Abstract
A common theme in bacterial pathogenesis is the manipulation of eukaryotic cells by targeting the cytoskeleton. This is in most cases achieved either by modifying actin, or indirectly via activation of key regulators controlling actin dynamics such as Rho-GTPases. A novel group of bacterial virulence factors termed the WXXXE family has emerged as guanine nucleotide exchange factors (GEFs) for these GTPases. The precise mechanism of nucleotide exchange, however, has remained unclear. Here we report the structure of the WXXXE-protein IpgB2 from Shigella flexneri and its complex with human RhoA. We unambiguously identify IpgB2 as a bacterial RhoA-GEF and dissect the molecular mechanism of GDP release, an essential prerequisite for GTP binding. Our observations uncover that IpgB2 induces conformational changes on RhoA mimicking DbI- but not DOCK family GEFs. We also show that dissociation of the GDP·Mg2+ complex is preceded by the displacement of the metal ion to the α-phosphate of the nucleotide, diminishing its affinity to the GTPase. These data refine our understanding of the mode of action not only of WXXXE GEFs but also of mammalian GEFs of the DH/PH family.
Keywords: Crystal Structure, Cytoskeleton, Enzyme Mechanisms, G Proteins, Protein Metal Ion Interaction, Protein Structure, IpgB2, RhoA-GEF Complex, Shigella, Actin Dynamics
Introduction
Remodeling of the actin cytoskeleton is essential for eukaryotic life, taking place in processes as diverse as division, motility, or cell-cell communication. The actin cytoskeleton is composed of actin monomers and helical filaments constantly assembling and disassembling at their ends, which is tightly regulated by multiple signaling pathways employing hundreds of actin-binding proteins. Most if not all signaling pathways converge on small GTPases of the Rho family, which are long recognized as key signaling switches inducing different actin filament structures (1–3). As effector interaction occurs in the GTP-bound state, their intrinsic GTPase activity is counterbalanced by guanine nucleotide exchange factors (GEFs)2 driving GTP re-loading by increasing the intrinsic nucleotide dissociation rate (4). Activation of the name-giving family member RhoA induces cell contraction. This drives the formation of stress fiber bundles and focal adhesions in fibroblasts or endothelial cells, exerting force onto the substratum or neighboring cells (5). In neurons for instance, RhoA activation induces growth cone retraction (6). Other Rho family members trigger the formation of cell protrusions antagonistic to RhoA, with Rac1 and Cdc42 inducing lamellipodia and filopodia, respectively (7, 8).
Given their importance in cell physiology, Rho-GTPases have also emerged as key targets of pathogens modifying cell signaling or actin remodeling for their own needs (9). For instance, multiple bacterial toxins inhibiting RhoA, such as C3 exoenzyme of Clostridium botulinum, have served as invaluable tools to study RhoA function in vivo (10, 11). As an alternative to inhibition, Rho-GTPase signaling pathways driving different types of actin reorganization are also frequently bypassed or activated by pathogens, at least transiently, allowing them to induce their tight adherence to or entry into eukaryotic cells (12–14). Pathogens often modulate host protein functions by virulence factors acting as perfect mimics of mammalian proteins. For example, the Salmonella toxin SopE can activate Cdc42 in a similar fashion as Dbl-like mammalian GEFs (15). Interestingly, SopE does not display any significant structural or sequence similarity to the mimicked mammalian protein.
An intriguing novel family of bacterial effectors shares a WXXXE sequence motif, the members of which were originally classified as GTPase mimics, despite the lack of any sequence homology or guanine nucleotide binding (16, 17). Prototypic members of the family include IpgB1 and IpgB2 from Shigella flexneri and Map from pathogenic Escherichia coli, which are all translocated into the host by type III secretion. Interestingly, these proteins were reported to induce filopodia (for Map) or lamellipodia and stress fiber formation, indicative of Rac1 and RhoA activity in the case of IpgB1 and IpgB2, respectively (16, 18). In another study, IpgB1 was described to drive Rac1 and Cdc42 activation (19). In addition, more recently identified family members such as EspM and EspT have been interpreted to activate rather than mimic RhoA and Rac1/Cdc42, respectively (20, 21), which was recently verified in the case of EspM2 (22). The finding that the WXXXE protein SifA displays a fold similar to the bacterial GEF SopE (15, 23) provided the first structural evidence for WXXXE proteins acting as GEFs. Finally, Map has recently been defined as Cdc42-GEF (24), refuting the mimicry hypothesis and questioning the relevance of IpgB2 binding to downstream effectors of RhoA (16).
Here we provide crystal structures of IpgB2 both in its free form and in complex with human RhoA. Our data reveal the mechanism and structurally decipher the sequence of events inducing GDP release catalyzed by this novel family of bacterial GEFs, constituting yet another remarkable example of convergent evolution, a common theme in bacterial pathogenesis (12, 25, 26).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
IpgB2 (residues 1–188, NCBI gene ID 56404000 was cloned into the expression vector pET-M41 (kindly provided by Gunter Stier, EMBL, Heidelberg). This pET-derived vector allows expressing the protein of interest as a fusion to the C terminus of His-tagged maltose binding protein (MBP) with a linker sensitive to tobacco etch virus protease. RhoA (residues 2–181, NCBI gene ID 10835049) and ROCK I were cloned into a modified pET-28c vector with an His8 tag and a linker sensitive to tobacco etch virus protease fused to the N terminus. The original ROCK I construct was purchased from GENEART and was optimized for protein production in E. coli. It encoded for amino acid residues 947–1015 according to NCBI gene ID 4885583. IpgB2 and RhoA were produced in E. coli Tuner (DE3) cells in LB medium containing additional 2 g/liter glucose after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The expression temperature was 15 °C for IpgB2 and 27 °C for RhoA. ROCK I was produced in E. coli Tuner (DE3) cells at 22 °C in 2×YT medium after induction with 0.8 mm isopropyl 1-thio-β-d-galactopyranoside. Selenomethionine-substituted IpgB2 was produced in minimal medium (1 g/liter NH4Cl, 3 g/liter KH2PO4, 6 g/liter Na2HPO4·7H2O) mixed 10:1 with supplement medium (21.82 g/liter d-glucose monohydrate, 0.45 g MgSO4·7H2O, 10.45 mg/liter Fe2(SO4)3·H2O, 10 mg/liter thiamine) and 50 mg/liter l-selenomethionine, with the pH adjusted to pH 7.4. Harvested cells containing IpgB2 were resuspended in buffer (50 mm HEPES, pH 7.5, 250 mm NaCl, 8.7% (v/v) glycerol, 7.5 mm imidazole, 250 mm trisodium citrate, and 10 mm β-mercaptoethanol). In the case of RhoA, the buffer composition was 30 mm Tris/HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 5% (v/v) glycerol, 10 mm β-mercaptoethanol, and 0.05 mm GDP, and in the case of ROCK I the buffer contained 50 mm Tris/HCl, pH 8.5, 10% (v/v) glycerol, 20% (w/v) sucrose, 0.2 mm Na2S2O4, 2 mm MgCl2, and2 mm dithiothreitol. To all resuspension buffers, a protease inhibitor mixture (Roche Applied Science, complete EDTA free) and a small amount of DNase I (<1 mg) was added. The suspended cells were mechanically disrupted using a TS series bench top cell homogenizer (Constant Systems Ltd.), and the soluble cell extract after centrifugation (60 min, 37,000 × g, 277 K) was purified by affinity chromatography using nickel-nitrilotriacetic acid-agarose. During affinity chromatography, the buffer of ROCK I was changed to a buffer containing 50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 0.1% (v/v) Triton X-100, 10 mm MgCl2, and 0.2 mm dithiothreitol. All elution buffers additionally contained 250 mm imidazole. IpgB2 and RhoA were mixed with tobacco etch virus protease in a molar ratio of 100:1 and dialyzed against buffer for 12 h at 277 K. ROCK I was first purified by anion chromatography on a Mono Q 5/5 column (Amersham Biosciences) and then treated equally. During dialysis, the imidazole concentration was reduced, and the proteins were digested to cleave off the fusion tags. The tags were removed by (negative) affinity chromatography using nickel-nitrilotriacetic acid-agarose, and IpgB2 and RhoA were further purified by gel-filtration chromatography on a Superdex 75 column (Amersham Biosciences). During gel-filtration chromatography, the buffer was exchanged against 200 mm trisodium citrate/citric acid, pH 6.0, and 10 mm dithiothreitol in the case of IpgB2 and against 30 mm Tris/HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 5% (v/v) glycerol, 5 mm dithiothreitol in the case of RhoA.
Nucleotide Exchange and GTPase and GEF Assays
GTPase and GEF assays were performed according to methods described by Eberth and Ahmadian (27). RhoA loaded with different nucleotides was prepared by a modified method similar as described by Hutchinson and Eccleston (28), utilizing the enhanced nucleotide dissociation rate of RhoA in buffers lacking Mg2+ (29).
Recombinant Protein Pulldown Experiments
His-MBP-fused IpgB2 was incubated with 5 μl of nickel-nitrilotriacetic acid-agarose (Qiagen) per milligram of fusion protein for 12 h at 4 °C in buffer containing 50 mm HEPES, pH 7.5, 250 mm NaCl, 8.7% (v/v) glycerol, 250 mm trisodium citrate, 10 mm β-mercaptoethanol. The resin was washed in a gravity flow column with 5 column volumes of buffer to remove unbound protein. Aliquots of 35 μl of packed resin mixed 1:1 (v/v) with glycerol were used in a typical binding assay with a 1-ml total reaction volume or stored at −70 °C until usage. The resin complexed with His-MBP-IpgB2 was incubated with recombinant tagless test protein (RhoA, ROCK I (Rho-binding domain (RBD))) for 10 min. The agarose was pelleted at 1000 × g, the supernatant was removed, and the agarose was washed three times to remove unbound test protein. Proteins were eluted with buffer additionally containing 250 mm imidazole, separated by SDS-PAGE, and visualized by Coomassie staining. To exclude unspecific binding of the test protein to the resin, all experiments were repeated with resin without complexed His-MBP-IpgB2.
Preparation of Soluble Bacterial WXXXE Effector Proteins
As untruncated wild-type IpgB2 was not soluble in standard buffers after cleavage of the MBP fusion tag, buffer conditions were optimized to obtain soluble, untagged IpgB2. 96 conditions from a standard crystallization screen (Classics Suite, Qiagen) were tested for optimization. The MBP-tagged IpgB2 was concentrated to ∼10 mg/ml, and 1.2 μl of protein solution was mixed with 1.2 μl of H2O, 0.3 μl of tobacco etch virus Protease (∼2.5 mg/ml), and 1.2 μl of reservoir solution from the crystallization screen in batch experiments. After incubation at 4 °C for 3 days, the solution was mixed with 10 μl of H2O, and insoluble particles were pelleted by centrifugation. 10 μl of supernatant of each experiment was separated by SDS-PAGE and visualized by Coomassie staining. In almost all tested conditions, complete cleavage of the MBP tag was observed, and approximately one-third of all conditions contained varying quantities of soluble IpgB2. Buffer components that repetitively were present in conditions leading to soluble IpgB2 were individually tested in further cleavage experiments. Citrate proved to be the most successful buffer component at concentrations of 100–200 mm, pH 5–7. Fresh IpgB2 was produced as described above using buffer systems based on this finding, which yielded >100 mg of soluble MBP-fused IpgB2 per liter bacterial culture and >10 mg of untagged IpgB2 per liter after purification to apparent homogeneity. Purified, untagged IpgB2 can also be handled in buffers without citrate (e.g. 10 mm Tris/HCl, pH 7.0) at 4 °C as long as no mechanical or oxidizing stress is applied to the protein. The proper folding state of purified IpgB2 was tested by 15N-, 13C-NMR spectroscopy on appropriately labeled IpgB2 (data not shown), indicating that the protein accommodates a compact, folded state in solution.
The buffer systems found for IpgB2 also allowed purification of the homologous WXXXE protein Map in full length, and allowed production of MBP-tagged IpgB1 (data not shown). IpgB1, however, slowly precipitates upon cleavage of the fusion tag and might require further buffer optimization.
Cell Assays
cDNA of the constitutively active variant Q14V of human RhoA was a kind gift from Laura Machesky (Glasgow, UK), cDNA of IpgB2 was amplified by PCR from genomic DNA of S. flexneri (serotype 5), and the mutants of IpgB2 were custom-synthesized (Genescript, Piscataway, NJ). All cDNAs were subcloned into pEGFP-C1 (Clontech, Mountain View, CA). The fidelity of all constructs was verified by sequencing. For transfections and fluorescence microscopy, SV40 large T-antigen-immortalized mouse embryonic fibroblasts were grown in Dulbecco's modified Eagle's medium, 4.5 g/liter glucose (Invitrogen) supplemented with 10% fetal bovine serum (Sigma) and 2 mm glutamine. Transfections were carried out with FuGENE 6 (Roche Applied Science) according to the manufacturer's protocols. Cells were plated on glass coverslips and fixed 16–24 h post transfection. Cells were fixed with 4% formaldehyde in phosphate-buffered saline for 20 min, permeabilized with a mixture of 0.1% Triton X-100 and formaldehyde for 45 s, and stained for the actin cytoskeleton using Alexa594-labeled phalloidin (Molecular Probes, Invitrogen). Epifluorescence images were acquired using an inverted microscope (Axiovert 100TV, Carl Zeiss, Jena, Germany) equipped with a 100×/1.3 numerical aperture PlanNeofluar objective and a back-illuminated charge-coupled device camera (CoolSNAP K4, Photometrics, Tucson, AZ) driven by MetaMorph software (Molecular Devices, Downingtown, PA), and further processed using Adobe Photoshop CS and Illustrator software (Adobe Systems, Mountain View, CA).
Crystallization
Native and selenomethionine-substituted IpgB2 and its complex with RhoA were crystallized using the hanging drop, vapor-diffusion technique at 4 °C. Initial spheroidal precipitate of free IpgB2 could be optimized by adding 10 mm FeCl3 to the protein in gel-filtration chromatography buffer, which was sufficient to obtain crystal growth without any additional precipitate. The crystal quality could be improved with low concentrations of polyethylene glycol 3350 and isopropanol. The final composition was 1 μl of 89 mg/ml IpgB2 in gel-filtration chromatography buffer mixed with 1 μl of reservoir containing 100 mm trisodium citrate/citric acid, pH 5.5, 10 mm NaCl, 4–7% isopropanol, 3–5% polyethylene glycol 3350, and 10 mm FeCl3. The crystal morphology was optimized by streak-seeding from initial crystallization conditions containing micro crystals. Crystals of IpgB2 in complex with RhoA were obtained from a (stoichiometric) mixture of 25 μl of RhoA (19.3 mg/ml) with 5.7 μl of IpgB2 (89.6 mg/ml). 1 μl of this protein solution was mixed with 1 μl of reservoir solution containing 100 mm Hepes, pH 7.5, and 20% (w/v) polyethylene glycol 3350 for crystallization of complex A. To obtain complex B, excess Mg2+ was removed from the mixture prior to crystallization by five cycles of 20-fold dilution with buffer containing 30 mm Tris/HCl, pH 7.5, 50 mm NaCl, 5% (v/v) glycerol, 20 mm EDTA, 0.05 mm GDP, and 5 mm dithiothreitol following concentration to the original volume using a Vivaspin concentrator unit (Sartorius, 5-kDa cutoff). The final concentration of the IpgB2·RhoA complex was 20.9 mg/ml. Crystals of complex A and B were cryoprotected in reservoir buffer with an additional 20% (v/v) glycerol and frozen in liquid nitrogen. Complex C was obtained by treatment of complex B crystals with a buffer containing 100 mm LiSO4, 100 mm HEPES, pH 7.5, 30 mm Tris, pH 7.0, 22.5% (w/w) polyethylene glycol 3350, 100 mm NaCl, 12% (v/v) glycerol, and 40 mm EDTA. Complex C crystals were directly frozen from this solution after 2–5 min of incubation time (30).
Free IpgB2 crystallized as cuboids with dimensions of up to 0.2 × 0.2 × 0.2 mm3 belonging to space group P4212. IpgB2 in complex with RhoA crystallized as rhomboids with dimensions of ∼0.2 × 0.2 × 0.08 mm3. Complex A crystals belonged to space group P21. Despite similar crystal morphology, complex B crystals belonged to space group P212121. Complex C crystals had a gel-like consistency and melted within ∼10 min after preparation. When frozen prior to complete disintegration, they showed an improved diffraction behavior compared with complex B. Changes in the crystal lattice led to the new space group P21212.
Data Collection, Structure Determination, and Refinement
X-ray diffraction data were collected at the beamlines ID23-1 and ID23-2 of the European Synchrotron Radiation Facility and by using a Rigaku MicroMax 007HF (home-source) x-ray generator equipped with a Saturn 944+ charge-coupled device detector. Data were processed and scaled using the XDS program package (31, 32). SHELXC, -D, and -E (33, 34) were used for structure solution and phasing of free selenomethionine-substituted IpgB2, and an initial model was built with AutoBuild of PHENIX (35). The structures of IpgB2 in complex with RhoA were solved by molecular replacement with MOLREP (36) using the partially refined structure of free IpgB2 and the structure of RhoA (37) as search models. The MOLREP solution was automatically corrected using ARP/wARP (38), manually completed in COOT (39), and refined with REFMAC5 (40). The figures were prepared using PyMOL (41). Data statistics are summarized in Table 1. Similarities between different structures were evaluated based on the r.m.s.d. of comparable residues using the DaliLite server (42).
TABLE 1.
Crystallographic data and refinement statistics
| Selenomethionine-labeled free IpgB2 | free IpgB2 | IpgB2/RhoA complex A | IpgB2/RhoA complex B | IpgB2/RhoA complex C | |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Beamline | ESRF (ID23-1) | ESRF (ID23-1) | ESRF (ID23-2) | Rigaku MicroMax 007HF | ESRF (ID23-1) |
| Space group | P4212 | P4212 | P21 | P212121 | P21212 |
| Unit cell dimensions | |||||
| a, b, c (Å) | 116.2 | 114.1 | 82.6 | 75.5 | 77.6 |
| 116.2 | 114.1 | 101.6 | 95.7 | 100.8 | |
| 90.8 | 88.8 | 97.0 | 103.0 | 50.9 | |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 96.4, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.900 | 0.979 | 0.873 | 1.54 | 0.972 |
| Resolution (Å)a | 48.9-2.9 (3.0-2.9) | 22.9-2.3 (2.36-2.3) | 48.2-1.85 (1.9-1.85) | 37.8-2.28 (2.34-2.28) | 42.5-1.68 (1.72-1.68) |
| Unique reflectionsa | 26125 (1950) | 25363 (1205) | 133487 (9777) | 34100 (2183) | 45588 (3248) |
| Multiplicitya | 6.3 (6.4) | 18.2 (6.4) | 3.5 (2.6) | 5.3 (3.5) | 4.7 (4.0) |
| Completeness (%)a | 98.9 (99.9) | 95.4 (63) | 98.4 (97.8) | 98.7 (86.3) | 98.6 (96.9) |
| I/σIa | 26.2 (3.35) | 33.35 (2.56) | 15.11 (2.49) | 9.81 (2.06) | 23.76 (2.64) |
| Rmerge (%)a,b | 4.5 (55.7) | 5.6 (67.5) | 5.2 (49.2) | 13.4 (65.4) | 4.1 (58.8) |
| Wilson B-factor (Å2) | 53.0 | 49.2 | 26.4 | 31.2 | 22.5 |
| Solvent content (%) | 64.9 | 63.8 | 48.0 | 44.5 | 47.4 |
| Refinement statistics | |||||
| Rcrys (%)c | 22.9 | 17.7 | 20.4 | 17.0 | |
| Rfree (%)d | 29.2 | 23.3 | 29.1 | 21.9 | |
| Number of atoms | 3117 | 13880 | 6381 | 3496 | |
| Monomers of IpgB2/RhoA per asymmetric unit r.m.s.d. from ideale | 2/0 | 4/4 | 2/2 | 1/1 | |
| Bond lengths (Å) | 0.014 | 0.014 | 0.016 | 0.023 | |
| Bond angle (°) | 1.589 | 1.423 | 1.661 | 1.975 | |
| Average B-factor (Å2) | 45.8 | 21.6 | 16.8 | 17.3 | |
| Ramachandran plot (%) (most favored regions/allowed/generously allowed/disallowed) | 90.8/8.9/0.3/0 | 94.4/5.6/0.0/0 | 92.8/7.0/0.2/0 | 93.6/6.4/0/0 | |
a Values in parentheses refer to statistics in the highest resolution shell.
b Rmerge = Σ|Iobs − 〈I〉|/ΣIobs.
c Rcrys = (Σ|Fo − Fc|/ΣFo) × 100, where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d Rfree was computed using 5% of the data assigned randomly.
e r.m.s.d., root mean square deviation.
Protein Data Bank Accession Numbers
The atomic coordinates and structure factors (code 3LYQ for free IpgB2, 3LW8 for IpgB2·RhoA complex A, 3LWN for complex B, and 3LXR for complex C) have been deposited in the RCSB Protein Data Bank.
RESULTS
IpgB2 Is a GEF for Human RhoA
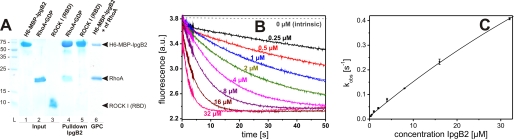
Consistent with the proposed function of IpgB2 acting as a GEF for RhoA, a weak but detectable interaction was found with GDP-bound RhoA in pulldown assays. Furthermore, a complex containing nucleotide-free RhoA in approximately stoichiometric ratio could be purified using gel-filtration chromatography (Fig. 1A). As opposed to previous suggestions (16), no interaction was detected for the RBD of the RhoA-effector Rho kinase (ROCK I) with IpgB2, although it readily interacted with RhoA (data not shown). This is consistent with a biological function as a RhoA-GEF rather than a RhoA-mimic. The catalytic activity of purified IpgB2 was confirmed using fluorescence-based GEF assays (Fig. 1, B and C). A nucleotide dissociation rate of 0.41 ± 0.005 s−1 was measured at the highest molar excess of IpgB2 over RhoA (128-fold), corresponding to a ∼104-fold acceleration of nucleotide exchange by IpgB2 compared with the intrinsic rate of 3.8 ± 0.2 × 10−5 s−1. This activity is well in the range observed for the bacterial GEF SopE (15, 43) and more than an order of magnitude higher when compared with mammalian GEFs like p115RhoGEF and p190RhoGEF (44).
FIGURE 1.
GEF activity of IpgB2 for RhoA. A, His6-MBP-IpgB2 binds RhoA but not ROCK I (RBD) in vitro. Lanes 1–5, SDS-PAGE analysis of pulldown experiments using immobilized His6-MBP-IpgB2 incubated with RhoA·GDP or ROCK I (RBD); lanes 1–3, pure samples; lanes 4–5, coelution of RhoA·GDP but not ROCK I with His6-MBP-IpgB2; lane 6, SDS-PAGE showing His6-MBP-IpgB2 in complex with nucleotide-free RhoA after purification by gel-permeation chromatography. B, time course of the dissociation of the RhoA·mantGDP complex, as followed by the decrease in mantGDP fluorescence after excitation at 335 ± 5 nm at 25 °C. The reaction mixture contained 0.25 μm RhoA in gel-filtration chromatography buffer (30 mm Tris/HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 5% (v/v) glycerol, 5 mm dithiothreitol), and the exchange was started by addition of 200 μm GDP and the indicated concentrations of (untagged) IpgB2 in a stopped flow apparatus. The intrinsic dissociation rate was measured with a conventional fluorescence spectrometer over a time frame of 50,000 s (gray dashed line). C, kinetic data as shown in B were analyzed by fitting single exponentials to them assuming pseudo-first-order conditions. The calculated rate constants (kobs) were plotted against the IpgB2 concentration used in the assay.
Structure of IpgB2
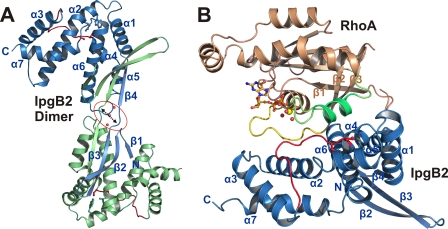
IpgB2 consists of two distinct domains: an extended N-terminal antiparallel β-sheet domain comprising the first 39 residues, followed by a C-terminal V-shaped α-helical domain (residues 40–188) divided into a four-helix bundle and a three-helix bundle. The N-terminal domain does not interact with its own C-terminal domain but is instead engaged via domain-swapping in interactions with the C-terminal domain of a neighboring IpgB2 molecule in the crystal, leading to a Z-shaped IpgB2 dimer (Fig. 2A). The absence of an IpgB2 dimer in solution (data not shown) and in the complex with RhoA (see below) indicates that the domain-swapping most likely represents a crystallographic feature. This is further supported by a disrupted hydrogen bonding pattern of the formed intermolecular antiparallel β-sheet at residues 31–33 (Fig. 2A), which is unlikely to occur in a physiologically relevant dimer.
FIGURE 2.
Structures of dimeric IpgB2 and IpgB2·RhoA complex A in schematic representation. The conserved residues of the WXXXE motif, the coordinating double serine motif of the catalytic loop, and GDP are shown in ball and stick representation. A, domain-swapped dimer in free IpgB2. The two monomers are shown in blue and green, respectively, the catalytic loops are marked in red. Each N-terminal β-sheet domain separates from its C-terminal helical domain and is flipped over to the C-terminal domain of the respective other monomer. The hydrogen-bonding network at the interface between strands β3 and β4 of each monomer is disrupted by three water molecules (red spheres, marked by an arrow and a red circle), indicating that this dimer architecture is not physiologically relevant (see text). B, IpgB2·RhoA·GDP (complex A). IpgB2 is shown in blue, RhoA in brown. GDP is shown in orange and the GDP-coordinating Mg2+ is a yellow sphere. Red spheres are water molecules coordinating Mg2+. The switch I region is shown as a yellow loop, and the switch II is green. The catalytic loop of IpgB2 is shown as a red loop. The IpgB2 monomer in complex A corresponds to β2 and β3 from one monomer of free IpgB2 as shown in A plus β3 to the end of the second monomer.
The rather loosely packed α-helical domain is further stabilized by a loop linking helices α3 and α4 that we refer to as the “catalytic loop” (residues Ile108–Ser118) herein, in line with previous nomenclature (15, 24). The loop winds around helix α2 and is oriented toward the N terminus of this helix. The conserved WXXXE motif is located close to the N-terminal end of helix α2. Trp62 and Glu66 of this motif form hydrogen bonds with Ser118 and Ser117 in the catalytic loop, respectively.
Both IpgB2 monomers in the asymmetric unit are structurally similar, but show a marked difference at the interface of helices α1 and α2. Helix α2 is disrupted between residue pairs 67/68 and 71/72 in one of the monomers, while forming a continuous helix in the other monomer. This conformational flexibility is important for binding to RhoA by induced fit (see below).
Structures of RhoA in Complex with IpgB2
The recently solved structure of the WXXXE protein Map in complex with Cdc42 (24) provided information about a GEF/GTPase complex after dissociation of the nucleotide. Initial complex formation, however, requires interaction with the nucleotide-bound GTPase, because the nucleotide-free state does not exist in significant amounts in vivo (45). Three different structures of IpgB2 in complex with RhoA·GDP were solved here, showing for the first time how bacterial GEFs interact with their nucleotide-bound human GTPase targets. One, representing the initial step of the nucleotide exchange reaction, contains RhoA GTPase with Mg2+-bound GDP (termed complex A). In the second complex structure (complex B), the Mg2+ is displaced from its original β-phosphate position of the nucleotide to a secondary binding site, thus representing the next step of the exchange reaction. In a third structure (complex C), the Mg2+ is completely removed from the complex.
The obtained IpgB2·RhoA complexes crystallized in different space groups, with four, two, and one heterodimers per asymmetric unit for complex A, B, and C, respectively. All complex structures show similar overall folds. The interaction surface between IpgB2 and RhoA buries roughly 1220 Å2. It is formed by extensive interactions between the helical bundles of IpgB2 and the flexible switch regions I and II, as well as a region involving residues of sheets β1, β2, and β3 of RhoA (in the following called the “β123 region”) (Fig. 2B). Although the overall fold of RhoA and the conformation of its bound GDP ligand remained essentially unchanged when compared with the free enzyme structure (46), the N-terminal domain of IpgB2 now packs against the four-helix bundle of the helical domain of the same protomer, without interacting with RhoA. This is in line with the observation that the first 19 residues of IpgB2 are dispensable for exerting GEF activity (24). The surface of this β-domain shows a pronounced positive charge, indicating potential interactions with lipids or other proteins.
Complex A
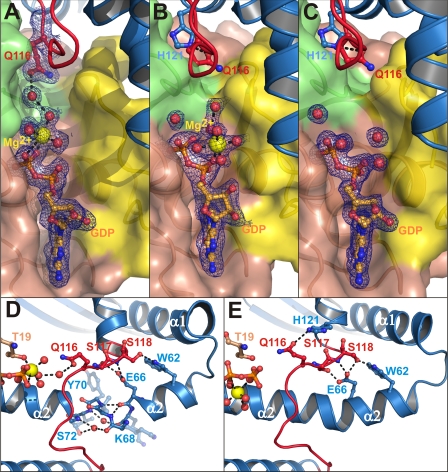
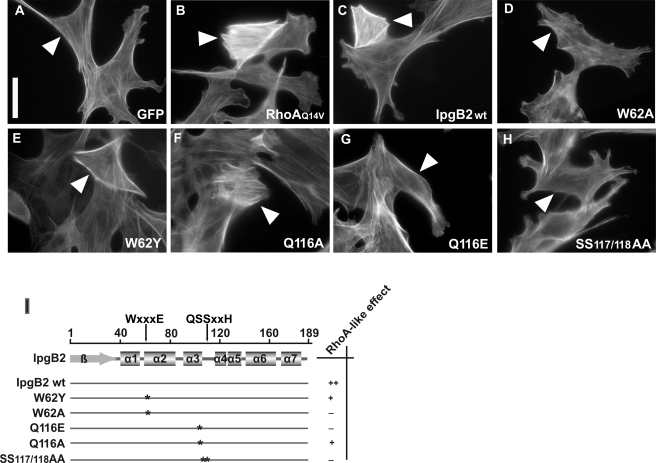
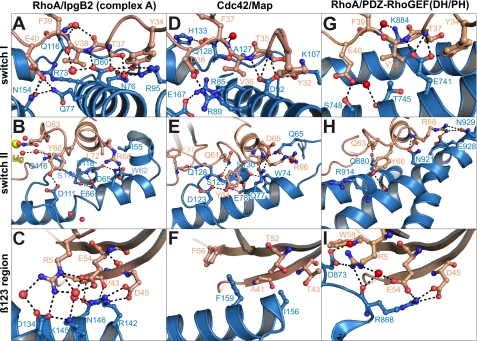
Upon complex formation, the catalytic loop of IpgB2 reorients along helix α2 toward its C terminus (8.4-Å movement) and is now cradled between the RhoA switch regions I and II. In complex A, it interacts with the Mg2+ hydration shell via Gln116 (Fig. 3A). Interestingly, replacement of this residue by alanine (Q116A) partially preserved stress fiber induction in cells upon expression of the IpgB2 mutant when compared with the respective positive and negative controls (Fig. 4, A–C and F, and supplemental Fig. 1). In contrast, mutation of Gln116 to glutamate (Q116E) completely abolished IpgB2-mediated stress fiber formation (Fig. 4G). This suggests that Gln116 is not essential for complex formation, but instead might have a supportive role for the nucleotide release induced by IpgB2. Substitution by glutamate may abolish this function due to electrostatic repulsions with Asp59 in RhoA, whereas mutation to alanine only results in a lowered catalytic efficiency of IpgB2.
FIGURE 3.
Nucleotide binding site and catalytic loop region in IpgB2·RhoA structures. IpgB2 is shown as a blue schematic, with the catalytic loop marked in red. GDP is shown as orange balls and sticks, and Mg2+ and water molecules are shown as yellow and red spheres, respectively. A, GDP-binding pocket and interaction of IpgB2 residue Gln116 in complex A. The [2Fo − Fc] electron density map contoured at 1 σ for GDP, Mg2+, the Mg2+ coordination sphere, and Gln116 is colored as blue mesh. B and C, similar representation as in A for complexes B and C, respectively. For A–C, RhoA is shown as a surface representation, with switch I, switch II, and other regions of RhoA as yellow, green, and brown surfaces, respectively (compare also supplemental Fig. 2 for a more detailed view in stereo representation). D, helix α2 in complex A, showing a conformation that is disrupted by two water molecules. Such a disrupted helix conformation is not observed in crystals of complex B (E) or of complex C (not shown). RhoA residues are shown as brown balls and sticks in D and E.
FIGURE 4.
Biological activity of IpgB2 and verification of critical residues in the putative catalytic loop. A–H, fibroblast cells expressing green fluorescent protein (GFP) alone (negative control), GFP-tagged constitutively active RhoAQ14V (positive control), wild-type IpgB2, and different point mutants as indicated were fixed and stained for the actin cytoskeleton. Transfected cells are marked by white arrowheads (compare also with supplemental Fig. 1). RhoA and wild-type IpgB2 strongly induced the formation of parallel actin stress fibers. IpgB2 mutants displayed no (D, G, and H) or moderate (E and F) biological activity. The bar equals 25 μm. I, schematic overview and summary of the IpgB2 mutants studied. Of note, the biological activity of none of the mutants was as strong as that seen with wild-type IpgB2, which caused effects virtually identical to RhoAQ14V already at low expression levels of the construct, as judged by GFP fluorescence (see also supplemental Fig. 1) (categorized as strong, ++). Mutants that lacked any notable activity even at high expression levels were classified as negative (−), whereas the responses observed with positive mutants were generally more variable than wild type and required strong overexpression to induce wild-type-like effects (categorized moderate, +). At least 25 individual cells were categorized for each construct.
Switch I is partially reoriented upon IpgB2 binding. Thr37, one of two threonine residues coordinating Mg2+ in free RhoA (46), is no longer involved in forming the octahedral Mg2+ coordination sphere. Instead, Asp80 of IpgB2 pulls switch I toward IpgB2 by forming hydrogen bonds to Tyr34, Thr37, and Val38. Additional hydrogen bonds are formed between IpgB2 residues Arg73, Gln77, and Asn154 and Glu40 of RhoA (Fig. 5A). In contrast to switch I, the conformation of switch II remained essentially unaffected upon formation of complex A. Minor rearrangements within switch II allow a closer approach of the catalytic loop of IpgB2 to the active site of RhoA (Fig. 5B). RhoA residues Arg5, Asp45, and Glu54 of the β123 region are the main determinants of specificity of RhoA-GEFs (47) and are involved in extensive interactions with complementary charged IpgB2 residues (Fig. 5C).
FIGURE 5.
Comparison of interactions of guanosine nucleotide exchange factors IpgB2 (A–C), Map (D–F (24)), and the DH/PH domain of PDZ-RhoGEF (G–I (52)) (in blue color) with the GTPases RhoA and Cdc42 (in brown color) as indicated. The interactions are shown for three characteristic regions of the GTPase (switch I and II and the β123 region). Water molecules are red spheres; Mg2+ is shown in yellow.
Complex B
After binding to a GTPase, GEFs destabilize the GDP-bound state to allow GDP dissociation and binding of GTP for activation. This is thought to be facilitated by a stepwise ligand release: First, Mg2+ is released, followed by dissociation of GDP (48–50). To obtain a structure of the RhoA·IpgB2 complex in the magnesium-free state, the metal ion was sequestered by repeated cycles of EDTA treatment prior to complex formation. Upon magnesium ion depletion, RhoA in the structure of complex B undergoes changes in switch II that are virtually identical to those described in the structure of magnesium-free RhoA (49). A flip of switch II residue Ala61 toward the GDP-binding region occludes the primary Mg2+ binding site at the β-phosphate. At the same time, Glu64 is repositioned and forms a hydrogen bond with Lys18 from the phosphate-binding loop (P-loop), thus lowering the affinity to the nucleotide (supplemental Fig. 2). However, while the Mg2+ is completely absent in free RhoA in complex with GDP (49), the complex with IpgB2 strikingly reveals a new, secondary Mg2+ binding site at the α-phosphate group of the nucleotide (Fig. 3B). Although the metal ion is well defined in the secondary binding site, it is no longer coordinated by protein atoms, but instead by five water molecules and an α-phosphate oxygen. Indirect interactions of this Mg2+(α) with RhoA residues are mediated by two magnesium-coordinating water molecules to the side chain of Thr19 and to the carbonyl oxygens of Pro36 and Thr37 of switch I, respectively (supplemental Fig. 3).
Complex C
Shimizu and coworkers (49) observed that high concentrations of LiSO4 are required to obtain the GDP-loaded, magnesium-free state of RhoA. In the current study, a milder method of Mg2+ depletion by EDTA treatment was utilized, which allowed remaining Mg2+ to bind to the α-phosphate. Indeed, LiSO4 can be used to completely remove the Mg2+ ion from the active site: treatment of complex B crystals with a buffer containing LiSO4 leads to significant rearrangements of the crystal lattice, an improved diffraction behavior, and quantitative Mg2+ elimination from the newly observed secondary binding site. The protein fold in this complex (complex C) essentially remains identical, showing virtually no conformational changes in the environment of the depleted site (Fig. 3C and supplemental Fig. 2). Switch I in complexes B and C is similar to the conformation observed in complex A, whereas in the free RhoA·GDP structure (49) it is moved away from the (magnesium-free) GDP binding pocket.
In addition to the conformational changes in RhoA, differences were also observed in helix α2 of IpgB2, which contacts the switch II region. In complex A, helix α2 preferentially adopts a conformation with two water molecules disrupting the helix between residues Lys67/Lys68 and Ser72/Val71 (Fig. 3D). In contrast, it adopts a well defined continuous α-helical arrangement in all IpgB2 molecules in complexes B and C (Fig. 3E). This suggests that the conformational rearrangements in switch II at low Mg2+ concentrations favor the undisrupted helix conformation.
DISCUSSION
Different Bacterial GEFs Induce GDP Release in a Similar Manner
IpgB2 shows a close structural homology to Map, the C-terminal region of SifA, and a less pronounced homology to SopE (r.m.s.d. of 2.6 Å, 3.5 Å, or 4.3 Å for 150, 158, or 140 common Cα positions, respectively). All four proteins share a V-shaped helical arrangement and a catalytically important loop connecting both lobes of the V. This loop winds around a helix that contains the conserved WXXXE motif in IpgB2, Map, and SifA.
Map in complex with the small GTPase Cdc42 (24) is the only other WXXXE family member co-crystallized with its cognate GTPase, allowing for a close comparison with the present IpgB2·RhoA complex. Both complexes share a number of common features. Structurally, the V-shaped C-terminal helical domain of Map (residues 51–203) and IpgB2 is basically conserved (Fig. 5, B and E). The interactions of IpgB2 and Map with switch I of their cognate GTPases are also remarkably similar (Fig. 5, A and D). As expected, the region involved in binding to the β123 region displays strong differences between IpgB2 and Map. Although the GTPase interaction surface of Map is predominantly apolar, IpgB2 interacts with RhoA via charged or polar interactions (Fig. 5, C and F). Both Map and IpgB2 contain the stabilizing double serine motif (Ser117 and Ser118 in IpgB2). The two adjacent serines form hydrogen bonds with the conserved residues of the WXXXE motif (Fig. 3, D and E) and are part of a sequence motif AQ(S/T)S(I/V) in the catalytic loop that is conserved throughout most currently known members of the WXXXE family. Substitution of both serines by alanine completely abolished induction of stress fiber formation by IpgB2 in vivo (Fig. 4H), consistent with the abrogation of Cdc42 binding observed for the S130H mutant of Map (24). This shows that the serines are crucial for biological activity and underscores their indirect function in catalyzing nucleotide exchange. Because the catalytic loop is directly involved in GTPase interactions whereas the WXXXE motif is not, mutations of the critical tryptophan in the WXXXE motif were tested for biological activity. A substitution of Trp62 for alanine led to a complete loss of cellular response (Fig. 4D), as described (16). However, a conservative change to tyrosine partially preserved IpgB2 activity (Fig. 4E), demonstrating that the tryptophan itself is not strictly required for biological function. This is also in line with the data on the novel family member EspM3 expressed in certain pathogenic E. coli strains, because the corresponding mutant W66Y also induced stress fibers when translocated into host cells (51).
The structure of the Map·Cdc42 complex represents a rather late state of the nucleotide exchange reaction where GDP is already released. Switch II of Cdc42 shows a conformation that is also observed in the complex with SopE, another bacterial GEF lacking a WXXXE motif (15). This switch II conformation is highly similar to the conformation of RhoA in IpgB2·RhoA complex B, but not in complex A (Fig. 5, B and E). Similarly, the contact surfaces of Map with switch II show a continuous conformation of helix α2, as seen in complex B. This demonstrates that Map, SopE, and IpgB2 induce very similar changes in their cognate GTPases, despite their different specificities. The remarkable similarity to complex B allows an allocation of the different complexes as consecutive snapshots of states that lead from complex A via complex B to a nucleotide-free state as represented by the Map·Cdc42 complex (Fig. 5, B and E).
Convergent Evolution of Bacterial and Human GEFs for RhoA
The mode of interaction with RhoA is largely conserved between IpgB2 and human GEFs such as the Dbl homologous/pleckstrin homologous fragments of leukemia-associated Rho guanine-nucleotide exchange factor (LARG) and PDZ-RhoGEF (in the following called “DH/PH”) (52, 53) despite very different tertiary structures. In contrast to the V-shaped helical arrangement in IpgB2, the DH domain of Dbl-like proteins forms a single elongated helical bundle in a “chaise longue” shape, with all the long helices packed into the “seat” and the short helices forming the “seatback” (52, 54). Interactions with the β123 region of RhoA are the main determinants for specificity of RhoA-GEFs (47). IpgB2 and DH/PH interact with this region in a remarkably similar fashion. Asp45 and Glu54 in RhoA form salt bridges with Arg142 of IpgB2 and Arg868 of DH/PH, while Arg5 forms hydrogen bonds with IpgB2-Asp134 and DH/PH-Asp873, respectively (Fig. 5, C and I). In both the (DH/PH)·RhoA and the IpgB2·RhoA complexes, switch I adopts a similar conformation. Asp80 of IpgB2 and Glu741 of DH/PH occupy similar positions in the RhoA interface and participate in essentially the same interactions with the switch I residues Tyr34, Thr37, and Val38 (Fig. 5, A and G). In addition, RhoA residue Glu40 forms hydrogen bonds with Asn154 and Gln77 of IpgB2 and with Ser748 of DH/PH (Fig. 5, A and G). The switch II conformation in (DH/PH)·RhoA resembles the state in complexes B and C of IpgB2·RhoA. Gln63 of RhoA is repositioned by >10 Å compared with complex A (Fig. 5, B and H) and now forms a water-mediated interaction with DH/PH residue Gln880 or a hydrogen bond to the main-chain carbonyl of IpgB2-Gln116 (supplemental Fig. 2), respectively. Additional hydrogen bonds with both GEFs are formed by RhoA residues Tyr66 and Arg68, although they occur with different binding modes (Fig. 5, B and H).
This structural comparison suggests that bacterial WXXXE GEFs and mammalian GEFs of the DbI family are the result of convergent evolution. In addition it ranks complexes B and C at a late stage of the nucleotide exchange reaction, strongly resembling the state in (DH/PH)·RhoA in which the GDP nucleotide is already released.
Role of the WXXXE Motif in Bacterial GEFs
The crystal structures of SifA and a Map·Cdc42 complex already revealed that WXXXE proteins are GEFs instead of GTPase mimics (23, 24, 30). Information on mechanistic details and GTPase selectivity of WXXXE proteins, however, is still limited. The structures of free and RhoA-complexed IpgB2 presented in this work together with the structures of SifA and Map·Cdc42 now provide a framework for the mode of action of bacterial WXXXE proteins. Although the WXXXE motif is only indirectly required for catalytic activity, residues of the catalytic loop form direct interactions with the cognate GTPase. For this, one important function of the WXXXE motif is to stabilize the catalytic loop by forming interactions with the double serine motif (Ser117/Ser118). This allows productive interactions with the cognate GTPase, e.g. of the catalytically important catalytic loop residue Gln116 with the Mg2+ hydration shell. The double serine motif is strictly required for activity, and interactions involving the WXXXE motif are strikingly conserved in IpgB2 and Map. SifA most likely utilizes a similar stabilization, despite harboring a more extended catalytic loop that is stabilized by several proline residues, leading to a different overall loop conformation (supplemental Fig. 4A). His255 of the catalytic loop in SifA is oriented toward Trp197 and Glu201 of the WXXXE motif, suggesting that it substitutes the hydrogen bonding potential of the two serine residues present in IpgB2, Map, and numerous other WXXXE proteins (supplemental Fig. 4B). However, the question arises whether the significant structural differences in the catalytic loop of SifA compared with IpgB2 suggest that RhoA might not be the main target of SifA, as recently proposed by Ohlsen et al. (23). An overlay with IpgB2·RhoA reveals highly unfavorable interactions between SifA and RhoA, particularly within the specificity determining regions (supplemental Fig. 4C). Interestingly, phenotypes induced by SifA in host cells as the formation of Salmonella-induced filaments (Sifs) do not typically coincide with RhoA activation but could derive, rather, from interference with Rab-GTPase signaling.
We conclude that the WXXXE motif in all family members forms an essential part of a conserved structural motif, which is complemented by the catalytic loop motif AQ(S/T)S(I/V) or related residues in the distinct sequence of SifA. Interactions between both motifs promote a functionally competent conformation of the catalytic loop that is required for productive interactions with the cognate GTPase. The role of the different catalytic loop in SifA still requires further analysis and might reveal other GTPases as targets of SifA which might not belong to the Rho family.
GEF Mechanism of Bacterial WXXXE Proteins
Various families of mammalian Rho-GTPase specific GEFs exist that activate their cognate GTPases by different mechanisms. The largest of those is the Dbl family of GEFs containing a DH/PH domain. In contrast to other Rho-specific GEFs like proteins from the DOCK family (55), DbI proteins do not directly insert residues into the active site of their cognate GTPase (4). Bacterial WXXXE GEFs utilize a similar mechanism for GTPase activation, despite having a different overall fold. By combining structural information about free and RhoA-complexed IpgB2 with knowledge about RhoA·GDP (with and without Mg2+ (46, 49)), and the structurally related complex of nucleotide-free Map·Cdc42 (24), it is now possible to outline the sequence of events finally leading to nucleotide release, the essential prerequisite of GTP loading (45). At least four distinct states, States 1–4 as follows, can now be discriminated (Fig. 6).
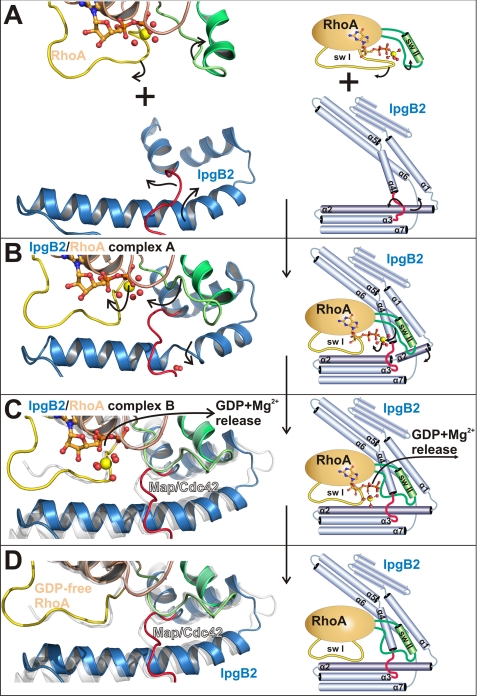
FIGURE 6.
Proposed mechanism of IgpB2 GEF function on RhoA. Crystal structures are shown on the left, and in a corresponding schematic drawing on the right hand side of each state. A, State 1: free RhoA·GDP·Mg2+ (46) and free IpgB2. B, State 2: IpgB2·RhoA complex A, with Mg2+ coordinated to the β-phosphate of the GDP nucleotide. C, State 3: complex B, with Mg2+ coordinated to the α-phosphate. D, State 4: proposed architecture of the nucleotide-free IpgB2·RhoA complex. The conformation of GDP-free RhoA was obtained from the complex of RhoA with the DH/PH fragment of PDZ-RhoGEF (52). Switch I and switch II of RhoA and the catalytic loop of IpgB2 are shown as yellow, green, and red schematics, respectively. GDP is shown as orange balls and sticks, and the Mg2+ ion and water molecules are yellow and red spheres, respectively. In C and D, the complex of (nucleotide-free) Map·Cdc42 (24) is shown in white for comparison. All GEF and GTPase structures were individually superposed with the corresponding structures of complex A as shown in B.
State 1: Uncomplexed Proteins
The structures of free IpgB2 and RhoA·GDP·Mg2+, respectively, represent the state before an interaction occurs (Fig. 6A). The catalytic loop of IpgB2 is oriented toward the N terminus of helix α2. The octahedral Mg2+ coordination sphere in RhoA includes switch I residue Thr37 (46).
State 2: Complex Formation
In a first step, IpgB2 binds to GDP-bound RhoA (Fig. 6B). Upon complex formation, the catalytic loop reorients along helix α2 toward its C terminus. In contrast to later states, the α-helical hydrogen-bonding pattern in helix α2 is disrupted by two water molecules. This way, a helical tilt is induced that bends the N-terminal residues of IpgB2 helix α2 toward switch II. While switch II stays mostly unchanged at this stage and IpgB2 reorients to form favorable interactions, the situation is opposite for switch I. Here, the contacting regions of IpgB2 remain similar to the uncomplexed protein, with switch I of RhoA undergoing a large reorientation. It adopts a conformation that is common to other GEF/GTPase structures, including Map·Cdc42 (24) and (DH/PH)·RhoA (52). Upon complex formation, Thr37 is removed from the octahedral coordination sphere of Mg2+ and is replaced by a fourth Mg2+-coordinating water molecule. This way, the GDP binding pocket becomes more solvent-exposed to initiate dissociation of GDP and Mg2+.
State 3: Change of Switch II and Release of Mg2+ from the β-Phosphate of GDP
In a second step, conformational changes are induced within switch II that displace Mg2+ and prepare the dissociation of GDP (Fig. 6C). This coincides with a conformation of helix α2 being no longer disrupted by water molecules, indicating that IpgB2 utilizes changes in this helix to adopt to different switch II conformations. Consistent with a subsequent release of Mg(β) and GDP, the Mg2+ ion is removed from its primary binding site at the β-phosphate. Such a stepwise mechanism has already been proposed based on a structure of GDP-loaded, magnesium-free RhoA in the absence of a GEF (49). It was discussed that the depletion of the Mg(β) ion reduces the affinity of GDP to RhoA to facilitate GDP release (49, 50), but the question remained as to whether a magnesium-free state would be stable under physiological conditions. The structure of complex B provides an intriguing answer to this question: the negative charge at the nucleotide is compensated by Mg2+ at a secondary, previously not described α-phosphate binding site. This secondary site can be depleted by soaking complex B crystals in buffers containing LiSO4 (Fig. 3C), which was also used to obtain the magnesium-free state of RhoA (49). This demonstrates that Mg2+ can bind to the α-phosphate of GTP without inducing any notable conformational rearrangements of the protein. The switch II conformation in complexes B and C is similar to free RhoA·GDP (49) and to nucleotide-free (DH/PH)·RhoA (52). Remarkably, even though an magnesium-bound α-phosphate binding site has never been described before, all analyzed structures of GEF·GTPase complexes in the presence as well as in the absence of GDP (15, 24, 50, 52, 54, 55) contain a cavity that can fit Mg2+ in a similar binding mode as observed in the IpgB2·RhoA complex B. This might indicate a broader relevance of this secondary site also for other GEF families such as mammalian Dbl-like GEFs. Because high concentrations of Mg2+ are present in vivo, complex B might represent a relevant intermediate in GTPase activation that was not structurally characterized before. The secondary Mg2+ binding site can be rationalized by the requirement to compensate the phosphate charge without interference with a switch II conformation blocking the primary Mg2+ binding site at the β-phosphate. This switch II reorientation is an essential step in the GEF-catalyzed nucleotide release and is required for a lowered GDP affinity. By binding to the α-phosphate, Mg2+ is able to compensate the phosphate charge without formation of additional interactions that could stabilize the binding of GDP to RhoA.
State 4: Release of GDP·Mg2+
In a final step, GDP and Mg(α) are released from the complex (Fig. 6D). The remarkable similarity of complex B with the GDP-free Map·Cdc42 complex (24) indicates that the latter can serve as a good model for a nucleotide-free IpgB2·RhoA complex, which is the final state in the nucleotide exchange reaction. As indicated by the comparison of both complexes, the dissociation of GDP does not require significant conformational changes. Under physiological conditions, a simultaneous release of Mg(α)·GDP seems probable. However, a stepwise dissociation of Mg2+ and GDP cannot be excluded, which would place the magnesium-depleted complex C crystals as an additional intermediate between states 3 and 4.
The presented scheme provides the first example of a fully deduced mechanism of a bacterial GEF in action. The conformational changes induced in the GTPase are comparable to those observed upon interaction with mammalian GEFs containing a DH/PH domain (e.g. Ref. 52), despite completely distinct GEF-folds. Based on structural comparisons of the PRONE domain of RopGEF8 with Rho of plants (Rop), Thomas and colleagues (50) concluded that mammalian and plant GEFs utilize common intermediate states in small G protein/GEF reactions. Our work shows that this notion can now be extended to bacterial WXXXE GEFs.
Conclusions
We report the first crystal structure of a free WXXXE family member. The structure of IpgB2 from S. flexneri combined with its complexes with RhoA allows a structural description of the sequence of events accompanying GDP release induced by this bacterial GEF, which is an essential prerequisite for GTP loading. A function as a GTPase mimicry protein, as proposed previously for IpgB2, can now be clearly ruled out. In addition, our data show the formation of an intermediate state with a magnesium ion relocalized from the β-phosphate to the α-phosphate of GDP. This way, the phosphate charge gets compensated without interference with a low GDP-affinity state, induced by switch II blocking the Mg2+-binding site at the β-phosphate. It is plausible that the molecular mechanism of GDP release involves a simultaneous dissociation of GDP·Mg2+(α) from the GEF·GTPase complex. In such a scenario, stable association of a complex containing GDP but lacking Mg2+ is not required to explain the GEF-induced nucleotide release. The depicted mechanism might also be considered as a model for mammalian RhoA-GEFs, which have not been crystallized as GEF·RhoA·GDP complexes so far.
Supplementary Material
Acknowledgments
We thank Thorsten Lührs for performing NMR experiments on 15N- and 13C-labelled IpgB2, Manfred Nimtz for mass spectrometry, Christiane Ritter for technical support during the stopped flow experiments, and Joachim Reichelt and Kevin Walkling for technical assistance. We also thank Gunhild Layer (Technical University Braunschweig (TU)) for sharing equipment and Johannes Walther (TU) for technical support during high-performance liquid chromatography experiments. We express our gratitude to the staff of beamlines ID14-4, ID23-1, ID23-2 and ID29 of the European Synchrotron Radiation Facility, Grenoble, France, of BL14.1 and BL14.2, Berliner Elektronen-Speicherring Gesellschaft für Synchrotronstrahlung (BESY), Berlin, Germany, and of Deutsches Elektronen Synchrotron (DESY), Hamburg, Germany, for access to beam time and assistance during data collection.
This work was funded in part by the Deutsche Forschungsgemeinschaft (Grants SFB621 to K. R. and SPP1150 to T. E. B. S.).
The atomic coordinates and structure factors (codes 3LYQ, 3LW8, 3LWN, and 3LXR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- GEF
- guanine nucleotide exchange factor
- MBP
- maltose-binding protein
- PFA
- formaldehyde
- RBD
- Rho-binding domain
- P-loop
- phosphate-binding loop
- Sif
- Salmonella-induced filament
- DH/PH
- Dbl homologous/pleckstrin homologous fragment of PDZ-RhoGEF
- r.m.s.d.
- root mean square deviation
- ROCK
- RhoA-effector Rho kinase.
REFERENCES
- 1.Ladwein M., Rottner K. (2008) FEBS Lett. 582, 2066–2074 [DOI] [PubMed] [Google Scholar]
- 2.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 3.Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 4.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell. Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 5.Chrzanowska-Wodnicka M., Burridge K. (1996) J. Cell Biol. 133, 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo G., Letourneau P. C. (2004) J. Neurobiol. 58, 92–102 [DOI] [PubMed] [Google Scholar]
- 7.Nobes C. D., Hall A. (1995) Biochem. Soc. Trans. 23, 456–459 [DOI] [PubMed] [Google Scholar]
- 8.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 9.Aktories K., Barbieri J. T. (2005) Nat. Rev. Microbiol. 3, 397–410 [DOI] [PubMed] [Google Scholar]
- 10.Vogelsgesang M., Pautsch A., Aktories K. (2007) Naunyn Schmiedebergs Arch. Pharmacol. 374, 347–360 [DOI] [PubMed] [Google Scholar]
- 11.Ridley A. J., Hall A. (1992) Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 12.Galán J. E. (2009) Cell Host. Microbe 5, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rottner K., Stradal T. E., Wehland J. (2005) Dev. Cell 9, 3–17 [DOI] [PubMed] [Google Scholar]
- 14.Boquet P., Lemichez E. (2003) Trends Cell Biol. 13, 238–246 [DOI] [PubMed] [Google Scholar]
- 15.Buchwald G., Friebel A., Galán J. E., Hardt W. D., Wittinghofer A., Scheffzek K. (2002) EMBO J. 21, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., Mattoo S., McMahon S. A., Ghosh P., Hughes T. R., Boone C., Dixon J. E. (2006) Cell 124, 133–145 [DOI] [PubMed] [Google Scholar]
- 17.Hayward R. D., Koronakis V. (2006) Cell 124, 15–17 [DOI] [PubMed] [Google Scholar]
- 18.Handa Y., Suzuki M., Ohya K., Iwai H., Ishijima N., Koleske A. J., Fukui Y., Sasakawa C. (2007) Nat. Cell Biol. 9, 121–128 [DOI] [PubMed] [Google Scholar]
- 19.Ohya K., Handa Y., Ogawa M., Suzuki M., Sasakawa C. (2005) J. Biol. Chem. 280, 24022–24034 [DOI] [PubMed] [Google Scholar]
- 20.Arbeloa A., Blanco M., Moreira F. C., Bulgin R., Lüpez C., Dahbi G., Blanco J. E., Mora A., Alonso M. P., Mamani R. C., Gomes T. A., Blanco J., Frankel G. (2009) J. Med. Microbiol. 58, 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulgin R. R., Arbeloa A., Chung J. C., Frankel G. (2009) Cell Microbiol. 11, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arbeloa A., Garnett J., Lillington J., Bulgin R. R., Berger C., Lea S. M., Matthews S., Frankel G. (2009) Cell Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohlson M. B., Huang Z., Alto N. M., Blanc M. P., Dixon J. E., Chai J., Miller S. I. (2008) Cell Host. Microbe 4, 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Z., Sutton S. E., Wallenfang A. J., Orchard R. C., Wu X., Feng Y., Chai J., Alto N. M. (2009) Nat. Struct. Mol. Biol. 16, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stebbins C. E., Galán J. E. (2001) Nature 412, 701–705 [DOI] [PubMed] [Google Scholar]
- 26.Shames S. R., Auweter S. D., Finlay B. B. (2009) Int. J. Biochem. Cell Biol. 41, 380–389 [DOI] [PubMed] [Google Scholar]
- 27.Eberth A., Ahmadian M. R. (2009) Curr. Protoc. Cell Biol., Chapter 14, 14.9.1–14.9.25 [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson J. P., Eccleston J. F. (2000) Biochemistry 39, 11348–11359 [DOI] [PubMed] [Google Scholar]
- 29.Zhang B., Zhang Y., Wang Z., Zheng Y. (2000) J. Biol. Chem. 275, 25299–25307 [DOI] [PubMed] [Google Scholar]
- 30.Diacovich L., Dumont A., Lafitte D., Soprano E., Guilhon A. A., Bignon C., Gorvel J. P., Bourne Y., Méresse S. (2009) J. Biol. Chem. 284, 33151–33160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 32.Kabsch W. (1988) J. Appl. Crystallogr. 21, 916–924 [Google Scholar]
- 33.Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 34.Sheldrick G. M. (2008) Acta Crystallogr. Sect. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 35.Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) J. Synchrotron. Radiat. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 37.Dvorsky R., Blumenstein L., Vetter I. R., Ahmadian M. R. (2004) J. Biol. Chem. 279, 7098–7104 [DOI] [PubMed] [Google Scholar]
- 38.Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 42.Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 43.Rudolph M. G., Weise C., Mirold S., Hillenbrand B., Bader B., Wittinghofer A., Hardt W. D. (1999) J. Biol. Chem. 274, 30501–30509 [DOI] [PubMed] [Google Scholar]
- 44.Häusler L. C. (2004) Aktivierung von GTPasen der Rho-Familie. Ph.D. Thesis, Bochum, Ruhr-Universität Bochum [Google Scholar]
- 45.Cherfils J., Chardin P. (1999) Trends Biochem. Sci. 24, 306–311 [DOI] [PubMed] [Google Scholar]
- 46.Wei Y., Zhang Y., Derewenda U., Liu X., Minor W., Nakamoto R. K., Somlyo A. V., Somlyo A. P., Derewenda Z. S. (1997) Nat. Struct. Biol. 4, 699–703 [DOI] [PubMed] [Google Scholar]
- 47.Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., Sondek J. (2002) Nat. Struct. Biol. 9, 468–475 [DOI] [PubMed] [Google Scholar]
- 48.Freymann D. M., Keenan R. J., Stroud R. M., Walter P. (1999) Nat. Struct. Biol. 6, 793–801 [DOI] [PubMed] [Google Scholar]
- 49.Shimizu T., Ihara K., Maesaki R., Kuroda S., Kaibuchi K., Hakoshima T. (2000) J. Biol. Chem. 275, 18311–18317 [DOI] [PubMed] [Google Scholar]
- 50.Thomas C., Fricke I., Scrima A., Berken A., Wittinghofer A. (2007) Mol. Cell 25, 141–149 [DOI] [PubMed] [Google Scholar]
- 51.Arbeloa A., Bulgin R. R., MacKenzie G., Shaw R. K., Pallen M. J., Crepin V. F., Berger C. N., Frankel G. (2008) Cell Microbiol. 10, 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. (2004) Structure 12, 1955–1965 [DOI] [PubMed] [Google Scholar]
- 53.Kristelly R., Gao G., Tesmer J. J. (2004) J. Biol. Chem. 279, 47352–47362 [DOI] [PubMed] [Google Scholar]
- 54.Worthylake D. K., Rossman K. L., Sondek J. (2000) Nature 408, 682–688 [DOI] [PubMed] [Google Scholar]
- 55.Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D. (2009) Science 325, 1398–1402 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.