Abstract
To maintain telomeres, telomerase evolved a unique biochemical activity: the use of a single-stranded RNA template for the synthesis of single-stranded DNA repeats. High repeat addition processivity (RAP) of the Tetrahymena telomerase holoenzyme requires association of the catalytic core with the telomere adaptor subcomplex (TASC) and an RPA1-related subunit (p82 or Teb1). Here, we used DNA binding and holoenzyme reconstitution assays to investigate the mechanism by which Teb1 and TASC confer high RAP. We show that TASC association with the recombinant telomerase catalytic core increases enzyme activity. Subsequent association of the Teb1 C-terminal domain with TASC confers the capacity for high RAP even though the Teb1 C-terminal domain does not provide a high-affinity DNA interaction site. Efficient RAP also requires suppression of nascent product folding mediated by the central Teb1 DNA-binding domains (DBDs). These sequence-specific high-affinity DBDs of Teb1 can be functionally substituted by the analogous DBDs of Tetrahymena Rpa1 to suppress nascent product folding but only if the Rpa1 high-affinity DBDs are physically tethered into holoenzyme context though the Teb1 C-terminal domain. Overall, our findings reveal multiple mechanisms and multiple surfaces of protein-DNA and protein-protein interaction that give rise to elongation processivity in the synthesis of a single-stranded nucleic acid product.
Keywords: DNA-binding Protein, DNA Polymerase, DNA-Protein Interaction, Reverse Transcription, Telomere, Processivity, Telomerase
Introduction
The eukaryotic reverse transcriptase telomerase elongates chromosome ends by addition of telomeric repeats, compensating for incomplete end replication and stochastic terminal sequence loss (1). Telomerase is specialized for this cellular task by its function as a ribonucleoprotein (RNP),2 with the integral telomerase RNA (TER) subunit providing a repeat complementary internal template to the telomerase reverse transcriptase (TERT) active site (2, 3). Expression of TER and TERT in a heterologous cell extract such as rabbit reticulocyte lysate (RRL) is sufficient to reconstitute the telomerase catalytic core. Minimal recombinant TERT-TER enzymes from several species show biological fidelity in copying the correct internal template, including a halt of synthesis at the 5′-template boundary. With a canonical reverse transcriptase, the formation of a template-product hybrid would restrict synthesis to a single round of template copying. For telomerase, however, as successively more 5′-template positions transit the active site, there is an accompanying unpairing of hybrid at the template 3′-end (4). Product release from the template 5′-end is generally rate-limiting for elongation in vitro, but most telomerases are capable of multiple-turnover repeat synthesis evident as an increase in product length and/or yield over time.
Telomerase is unique in its reiterative use of a single-stranded template for the synthesis of a single-stranded product. In vivo, telomerase repeat addition processivity (RAP) expedites telomere length homeostasis (2, 5). In vitro, uncoupled from regulation by telomere-associated factors, a ciliate or human telomerase holoenzyme can add hundreds of repeats to a single primer (6, 7). In contrast to the high RAP of an endogenously assembled telomerase holoenzyme, RAP of the recombinant Tetrahymena thermophila catalytic core has <50% frequency of next-repeat synthesis (8). Even low-RAP activity requires the coordination of at least two enzyme sites for single-stranded DNA interaction: the DNA 3′-end is engaged in the active site, whereas adjacent single-stranded DNA (ssDNA) contacts a surface of the TERT N-terminal domain (9, 10). The low-RAP products of the T. thermophila recombinant telomerase catalytic core or a telomerase holoenzyme subpopulation in cell extract continue to accumulate over reaction time, suggesting that the difference between low-RAP and high-RAP activities does not derive from a failure of product to release from the template (which limits the in vitro activity of Saccharomyces cerevisiae telomerase) but instead derives from reduced stability of the enzyme-product interaction(s) necessary to maintain an elongation complex during dissociation of the template hybrid (3, 5).
Structures that can form on G-rich ssDNA repeats affect primer use by telomerase (11). The folding of nascent product as a G-hairpin structure was originally suggested to promote RAP of telomerase from the ciliate Euplotes (12, 13). More recently, studies of human telomerase have demonstrated an in vitro activity of telomere proteins in promoting telomerase RAP by unfolding structure(s) formed in the substrate primer. The human ssDNA-binding telomere protein POT1 promotes unfolding of an intramolecular G-quadruplex (14). Depending on the position of its binding within a substrate, POT1 either inhibits primer use or modestly increases RAP (15). The combination of POT1 and its interaction partner TPP1 can synergistically enhance RAP, potentially involving an association of TPP1 and TERT (16, 17). One report describes a stimulation of human telomerase RAP by the general eukaryotic ssDNA-binding factor replication protein A (RPA), although high concentrations of human RPA or Escherichia coli ssDNA-binding protein inhibit human and T. thermophila telomerase activities (18, 19). Notably, human RPA can unfold an intramolecular G-quadruplex (20, 21).
Telomerase RAP can also be increased through the function of telomerase holoenzyme subunits. Different forms of the Euplotes crassus telomerase holoenzyme have dramatically different RAP, which could reflect changes in TERT and/or other holoenzyme subunits (22, 23). The T. thermophila telomerase holoenzyme has numerous telomerase-specific subunits beyond the catalytic core that are each biologically essential for telomere maintenance (24–26). The TER-binding protein p65 initiates the hierarchical steps of RNA and protein folding that are required for biogenesis of an endogenously assembled p65-TER-TERT catalytic core (27). The three subunits p75, p45, and p19 form the telomere adaptor subcomplex (TASC), which is recruited to the catalytic core in a manner regulated by the substoichiometric factor p50 (26). The final telomerase holoenzyme subunit p82 has direct telomeric repeat ssDNA-binding activity; for ease of reference, we will designate it as Teb1 (telomeric repeat-binding subunit 1). The presence or absence of Teb1 distinguishes high-RAP versus low-RAP holoenzyme activities (26). Teb1 can interact directly with TASC, suggesting that TASC may bridge Teb1 to the catalytic core.
Here, we investigated the mechanism of high RAP conferred by telomerase holoenzyme subunits beyond the catalytic core. To address the role of sequence-specific DNA-binding sites and their physical connectivity with the active site, we exploited the paralogous relationship of telomerase-specific Teb1 and T. thermophila Rpa1. Only Teb1 shows sequence-specific interaction with ssDNA, dependent on both the permutation and number of telomeric repeats, and only Teb1 affects RAP. With a newly developed telomerase holoenzyme reconstitution system, we used Teb1 domain truncations and a Teb1/Rpa1 chimera to uncover a combination of mechanisms that underlie high RAP. The C-terminal domain of Teb1 lacks independent DNA-binding activity but confers an inherent capacity for high RAP through association with TASC. However, only a fraction of holoenzyme with the isolated C-terminal domain of Teb1 has high RAP due to enhanced dissociation of products with a length of five telomeric repeats. Two central domains of Teb1 bind sequence specifically to telomeric repeats. When bridged to holoenzyme through the Teb1 C-terminal domain, these sequence-specific DNA-binding domains increase RAP through suppression of nascent product folding. The central DNA-binding domains of Teb1 can be replaced by the analogous Rpa1 DNA-binding domains to promote RAP. These findings uncover several mechanisms required for the high processivity of a telomerase holoenzyme.
EXPERIMENTAL PROCEDURES
Protein and Subcomplex Expression and Purification
All recombinant proteins produced in E. coli were expressed from pET28a with an N-terminal His6 tag using BL21(DE3) cells as described previously for full-length Teb1 (26). The proteins were eluted in 20 mm Tris-HCl (pH 8), 1 mm MgCl2, 10% glycerol, 300 mm NaCl, 0.1% IGEPAL, and 300 mm imidazole and supplemented with 5 mm dithiothreitol (DTT) before storage at negative 80 °C. Proteins were supplemented with an additional 2 mm DTT upon thawing.
The recombinant telomerase catalytic core was made using Promega TnT RRL following the manufacturer's instructions, with protein expression from pCITE4a. In vitro transcribed TER was added to the RRL reaction during protein synthesis at 1 ng/μl. Scaled up for the total number of assays, 25 μl of an RRL reaction was immunopurified with 7 μl of a resin slurry of FLAG M2 resin (Sigma) and washed with buffer A (20 mm Tris-HCl (pH 8), 1 mm MgCl2, and 10% glycerol) supplemented with 5 mm β-mercaptoethanol.
TASC was prepared by purifying telomerase from cells expressing tagged p45 (26). After a two-step purification of the tagged protein 45fzz, telomerase holoenzyme immobilized on FLAG M2 resin was treated with 10 μl/ml micrococcal nuclease in buffer B (20 mm Tris-HCl (pH 8), 1 mm MgCl2, 10% glycerol, 50 mm NaCl, and 0.1% IGEPAL) supplemented with 2 mm CaCl2 and incubated at room temperature for 30 min. Bound TASC was washed for 30 min with 4 changes of buffer B and eluted with 0.2 mg/ml 3XFLAG peptide (Sigma) in buffer B with 2 mm DTT. Typical purifications contained ∼ 0.2–0.5 ng/μl p75 estimated relative to a protein ladder following SDS-PAGE and silver staining.
Binding and Activity Assays
Electrophoretic mobility shift assays and activity assays with native holoenzyme were done as described previously (26). Binding affinities were calculated based on free probe signal using ImageQuant software (26). For holoenzyme reconstitution, the catalytic core was provided in 7 μl, TASC in 3.5 μl, and recombinant protein from E. coli in 2 μl. All reactions were matched to contain the appropriate volume of each buffer, so the final reaction contained ∼40 mm NaCl, ∼0.03% IGEPAL detergent, ∼25 ng/μl 3XFLAG peptide, and ∼50 mm imidazole regardless of protein content. All assays were carried out by incubating the catalytic core, all proteins, and primer in a 15-μl volume for 10–15 min before the addition of 5 μl of buffer and nucleotides to start primer extension. The assay buffer contained final concentrations of 50 mm Tris-HCl (pH 8), 1.25 mm MgCl2, 5 mm DTT, 0.1 mm dTTP, 10 μm dGTP, and 0.5 μl of [α-32P]dGTP (3000 Ci/mmol, 10 mCi/ml; PerkinElmer Life Sciences). Incubation was carried out at room temperature for 20–30 min unless noted otherwise. Products were purified, resolved by denaturing PAGE, imaged by Typhoon Trio, and quantified using ImageQuant software. For assays shown in Fig. 5, 2 μl of RRL RNP reconstitution reaction was used directly without purification; assays also contained 5 mm DTT instead of β-mercaptoethanol, and the final dGTP concentration was 0.3 μm instead of 10 μm.
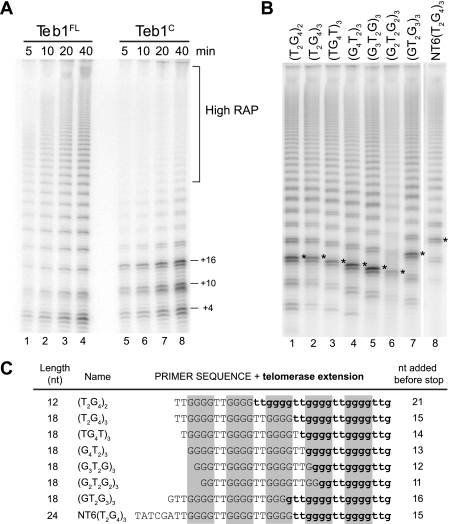
FIGURE 5.
Length-dependent product dissociation from Teb1C holoenzyme. A, time course activity assays of Teb1FL and Teb1C holoenzymes. Reactions contained final concentrations of 0.3 μm dGTP, 200 nm (GT2G3)3, and the indicated form of Teb1 at 200 nm. B, primer sequence dependence of short-product synthesis by Teb1C holoenzyme. Reactions contained final concentrations of 0.3 μm dGTP, 200 nm primer, and 1 μm Teb1C. Asterisks indicate the products dissociated when the product length reached five telomeric repeats. C, summary of predominant products from assays shown in B. Primer sequence is in uppercase letters, whereas sequence added by telomerase is in lowercase boldface letters. Gray boxes highlight the product G-tracts. nt, nucleotides.
RESULTS
Functional Comparison of Teb1 and Rpa1
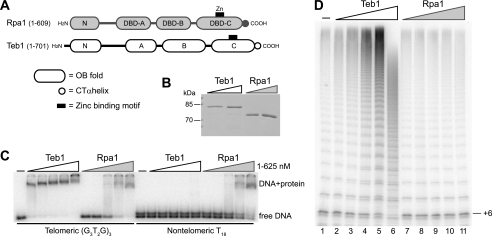
Teb1 is a telomerase-specific protein with a primary sequence and predicted tandem OB-fold domain organization similar to those of RPA1, the largest subunit of the RPA heterotrimer (26). BLAST searches identified two T. thermophila RPA1-like predicted proteins other than Teb1. One of these, Rlp1 (RPA1-like protein 1) was not abundantly expressed, and its gene could be eliminated without phenotype (supplemental Fig. 1). In contrast, the T. thermophila Rpa1 protein most homologous to RPA1 subunits of other species was highly expressed (26), and even modest gene knockdown was strongly inhibitory for culture growth (data not shown). Thus, Teb1 and Rpa1 are the essential RPA1-like proteins of T. thermophila. For purposes of domain analysis below, we designate the predicted tandem OB-fold domains of T. thermophila Teb1 and Rpa1 according to RPA1 convention (Fig. 1A). The RPA heterotrimer has four DNA-binding domain (DBD) OB-folds, three of which (DBD-A, DBD-B, and DBD-C) are found within RPA1 (28). DBD-A and DBD-B have the highest affinity for ssDNA and are the initial sites of ssDNA interaction, whereas DBD-C and, in particular, its C-terminal α-helical extension are critical for heterotrimer association (29).
FIGURE 1.
Comparison of DNA binding and processivity stimulation by Teb1 and Rpa1. A, comparison of predicted T. thermophila Rpa1 and Teb1 domain architectures. B, recombinant Teb1 and Rpa1. Purified recombinant proteins were resolved by SDS-PAGE and Coomassie Blue-stained. C, DNA-binding activity of Teb1 and Rpa1. End-radiolabeled oligonucleotides (∼10 pm) were used with 5-fold concentration steps of protein over the range of 1–625 nm. D, processivity factor activity of Teb1. Affinity-purified telomerase holoenzyme was assayed with 200 nm (G3T2G)3 primer, 0.3 μm dGTP, and 5-fold concentration steps of protein over the range of 0.32–200 nm. CT, C-terminal.
To investigate the biochemical mechanisms that underlie Teb1 function as a telomerase processivity factor, we first assessed its ssDNA-binding activity in comparison with that of the paralogous Rpa1 protein. Like Teb1, T. thermophila Rpa1 was readily purified as a soluble protein following expression in E. coli (Fig. 1B). This allowed us to directly compare the ssDNA binding properties of Teb1 and Rpa1 by electrophoretic mobility shift assay. Teb1 was shown previously to bind to ssDNAs containing the Tetrahymena telomeric repeat sequence (T2G4)3, with or without additional non-telomeric 5′- or 3′-sequence (26). Therefore, we compared the binding of Teb1 and Rpa1 to telomeric or non-telomeric 18-nucleotide ssDNA oligonucleotides. Teb1 bound the telomeric repeat ssDNA with nanomolar affinity but did not detectably bind polythymidine, whereas Rpa1 bound both ssDNAs with approximately equal affinity (Fig. 1C). We conclude that Teb1 differs from Rpa1 in its increased affinity for tandem telomeric repeats and also its decreased affinity for non-telomeric sequence. Both proteins showed dissociation rates too rapid to measure reliably by electrophoretic mobility shift assay (data not shown), consistent with a dynamic binding rather than stable capping function for both proteins in vivo. For comparison, we attempted to purify the T. thermophila RPA complex using a strain with Rpa1 C-terminally tagged at its endogenous locus, but subunit tagging was deleterious for its biological function (data not shown).
The addition of recombinant Teb1 to a total holoenzyme population purified by tagged TERT converts the percentage of holoenzyme with low-RAP activity to additional high-RAP activity (26). The increase in RAP was particularly evident in the long products with high specific activity of radiolabeled dGTP incorporation (Fig. 1D, lanes 2–5). At high concentration, Teb1 competed with holoenzyme for substrate and product binding, leading to inhibition of RAP and activity overall (Fig. 1D, lane 6). Over the same concentration range, the addition of purified Rpa1 had no impact on the low-RAP or high-RAP activities of telomerase holoenzyme (Fig. 1D, lanes 7–11). Much higher concentrations of Rpa1 caused overall inhibition of telomerase activity, consistent with weak competition for substrate (data not shown). We conclude that Teb1 function as a telomerase processivity factor depends on properties of Teb1 that are distinct from those of Rpa1, such as its sequence specificity of DNA binding and/or its physical association with telomerase holoenzyme.
Reconstitution of Holoenzyme RAP Fully Dependent on Recombinant Teb1
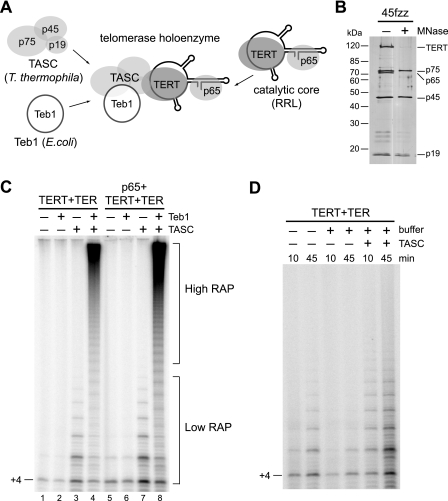
To study the mechanism of Teb1-dependent processivity, we first established a holoenzyme reconstitution system entirely dependent on recombinant Teb1 for high RAP (Fig. 2A). The minimal catalytic core (TERT-TER) and the endogenous RNP catalytic core (p65-TER-TERT) produced in RRL have low RAP (8, 30). TASC may be required to recruit Teb1 to holoenzyme (26), but we have not been able to reconstitute TASC from recombinant p75, p45, and p19. Instead, we isolated TASC from holoenzyme purified using tagged p45 by treating resin-immobilized holoenzyme with nuclease to degrade TER, release p65, and induce unfolding of TERT (26). After extensive washing and elution, highly purified TASC was recovered (Fig. 2B). Recombinant Teb1 was purified from E. coli. Purified TASC or Teb1 alone had no background of associated enzyme activity (data not shown). We therefore used the combination of the catalytic core, TASC, and Teb1 from three systems (Fig. 2A) to determine whether Teb1 could convert the low-RAP activity of the recombinant telomerase catalytic core to the high-RAP activity of a native holoenzyme.
FIGURE 2.
Reconstitution of telomerase holoenzyme. A, diagram of the holoenzyme reconstitution strategy. B, TASC purification. Telomerase holoenzyme and micrococcal nuclease (MNase)-treated TASC purified by 45fzz were resolved by SDS-PAGE and silver-stained. C, requirements for high RAP. Activity assays of the recombinant minimal (TERT-TER) or endogenous (p65-TERT-TER) catalytic core were performed with or without TASC and/or 40 nm Teb1 using 200 nm (GT2G3)3 primer at 30 °C. D, effect of TASC on the activity of the minimal recombinant enzyme (TERT-TER). Activity assays were performed using 1 μm (GT2G3)3 primer at room temperature with or without TASC and with or without the buffer components present in a purified recombinant Teb1 preparation (indicated as + or − buffer; no Teb1 protein was added).
The activity of an RRL-reconstituted T. thermophila minimal or endogenous RNP catalytic core had low RAP (Fig. 2C, lanes 1 and 5). The addition of Teb1 alone had no impact on the activity of either RNP (Fig. 2C, lanes 2 and 6). The addition of TASC alone increased activity without increasing RAP above that characteristic of the recombinant catalytic core (Fig. 2C, lanes 3 and 7). (Quantification of RAP indicated a 30–35% frequency of next-repeat synthesis.) Activity stimulation by TASC derived in part from its ability to reduce enzyme inhibition by buffer components required for recombinant Teb1 purification (Fig. 2D). In striking contrast to the addition of either Teb1 or TASC alone, their combination resulted in a dramatic conversion of low-RAP to high-RAP activity (Fig. 2C, lanes 4 and 8). The presence of p65 was not essential for high RAP in vitro, and thus, subsequent reconstitutions were performed with the minimal catalytic core of TERT-TER. These results demonstrate the first high-RAP holoenzyme reconstitution of a minimal recombinant telomerase catalytic core.
Permutation and Length Dependence of Teb1 DNA Binding
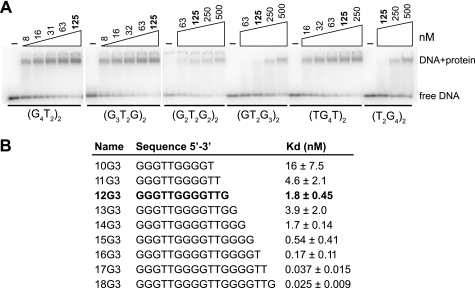
Teb1 binding to the 18-nucleotide oligonucleotide (T2G4)3 was poorly competed by the 12-nucleotide oligonucleotide (T2G4)2 (26). Because repeat permutation could have a major impact on Teb1 binding, previous assays using oligonucleotides with a single repeat permutation may have overestimated the significance of the three-repeat rather than two-repeat ssDNA length. We therefore examined Teb1 binding to all possible two-repeat oligonucleotide sequence permutations. Teb1 demonstrated a dramatic permutation dependence of binding affinity, with an ∼50-fold preference for (G3T2G)2 compared with (T2G4)2 (Fig. 3A, note the different Teb1 concentrations in each panel). We then tested an oligonucleotide series maintaining the 5′-permutation of the two-repeat ssDNA (G3T2G)2 with truncation or extension of the oligonucleotide 3′-end (Fig. 3B). Loss of 3′-nucleotides progressively reduced binding affinity, whereas affinity increased by a total of ∼100-fold as the ssDNA gained another full repeat (Fig. 3B). These results support the likelihood of an extended Teb1 contact surface for ssDNA that can engage multiple telomeric repeats.
FIGURE 3.
Teb1 binding dependence on repeat permutation and ssDNA length. A, permutation-dependent Teb1 DNA-binding activity. End-radiolabeled oligonucleotides (∼10 pm) were used with 2-fold concentration steps of protein over different final concentration ranges (the common 125 nm concentration is indicated in boldface). B, length-dependent Teb1 DNA-binding activity. Kd calculations were made from three independent experiments performed using end-radiolabeled oligonucleotides.
Teb1 Domain Requirements for DNA Binding and High RAP
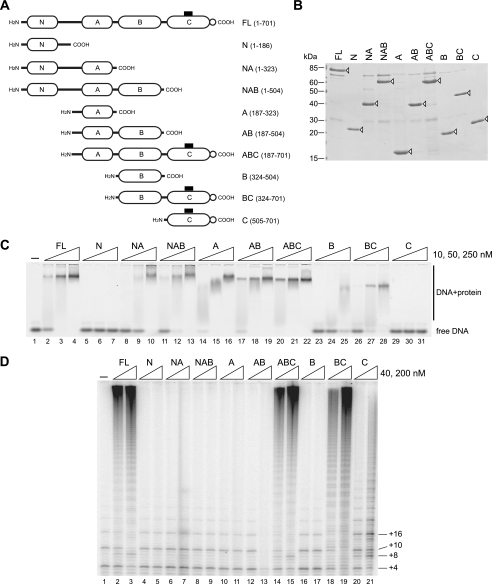
Because Teb1 binds preferentially to an at least three-repeat length of ssDNA, we next explored whether multiple domains of Teb1 associate with ssDNA. Truncation from the Teb1 N terminus and/or C terminus created domain combinations designated as N, NA, NAB, A, AB, ABC, B, BC, and C (Fig. 4A). Each protein was purified following expression in E. coli (Fig. 4B) and used for DNA interaction assays (Fig. 4C). Only Teb1N and Teb1C lacked detectable DNA-binding activity (Fig. 4C, lanes 5–7 and 29–31). Teb1A and Teb1B each bound ssDNA, with Teb1A showing the highest binding affinity of the individual domains (Fig. 4C, lanes 14–16 and 23–25). All proteins with at least one of the central Teb1 DNA-binding domains showed robust ssDNA interaction, and the combination of both of the central DNA-binding domains was sufficient for binding affinity comparable with that of full-length Teb1 (Teb1FL). These results demonstrate that Teb1 has two domains with autonomous ssDNA-binding activity, which are analogous to RPA1 DBD-A and DBD-B in overall protein architecture.
FIGURE 4.
DNA binding and processivity factor function of Teb1 domains. A, diagram of Teb1 domain truncations with amino acid endpoints indicated to the right. B, truncated Teb1 proteins. Purified recombinant proteins were resolved by SDS-PAGE and Coomassie Blue-stained. Open arrowheads mark the Teb1 polypeptides. C, DNA-binding activity of truncated Teb1 proteins. Proteins were used in 5-fold concentration steps over the range of 10–250 nm with 10 nm 6-carboxyfluorescein-(T2G4)3. D, processivity factor function of truncated Teb1 proteins. Proteins were assayed in reconstituted telomerase holoenzyme reactions using 200 nm (GT2G3)3 primer and 40 or 200 nm each Teb1 polypeptide.
The panel of Teb1 truncations was also assayed for ability to confer high RAP to reconstituted telomerase holoenzyme. The low RAP of the catalytic core with TASC was converted to high RAP by the addition of Teb1FL (Fig. 4D, lanes 1–3). High RAP was also induced by Teb1ABC or Teb1BC (Fig. 4D, lanes 14 and 15 and lanes 18 and 19). Teb1 truncations missing the C-terminal domain did not increase RAP. Furthermore, at high concentration, Teb1AB inhibited activity overall (Fig. 4D, lanes 12 and 13). These results suggest that Teb1C functions to link the ssDNA-binding activity of the central Teb1 domains to the rest of the holoenzyme complex. Without the Teb1C bridge, the central ssDNA-binding domains of Teb1 compete for substrate rather than conferring high RAP to holoenzyme.
Teb1C alone supported some high-RAP product synthesis (Fig. 4D, lanes 20 and 21). Curiously, the product profile of Teb1C holoenzyme is distinct from that of any other T. thermophila telomerase preparation. In assays of the catalytic core with TASC (Fig. 4D, lane 1), products of progressively increasing length had progressively decreasing intensity due to the limited frequency of next-repeat synthesis. In assays of Teb1FL holoenzyme with high RAP (Fig. 4D, lanes 2 and 3), long products had the highest signal intensity. The products of Teb1C holoenzyme had components of low-RAP and high-RAP activity but also a predominance of distinct short products (Fig. 4D, lanes 20 and 21). Overall, we conclude that Teb1C holoenzyme has a mechanism for high RAP that was lacking in the catalytic core, but without at least one of the central Teb1 DNA-binding domains, high-RAP activity is severely limited.
RAP Inhibition by Product Folding
To investigate the short products that limit high RAP of Teb1C holoenzyme, we first compared the time courses of product synthesis by Teb1FL and Teb1C holoenzymes. High-RAP products in reactions with a standard dGTP concentration (10 μm) were already too long to resolve by gel electrophoresis at early time points, but reactions with a lower dGTP concentration (0.3 μm) reduced the rate of product synthesis enough to visualize the increase in high-RAP product length over time. Teb1FL holoenzyme and, to a lesser extent, Teb1C holoenzyme both generated some high-RAP products that increased in length over time (Fig. 5A). For the high-RAP reaction products, normalization of product intensities for the content of radiolabeled dGTP suggested that Teb1C holoenzyme approached the same RAP as Teb1FL holoenzyme (representative quantification is shown in supplemental Fig. 2A). Teb1C holoenzyme differed from Teb1FL holoenzyme in the enhanced accumulation of short products of specific lengths (product quantification is shown in supplemental Fig. 2A). The unique short-product profile of Teb1C holoenzyme did not change over time as product abundance increased, suggesting multiple rounds of product synthesis and dissociation (Fig. 5A, lanes 5–8). Chase assays with unlabeled dGTP confirmed that the predominant short products of Teb1C holoenzyme were not intermediates in the synthesis of high-RAP products (data not shown).
To probe whether short-product dissociation from Teb1C holoenzyme was a consequence of total product length, number of repeats synthesized, and/or product sequence, we compared the Teb1C holoenzyme products generated from different primers. The two-repeat primer (T2G4)2 was extended by about three repeats to generate the most intense product (Fig. 5B, lane 1), representing +21 nucleotides of synthesis beyond the 12-nucleotide primer (Fig. 5C). A three-repeat primer of the same permutation, (T2G4)3, was extended by about two repeats to generate the most intense product, representing +15 nucleotides of synthesis beyond the 18-nucleotide primer (Fig. 5, B, lane 2, and C). Three-repeat primers of other permutations were also extended by about two repeats to generate the most intense product (Fig. 5B, lanes 3–6), with the exception of the primer with permutation (GT2G3)3, which was extended by about three repeats (lane 7). These profiles suggest that product dissociation occurs preferentially when a product harbors five consecutive G-tracts, with the most 5′-tract at least G2 but optimally G3 or G4 (Fig. 5C). As predicted from this pattern, the most intense product of a primer with non-telomeric 5′-sequence had a longer total length (Fig. 5, B, lane 8, and C). Product quantifications normalized for the content of radiolabeled dGTP confirmed the increase in product dissociation after synthesis of the fifth telomeric repeat (supplemental Fig. 2B). These findings suggest that G-quadruplex folding of the first four repeats released from the template hybrid limits the high-RAP activity of Teb1C holoenzyme by inducing product dissociation (see “Discussion”).
High RAP from Fusion of Rpa1 Domains to Teb1C
The above results suggest that Teb1FL confers high RAP by a combination of two mechanisms, one mediated by Teb1C and another involving the suppression of product folding by the central Teb1 DNA-binding domains. The central DNA-binding domains of Teb1 differ from those of Rpa1 in their sequence specificity of DNA binding and in their physical association with telomerase holoenzyme through Teb1C. We addressed whether both of these features were required for Teb1 suppression of product folding and efficient high RAP. We exploited the parallel modularity of Teb1 and Rpa1 to design a chimeric protein with the major DNA-binding domains of Teb1 replaced by those of Rpa1 (Rpa1NAB-Teb1C) (Fig. 6A). This protein should associate with holoenzyme through the Teb1 C-terminal domain to position general ssDNA-binding activity in the vicinity of nascent product. Also, to investigate the requirements for Teb1C association with holoenzyme, we designed a variant that removed a predicted α-helical C-terminal extension from the Teb1 C-terminal OB-fold (Teb1ΔCTαh) (Fig. 6A). In the structure of the RPA trimerization core, the C-terminal α-helical peptide of RPA1 interacts with RPA2 and RPA3 (29). If Teb1C association with holoenzyme occurred through an association similar to that of RPA1 interaction with its heterotrimer partners, we would expect the Teb1 C-terminal peptide truncation to preclude Teb1-dependent RAP. The domain chimera and the Teb1 C-terminal peptide truncation variant were both purified as soluble protein following expression in E. coli (Fig. 6B). As expected, each protein bound to ssDNA (Fig. 6C).
FIGURE 6.
High RAP from general ssDNA-binding activity tethered to telomerase holoenzyme. A, diagram of the domain chimera and Teb1 C-terminal peptide truncation relative to wild-type Teb1 and Rpa1, with amino acid endpoints indicated. B, purified recombinant proteins resolved by SDS-PAGE and Coomassie Blue-stained. C, DNA-binding activity of Teb1 variants. End-radiolabeled oligonucleotide (∼10 pm) was used with 5-fold protein concentration steps over the indicated ranges. D, processivity stimulation by Teb1 variants as assayed in reconstituted telomerase holoenzyme reactions using 200 nm (GT2G3)3 primer and the indicated proteins at 40 or 100 nm. E, DNA-binding activity of Rpa1 and the domain chimera. End-radiolabeled oligonucleotides (∼10 pm) were used with 10-fold protein concentration steps over the indicated ranges.
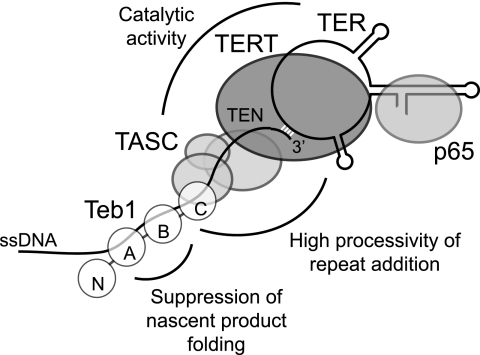
The Rpa1NAB-Teb1C and Teb1ΔCTαh proteins were compared with Rpa1FL, Teb1FL, and Teb1C for ability to induce high RAP. Teb1 C-terminal peptide truncation did not prevent high RAP of the reconstituted holoenzyme, suggesting that the Teb1C putative OB-fold itself interacts with TASC (Fig. 6D, compare lanes 2 and 3 with lanes 4 and 5). Notably, the domain chimera was also permissive for high RAP: unlike Rpa1FL, Rpa1NAB-Teb1C stimulated high RAP without the tendency to short-product dissociation evident for holoenzyme with Teb1C alone (Fig. 6D, lanes 6–11). These results indicate that the sequence specificity of Teb1AB-DNA interaction is not required for high RAP of telomeric repeat synthesis in vitro. Curiously, unlike Rpa1, the domain chimera showed a preference for binding to telomeric versus non-telomeric sequence (Fig. 6E). Overall, our dissection of telomerase processivity reveals that the catalytic core with TASC gains an inherent capacity for high RAP through association of Teb1C and gains additional efficiency of high RAP through Teb1AB suppression of product folding. Numerous protein-protein interactions establish the holoenzyme architecture required for processive repeat synthesis, and numerous sites of protein-DNA interaction engage the single-stranded product (Fig. 7).
FIGURE 7.
Model for telomerase holoenzyme protein-protein and protein-DNA interactions that contribute to catalytic activity and RAP. Individual Teb1 domains are designated N, A, B, and C. TEN, TERT N-terminal domain.
DISCUSSION
In this study, we have demonstrated the first telomerase holoenzyme reconstitution from a recombinant catalytic core. We then exploited this reconstitution system to define mechanisms that confer the unique ssDNA repeat synthesis processivity of telomerase. RAP requires both product dissociation from the template 5′-end and retention of an enzyme-product complex while the released template returns to its default position in the active site (3). Here, we have shown that multiple mechanisms contribute to the high processivity of repeat synthesis by a telomerase holoenzyme.
At the level of holoenzyme architecture, the ability to reconstitute telomerase holoenzyme catalytic activity from a recombinant catalytic core bypasses previous limitations imposed by endogenous RNP accumulation and proteolytic truncation of Teb1 in cell extract. We found that the p65 subunit essential for TER folding and TERT RNP assembly in vivo does not play additional roles as a platform for assembly of TASC or Teb1. Also, our findings demonstrate that ciliate TERT translated in RRL and ciliate TER transcribed by T7 RNA polymerase are competent for high RAP. As proposed based on Teb1 and TASC physical interaction (26), we have shown here that TASC functionally links Teb1 to the catalytic core. The C-terminal domain of Teb1 interacts with TASC without a requirement for the peptide extension that mediates RPA1 association with RPA2 and RPA3 (29). It remains possible that Teb1 forms an RPA-like interface with TASC subunits p75, p45, and/or p19, although these three subunits themselves could form an RPA-like complex.
Biochemical analysis of Teb1 truncations supports the overall similarity of Teb1 and Rpa1 domain architectures but also highlights some divergent properties of individual domains. The Teb1 N-terminal domain makes no evident contribution to DNA binding. Instead, like the RPA1 N-terminal domain, Teb1N could mediate cellular regulations such as Teb1 turnover by the proteasome (26). Teb1 and RPA1 each have two autonomously functional DNA-binding domains. However, unlike RPA1 DBD-A and DBD-B, the major Teb1 DNA-binding domains specifically recognize telomeric repeat ssDNA. In addition to increased affinity for telomeric repeats, Teb1 has decreased affinity for non-telomeric sequence compared with T. thermophila Rpa1. Because Teb1 has a much lower endogenous expression level than Rpa1 and does not physically associate with Rpa1 interaction partners (26), it seems unlikely that general ssDNA binding by Teb1 would be counterselected due to functional interference with RPA. Instead, Teb1AB loss of general ssDNA-binding activity could give an advantageous increase in specificity of telomerase holoenzyme recruitment to established telomeres. Finally, similarities and differences are also evident upon comparison of the Teb1 C-terminal domain with RPA1 DBD-C. Teb1C seems likely to contact ssDNA because Teb1BC has higher affinity for ssDNA than does Teb1B alone (Fig. 4C), and Teb1C increases the preference of Rpa1NAB for telomeric repeat ssDNA (Fig. 6E). Teb1C also plays unique roles as a telomerase holoenzyme subunit, bridging TASC to the Teb1AB domains that suppress nascent product folding.
Multiple telomerase holoenzyme sites of ssDNA interaction contribute to high RAP (Fig. 7). Within the catalytic core, TER and TERT both provide sites of DNA interaction. The template binds a ssDNA 3′-end by hybridization, and the TER pseudoknot has been suggested to position a template hybrid in the active site (31). TERT contacts the template hybrid and also immediately adjacent ssDNA in a nascent product-binding site that supports low RAP (8, 32, 33). A surface of the TERT N-terminal domain interacts with additional unpaired ssDNA, improving the Km of primer elongation (8–10). TASC does not substantially enhance RAP, but it does enhance the activity of the recombinant catalytic core (Fig. 7). Only upon the addition of Teb1C to TASC and the catalytic core does the nature of telomerase association with ssDNA change to support high RAP. The weak ssDNA-binding activity of Teb1C could contribute to RAP, but high RAP conferred by Teb1C does not appear to reflect the consequence of a simple increase in ssDNA-binding affinity. Instead, in addition to protein-DNA interactions, a network of protein-protein interactions between Teb1C, TASC, and the catalytic core provides a critical part of the mechanism for high RAP (Fig. 7).
Teb1AB domains provide an additional mechanism for increase in RAP. The Teb1AB interactions with ssDNA that suppress short-product dissociation do not need to be sequence-specific, but they do need to occur in holoenzyme context (Fig. 7). Short-product dissociation from the Teb1C holoenzyme occurs preferentially after the accumulation of four telomeric repeats beyond the template hybrid. We suggest that intramolecular G-quadruplex formation can out-compete the protein-DNA interactions required to stabilize enzyme-product association upon product release from the template. If the product reaches a sufficient length, competition of protein-DNA interaction by nascent product folding appears reduced. G-quadruplex folding may still occur on long products but would not absolutely require the participation of the nascent product region capable of protein interaction. Consistent with this scenario, Teb1C holoenzyme reactions with a faster rate of repeat synthesis yield more high-RAP products (for example, compare Fig. 4 reactions using a standard dGTP concentration with Fig. 5 reactions using a reduced dGTP concentration).
Overall, our work reveals an unexpected strategy for the high RAP of a telomerase holoenzyme. The protein-DNA interactions that confer high RAP appear to function as an extensive surface of dynamic toeholds rather than a coordinated set of alternately locked binding sites. The combination of individually dynamic protein-DNA interactions could enable the T. thermophila telomerase holoenzyme to have high RAP and yet thread forward on chromosome ends in vivo, reducing the topological challenge for lagging strand synthesis and potentially accelerating the assembly of telomere proteins for feedback control of telomere length homeostasis.
Related principles of telomerase interaction with ssDNA may apply in other organisms. Budding yeast Cdc13 binds sequence-specifically to G-rich telomeric repeat ssDNA and promotes telomere elongation by association with telomerase holoenzyme subunit(s) beyond the catalytic core (34). This role of Cdc13 requires its release from Stn1 and Ten1, two smaller subunits of an RPA-like CST complex generally involved in lagging strand synthesis at replication-challenged regions of the genome (35). Most eukaryotes except budding yeast have a G-rich telomeric repeat ssDNA-binding protein, POT1, which is recruited to telomeres through interactions with both ssDNA and other telomere-bound proteins (36, 37). Human POT1 can unfold an intramolecular G-quadruplex and, depending on its binding register, either prevent telomerase elongation or enhance elongation processivity (14, 15). The POT1-TPP1 complex increases human telomerase RAP more than POT1 alone, and TPP1 itself shows some stimulation of RAP potentially linked to an association with TERT (16, 17). It is possible that TPP1 acts analogously to Teb1C, whereas POT1 provides the function of Teb1AB. In addition to increasing telomerase processivity, both Teb1 and POT1-TPP1 are likely to play key roles in the recruitment of telomerase to telomeres.
Supplementary Material
Acknowledgments
We thank Kyungah Hong for expression and purification of the Teb1ΔCTαh and Rpa1NAB-Teb1C proteins and Richard Sun for cloning Teb1 truncation constructs.
This work was supported, in whole or in part, by National Institutes of Health GM54198. This work was also supported by a University of California Berkeley Cancer Research Laboratory training grant.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- RNP
- ribonucleoprotein
- TER
- telomerase RNA
- TERT
- telomerase reverse transcriptase
- RRL
- rabbit reticulocyte lysate
- RAP
- repeat addition processivity
- ssDNA
- single-stranded DNA
- RPA
- replication protein A
- TASC
- telomere adaptor subcomplex
- DTT
- dithiothreitol
- DBD
- DNA-binding domain
- FL
- full-length.
REFERENCES
- 1.Blackburn E. H., Greider C. W., Szostak J. W. (2006) Nat. Med. 12, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 2.Sekaran V. G., Soares J., Jarstfer M. B. (2010) Biochim. Biophys. Acta 1804, 1190–1201 [DOI] [PubMed] [Google Scholar]
- 3.Collins K. (2009) in Non-Protein Coding RNAs (Walter N. G., Woodson S. A., Batey R. T. eds) pp. 285–301, Springer-Verlag, Berlin [Google Scholar]
- 4.Förstemann K., Lingner J. (2005) EMBO Rep. 6, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lue N. F. (2004) BioEssays 26, 955–962 [DOI] [PubMed] [Google Scholar]
- 6.Greider C. W. (1991) Mol. Cell. Biol. 11, 4572–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maine I. P., Chen S. F., Windle B. (1999) Biochemistry 38, 15325–15332 [DOI] [PubMed] [Google Scholar]
- 8.Hardy C. D., Schultz C. S., Collins K. (2001) J. Biol. Chem. 276, 4863–4871 [DOI] [PubMed] [Google Scholar]
- 9.Jacobs S. A., Podell E. R., Cech T. R. (2006) Nat. Struct. Mol. Biol. 13, 218–225 [DOI] [PubMed] [Google Scholar]
- 10.Romi E., Baran N., Gantman M., Shmoish M., Min B., Collins K., Manor H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8791–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oganesian L., Moon I. K., Bryan T. M., Jarstfer M. B. (2006) EMBO J. 25, 1148–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shippen-Lentz D., Blackburn E. H. (1990) Science 247, 546–552 [DOI] [PubMed] [Google Scholar]
- 13.Jarstfer M. B., Cech T. R. (2002) Biochemistry 41, 151–161 [DOI] [PubMed] [Google Scholar]
- 14.Zaug A. J., Podell E. R., Cech T. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei M., Zaug A. J., Podell E. R., Cech T. R. (2005) J. Biol. Chem. 280, 20449–20456 [DOI] [PubMed] [Google Scholar]
- 16.Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 17.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007) Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 18.Cohen S., Jacob E., Manor H. (2004) Biochim. Biophys. Acta 1679, 129–140 [DOI] [PubMed] [Google Scholar]
- 19.Rubtsova M. P., Skvortsov D. A., Petruseva I. O., Lavrik O. I., Spirin P. V., Prasolov V. S., Kisseljov F. L., Dontsova O. A. (2009) Biochemistry 74, 92–96 [DOI] [PubMed] [Google Scholar]
- 20.Salas T. R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J. L., Favre A., Saintomé C. (2006) Nucleic Acids Res. 34, 4857–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J. H., Bochkareva E., Bochkarev A., Gray D. M. (2009) Biochemistry 48, 1099–1111 [DOI] [PubMed] [Google Scholar]
- 22.Greene E. C., Shippen D. E. (1998) Genes Dev. 12, 2921–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karamysheva Z., Wang L., Shrode T., Bednenko J., Hurley L. A., Shippen D. E. (2003) Cell 113, 565–576 [DOI] [PubMed] [Google Scholar]
- 24.Witkin K. L., Collins K. (2004) Genes Dev. 18, 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witkin K. L., Prathapam R., Collins K. (2007) Mol. Cell. Biol. 27, 2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min B., Collins K. (2009) Mol. Cell 36, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone M. D., Mihalusova M., O'Connor C. M., Prathapam R., Collins K., Zhuang X. (2007) Nature 446, 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochkareva E., Korolev S., Lees-Miller S. P., Bochkarev A. (2002) EMBO J. 21, 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prathapam R., Witkin K. L., O'Connor C. M., Collins K. (2005) Nat. Struct. Mol. Biol. 12, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao F., Cech T. R. (2008) Nat. Struct. Mol. Biol. 15, 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baran N., Haviv Y., Paul B., Manor H. (2002) Nucleic Acids Res. 30, 5570–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie M., Podlevsky J. D., Qi X., Bley C. J., Chen J. J. (2010) Nucleic Acids Res. 38, 1982–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shore D., Bianchi A. (2009) EMBO J. 28, 2309–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellinger R. J. (2009) Mol. Cell 36, 168–169 [DOI] [PubMed] [Google Scholar]
- 36.Palm W., de Lange T. (2008) Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 37.Oganesian L., Karlseder J. (2009) J. Cell Sci. 122, 4013–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.