Abstract
MicroRNAs are a class of small non-coding RNAs and participate in the regulation of apoptotic program. Although miR-21 is able to inhibit apoptosis, its expression regulation and downstream targets remain to be fully elucidated. Here we report that the transcriptional factor Foxo3a initiates apoptosis by transcriptionally repressing miR-21 expression. Our results showed that doxorubicin could simultaneously induce the translocation of Foxo3a to the cell nuclei and a reduction in miR-21 expression. Knockdown of Foxo3a resulted in an elevation in miR-21 levels, whereas enforced expression of Foxo3a led to a decrease in miR-21 expression. In exploring the molecular mechanism by which Foxo3a regulates miR-21, we observed that Foxo3a bound to the promoter region of miR-21 and suppressed its promoter activity. These results indicate that Foxo3a can transcriptionally repress miR-21 expression. In searching for the downstream targets of miR-21 in apoptosis, we found that miR-21 suppressed the translation of Fas ligand (FasL), a pro-apoptotic factor. Furthermore, Foxo3a was able to up-regulate FasL expression through down-regulating miR-21. Our data suggest that Foxo3a negatively regulates miR-21 in initiating apoptosis.
Keywords: Apoptosis, Cancer Therapy, MicroRNA, RNA, Signal Transduction
Introduction
miRNAs2 are a class of small non-coding RNAs that mediate post-transcriptional gene silencing. Recently, the work on miRNAs renovates our understanding about apoptotic regulation. They can be classified as either pro- or anti-apoptotic miRNAs (1, 2). For example, miR-1 participates in the initiation of apoptosis (3), whereas miR-21 is able to inhibit apoptosis (4). miRNAs are expressed at a constant level under physiological condition, and their functions depend on their expression levels. It remains a challenging question as to how their expression is regulated in the apoptotic program.
The forkhead family of transcription factors is characterized by the presence of a conserved 100-amino acid DNA binding domain and participate in regulating diverse cellular functions such as apoptosis, differentiation, metabolism, proliferation, and survival (5). Foxo3a is a substrate of protein kinase Akt, and its transcriptional output is controlled via phosphorylation. In the absence of cellular stimulation and when Akt is inactive, Foxo3a is localized within the nucleus where it performs transcription of target genes. However, upon phosphorylation by Akt at Thr-32, Ser-253, and Ser-315, it binds to 14-3-3 and cannot exert the transcriptional function (6).
Fas ligand (FasL) is a potential transcriptional target of Foxo3a, but the transcriptional output can be either activation or suppression. It has been reported that Foxo3a can stimulate FasL expression, thereby triggering apoptosis (6). However, there is also evidence showing that Foxo3a decreases the expression of FasL (7). The molecular mechanism by which Foxo3a regulates FasL expression remains further elusive.
miR-21 has been shown to regulate apoptosis by targeting a variety of apoptotic factors. It contributes to glioma malignancy by down-regulating matrix metalloproteinase inhibitors, thereby leading to the activation of matrix metalloproteinases and promoting invasiveness of cancer cells (8). It promotes cell transformation by targeting the programmed cell death 4 gene (9). miR-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and the tissue inhibitor of metalloproteinase 3 (10). Given the important role of FasL in apoptosis, it is not yet clear whether FasL can be a target of miR-21 in the apoptotic machinery.
The expression levels of miRNAs remain constant under physiological condition. Their alterations may cause the pathological disorders. miRNA expression can be regulated by transcriptional factors. For example, miR-34a causes dramatic reprogramming of gene expression and promotes apoptosis, and it is transactivated by the tumor suppression gene p53 (11). Serum response factor can directly bind to the promoter of miR-1-1 and miR-1-2 genes and activate their expression (12). Our recent work reveals that miR-23a is a transcriptional target of nuclear factor of activated T cells c3 (13). Because the functions of miRNAs are closely related to their expression levels, it is important to explore the molecular mechanism governing their expression. Foxo3a and miR-21 are involved in apoptosis, but it is not yet clear whether there is an impact between these two factors in the apoptotic machinery.
Lung cancer is the most common cause of cancer-related death. Chemotherapy plays an important role for the treatment of lung cancer, and doxorubicin is commonly used for lung cancer therapy. However, a major obstacle of chemotherapy is the cancer resistance. It is critically important to identify the factors that participate in the regulation of cancer cell apoptosis, so that novel approaches can be developed for cancer therapy.
Our present work aimed at elucidating the molecular mechanism by which Foxo3a regulates the apoptotic program in A549 human lung cancer cells. Our results revealed that Foxo3a transcriptionally represses the expression of miR-21. Furthermore, we identified FasL as a target of miR-21. In addition, we found that Foxo3a regulates FasL through miR-21. Our data shed new light on understanding the regulation of miRNA expression and the relationship among Foxo3a, miR-21, and FasL in the apoptotic cascades.
EXPERIMENTAL PROCEDURES
Cell Cultures and Treatment
Human lung cancer cells (A549) and human neuroblastoma cells (SH-EP1) were grown in Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. The treatment with doxorubicin (Sigma) was performed as we described (14). The culture of cardiac fibroblasts was performed as we described (15).
Cell Viability and Apoptosis Assays
Cell death was determined by trypan blue exclusion, and the numbers of trypan blue-positive and -negative cells were counted on a hemocytometer. Apoptosis was determined by the terminal deoxyribonucleotidyltransferase-mediated TUNEL using a kit from Roche Applied Science. The detection procedures were in accordance with the kit instructions.
Constructions of Foxo3a and FasL RNA Interference (RNAi)
The Foxo3a RNAi sense sequence is 5′-GAGCTCTTGGTGGATCATC-3′; the antisense sequence is 5′-GATGATCCACCAAGAGCTC-3′. The scramble Foxo3a RNAi sense sequence is 5′-GGCGTAGTCGTAGTTCTCA-3′; the scramble antisense sequence is 5′-TGAGAACTACGACTACGCC-3′. FasL RNAi sense sequence is 5′-AAAGGAGCTGAGGAAAGTG-3′; the antisense sequence is 5′-CACTTTCCTCAGCTCCTTT-3′. The scramble FasL RNAi sense sequence is 5′-TGGAAGAGACAGTAGAGAG-3′; the scramble antisense sequence is 5′-CTCTCTACTGTCTCTTCCA-3′. They were cloned into pSilencer adeno 1.0-CMV vector (Ambion) according to the manufacturer's instructions.
Constructions of miR-21 and FasL Promoters
Human mir-21 promoter region was amplified from human genomic DNA using PCR to generate wild type promoter. The large fragment containing two Foxo3a potential binding sites (wild type promoter-1, wt-1) was amplified using the forward primer, 5′-AAACCAAGGCTCTTACCATAGC-3′. The short fragment containing one Foxo3a potential binding (wild type promoter-2 (wt-2)) was amplified using the forward primer, 5′-GCATGAGAGAGCCACTACCAAG-3′. Both fragments were amplified using the reverse primer, 5′-TGGTACAGCCATGGAGATGTCA-3′. The promoters were cloned into the reporter plasmid, pGL4.17 (Promega). The introduction of mutations in the putative Foxo3a binding site in wt-1 fragment (−197 to −191 wild type, 5′-ATAAACA-3′; mutant, 5′-ACGGCCA-3′) was generated using QuikChange II XL site-directed mutagenesis kit (Stratagene). Human FasL promoter region (from −3157 to +2) was amplified using the same protocol as described for miR-21. The forward primer was 5′-GTGACCTGTCCAGTTCACACAG-3′; the reverse primer was 5′-TGCATGGCAGCTGGTGAGTCAG-3′. The constructs were sequence-verified.
Preparation of miR-21 Expression Construct
miR-21 was synthesized by PCR using human genomic DNA as the template. The upstream primer was 5′-GCATTATGAGCATTATGTCAGA-3′; the downstream primer was 5′-CATACAGCTAGAAAAGTCCCTG-3′. The PCR fragment was finally cloned into the Adeno-XTM Expression System (Clontech) according to the manufacturer's instructions.
Preparation of the Luciferase Construct of FasL 3′-UTR
FasL with 3′-UTR was amplified by PCR. The forward primer was 5′-GAGAAGCACTTTGGGATTCTTTC-3′. The reverse primer was 5′-CCCTACAATTGCACTGGAAATAC-3′. The PCR fragment was subcloned into the pGL3 vector (Promega) immediately downstream of the stop codon of the luciferase gene. To produce mutated 3′-UTR, the mutations (wild type 3′-UTR, 5′-ATAAGCTA-3′; mutated 3′-UTR, 5′-ATCCATTA-3′) was generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene). The constructs were sequence-verified.
Preparation of FasL with 3′-UTR
FasL with the 3′-UTR was from Origene. To produce FasL with mutated 3′-UTR, the mutations (wild type 3′-UTR, 5′-ATAAGCTA-3′; mutated, 3′-UTR: 5′-ATCCATTA-3′) was generated using QuikChange II XL site-directed mutagenesis kit (Stratagene). The constructs were sequence-verified. They were cloned into the Adeno-XTM Expression System (Clontech) according to the manufacturer's instructions.
Immunoblot and Immunofluorescence
Immunoblotting was carried out as we previously described (16). Cells were lysed for 1 h at 4 °C in a lysis buffer (20 mm Tris, pH 7.5, 2 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, 250 mm sucrose, 0.1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100) containing a protease inhibitor mixture. Samples were subjected to 12% SDS-PAGE and transferred to nitrocellulose membranes. Equal protein loading was controlled by Ponceau Red staining of membranes. Blots were probed using the primary antibodies. The anti-Foxo3a and the anti-phospho Foxo3a antibody (Thr-32) were from Cell Signaling. The anti-FasL antibody was from Abcam. The anti-PCNA (proliferating cell nuclear antigen) antibody and the anti-actin antibody were from Santa Cruz Biotechnology. After four washes with phosphate-buffered saline, Tween 20, the horseradish peroxidase-conjugated secondary antibodies were added. Antigen-antibody complexes were visualized by enhanced chemiluminescence. Immunofluorescence was performed as we described (17). The samples were imaged using a laser scanning confocal microscope (Zeiss LSM 510 META).
Preparation of Subcellular Fractions
Subcellular fractions were prepared as described with modifications (18). In brief, cells were washed twice with phosphate-buffered saline, and the pellets were suspended in 0.2 ml of ice-cold buffer (20 mm potassium-HEPES, pH 7.8, 5 mm potassium acetate, 0.5 mm MgCl2, and 0.5 mm dithiothreitol) containing a protease inhibitor mixture. The cells were homogenized by 12 strokes in a Dounce homogenizer. The homogenates were centrifuged at 750 × g for 5 min at 4 °C to collect nuclei. The resulting supernatants were centrifuged at 20,000 × g for 5 min at 4 °C to collect the cytosolic fractions.
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP was performed as we and other described (19, 20). In brief, cells were washed with phosphate-buffered saline and incubated for 10 min with 1% formaldehyde at room temperature. The cross-linking was quenched with 0.1 m glycine for 5 min. Cells were washed twice with phosphate-buffered saline and lysed for 1 h at 4 °C in a lysis buffer. The cell lysates were sonicated into chromatin fragments with an average length of 500–800 bp as assessed by agarose gel electrophoresis. The samples were precleared with Protein A-agarose (Roche Applied Science) for 1 h at 4 °C on a rocking platform, and 5 μg of specific antibodies were added and rocked for overnight at 4 °C. The anti-NFAT4 antibody was from Santa Cruz Biotechnology. Immunoprecipitates were captured with 10% (v/v) Protein A-agarose for 4 h. Before use, Protein A-agarose was blocked twice at 4 °C with salmon sperm DNA (2 μg/ml) overnight. For the analysis of Foxo3a binding to the promoter of FasL, PCRs were performed with the primers that encompass Foxo3a BS1 or BS2 of the human FasL promoter. The oligonucleotides were as follows: BS1 (forward, 5′-GGATGGGCAGAATGTATCAGAG-3′; reverse, 5′-GCCAATAACTTCCAAGTAGTTA-3′); BS2 (forward 5′-CAAGGCAAGAGGATTGCTTGAG-3′; reverse, 5′-ACCTGCTACACCCACTTTAGAA-3′). For the analysis of Foxo3a binding to the promoter region of miR-21, the oligonucleotides were as follows: BS1 (forward 5′-AAACCAAGGCTCTTACCATAGC-3′ and reverse, 5′-CATTGCACTCCAACTTGGGCAA-3′); BS2 (forward, 5′-CTCTGGTTTCAACAGACACAAA-3′; reverse, 5′-TCTGGCCTGTTAAGATCGAACC-3′). For the analysis of NFAT4 binding to the promoter region of FasL, the oligonucleotides were as follows: forward, 5′-GGTATCCAGCGCTGATTTGCT-3′; reverse, 5′-ACCTCTCTCCAGTTCTCTTCT-3′.
Luciferase Assay
Luciferase assay was performed as we described (13, 21). Briefly, for miR-21 and FasL promoters luciferase assays cells were seeded in 24-well plates. They were transfected with the plasmid constructs using the Lipofectamine 2000 (Invitrogen). Each well contained 0.2 μg of luciferase reporter plasmids and 2.5 ng of SV-Renilla luciferase plasmids as the internal control. Cells were harvested at the indicated times after transfection for the detection of luciferase activity using the Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions. 20 μl of protein extracts were analyzed in a luminometer. Firefly luciferase activities were normalized to Renilla luciferase activity.
For FasL 3′-UTR luciferase assay, cells were co-transfected with the plasmid constructs of 200 ng/well pGL3-FasL-3′-UTR, 400 ng/well miR-21, 20 pmol of either miR-21 antagomir or antagomir negative control (antagomir-NC) using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, cells were lysed, and luciferase activity was measured.
Analysis of miR-21, miR-670, and FasL by Quantitative Reverse Transcription (qRT)-PCR
Stem-loop qRT-PCR for mature miR-21 and miR-670 was performed as described (22) on an Applied Biosystems AB 7000 Real Time PCR system. Total RNA was extracted using Trizol reagent. After DNase I (Takara, Japan) treatment, RNA was reverse-transcribed with reverse transcriptase (ReverTra Ace, Toyobo). The levels of miR-21 and miR-670 analyzed by qRT-PCR were normalized to that of U6. U6 primers were: forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′. qRT-PCR analysis for FasL was standardized to control values of glyceraldehyde-3-phosphate dehydrogenase. The sequences of FasL primers were: forward, 5′-CTGAAGAAGAGAGGGAACCACA-3′; reverse, 5′-AGCTCCTTCTGTAGGTGGAAGA-3′. The sequences of glyceraldehyde-3-phosphate dehydrogenase primers were: forward primer, 5′-CCAAAAGGGTCATCATCTCTGC-3′; reverse, 5′-TGCTAAGCAGTTGGTGGTGCAG-3′.
Transfection of Antagomir
Chemically modified antagomir complementary to miR-21 designed to inhibit endogenous miR-21 expression and antagomir-NC were obtained from GenePharma Co. Ltd. The antagomir sequence was 5′-UCAACAUCAGUCUGAUAAGCUA-3′. The antagomir-NC sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. Cells were transfected with the antagomirs or the antagomir-NC using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Statistical Analysis
The results are expressed as means ± S.E. of at least three independent experiments. The statistical comparison among different groups was performed by one-way analysis of variance. Paired data were evaluated by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Foxo3a Relocalization to the Nucleus and miR-21 Down-regulation Simultaneously Occur upon Doxorubicin Treatment
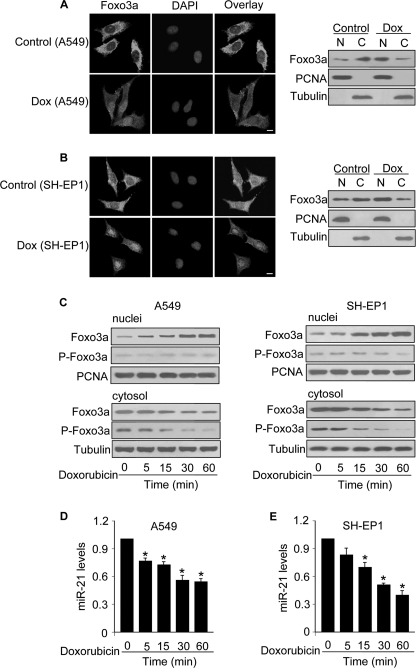
We analyzed whether Foxo3a and miR-21 are involved in the apoptotic program of doxorubicin. The immunofluorescence of A549 cells revealed that Foxo3a was predominantly distributed in the cytoplasm in the control cells without treatment. In contrast, Foxo3a was accumulated in the nuclei in response to doxorubicin treatment. The immunoblotting of subcellular fractions also revealed that Foxo3a was translocated from the cytoplasm to the nuclei upon doxorubicin treatment (Fig. 1A). Because doxorubicin is a component of the standard chemotherapeutic protocol to treat neuroblastoma, we tested whether it can also influence Foxo3a in the human neuroblastoma cell line SH-EP1. A similar result was obtained in SH-EP1 cells (Fig. 1B). The subcellular distributions of Foxo3a are controlled by its phosphorylation status (6). Accordingly, we detected the subcellular distributions of the phosphorylated and nonphosphorylated Foxo3a upon doxorubicin treatment. We observed a time-dependent elevation of nonphosphorylated Foxo3a in the nuclei and a reduction of phosphorylated Foxo3a in the cytoplasm in both A549 and SH-EP1 cells (Fig. 1C). These data suggest that doxorubicin can induce the relocalization of Foxo3a to the nuclei.
FIGURE 1.
Doxorubicin induces a redistribution of Foxo3a from the cytoplasm to nuclei and a reduction of miR-21 levels. A, doxorubicin induces Foxo3a redistributions from the cytoplasm to nuclei in A549 cells. A549 cells were treated with 2 μm doxorubicin (Dox). 1 h after treatment, cells were collected for the analysis of Foxo3a by immunofluorescent staining (left panel) or by immunoblotting with the cellular fractions of nuclei (N) or cytosol (C) (right panel). Cell nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI). Bar = 10 μm. Proliferating cell nuclear antigen (PCNA) is a nucleic marker. Tubulin is a cytosolic marker. B, doxorubicin induces Foxo3a redistributions from the cytoplasm to nuclei in SH-EP1 cells. The experiments were performed as described for A except that SH-EP1 cells were used. C, doxorubicin induces a time-dependent redistribution of Foxo3a. A549 (left panel) and SH-EP1 cells (right panel) were treated with 2 μm doxorubicin. Cells were harvested at the indicated times for the analysis of phosphorylated Foxo3a (P-Foxo3a) and total Foxo3a in the cytosolic and nuclear fractions by immunoblotting. D and E, miR-21 is down-regulated by doxorubicin. A549 (D) and SH-EP1 cells (E) were treated with 2 μm doxorubicin. Cells were harvested at the indicated times for the analysis of miR-21 by qRT-PCR. The levels of miR-21 analyzed by qRT-PCR were normalized to that of U6. *, p < 0.05 versus control. Data are expressed as the mean ± S.E. of three independent experiments.
We detected the levels of miR-21 in cells upon doxorubicin treatment. Doxorubicin induced a reduction in miR-21 levels in A549 (Fig. 1D) and SH-EP-1 cells (Fig. 1E). To know if the alterations of miR-21 is specific or not, we detected miR-670, and no significant alterations in miR-670 levels were observed (data not shown). Thus, it appears that miR-21 can be down-regulated by doxorubicin.
Foxo3a Can Negatively Regulate miR-21 Expression
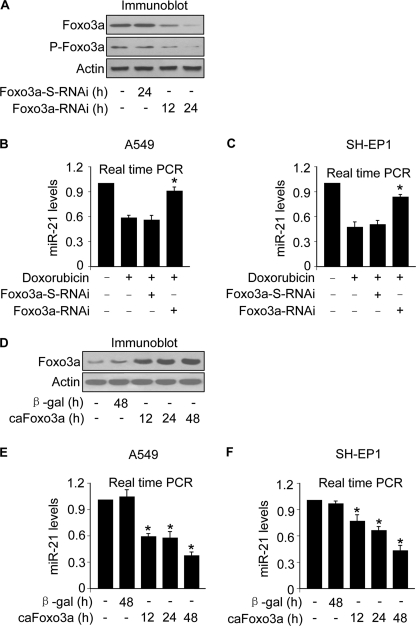
The simultaneous alterations in Foxo3a and miR-21 led us to consider whether these two events are related. To address this issue, we produced Foxo3a RNAi constructs and tested whether knockdown of Foxo3a can influence miR-21 levels. The RNAi construct of Foxo3a but not its scramble form was able to suppress Foxo3a expression (Fig. 2A). Surprisingly, knockdown of Foxo3a could attenuate the reduction of miR-21 induced by doxorubicin in A549 (Fig. 2B) and SH-EP1 cells (Fig. 2C). To further understand whether Foxo3a is able to influence miR-21 expression, we detected miR-21 levels in cells expressing the constitutively active form of Foxo3a (caFoxo3a) (Fig. 2D). Enforced expression of caFoxo3a led to an elevation in miR-21 levels in A549 (Fig. 2E) and SH-EP1 cells (Fig. 2F). These data suggest that Foxo3a can negatively influence the expression of miR-21.
FIGURE 2.
Foxo3a can negatively regulate miR-21 expression. A, Foxo3a RNAi induces a reduction of Foxo3a expression levels. A549 cells were infected with the RNAi constructs of Foxo3a or its scramble form (Foxo3a-S-RNAi). Cells were harvested at the indicated times for the analysis of Foxo3a and P-Foxo3a by immunoblotting. B and C, knockdown of Foxo3a attenuates the reduction of miR-21 levels induced by doxorubicin. A549 (B) and SH-EP1 cells (C) were infected with the RNAi constructs of Foxo3a or its scramble form (Foxo3a-S-RNAi). 24 h after infection cells were treated with 2 μm doxorubicin. 1 h after doxorubicin treatment miR-21 was analyzed by qRT-PCR. The levels of miR-21 analyzed by qRT-PCR were normalized to that of U6. *, p < 0.05 versus doxorubicin alone. D, enforced expression of the constitutively active form of Foxo3a (caFoxo3a) is shown. A549 cells were infected with the adenoviral construct of caFoxo3a. The adenovirus containing β-galactosidase (β-gal) was used as a control. Cells were harvested at the indicated times for the analysis of caFoxo3a expression by immunoblot. E and F, enforced expression of caFoxo3a leads to a reduction of miR-21 levels. A549 (E) and SH-EP1 cells (F) were infected with the adenoviral construct of caFoxo3a. The adenovirus containing β-galactosidase was used as a control. Cells were harvested at the indicated times for the analysis of miR-21 expression by qRT-PCR. The levels of miR-21 analyzed by qRT-PCR were normalized to that of U6. *, p < 0.05 versus control. Data are expressed as the means ± S.E. of three independent experiments.
Foxa3a and miR-21 Cross-talk in the Apoptotic Program
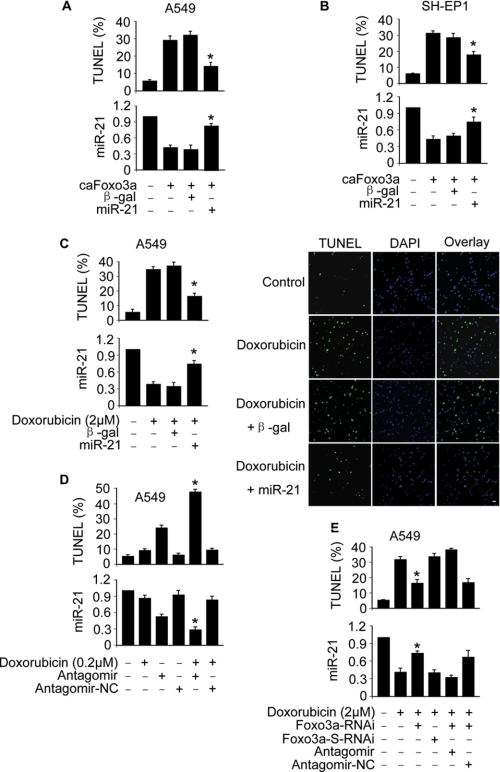
We tested whether Foxo3a and miR-21 are functionally related in apoptosis. Enforced expression of miR-21 could attenuate apoptosis induced by caFoxo3a in A549 (Fig. 3A) and SH-EP1 cells (Fig. 3B). miR-21 also could inhibit apoptosis induced by doxorubicin (Fig. 3C). We characterized the role of endogenous miR-21 in the apoptotic program of doxorubicin. Doxorubicin at a dose of 0.2 μm led to a less amount of cells to undergoing apoptosis. However, knockdown of miR-21 resulted in a significant amount of cells undergoing apoptosis in response to the same dose of doxorubicin treatment (Fig. 3D). We tested whether Foxo3a is involved in mediating the apoptotic signal of doxorubicin. Knockdown of Foxo3a was able to attenuate apoptosis induced by doxorubicin, but this effect could be abolished by miR-21 antagomir (Fig. 3E). Collectively, Foxo3a and miR-21 are functionally related in the apoptotic cascades.
FIGURE 3.
Foxa3a and miR-21 cross-talk in apoptosis. A and B, enforced expression of miR-21 attenuates apoptosis induced by caFoxo3a. A549 (A) or SH-EP1 cells (B) were co-infected with the adenoviral construct of caFoxo3a or miR-21. The adenovirus containing β-galactosidase (β-gal) was used as a control. Apoptosis was analyzed by TUNEL assay 48 h after infection. TUNEL-positive cells were counted under the fluorescent microscope according to the kit instructions. *, p < 0.05 versus caFoxo3a alone. C, enforced expression of miR-21 inhibits apoptosis induced by doxorubicin. A549 cells were infected with the adenoviral construct of miR-21 or β-gal. Cells were treated with 2 μm doxorubicin 24 h after infection. A TUNEL assay was performed 12 h after doxorubicin treatment. The percentages of TUNEL-positive cells are shown in the left panel. *, p < 0.05 versus doxorubicin alone. Representative photos of TUNEL staining are shown in the right panel. Bar = 50 μm. The nuclei with the green color are TUNEL-positive. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, blue). D, knockdown of miR-21 sensitizes doxorubicin to inducing apoptosis is shown. A549 cells were transfected with miR-21 antagomir or the antagomir-NC. 24 h after transfection cells were treated with 0.2 μm doxorubicin. A TUNEL assay was performed 12 h after doxorubicin treatment. *, p < 0.05 versus doxorubicin alone or antagomir alone. E, knockdown of endogenous Foxo3a attenuates apoptosis induced by doxorubicin is shown. A549 cells were infected with the RNAi constructs of Foxo3a or its scramble form (Foxo3a-S-RNAi). Cells were then transfected with miR-21 antagomir or the antagomir-NC. 24 h after transfection cells were treated with 2 μm doxorubicin. Apoptosis was analyzed by TUNEL assay. *, p < 0.05 versus doxorubicin alone. Data are expressed as the mean ± S.E. of three independent experiments.
miR-21 Is a Transcriptional Target of Foxo3a
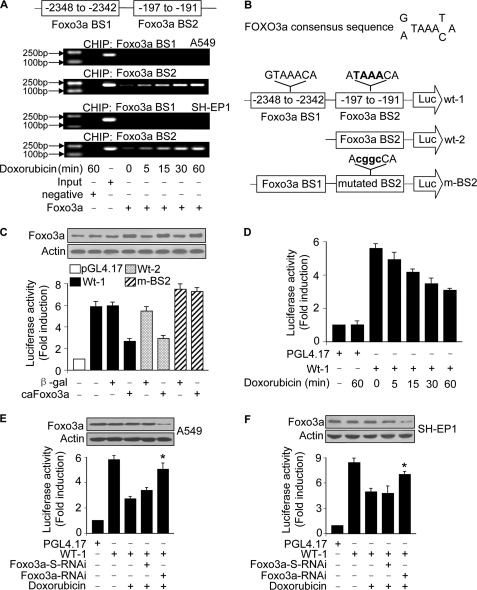
The functional correlation between Foxo3a and miR-21 necessitates the elucidation of the mechanism by which miR-21 is regulated by Foxo3a. Foxo3a is a transcriptional factor. Accordingly, we tested whether the regulation of miR-21 by Foxo3a occurs through a transcriptionally dependent or independent manner. To this end, we analyzed the promoter region of miR-21. There are two optimal Foxo3a consensus binding sites (Fig. 4, A and B). ChIP assay revealed an increase in the association levels of Foxo3a with BS2 in response to doxorubicin treatment. However, an association of Foxo3a with BS1 was not detectable (Fig. 4A). We tested whether Foxo3a can influence miR-21 promoter activity. Wild type miR-21 promoter (wt-1) demonstrated a low activity in the presence of caFoxo3a. Also, the truncated form of wild type miR-21 promoter containing only the binding site-2 (wt-2) showed a low activity in the presence of caFoxo3a. However, mutations in the Foxo3a consensus binding site-2 (BS2) could abolish the inhibitory effect of Foxo3a on miR-21 promoter activity (Fig. 4C). These data suggest that BS2 is the Foxo3a binding site.
FIGURE 4.
miR-21 is a transcriptional target of Foxo3a. A and B, Foxo3a binds to the miR-21 promoter. The promoter region of miR-21 contains two optimal Foxo3a binding sites. Cells were treated with 2 μm doxorubicin and harvested at the indicated times for the ChIP analysis. Chromatin-bound DNA was immunoprecipitated with the anti-Foxo3a antibody. Anti-FasL antibody was used as a negative control (A). The wild type miR-21 promoters (wt-1 and wt-2) were linked to luciferase (Luc) reporter gene. The mutations were introduced to the consensus binding site-2 (m-BS2) (B). C, Foxo3a inhibits miR-21 promoter activity. HEK293 cells were infected with adenoviral caFoxo3a or (β-galactosidase (β-gal)). 24 h after infection cells were transfected with the constructs of the empty vector (pGL-4.17), wt-1, wt-2, or the mutated promoter (m-BS2), respectively. The expression levels of Foxo3a were detected by immunoblotting (upper panel). Firefly luciferase activities are shown in the lower panel. D, doxorubicin induces a reduction of miR-21 promoter activity. A549 cells were transfected with the constructs of the empty vector (pGL-4.17) or miR-21 wt-1. 24 h after transfection cells were treated with 2 μm doxorubicin and collected at the indicated times for luciferase assay. E and F, knockdown of Foxo3a attenuates the reduction of the promoter activity upon treatment with doxorubicin. A549 (E) and SH-EP1 cells (F) were infected with the adenoviral RNAi constructs of Foxo3a or its scramble form (Foxo3a-S-RNAi) and then transfected with the constructs of the empty vector (pGL-4.17) or miR-21 wt-1 promoter. Cells were treated with 2 μm doxorubicin. Firefly luciferase activities are shown in the lower panel. The expression levels of Foxo3a were detected by immunoblot (upper panel). *, p < 0.05 versus wt-1 plus doxorubicin. Data are expressed as the mean ± S.E. of three independent experiments.
Doxorubicin could induce a time-dependent reduction in miR-21 promoter activity (Fig. 4D). Concomitantly, knockdown of Foxo3a could attenuate the reduction of miR-21 promoter activity induced by doxorubicin in A549 (Fig. 4E) and SH-EP-1 cells (Fig. 4F). These data indicate that miR-21 can be transcriptionally repressed by Foxo3a.
The Regulation of FasL by Foxo3a Is Not in a Transcription Manner
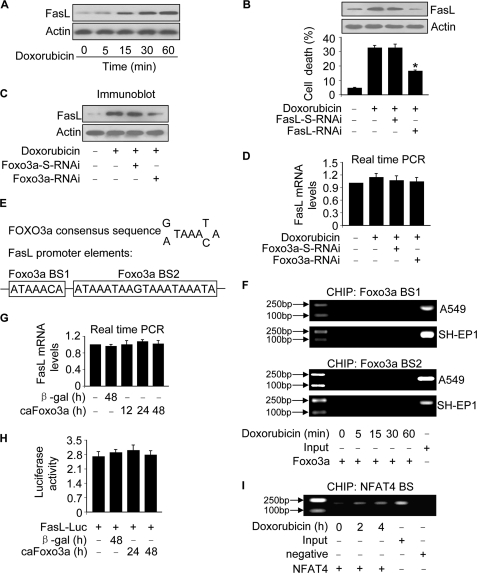
Foxo3a and miR-21 cannot directly execute apoptosis. Which molecules are their downstream mediators? Foxo3a has been shown to either activate or suppress FasL transcription depending on the cellular context (6, 7). We detected the levels of FasL in response to doxorubicin treatment. An up-regulation of FasL could be observed upon doxorubicin treatment (Fig. 5A). Knockdown of FasL attenuated cell death (Fig. 5B), suggesting that FasL participates in conveying the death signal of doxorubicin.
FIGURE 5.
Foxo3a regulates FasL expression independent of the transcriptional manner. A, doxorubicin stimulates FasL expression. A549 cells were treated with 2 μm doxorubicin and harvested at the indicated times for the analysis of FasL by immunoblotting. B, knockdown of FasL attenuates cell death induced by doxorubicin. A549 cells were infected with the adenoviral RNAi constructs of FasL or its scramble form (FasL-S-RNAi) and then treated with 2 μm doxorubicin. FasL was detected by immunoblot. Cell death was analyzed by trypan blue exclusion 24 h after doxorubicin treatment. *, p < 0.05 versus doxorubicin alone. C and D, knockdown of Foxo3a reduces FasL protein but not mRNA expression levels. A549 cells were infected with the adenoviral RNAi constructs of Foxo3a or its scramble form (Foxo3a-S-RNAi) and then treated with 2 μm doxorubicin. FasL protein levels were analyzed by immunoblotting (C), and its mRNA levels were detected by qRT-PCR (D). The values of FasL mRNA analyzed by qRT-PCR were normalized to that of glyceraldehyde-3-phosphate dehydrogenase. E, the promoter region of human FasL contains two Foxo3a consensus binding sites. F, Foxo3a does not bind to FasL promoter, analyzed by ChIP assay. Cells were treated with 2 μm doxorubicin and harvested at the indicated times for the ChIP assay. PCR primers encompassing Foxo3a potential binding site-1 (BS1) and binding site-2 (BS2) were used. G, caFoxo3a induces no significant alterations in FasL mRNA levels. A549 cells were infected with the adenoviral caFoxo3a. The adenoviral β-galactosidase (β-gal) was used as a control. Cells were harvested at the indicated times for the analysis of FasL mRNA by qRT-PCR. The values of FasL mRNA analyzed by qRT-PCR were normalized to that of glyceraldehyde-3-phosphate dehydrogenase. H, caFoxo3a induces no significant changes in FasL promoter activity. A549 cells were infected with the adenoviral caFoxo3a or β-galactosidase and then transfected with the luciferase construct of FasL (FasL-Luc) or the empty vector pGL4.17. Firefly luciferase activities were normalized to Renilla luciferase activities. Data are expressed the ratios of FasL-Luc/the empty vector pGL4.17. I, NFAT4 binds to FasL promoter analyzed by ChIP assay. The primary cardiac fibroblasts were treated with 2 μm doxorubicin and harvested at the indicated time for the ChIP assay. The anti-actin antibody was used as a negative control. The results are expressed as the mean ± S.E. of three independent experiments.
We tested whether the up-regulation of FasL is related to Foxo3a. Knockdown of Foxo3a attenuated FasL protein levels upon doxorubicin treatment (Fig. 5C). Surprisingly, FasL mRNA levels were not significantly altered by knockdown of Foxo3a (Fig. 5D). Human FasL promoter region contains 2 Foxo3a consensus binding sites (Fig. 5E). We performed a ChIP assay to detect whether Foxo3a binds to these sites upon doxorubicin treatment. The ChIP assay revealed that there was no detectable association between Foxo3a and the consensus binding site-1 (BS1) or binding site-2 (BS2) (Fig. 5F). We tested whether caFoxo3a can influence FasL mRNA levels and promoter activity. caFoxo3a led to no significant alterations in FasL mRNA levels (Fig. 5G) as well as FasL promoter activity (Fig. 5H). We sequenced the promoter region of FasL (3157 bp) and found no mutations in this region. We used NFAT4 as a positive control because FasL promoter contains NFAT4 binding sites (23). We tested whether NFAT4 can bind to the promoter region of FasL in the primary cardiac fibroblasts. NFAT4 association with FasL promoter could be detectable (Fig. 5I). Taken together, these data suggest that Foxo3a does not transcriptionally regulate FasL expression upon doxorubicin treatment.
miR-21 Is Able to Regulate FasL
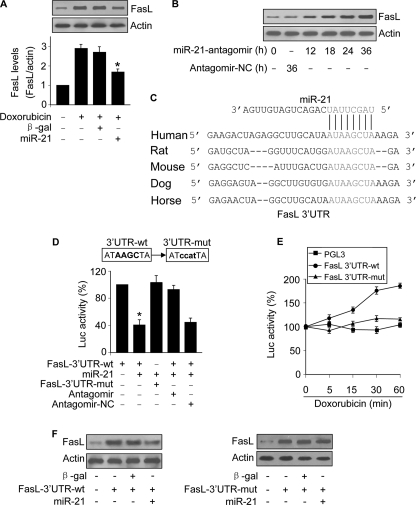
The deficiency of Foxo3a to transcriptionally regulate FasL led us to search for other mechanisms that account for the up-regulation of FasL upon doxorubicin treatment. miRNAs can suppress gene expression, whereas doxorubicin can induce a reduction of miR-21 expression. These lines of evidence encouraged us to test whether miR-21 participates in the regulation of FasL. Enforced expression of miR-21 could attenuate the elevation of FasL levels induced by doxorubicin (Fig. 6A). Knockdown of endogenous miR-21 augmented FasL expression (Fig. 6B). To understand whether miR-21 can directly target FasL, we analyzed the 3′-UTR region of FasL and observed that miR-21 is a potential target of FasL (Fig. 6C). A luciferase assay revealed that miR-21 could suppress the translational activity of FasL. Introduction of mutations in the 3′-UTR region of FasL led to the failure of miR-21 to repress FasL translation (Fig. 6D). These data suggest that FasL is a direct target of miR-21.
FIGURE 6.
FasL is a target of miR-21. A, enforced expression of miR-21 attenuates FasL elevations induced by doxorubicin. A549 cells were infected with the adenoviral miR-21 or β-gal. 24 h after infection cells were treated with 2 μm doxorubicin. Cells were harvested 1 h after doxorubicin treatment for FasL analysis by immunoblotting. The upper panel shows a representative blot. The lower panel shows the quantitative analysis of FasL levels. The films were densitometrically scanned using NIH ImageJ. The ratios of FasL to actin are shown. *, p < 0.05 versus doxorubicin alone. Data are expressed as the mean ± S.E. of three independent experiments. β-gal, β-galactosidase. B, knockdown of miR-21 leads to an elevation in FasL levels. A549 cells were transfected with miR-21 antagomir or the antagomir-NC. Cells were harvested at the indicate times for FasL analysis by immunoblotting. C, the miR-21 targeting sites in FasL 3′-UTR are conserved. D, miR-21 suppresses FasL translation. HEK293 cells were transfected with the luciferase constructs of wild type FasL-3′-UTR (FasL-3′-UTR-wt) or the mutated FasL-3′-UTR (FasL-3′-UTR-mut) along with the expression plasmids for miR-21, miR-21 antagomir or antagomir-NC. *, p < 0.05 versus FasL-3′-UTR-wt. E, doxorubicin can promote the translational activity of wild type FasL-3′-UTR but not the mutated FasL-3′-UTR (FasL-3′-UTR-mut). A549 cells were transfected with the luciferase constructs of wild type FasL-3′-UTR or the FasL-3′-UTR-mut and then treated with 2 μm doxorubicin and harvested at the indicated times for the luciferase assay. The luciferase activity (Luc) of each construct before doxorubicin treatment was taken as 100%. F, miR-21 can suppress the expression of FasL with wild type but not mutated 3′-UTR. A549 cells were co-infected with the adenoviral miR-21, β-galactosidase, wild type FasL-3′-UTR (FasL-3′-UTR-wt) or the mutated FasL-3′-UTR (FasL-3′-UTR-mut). FasL expression was detected by immunoblotting.
To know whether the endogenous miR-21 can regulate FasL translation, the cells were transfected with the constructs of FasL 3′-UTR and then exposed to doxorubicin. Treatment with doxorubicin led to an elevation in the luciferase activity of wild type 3′-UTR but not the mutated 3′-UTR (Fig. 6E), suggesting that endogenous miR-21 can target FasL 3′-UTR.
We finally tested whether miR-21 regulates FasL expression via targeting its 3′-UTR. FasL with wild type 3′-UTR expressed at a low level in the presence of miR-21. Introduction of mutations in the miR-21 binding site abolished the inhibitory effect of miR-21 on FasL expression (Fig. 6F). Taken together, it appears that miR-21 can inhibit the translation of FasL.
DISCUSSION
The growing evidence has shown that miR-21 is an oncogenic miRNA and is highly expressed in a variety of malignant tumors (24, 25). It is still a challenging question as to how the expression of miRNAs is regulated. Our present work demonstrated that miR-21 can be transcriptionally suppressed by Foxo3a. We further identified FasL as a target of miR-21. In particular, we found that Foxo3a can up-regulate FasL through down-regulating miR-21. Our results provide novel evidence revealing that miR-21 can be a linker between Foxo3a and FasL, and Foxo3a may promote apoptosis by targeting the anti-apoptotic miRNA, miR-21.
Foxo3a participates in the regulation of apoptosis by targeting the apoptotic factors such as FasL (6) and Bim (26). However, it remains controversial as to the transcriptional output of Foxo3a in regulating FasL. There is evidence showing that Foxo3a can up-regulate FasL expression (6). In contrast, the opposite evidence demonstrates that the transcriptional output of Foxo3a is the suppression rather than activation of FasL (7). Such a discrepancy raises a question as to whether Foxo3a can directly target FasL. The results obtained from our present study demonstrate that Foxo3a can induce an elevation in FasL, but it does not regulate FasL expression through a transcriptional manner. Out data further revealed that Foxo3a inhibits miR-21 expression and suppresses FasL translation. Thus, it appears that miR-21 is a linker between Foxo3a and FasL. Our results shed new light on understanding the relationship between Foxo3a and FasL.
The growing evidence has shown that miR-21 is highly expressed in a variety of malignant tumors (25). It is up-regulated in laryngeal carcinoma tissues (27) and human cholangiocarcinoma (10). miR-21 is higher in estrogen receptor-α-positive tumors, and estradiol inhibits miR-21 expression in MCF-7 human breast cancer cells (28). miR-21 is abundantly expressed and is a putative oncogenic miRNA in head and neck cancer (29). Overexpression of miR-21 can be observed in multiple myeloma and is associated with its pathogenesis (30). Elevated expression of miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype (31). Foxo3a function is dependent on its phosphorylation by Akt with the nonphosphorylated but not the phosphorylated form to elicit transcriptional activity (6). Akt plays a key role in promoting cell survival and is highly active in many malignant tumors. Based on our present finding that Foxo3a can negatively regulate miR-21 expression, it is reasonable to speculate that the abundant expression of miR-21 in malignant tumors may be related to the high activity of Akt. Future studies are required to test this hypothesis.
It is of note that miR-21 contributes to cancer resistance to therapies. For example, miR-21 is overexpressed in glioblastoma and mediates chemoresistance to the chemotherapeutic agent VM-26 in glioblastoma cells (32). The tumor necrosis factor-related apoptosis-inducing ligand in combination with miR-21 suppression leads to a synergistic increase in caspase activity and a decrease in cell viability in human glioma cells (33). Our data that Foxo3a can transcriptionally down-regulate miR-21 may warrant future studies to develop a therapeutic approach to target miR-21 by modulating Foxo3a in cancer treatment.
Our present work revealed that the transcriptional factor Foxo3a can suppress miR-21 expression. There are reports showing that miR-21 can be transcriptionally up-regulated. The transcription factor AP-1 plays a key role in tumorigenesis, and it can induce miR-21 expression (34). Signal transducer and activator of transcription 3 (Stat3) is implicated in the pathogenesis of many malignancies and essential for interleukin-6-dependent survival and growth of multiple myeloma cells. Interleukin-6 can induce miR-21 expression through Stat3 (4). It is possible that miR-21 can also be directly or indirectly regulated by other transcriptional factors in the apoptotic cascades. For example, p53 activation leads to Foxo3a phosphorylation and subcellular localization changes that result in inhibition of Foxo3a transcription activity (35). It would be interesting to elucidate whether p53 can target miR-21 through Foxo3a.
Foxo3a is able to transcriptionally repress gene expression (7). Our present work revealed that Foxo3a could transcriptionally repress miR-21. Other transcriptional factors also can either activate and/or suppress gene expression. For example, in response to p53 transcriptional stimulation, 38 genes are up-regulated, and 24 genes are down-regulated (36). p53 can up-regulate the pro-apoptotic factors such as Bax (37), PUMA (38, 39), and Bad (40) but down-regulate the anti-apoptotic factors such as glutathione S-transferase-α (41) and Survivin (42).
Our present study demonstrated that FasL is a target of miR-21. miR-21 has been shown to suppress other apoptotic proteins. For example, it can suppress tissue inhibitor of metalloproteinases 3 (10). miR-21 targets the network of p53, transforming growth factor-β, and mitochondrial apoptosis tumor suppressor genes in glioblastoma cells (43). miR-21 promotes glioma invasion by targeting matrix metalloproteinase regulators (8). Programmed cell death 4 is an important functional target of miR-21 in breast cancer (44) and colorectal cancer (45). Apoptosis can be initiated through the extrinsic and intrinsic pathways. In particular, there is cross-talk between these two pathways (46–48). FasL can trigger the activation of extrinsic apoptotic pathway by promoting DISC formation (49). It is necessary to elucidate the consequences of miR-21 regulation on FasL in apoptotic cascades.
Doxorubicin is an important component for the treatment of lung cancer and neuroblastoma. Induction of cancer cells to undergo apoptosis is a cellular basis of doxorubicin effect. However, a major obstacle of doxorubicin treatment is cancer resistance. Our present work demonstrated that miR-21 is negatively regulated by Foxo3a in both A549 and SH-EP1 cells upon doxorubicin treatment. We identified FasL as a new target of miR-21. Furthermore, Foxo3a can regulate FasL through miR-21. Future studies are required to delineate whether doxorubicin has a similar effect in other types of cancer cells. Overall, based on our findings, combinatorial therapies for cancer treatment may be developed by targeting the Foxo3a-miR-21-FasL pathway.
This work was supported by National Natural Science Foundation of China Grants 30730045 and 30871243 and by the National Basic Research Program of China (973 Program, Grant 2007CB512000).
- miRNA
- microRNA
- FasL
- Fas ligand
- Foxo3a
- Forkhead bOX-containing protein O subfamily 3a
- NFAT4
- nuclear translocation of nuclear factor of activated T cells 4
- TUNEL
- terminal dUTP nick-end labeling
- ChIP
- chromatin immunoprecipitation
- qRT
- quantitative reverse transcription
- antagomir-NC
- antagomir negative control
- wt
- wild type
- UTR
- untranslated region.
REFERENCES
- 1.Park S. M., Schickel R., Peter M. E. (2005) Curr. Opin. Cell Biol. 17, 610–616 [DOI] [PubMed] [Google Scholar]
- 2.He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasser M. W., Datta J., Nuovo G., Kutay H., Motiwala T., Majumder S., Wang B., Suster S., Jacob S. T., Ghoshal K. (2008) J. Biol. Chem. 283, 33394–33405 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Löffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermüller J., Kretzschmar A. K., Burger R., Gramatzki M., Blumert C., Bauer K., Cvijic H., Ullmann A. K., Stadler P. F., Horn F. (2007) Blood 110, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 5.Accili D., Arden K. C. (2004) Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 6.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 7.Jonsson H., Allen P., Peng S. L. (2005) Nat. Med. 11, 666–671 [DOI] [PubMed] [Google Scholar]
- 8.Gabriely G., Wurdinger T., Kesari S., Esau C. C., Burchard J., Linsley P. S., Krichevsky A. M. (2008) Mol. Cell. Biol. 28, 5369–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Z., Liu M., Stribinskis V., Klinge C. M., Ramos K. S., Colburn N. H., Li Y. (2008) Oncogene 27, 4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selaru F. M., Olaru A. V., Kan T., David S., Cheng Y., Mori Y., Yang J., Paun B., Jin Z., Agarwal R., Hamilton J. P., Abraham J., Georgiades C., Alvarez H., Vivekanandan P., Yu W., Maitra A., Torbenson M., Thuluvath P. J., Gores G. J., LaRusso N. F., Hruban R., Meltzer S. J. (2009) Hepatology 49, 1595–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007) Mol. Cell 26, 731–743 [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y., Samal E., Srivastava D. (2005) Nature 436, 214–220 [DOI] [PubMed] [Google Scholar]
- 13.Lin Z., Murtaza I., Wang K., Jiao J., Gao J., Li P. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12103–12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J. X., Li Q., Li P. F. (2009) Cancer Res. 69, 492–500 [DOI] [PubMed] [Google Scholar]
- 15.Li P. F., Dietz R., von Harsdorf R. (1999) FEBS Lett. 448, 206–210 [DOI] [PubMed] [Google Scholar]
- 16.Li P. F., Dietz R., von Harsdorf R. (1999) EMBO J. 18, 6027–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P. F., Li J., Müller E. C., Otto A., Dietz R., von Harsdorf R. (2002) Mol. Cell 10, 247–258 [DOI] [PubMed] [Google Scholar]
- 18.Krude T., Jackman M., Pines J., Laskey R. A. (1997) Cell 88, 109–119 [DOI] [PubMed] [Google Scholar]
- 19.Li Y. Z., Lu D. Y., Tan W. Q., Wang J. X., Li P. F. (2008) Mol. Cell. Biol. 28, 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szak S. T., Mays D., Pietenpol J. A. (2001) Mol. Cell. Biol. 21, 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan W. Q., Wang K., Lv D. Y., Li P. F. (2008) J. Biol. Chem. 283, 29730–29739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latinis K. M., Norian L. A., Eliason S. L., Koretzky G. A. (1997) J. Biol. Chem. 272, 31427–31434 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Lee C. G. (2009) J. Cell. Mol. Med. 13, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krichevsky A. M., Gabriely G. (2009) J. Cell. Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dijkers P. F., Medema R. H., Lammers J. W., Koenderman L., Coffer P. J. (2000) Curr. Biol. 10, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 27.Liu M., Wu H., Liu T., Li Y., Wang F., Wan H., Li X., Tang H. (2009) Cell Res. 19, 828–837 [DOI] [PubMed] [Google Scholar]
- 28.Wickramasinghe N. S., Manavalan T. T., Dougherty S. M., Riggs K. A., Li Y., Klinge C. M. (2009) Nucleic Acids Res. 37, 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang S. S., Jiang W. W., Smith I., Poeta L. M., Begum S., Glazer C., Shan S., Westra W., Sidransky D., Califano J. A. (2008) Int. J. Cancer 123, 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichiorri F., Suh S. S., Ladetto M., Kuehl M., Palumbo T., Drandi D., Taccioli C., Zanesi N., Alder H., Hagan J. P., Munker R., Volinia S., Boccadoro M., Garzon R., Palumbo A., Aqeilan R. I., Croce C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly E., Melegari M., Landgraf P., Tchaikovskaya T., Tennant B. C., Slagle B. L., Rogler L. E., Zavolan M., Tuschl T., Rogler C. E. (2008) Am. J. Pathol. 173, 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Li W., Yang Y., Lu Y., He C., Hu G., Liu H., Chen J., He J., Yu H. (2009) Brain Res. 1286, 13–18 [DOI] [PubMed] [Google Scholar]
- 33.Corsten M. F., Miranda R., Kasmieh R., Krichevsky A. M., Weissleder R., Shah K. (2007) Cancer Res. 67, 8994–9000 [DOI] [PubMed] [Google Scholar]
- 34.Talotta F., Cimmino A., Matarazzo M. R., Casalino L., De Vita G., D'Esposito M., Di Lauro R., Verde P. (2009) Oncogene 28, 73–84 [DOI] [PubMed] [Google Scholar]
- 35.You H., Jang Y., You-Ten A. I., Okada H., Liepa J., Wakeham A., Zaugg K., Mak T. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14057–14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kannan K., Amariglio N., Rechavi G., Jakob-Hirsch J., Kela I., Kaminski N., Getz G., Domany E., Givol D. (2001) Oncogene 20, 2225–2234 [DOI] [PubMed] [Google Scholar]
- 37.Miyashita T., Reed J. C. (1995) Cell 80, 293–299 [DOI] [PubMed] [Google Scholar]
- 38.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 39.Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 40.Jiang P., Du W., Heese K., Wu M. (2006) Mol. Cell. Biol. 26, 9071–9082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faraonio R., Vergara P., Di Marzo D., Pierantoni M. G., Napolitano M., Russo T., Cimino F. (2006) J. Biol. Chem. 281, 39776–39784 [DOI] [PubMed] [Google Scholar]
- 42.Hoffman W. H., Biade S., Zilfou J. T., Chen J., Murphy M. (2002) J. Biol. Chem. 277, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 43.Papagiannakopoulos T., Shapiro A., Kosik K. S. (2008) Cancer Res. 68, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 44.Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008) J. Biol. Chem. 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 45.Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008) Oncogene 27, 2128–2136 [DOI] [PubMed] [Google Scholar]
- 46.Dhanasekaran D. N., Reddy E. P. (2008) Oncogene 27, 6245–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallach D., Kang T. B., Kovalenko A. (2008) Cell Death Differ. 15, 1533–1541 [DOI] [PubMed] [Google Scholar]
- 48.Galluzzi L., Zamzami N., de La Motte Rouge T., Lemaire C., Brenner C., Kroemer G. (2007) Apoptosis 12, 803–813 [DOI] [PubMed] [Google Scholar]
- 49.Yu J. W., Shi Y. (2008) Oncogene 27, 6216–6227 [DOI] [PubMed] [Google Scholar]