 | Zinc finger protein 36, C3H type-like 2 | 1.47 | 1.22 |
| 92777_at | 4.91 | Cyr61 | Cysteine-rich protein 61 | 5.53 | 4.19 |
| 102371_at | 4.41 | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 15.10 | 8.84 |
| 104647_at | 4.36 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 3.81 | 2.28 |
| 160829_at | 3.56 | Phlda1 | Pleckstrin homology-like domain, family A, member 1 | 2.19 | 1.68 |
| 93974_at | 3.50 | Errfi1 | ERBB receptor feedback inhibitor 1 | 3.81 | 3.45 |
| 93975_at | 3.30 | Errfi1 | ERBB receptor feedback inhibitor 1 | 3.81 | 3.45 |
| 94384_at | 3.12 | Ier3 | Immediate early response 3 | 3.51 | 2.75 |
| 102048_at | 3.04 | Ankrd1 | Ankyrin repeat domain 1 (cardiac muscle) | 1.69 | 1.35 |
| 92310_at | 2.92 | Plk2 | Polo-like kinase 2 (Drosophila) | 2.44 | 2.06 |
| 92830_s_at | 2.86 | Zfp36 | Zinc finger protein 36 | 1.48 | 1.12 |
| 102363_r_at | 2.75 | Junb | JunB oncogene | 4.17 | 2.97 |
| 94147_at | 2.60 | Serpine1 | Serine (or cysteine) proteinase inhibitor, clade E, member 1 | 1.63 | 1.27 |
| 102477_at | 2.43 | ··· | ··· | ||
| 93285_at | 2.37 | Dusp6 | Dual specificity phosphatase 6 | 1.85 | 1.89 |
| 104598_at | 2.33 | Dusp1 | Dual specificity phosphatase 1 | 2.26 | 2.09 |
| 97890_at | 2.23 | Sgk | Serum/glucocorticoid regulated kinase | 2.48 | 1.95 |
| 99942_s_at | 2.22 | Cnn1 | Calponin 1 | 1.05 | 1.09 |
| 103362_at | 2.18 | Ptger4 | Prostaglandin E receptor 4 (subtype EP4) | 1.61 | 1.18 |
| 101979_at | 2.17 | Gadd45g | Growth arrest and DNA damage-inducible 45γ | ||
| 162374_r_at | 2.14 | Myh8 | Myosin, heavy polypeptide 8, skeletal muscle, perinatal | ||
| 96532_at | 2.11 | Ddx50 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 | ||
| 101827_at | 2.11 | Hpvc2 | Human papillomavirus 18 E5 central sequence motif gene 2 | 0.69 | 0.73 |
| 100050_at | 2.08 | Idb1 | Inhibitor of DNA binding 1 | ||
| 98569_at | 2.08 | Slc25a25 | Solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | ||
| 161281_f_at | 2.05 | ··· | ··· | ||
| 94761_at | 2.04 | Ccl7 | Chemokine (C-C motif) ligand 7 | ||
| 101973_at | 2.04 | Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxyl-terminal domain, | 2 | |
| 92742_at | 2.04 | Ccl11 | Small chemokine (C-C motif) ligand 11 | ||
| 104712_at | 2.02 | Myc | Myelocytomatosis oncogene | ||
| 98318_at | 2.02 | Tnfsf7 | Tumor necrosis factor (ligand) superfamily, member 7 | NDa | ND |
| 101583_at | 2.01 | Btg2 | B-cell translocation gene 2, anti-proliferative | ||
| 99449_at | 0.50 | Kcnq2 | Potassium voltage-gated channel, subfamily Q, member 2 | ||
| 100921_at | 0.47 | Tnni3 | Troponin I, cardiac | ||
| 100061_f_at | 0.45 | Klk6 | Kallikrein 6 | ||
| 102009_at | 0.41 | Cyfip2 | Cytoplasmic FMR1 interacting protein 2 | ||
| 93265_at | 0.40 | Tpm3 | Tropomyosin 3 | 0.65 | 0.73 |
| 98789_at | 0.40 | Cdkn2a | Cyclin-dependent kinase inhibitor 2A | 1.18 | 1.12 |
a ND, not detected.
The pattern of gene expression changes was completely different after a 6-h IGF-1 stimulation: at this time point, IGF-1 induced a broader, but also weaker, regulation of gene expression, with a majority of the genes being down-regulated (Fig. 1A). Indeed, 1221 probesets were significantly regulated 6 h after IGF-1 treatment (p < 0.05), with 592 up-regulated and 630 down-regulated (Fig. 1 and supplemental Table S1). Of these, only 14 genes were up-regulated more than 2-fold (with a maximum fold-change of 2.6 for spermidine synthase), whereas 40 genes were down-regulated 2-fold or more with a maximum change of 3 for yippee-like 3 (Fig. 1 and Table 2).
TABLE 2.
Probesets significantly regulated more than 2-fold by a 6-h IGF-1 treatment in WT cells
54 probesets regulated more than 2-fold by 10 nm IGF-1 after 30 min in WT preadipocytes are listed with fold-change, gene symbol, full gene name, and fold-change measured by real time PCR after IGF-1 or insulin treatment (for selected genes).
| Probeset | Fold | Symbol | Name | Real time PCR |
|
|---|---|---|---|---|---|
| Fold-change IGF1 | Fold-change insulin | ||||
| 92540_f_at | 2.65 | Srm | Spermidine synthase | 2.22 | 1.41 |
| 96519_at | 2.59 | Pdxk | Pyridoxal (pyridoxine, vitamin B6) kinase | ||
| 104351_at | 2.47 | Amigo3 | Amphoterin-induced gene and ORF 3 | ||
| 160786_f_at | 2.45 | Actr1b | ARP1 actin-related protein 1 homolog B (yeast) | ||
| 162341_r_at | 2.45 | Akr1b3 | Aldo-keto reductase family 1, member B3 (aldose reductase) | ||
| 92483_g_at | 2.36 | Fmnl1 | Formin-like 1 | ||
| 95735_at | 2.24 | Nolc1 | Nucleolar and coiled-body phosphoprotein 1 | ||
| 160745_at | 2.21 | Gcn5l2 | GCN5 general control of amino acid synthesis-like 2 (yeast) | ||
| 160653_at | 2.21 | Tomm40 | Translocase of outer mitochondrial membrane 40 homolog (yeast) | ||
| 96594_at | 2.10 | Hspa4 | Heat shock protein 4 | ||
| 96072_at | 2.09 | Ldh1 | Lactate dehydrogenase 1, A chain | ||
| 93845_at | 2.08 | Abcf2 | ATP-binding cassette, subfamily F (GCN20), member 2 | ||
| 100566_at | 2.06 | Igfbp5 | Insulin-like growth factor-binding protein 5 | 3.68 | 2.24 |
| 101180_at | 2.06 | Atm | Ataxia telangiectasia mutated homolog (human) | ||
| 161070_at | 0.50 | Spred2 | Sprouty protein with EVH-1 domain 2, related sequence | ||
| 104210_at | 0.50 | Itga3 | Integrin α 3 | ||
| 94843_at | 0.49 | Pold4 | Polymerase (DNA-directed), δ4 | ||
| 98824_at | 0.49 | Irs1 | Insulin receptor substrate 1 | 0.18 | 0.36 |
| 102048_at | 0.49 | Ankrd1 | Ankyrin repeat domain 1 (cardiac muscle) | ||
| 162462_r_at | 0.49 | Pck2 | Phosphoenolpyruvate carboxykinase 2 | ||
| 160138_at | 0.49 | Mxi1 | Max interacting protein 1 | ||
| 93265_at | 0.48 | Tpm3 | Tropomyosin 3 | 0.83 | 0.81 |
| 99864_at | 0.48 | Adora2b | Adenosine A2b receptor | ||
| 98478_at | 0.48 | Ccng2 | Cyclin G2 | ||
| 94354_at | 0.48 | Abca1 | ATP-binding cassette, subfamily A (ABC1), member 1 | ||
| 92318_at | 0.48 | Aplf | Aprataxin and PNKP-like factor | ||
| 99845_at | 0.47 | Slc1a6 | Solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | ||
| 160948_at | 0.47 | Ppp3cc | Protein phosphatase 3, catalytic subunit, γ isoform | ||
| 160816_at | 0.47 | Fjx1 | Four jointed box 1 (Drosophila) | ||
| 102673_at | 0.47 | Creb1 | cAMP responsive element-binding protein 1 | ||
| 92777_at | 0.47 | Cyr61 | Cysteine-rich protein 61 | 0.54 | 0.62 |
| 94988_at | 0.46 | Pten | Phosphatase and tensin homolog | ||
| 160061_at | 0.46 | Il1rl1l | Interleukin 1 receptor-like 1 ligand | ||
| 161342_r_at | 0.45 | Eif4b | Eukaryotic translation initiation factor 4B | ||
| 161033_at | 0.45 | Papolb | Poly(A) polymerase beta (testis specific) | ||
| 95294_at | 0.45 | Agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | ||
| 92705_at | 0.44 | Tbx2 | T-box 2 | ||
| 160894_at | 0.44 | Cebpd | CCAAT/enhancer binding protein (C/EBP), δ | ||
| 102362_i_at | 0.44 | Junb | JunB oncogene | 0.71 | 0.82 |
| 102370_at | 0.44 | Dhrs8 | Dehydrogenase/reductase (SDR family) member 8 | ||
| 96623_at | 0.44 | Ugcg | UDP-glucose ceramide glucosyltransferase | 0.31 | 0.43 |
| 103536_at | 0.43 | Tmeff2 | Transmembrane protein with EGF-like and two follistatin-like domains 2 | ||
| 93294_at | 0.43 | Ctgf | Connective tissue growth factor | 0.33 | 0.46 |
| 96728_at | 0.43 | Wdrx1 | WD repeat domain, X-linked 1 | ||
| 160469_at | 0.41 | Thbs1 | Thrombospondin 1 | ||
| 102922_at | 0.40 | Pitpnc1 | Phosphatidylinositol transfer protein, cytoplasmic 1 | ||
| 160373_i_at | 0.39 | Sdpr | Serum deprivation response | 0.27 | 0.58 |
| 96494_at | 0.38 | Klhl24 | Kelch-like 24 | ||
| 104513_at | 0.38 | 2410004N09 | RIK EN cDNA 2410004N09 gene | ||
| 94113_at | 0.36 | ··· | ··· | ||
| 102798_at | 0.33 | Adm | Adrenomedullin | 0.27 | 0.48 |
| 102292_at | 0.33 | Gadd45a | Growth arrest and DNA damage-inducible 45α | ||
| 96615_at | 0.32 | Ypel3 | Yippee-like 3 (Drosophila) | ||
To confirm the microarray data, we measured the expression of 38 of the most significantly regulated genes using quantitative real time PCR using independent samples, from cells treated with insulin and IGF-1 for 30 min or 6 h. For more than 90% of the genes tested, the significant regulation by IGF-1 was confirmed (see Tables 1 and 2). Stimulation with 10 nm insulin was also able to regulate expression of all the IGF-1-regulated genes as assessed by quantitative PCR, although with a lower magnitude of response than IGF-1 (see Tables 1 and 2).
Insulin and IGF-1 Regulation of Gene Expression in Non-differentiated and Differentiated Brown Preadipocytes
Brown preadipocytes express high levels of IGF-1 receptors but also low levels of insulin receptors (21) (Fig. 2, A and B). Although insulin and IGF-1 have a higher affinity for their respective receptors, 10 nm insulin or IGF-1 is still a supra-physiological concentration that is high enough to allow cross-reaction of each ligand to the receptor of the other, making it almost impossible to find genes specifically regulated by one receptor or the other using this paradigm.
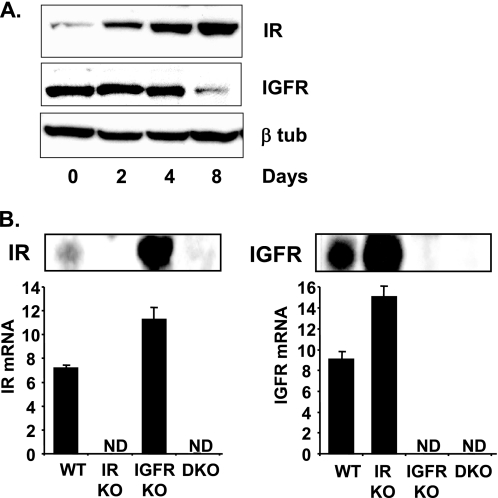
FIGURE 2.
IR and IGF1R expression in WT, IRKO, IGFRKO, and DKO cells. A, WT brown preadipocytes were differentiated for 2, 4, and 8 days as described under “Experimental Procedures” and IR, IGF1R, and β-tubulin protein levels were measured by Western blot analysis. A representative blot from 3 experiments is shown. B, IR and IGF1R immunoblots were performed on confluent WT, IRKO, IGFRKO, or DKO cells. mRNA was extracted from confluent WT, IRKO, IGFRKO, or DKO cells and IR and IGF1R mRNA expression were measured by real time PCR using specific oligonucleotide primers. The data were normalized to levels of TBP mRNA. ND, not detected. Results are mean ± standard error of the mean from 5 independent measurements.
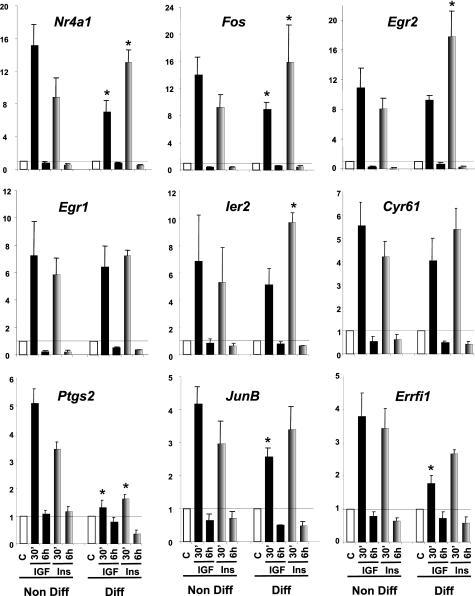
To see how the relative receptor levels affect the response to their ligand, we performed similar insulin and IGF-1 stimulation in differentiated WT brown adipocytes, where the balance between the expression of IR and IGF1R is changed. Over an 8-day course of differentiation into adipocytes, IR expression progressively increased over 5-fold, whereas IGF1R levels decreased by 60% (Fig. 2A). For all the genes studied, IGF-1 had a more potent effect than insulin on WT brown preadipocytes, consistent with the fact that preadipocytes have more IGF1R than IR. However, in differentiated cells, the pattern was the opposite with insulin having a stronger effect. Thus, insulin was now more potent in its ability to increase expression of Nr4a1, Fos, Egr2, Egr1, Ier2, Cyr61, Ptgs2, JunB, Errfi1 in differentiated brown adipocytes compared with IGF-1 (Fig. 3).
FIGURE 3.
Insulin and IGF-1 regulation of gene expression in nondifferentiated versus differentiated WT cells. WT cells were grown to confluence, and differentiation was induced as described under “Experimental Procedures” for 8 days. Nondifferentiated cells at day 0 (Non Diff) and differentiated cells at day 8 (Diff) were serum starved overnight and stimulated with 10 nm insulin (gray bars) or IGF-1 (black bars) during 30 min or 6 h. Gene expression was measured by real time PCR using specific oligonucleotides. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 6 independent experiments for nondifferentiated cells and 3 independent experiments for differentiated adipocytes. * indicates a significant difference compared with the similar treatment in nondifferentiated cells, p value <0.05 by Student's t test.
Insulin and IGF-1 Regulation of Gene Expression in WT, IRKO, and IGF1RKO Cells
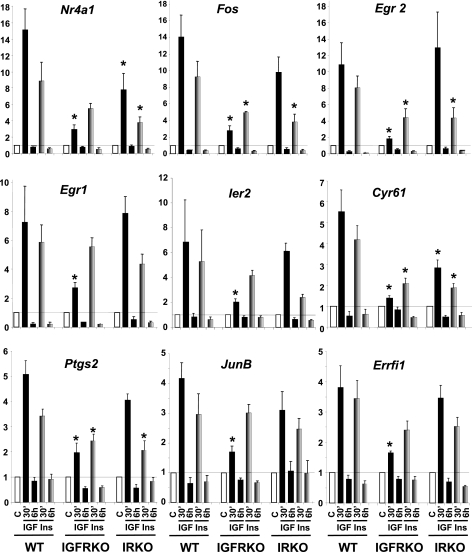
To be able to identify genes regulated by IGF-1 or insulin specifically through the IGF1R or IR, but not both, we used cells with genetically inactivated IGF-1 or insulin receptors. As expected, IGF1R, IR mRNA, and protein were not detected in IGFRKO and IRKO cells, respectively (Fig. 2B). Again, cells were stimulated with 10 nm IGF-1 or insulin for 30 min and 6 h and expression of the genes previously identified to be significantly regulated in WT cells were assessed by real time PCR. Genes regulated exclusively via the IGF1R should not be regulated anymore in IGFRKO cells and those regulated via the IR should not be regulated in IRKO cells, regardless of the ability of insulin or IGF-1 to regulate their expression in WT cells. Results of these studies are shown in Fig. 4.
FIGURE 4.
Insulin and IGF-1 regulation of gene expression in WT, IRKO, and IGFRKO preadipocytes. WT, IRKO, and IGFRKO cells were grown to confluence, serum starved overnight, and stimulated with 10 nm insulin (gray bars) or IGF-1 (black bars) for 30 min or 6 h. Gene expression was measured by real time PCR using specific oligonucleotides. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 5 to 6 independent experiments. * indicates a significant difference compared with a similar treatment in WT cells, p value <0.05 by Student's t test.
We found that Egr2 up-regulation after a 30-min IGF-1 treatment was highly IGF1R specific, because IGF-1 induced a 11-fold increase in WT cells, and this effect was almost completely lost in IGFRKO cells (1.8-fold increase). However, insulin was still able to induce a 4.4-fold increase in Egr2 expression in IGFRKO cells, indicating that the insulin receptor is also able to regulate Egr2 gene expression (Fig. 4). In IRKO cells, the insulin effect on Egr2 gene expression was reduced compared with WT cells (4.4- versus 8-fold increase) indicating that the IR contributes to the insulin effect on the regulation of Egr2 expression in WT cells. On the other hand, the IGF-1 effect was similar between IRKO and WT cells suggesting that the IR had no significant contribution in the regulation of Egr2 by IGF-1. This is consistent with the fact that the IGF-1 effect was almost completely abolished in IGFRKO cells. The up-regulation of Egr2 by insulin or IGF-1 was very transient; 6 h after stimulation, Egr2 levels returned to below basal levels.
Fos, the second most regulated gene by IGF-1 in WT cells showed a similar regulation by insulin and IGF-1 in the different KO cells. In IGFRKO cells, the IGF-1 effect was dramatically reduced (2.8-fold increased compared with 14-fold in WT cells), but not completely abolished due to the remaining effect from the IR. The effect of insulin was stronger compared with IGF-1 in IGFRKO cells (4.9- versus 2.8-fold increase) but was significantly decreased compared with WT cells (4.9- versus 9.2-fold increase), showing the contribution of the IR in the effect of insulin and IGF-1 regulating Fos expression. The contribution of the IGF1R could be visualized in IRKO cells: the IGF-1 and insulin effects on the regulation of Fos expression were both modestly decreased in IRKO cells compared with WT cells (9.8-fold increase in IRKO cells compared with a 14-fold increase in WT cells for IGF-1 and 3.8- versus 9.2-fold increase for insulin) indicating that both the IGF1R and IR to a lesser extent, were involved in mediating the effects of insulin and IGF-1 in the regulation of Fos expression. Other genes highly up-regulated by IGF-1 such as Nr4a1, Egr1, Ier2, Cyr61, Ptgs2, JunB, and Errfi1 or genes more modestly regulated by IGF-1 (between 3.5- and 1.5-fold change in WT cells) such as Ier3, Sgk, Dusp6, Dusp1, Ctgf, Plk2, Phlda1, and Serpine 1 all showed a similar pattern of regulation by IGF-1 and insulin in WT, IGFRKO, and IRKO cells (Fig. 4 and supplemental Fig. S1). We also measured the expression of genes regulated by IGF-1 at 6 h and found similar results. Expression of Igfbp5 and Srm are shown as an example in supplemental Fig. S2.
In conclusion, the effect of IGF-1 on the regulation of gene expression is mainly achieved through activation of the IGF1R, and to a lesser extent via the IR in WT cells. However, studies in single KO cells revealed that the IR mediates part of the IGF-1 effect in WT cells, as both insulin and IGF-1 were still able to regulate the expression of these genes in cells lacking IGF1R (IGFRKO cells), and that insulin and IGF-1 effects were significantly decreased in cells lacking IR (IRKO cells). Based on the combined analysis of microarray data identifying IGF-1-regulated genes and real time PCR data on the effects of IGF-1 and insulin in the regulation of more than 50 of these genes in WT, IGFRKO, and IRKO cells, we failed to identify one single gene that was regulated by IGF-1 or insulin specifically via the IGF1R or IR. These results indicate that differences in receptor expression, rather than receptor type, are more important for the specificity of insulin and IGF-1 effects.
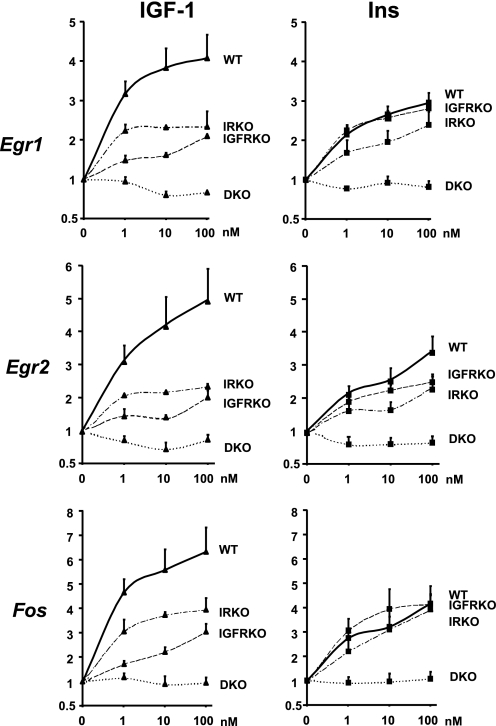
Insulin and IGF-1 Dose Response on the Regulation of Egr1, Egr2, and Fos Gene Expression in Cells Expressing IR, IGF1R, Both, or None
To further explore the relative roles of IR and IGF1R in insulin and IGF-1 action, we performed a dose-dependent stimulation of WT, IGFRKO, IRKO, and double IR/IGFR KO cells (DKO) with IGF-1 or insulin for 30 min and measured the regulation of three of the most highly up-regulated genes by IGF-1 and insulin, namely Egr1, Egr2, and Fos. Insulin and IGF-1 induced a significant up-regulation of Egr1 in WT preadipocyte cells at the lowest dose used (1 nm). As shown in Fig. 3, IGF-1 was more potent than insulin to up-regulate Egr1 gene expression in WT preadipocytes at all concentrations used from 1 to 100 nm (Fig. 5). Inactivation of IGF1R in IGFRKO cells had no effect on the insulin dose response, because insulin was as potent to up-regulate Egr1 gene expression in IGFRKO cells as in WT cells at all concentrations. By contrast, the IGF-1 effect on Egr1 expression was dramatically reduced in IGFRKO cells compared with WT cells at all doses. Both insulin and IGF-1 effects were reduced in IRKO cells compared with WT cells at all concentrations indicating that IR contributes to both the insulin and IGF1 effects on the regulation of Egr1 expression in WT cells. 1 nm IGF-1 or insulin was able to significantly up-regulate Egr1 expression in IGFRKO and IRKO cells, respectively, indicating that insulin and IGF-1 can efficiently cross-react with IGF1R and IR, even at this physiological concentration. As expected, Egr1 expression was not modified by an insulin or IGF-1 treatment in cells lacking both the IR and IGF1R (DKO), even at the highest concentration of 100 nm. Egr2 and Fos showed a similar pattern of regulation by insulin and IGF-1 than Egr1 in WT, IRKO, IGFRKO, and DKO cells (Fig. 5). These results indicate that both IR and IGF1R contribute to the effects of either insulin or IGF-1 in regulating gene expression in brown preadipocytes. However, results are confounded by the fact that these cells express different levels of receptors and that the KO of one receptor does result in some compensatory up-regulation of the other (Fig. 2B).
FIGURE 5.
Insulin and IGF-1 dose response on Egr1, Egr2, and Fos gene expression in WT, IRKO, IGFRKO, and DKO cells. WT, IRKO, and IGFRKO cells were grown to confluence, serum starved overnight, and stimulated with 1, 10, or 100 nm insulin or IGF-1 during 30 min. Egr1, Egr2, and Fos mRNA levels were measured by real time PCR using specific oligonucleotides. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 5 independent experiments.
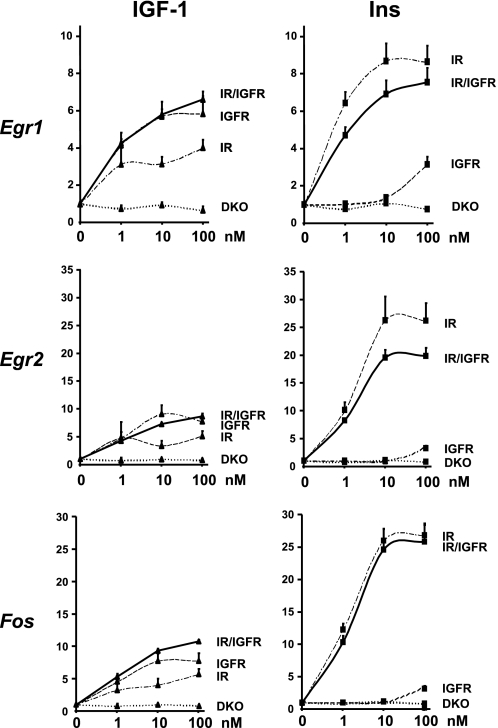
To overcome this problem, we performed a similar experiment in DKO cells stably overexpressing hIR, hIGF1R, or both, created as described under “Experimental Procedures.” 1, 10, or 100 nm insulin treatment for 30 min induced a 6.4-, 8.7-, and 8.7-fold increase in Egr1 gene expression in IR expressing cells, respectively (Fig. 6). Similar concentrations of IGF-1 induced a reduced, but significant, increase in Egr1 expression increasing Egr1 levels by 3.1-, 3.2-, and 4-fold, respectively, in agreement with the fact that the affinity of IGF-1 for the insulin receptor is reduced compared with its affinity for its own receptor. More surprisingly, even at the highest concentration used (100 nm), IGF-1 was less potent that even the lowest concentration of insulin (1 nm), indicating that even in excess, IGF-1 is unable to fully activate IR-mediated gene expression. In IGF1R expressing cells, IGF-1 increased Egr1 levels by 4.1-, 5.7-, and 5.9-fold at 1, 10, and 100 nm, respectively. Insulin, however, was ineffective at 1 and 10 nm and was only able to modestly increase Egr1 gene expression by 3.1-fold at 100 nm, indicating that insulin is less potent in regulating gene expression via the IGF1R than IGF-1 via the IR. There was no additive effect having both receptors in the same cells as insulin increased Egr1 mRNA levels by 4.7-, 6.9-, and 7.5-fold and IGF-1 by 4.3-, 5.8-, and 6.6-fold at concentrations of 1, 10, and 100 nm, respectively. Similar results were obtained when assessing regulation of expression of Egr2 and Fos (Fig. 6).
FIGURE 6.
Insulin and IGF-1 dose response on Egr1, Egr2, and Fos gene expression in DKO cells re-expressing IR, IGF1R, or both. DKO cells re-expressing IR, IGF1R, or both were grown to confluence, serum starved overnight, and stimulated with 1, 10, or 100 nm insulin or IGF-1 during 30 min. Egr1, Egr2, and Fos mRNA levels were measured by real time PCR using specific oligonucleotides. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 5 independent experiments.
IR and IGF1R Are Equally Potent to Induce Adipocyte Differentiation
The results thus far suggest that despite differences in receptor expression that modulate the amplitude of the signal created by the specific ligand-receptor interaction, IR and IGF1R have virtually identical roles in the regulation of gene expression by insulin and IGF-1. We then investigated if this is true for one of the physiological responses of insulin and IGF-1 in preadipocytes: stimulation of adipocyte differentiation. Insulin and IGF-1 are potent pro-adipogenic hormones, but it remains an open question whether this effect is mediated by the IR, IGF1R, or both. In culture, adipocyte differentiation is induced using high insulin concentrations that can activate the IGF1R. Furthermore, preadipocytes express a lot more IGF1R than IR. On the other hand, cells lacking IR display a defect in adipocyte differentiation (22) and mice with a specific KO of IR in adipose tissue display decreased adipose tissue mass (23), whereas mice with a specific KO of IGF1R in adipose tissue do not (24).
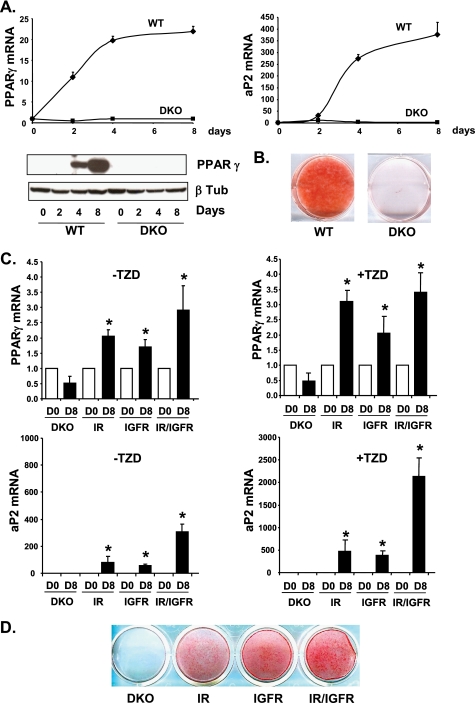
Eight days after induction of adipocyte differentiation, WT cells were fully differentiated as visualized by oil red O staining of accumulated lipids (Fig. 7B). DKO cells lacking both IR and IGF1R were unable to differentiate and showed no lipid accumulation. mRNA levels of transcription factor PPARγ, a central regulator of adipocyte differentiation, were increased 22-fold during adipocyte differentiation in WT cells between days 0 and 8 of differentiation, whereas they did not change in DKO cells, consistent with the lack of differentiation (Fig. 7A). Protein levels showed a similar change. Similarly, mRNA levels of aP2, a fatty acid-binding protein highly abundant in differentiated adipocytes, were increased 400-fold over the differentiation time course in WT cells, but not in DKO cells. Addition of the PPARγ agonist rosiglitazone, a potent inducer of adipocyte differentiation, also failed to stimulate differentiation of the DKO cells (Fig. 7D).
FIGURE 7.
Differentiation of WT and DKO cells or DKO cells re-expressing IR, IGF1R, or both. A, confluent WT and DKO brown preadipocytes were differentiated for 2, 4, and 8 days. PPARγ and aP2 mRNA levels were measured by real time PCR using specific oligonucleotide primers. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 3 independent experiments. PPARγ and β-tubulin protein levels were measured by Western blot analysis. A representative blot from 3 experiments is shown. B, oil red O staining from WT and DKO cells differentiated for 8 days. C, DKO cells re-expressing IR, IGF1R, or both were grown to confluence and differentiated with or without the thiazolidinedione (TZD) rosiglitazone (1 μm) for 8 days. PPARγ and aP2 mRNA levels were measured by real time PCR using specific oligonucleotide primers. The data were normalized to levels of TBP mRNA. Results are mean ± standard error of the mean from 3 independent experiments. D, oil red O staining from DKO cells re-expressing IR, IGF1R, or both after 8 days of differentiation in the presence of rosiglitazone.
We then investigated whether re-expressing IR, IGF1R, or both would rescue adipocyte differentiation. DKO cells re-expressing IR or IGF1R were partially rescued from their differentiation defect. Using the standard differentiation mixture without rosiglitazone, PPARγ mRNA levels were up-regulated 2.1-, 1.7-, and 2.9-fold after 8 days of differentiation in IR, IGF1R, and IR/IGF1R re-expressing cells, respectively (Fig. 7C). mRNA levels for the fatty acid-binding protein aP2 were up-regulated 80- and 60-fold between days 0 and 8 of differentiation in IR and IGF1R re-expressing cells, respectively. An additive effect was observed when both receptors were re-expressed with aP2 being up-regulated 300-fold after 8 days of differentiation. Addition of the PPARγ agonist rosiglitazone significantly improved the overall differentiation efficiency as visualized by lipid accumulation after oil red O staining (Fig. 7D). In these conditions, PPARγ mRNA levels were up-regulated 3.1-, 2.1-, and 3.4-fold and aP2 mRNA levels 470-, 380-, and 2100-fold after 8 days of differentiation in IR, IGF1R, and IR/IGF1R re-expressing cells, respectively (Fig. 7C). These results indicate that both IR and IGF1R are able to promote adipocyte differentiation to a similar extent, and that they can regulate the same sets of genes critical for adipocyte differentiation.
DISCUSSION
We have explored the key question of how IR and IGF1R signaling might differ by studying their role in regulation of gene expression. We performed studies on brown preadipocytes before and after differentiation and on IGFRKO, IRKO, and DKO cells with or without re-expression of IR, IGF1R, or both, to assess whether or not genes could be regulated specifically via the IR or IGF1R but not both. Among all genes identified by microarray analysis to be regulated more than 2-fold by IGF-1 in WT preadipocytes, we found that none are regulated strictly via one receptor or the other. IGF-1 was able to regulate the expression of 44 genes more than 2-fold after a 30-min treatment but most of the IGF-1 effect was gone when the same stimulation was performed in IGFRKO cells, indicating that the IGF-1 effect was predominantly IGF1R specific. However, insulin stimulation could strongly regulate the expression of the same genes indicating that the IR stimulated by its own ligand was able to have the same effect as IGF1R stimulation in the control of gene expression. The remaining IGF-1 effect in IGFRKO cells or insulin effect in IRKO cannot be explained by a residual expression of the receptors because the IGFRKO, IRKO, or DKO cell lines were generated from the corresponding IRlox, IGFRlox, or IRlox/IGFRlox cells lines after in vitro recombination with a Cre recombinase expressing adenovirus, and contain no detectable IR or IGF1R mRNA and protein or both as predicted by the gene inactivation. Furthermore, DKO cells show a complete absence of signaling after 100 nm insulin or IGF-1 treatment, and no regulation of Egr1, Egr2, or Fos gene expression after 30 min (Figs. 3 and 4).
The two time points used were chosen to look at short term (30 min) and long term (6 h) regulation of gene expression induced by IGF-1 and insulin. The list of genes identified to be regulated by IGF-1/insulin in WT cells at 30 min and 6 h probably only contains a subset of all genes regulated by IGF-1/insulin. It is possible that IR or IGF1R specifically regulate certain genes that have a different kinetic of regulation by IGF-1/insulin than the ones investigated here. We used an IGF-1 stimulation in WT cells to identify genes regulated by the activation of IGF-1 and insulin receptors. Although IGF-1 at the concentration used also activates the IR, we cannot exclude that insulin stimulation would have given us a slightly different list of genes to test. Furthermore, we only tested genes regulated 2-fold or more for their IR or IGF1R specificity. Thus, it is possible that differences exist among the genes more modestly regulated, or some with unusual kinetics.
For all genes identified in this study to be regulated by IGF-1 and measured by real time PCR, insulin also regulates their expression in WT cells but is less potent than IGF-1. Preadipocytes express many more IGF1R than IR (21). Indeed, in IRKO cells, the IGF-1 response on gene expression was only moderately decreased due to the low number of IR in brown preadipocytes. In adipocytes, however, the IR/IGF1R ratio is changed as IR expression increases with adipocyte differentiation (21, 25) (Fig. 5). As a consequence, insulin was more potent than IGF-1 in regulating gene expression in differentiated adipocytes compared with preadipocytes. Using the KO cell lines in the differentiated state was not possible as these cells show a defect in adipocyte differentiation (22). Not surprisingly, DKO preadipocyte cells were unable to differentiate into adipocytes even in the presence of the PPARγ agonist thiazolidinediones, highlighting the importance of IR and/or IGF1R in the control of brown adipocyte differentiation. However, the relative contribution of IR and IGF1R in that process is not clearly established. Re-expressing IR, IGF1R, or both receptors in these cells partially rescued adipocyte differentiation. Furthermore, IR and IGF1R rescued the adipocyte differentiation to the same extent, showing once again the redundancy between IR and IGF1R, this time controlling a network of genes required for adipocyte differentiation.
In agreement with our results, other studies have shown that IGF-1 and insulin receptors can mediate similar functions. Muscle-specific IRKO mice remain normoglycemic and develop only mild insulin resistance suggesting insulin and/or IGF-1 acting through the IGF1R can help control blood glucose (26, 27). IGF-1 has also been shown to have a direct insulin-like effect in human skeletal muscle cells (28, 29) and L6 myotubes (30) in inducing glucose uptake. Furthermore, IGF-1 induces glucose uptake and glycogen synthesis in IRKO myotubes with a potency close to that of insulin or IGF-1 in wild-type cells (31). In IR-deficient mouse fibroblasts, IGF-1 and insulin both stimulate glucose uptake, glycogen synthesis, and thymidine incorporation via phosphatidylinositol 3-kinase and mitogen-activated protein kinase (MAPK) pathways proving once again that the IGF1R is also capable of inducing metabolic responses (32). Furthermore, IGF-1 has been shown to play a major role in glucose metabolism in the brain. In ependymal cells residing in the central nervous system, IGF-1 is reported to be at least 10 times more efficient than insulin in stimulating glucose uptake, and it has been suggested that IGF-1 rather than insulin is the predominant metabolic regulator in this organ (33, 34). IGF-1 also induces lipogenesis and differentiation of preadipocytes, glucose uptake and protein synthesis in skeletal muscle, and decreases gluconeogenesis in the liver (reviewed in Ref. 35). Conversely, activation of the IR with insulin in brown preadipocytes lacking IGF1R leads to a strong mitogenic response (36).
Several studies have tried to identify genes specifically regulated by insulin or IGF-1. Dupont et al. (37) stimulated NIH-3T3 cells overexpressing hIR or hIGF1R with insulin and IGF-1, respectively, for 90 min and performed cDNA arrays containing 3899 probesets. They identified a few genes specifically regulated by IGF1/IGF1R or insulin/IR including the Egr1 gene that we found in our study. They found that Egr1 was up-regulated 4-fold by IGF-1/IGF1R but not by insulin/IR. In our hands, Egr1 can be equally regulated by the IR or IGF1R. This discrepancy could be due to the different cell type used, the different time chosen (30 versus 90 min), or the sensitivity and reproducibility of the method. In fact, it is possible that IR and IGF1R activation could induce regulation of the same genes but with different amplitudes and kinetics. In another study, Mulligan et al. (38) compared IR and IGF1R regulation of gene expression by microarray analysis using 3T3-L1 cells expressing either TrkC/IR or TrkC/IGF1R chimeric receptors. They found heparin-binding EGF-like growth factor (HB-EGF) to be regulated specifically by TrkC/IGF1R, but not TrkC/IR, following a 4-h stimulation by neurotrophin-3. However, they found that HB-EGF could be regulated by insulin in cells overexpressing non-chimeric IR, although to a lesser extent than by IGF-1 in cells overexpressing IGF1R. In our WT preadipocytes, HB-EGF was up-regulated 2.0-fold after a 30-min IGF-1 stimulation, an effect that was almost completely abolished in IGFRKO cells (1.2-fold increase). However, insulin was still able to significantly increase HB-EGF gene expression by 1.5-fold in these cells indicating that the IR can also increase HB-EGF gene transcription. HB-EGF mRNA levels were not different after a 6-h insulin or IGF-1 stimulation. Palsgaard et al. (39) performed a microarray analysis in nondifferentiated or differentiated primary human skeletal muscle cells stimulated with insulin or IGF-1. They found that IGF-1 was more potent than insulin in regulating gene expression, correlating with a higher IGF1R than IR level, in both undifferentiated and differentiated cells. However, as in our study, not a single gene regulated specifically by insulin or IGF-1 could be identified.
Taken together, our results indicate that in a given cell type, IR and IGF1R act as identical portals to the regulation of gene expression, with differences between insulin and IGF-1 effects due to a modulation of the amplitude of the signal created by the specific ligand-receptor interaction. This implies that the specificity in vivo of insulin and IGF-1 reflects at least in part the levels and timing of expression of IR and IGFR in target tissues, and ligand concentration and availability. Further studies will be required to determine the differences between IR and IGF1R signaling in vivo.
Supplementary Material
Acknowledgments
We thank the Joslin Genomics Core facility supported by Joslin Diabetes Center's Diabetes and Endocrinology Research Center (DERC) (supported by National Institutes of Health NIDDK Grant P30DK036836) and Jane Palsgaard for careful reading of the manuscript and scientific discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK31036, DK33201, and DK077097, Diabetes Genome Anatomy Project Grant DK60837, and Harvard Catalyst/The Harvard Clinical and Translational Science Center Grant UL1 RR 025758-01.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table 1.
- IGF-1
- insulin-like growth factor-1
- IR
- insulin receptor
- IGF1R
- IGF-1 receptor
- HB-EGF
- heparin-binding epidermal growth factor-like growth factor
- PPAR
- peroxisome proliferator-activated receptor
- DKO
- double knockout
- TBP
- TATA binding protein
- WT
- wild-type
- IRKO
- insulin receptor knockout
- IGF1RKO
- IGF-1 receptor knockout.
REFERENCES
- 1.Siddle K., Ursø B., Niesler C. A., Cope D. L., Molina L., Surinya K. H., Soos M. A. (2001) Biochem. Soc. Trans. 29, 513–525 [DOI] [PubMed] [Google Scholar]
- 2.Baker J., Liu J. P., Robertson E. J., Efstratiadis A. (1993) Cell 75, 73–82 [PubMed] [Google Scholar]
- 3.Accili D., Drago J., Lee E. J., Johnson M. D., Cool M. H., Salvatore P., Asico L. D., José P. A., Taylor S. I., Westphal H. (1996) Nat. Genet. 12, 106–109 [DOI] [PubMed] [Google Scholar]
- 4.Joshi R. L., Lamothe B., Cordonnier N., Mesbah K., Monthioux E., Jami J., Bucchini D. (1996) EMBO J. 15, 1542–1547 [PMC free article] [PubMed] [Google Scholar]
- 5.Duvillié B., Cordonnier N., Deltour L., Dandoy-Dron F., Itier J. M., Monthioux E., Jami J., Joshi R. L., Bucchini D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5137–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell-Braxton L., Hollingshead P., Warburton C., Dowd M., Pitts-Meek S., Dalton D., Gillett N., Stewart T. A. (1993) Genes Dev. 7, 2609–2617 [DOI] [PubMed] [Google Scholar]
- 7.Liu J. P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A. (1993) Cell 75, 59–72 [PubMed] [Google Scholar]
- 8.Laron Z. (2008) Arch. Physiol. Biochem. 114, 11–16 [DOI] [PubMed] [Google Scholar]
- 9.Di Cola G., Cool M. H., Accili D. (1997) J. Clin. Invest. 99, 2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moses A. C., Young S. C., Morrow L. A., O'Brien M., Clemmons D. R. (1996) Diabetes 45, 91–100 [DOI] [PubMed] [Google Scholar]
- 11.Simpson H. L., Umpleby A. M., Russell-Jones D. L. (1998) Growth Horm. IGF Res. 8, 83–95 [DOI] [PubMed] [Google Scholar]
- 12.Simpson H. L., Jackson N. C., Shojaee-Moradie F., Jones R. H., Russell-Jones D. L., Sönksen P. H., Dunger D. B., Umpleby A. M. (2004) J. Clin. Endocrinol. Metab. 89, 425–432 [DOI] [PubMed] [Google Scholar]
- 13.Clemmons D. R., Moses A. C., Sommer A., Jacobson W., Rogol A. D., Sleevi M. R., Allan G. (2005) Growth Horm. IGF Res. 15, 265–274 [DOI] [PubMed] [Google Scholar]
- 14.Clemmons D. R. (2006) Curr. Opin. Pharmacol. 6, 620–625 [DOI] [PubMed] [Google Scholar]
- 15.Dupont J., Dunn S. E., Barrett J. C., LeRoith D. (2003) Recent Prog. Horm. Res. 58, 325–342 [DOI] [PubMed] [Google Scholar]
- 16.Lammers R., Gray A., Schlessinger J., Ullrich A. (1989) EMBO J. 8, 1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tartare S., Mothe I., Kowalski-Chauvel A., Breittmayer J. P., Ballotti R., Van Obberghen E. (1994) J. Biol. Chem. 269, 11449–11455 [PubMed] [Google Scholar]
- 18.Kalloo-Hosein H. E., Whitehead J. P., Soos M., Tavaré J. M., Siddle K., O'Rahilly S. (1997) J. Biol. Chem. 272, 24325–24332 [DOI] [PubMed] [Google Scholar]
- 19.Ursø B., Cope D. L., Kalloo-Hosein H. E., Hayward A. C., Whitehead J. P., O'Rahilly S., Siddle K. (1999) J. Biol. Chem. 274, 30864–30873 [DOI] [PubMed] [Google Scholar]
- 20.Dupont J., LeRoith D. (2001) Horm. Res. 55, Suppl. 2, 22–26 [DOI] [PubMed] [Google Scholar]
- 21.Entingh-Pearsall A., Kahn C. R. (2004) J. Biol. Chem. 279, 38016–38024 [DOI] [PubMed] [Google Scholar]
- 22.Entingh A. J., Taniguchi C. M., Kahn C. R. (2003) J. Biol. Chem. 278, 33377–33383 [DOI] [PubMed] [Google Scholar]
- 23.Blüher M., Michael M. D., Peroni O. D., Ueki K., Carter N., Kahn B. B., Kahn C. R. (2002) Dev. Cell 3, 25–38 [DOI] [PubMed] [Google Scholar]
- 24.Klöting N., Koch L., Wunderlich T., Kern M., Ruschke K., Krone W., Brüning J. C., Blüher M. (2008) Diabetes 57, 2074–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bäck K., Arnqvist H. J. (2009) Growth Horm. IGF Res. 19, 101–111 [DOI] [PubMed] [Google Scholar]
- 26.Le Roith D., Kim H., Fernandez A. M., Accili D. (2002) Curr. Opin. Clin. Nutr. Metab. Care 5, 371–375 [DOI] [PubMed] [Google Scholar]
- 27.Brüning J. C., Michael M. D., Winnay J. N., Hayashi T., Hörsch D., Accili D., Goodyear L. J., Kahn C. R. (1998) Mol. Cell 2, 559–569 [DOI] [PubMed] [Google Scholar]
- 28.Ciaraldi T. P., Phillips S. A., Carter L., Aroda V., Mudaliar S., Henry R. R. (2005) J. Clin. Endocrinol. Metab. 90, 5551–5558 [DOI] [PubMed] [Google Scholar]
- 29.Henry R. R., Abrams L., Nikoulina S., Ciaraldi T. P. (1995) Diabetes 44, 936–946 [DOI] [PubMed] [Google Scholar]
- 30.Wilson C. M., Mitsumoto Y., Maher F., Klip A. (1995) FEBS Lett. 368, 19–22 [DOI] [PubMed] [Google Scholar]
- 31.Baudry A., Lamothe B., Bucchini D., Jami J., Montarras D., Pinset C., Joshi R. L. (2001) FEBS Lett. 488, 174–178 [DOI] [PubMed] [Google Scholar]
- 32.Lamothe B., Baudry A., Christoffersen C. T., De Meyts P., Jami J., Bucchini D., Joshi R. L. (1998) FEBS Lett. 426, 381–385 [DOI] [PubMed] [Google Scholar]
- 33.Cheng C. M., Reinhardt R. R., Lee W. H., Joncas G., Patel S. C., Bondy C. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verleysdonk S., Hirschner W., Wellard J., Rapp M., de los Angeles G. M., Nualart F., Hamprecht B. (2004) Neurochem. Res. 29, 127–134 [DOI] [PubMed] [Google Scholar]
- 35.LeRoith D., Yakar S. (2007) Nat. Clin. Pract. Endocrinol. Metab. 3, 302–310 [DOI] [PubMed] [Google Scholar]
- 36.Mur C., Valverde A. M., Kahn C. R., Benito M. (2002) Diabetes 51, 743–754 [DOI] [PubMed] [Google Scholar]
- 37.Dupont J., Khan J., Qu B. H., Metzler P., Helman L., LeRoith D. (2001) Endocrinology 142, 4969–4975 [DOI] [PubMed] [Google Scholar]
- 38.Mulligan C., Rochford J., Denyer G., Stephens R., Yeo G., Freeman T., Siddle K., O'Rahilly S. (2002) J. Biol. Chem. 277, 42480–42487 [DOI] [PubMed] [Google Scholar]
- 39.Palsgaard J., Brown A. E., Jensen M., Borup R., Walker M., De Meyts P. (2009) Growth Horm. IGF Res. 19, 168–178 [DOI] [PubMed] [Google Scholar]
- 40.Klein J., Fasshauer M., Ito M., Lowell B. B., Benito M., Kahn C. R. (1999) J. Biol. Chem. 274, 34795–34802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.