Abstract
Human African trypanosomiasis, caused by the eukaryotic parasite Trypanosoma brucei, is a serious health problem in much of central Africa. The only validated molecular target for treatment of human African trypanosomiasis is ornithine decarboxylase (ODC), which catalyzes the first step in polyamine metabolism. Here, we describe the use of an enzymatic high throughput screen of 316,114 unique molecules to identify potent and selective inhibitors of ODC. This screen identified four novel families of ODC inhibitors, including the first inhibitors selective for the parasitic enzyme. These compounds display unique binding modes, suggesting the presence of allosteric regulatory sites on the enzyme. Docking of a subset of these inhibitors, coupled with mutagenesis, also supports the existence of these allosteric sites.
Keywords: Cell/Division, Diseases, Enzymes/Decarboxylase, Enzymes/Inhibitors, Metabolism/Regulation, Organisms/Parasite, Polyamines, Ornithine Decarboxylase
Introduction
Human African trypanosomiasis, caused by the eukaryotic parasite Trypanosoma brucei, is a most neglected disease in central Africa, with at least 50,000 active cases and 17,000 new cases each year (1). The disease is fatal if left untreated (2). Current treatments require prolonged drug regimens with drugs that have unacceptable toxic side effects (3). Additionally, with one exception, current drugs have poorly understood modes of action (3). The only clinically validated molecular target for treatment of human African trypanosomiasis is ornithine decarboxylase (ODC),3 which catalyzes the decarboxylation of ornithine to produce putrescine, the first step in polyamine metabolism (Fig. 1). The polyamines putrescine, spermidine, and spermine are known to be necessary for cellular replication (4, 5). Increases in polyamine concentration have also been linked to carcinogenesis. Therefore, inhibitors of the polyamine biosynthetic pathway have been extensively investigated as potential chemotherapeutic and chemopreventative compounds (reviewed in Ref. 6).
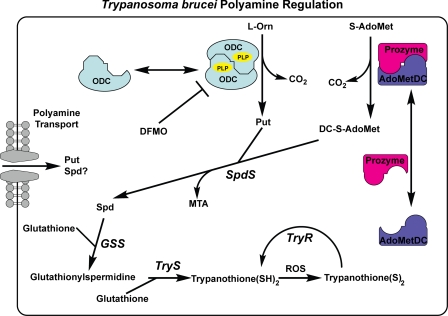
FIGURE 1.
Schematic of the polyamine biosynthetic pathway in T. brucei. The following abbreviations are used: S-adenosylmethionine decarboxylase (SAMDC), S-adenosylmethionine (S-AdoMet), 5′-methylthioadenosine (MTA), decarboxylated S-adenosylmethionine (DC-S-AdoMet), putrescine (Put), spermidine (Spd), spermidine synthase (SpdS), DFMO, glutathionyl spermidine synthase (GSS), trypanothione synthase (TryS), trypanothione reductase (TryR), and reactive oxygen species (ROS).
In mammalian systems, ODC is tightly controlled via transcriptional, translational, and post-translational mechanisms (6–8). It is active as a homodimer and is dependent upon binding to pyridoxal 5′-phosphate (PLP), a cofactor shared with many other enzymes (9). The mammalian ODC protein has one of the shortest known half-lives (∼10–20 min), which is primarily regulated by antizyme, a polyamine-inducible protein inhibitor that binds to ODC monomers and targets ODC for degradation (10). The levels of the polyamines are further regulated by inter-conversion of individual pools and by a highly efficient transport system allowing import and export of polyamines and intermediates (11). This highly redundant regulation means that mammalian cells are strongly resistant to changes in polyamine levels. The most widely used inhibitor of mammalian ODC is α-difluoromethylornithine (DFMO), a highly selective compound that alkylates Cys-360, a catalytic residue in the ODC active site (12, 13). DFMO is orally available but rapidly cleared (t½ of 1.5 h (intravenous dosing) to 4 h (oral dosing)) (14). It is relatively nontoxic and can be dosed to extremely high levels (up to 3.75 g/m2) with only minor side effects (15). Despite this, DFMO has largely been abandoned as a chemotherapeutic due to poor efficacy, which has been attributed to the robustness of the mammalian polyamine pool. Recently, interest has risen in the use of DFMO as a chemopreventative in combination with other agents (16–18).
In T. brucei, polyamine biosynthesis is much simpler (Fig. 1). There are no inter-conversion pathways, and transport of exogenous polyamines plays a lesser role (19). In addition, T. brucei ODC is much longer lived than its mammalian counterpart, and levels are not actively regulated (20, 21). The pathway is regulated in T. brucei apparently via activation of S-adenosylmethionine decarboxylase by heterodimer formation with a catalytically inactive homolog termed prozyme (22). Prozyme expression is in turn regulated in response to changes in polyamine levels (23). Depletion of polyamines either by inhibition of the biosynthetic enzymes or by gene knockdown leads to reduced trypanothione levels and to cell death (19, 23, 24). DFMO is clinically approved for the treatment of human African trypanosomiasis, and a new combination with nifurtimox is now the recommended frontline therapy for late stage Trypanosoma brucei gambiense (25). The mechanism of action of DFMO has been demonstrated to be the inhibition of polyamine biosynthesis (3). However, the poor pharmacokinetic behavior of DFMO is a major limiting factor in its use (26). Furthermore, DFMO is only effective against one of two subspecies of T. brucei causing human disease (27). As the parasite cannot increase ODC levels in response to polyamine depletion, the development of potent reversible inhibitors may allow inhibition of ODC to be fruitful in both subspecies (19).
To date, most discovery efforts directed toward ODC have been focused on analogs of ornithine (such as DFMO), putrescine, or PLP (28). None of these has proved as effective as DFMO for treatment of T. brucei infections. No large scale efforts to discover novel inhibitors have been reported. The lack of prior high throughput screening efforts directed at ODC is due in part to the difficulty in assaying its activity. Classical assay methods utilize capture of 14CO2 from radiolabeled ornithine or derivatization of putrescine followed by high pressure liquid chromatography analysis (29, 30). Neither of these techniques is tenable for a large scale screening effort.
Therefore, we have optimized an enzyme-linked assay suitable for high throughput screening of ODC. This assay links the production of CO2 to the consumption of NADH using phosphoenolpyruvate carboxylase and malate dehydrogenase (31). The CO2 produced by ODC is captured as bicarbonate by the basic reaction buffer and then used by phosphoenolpyruvate carboxylase to carboxylate phosphoenolpyruvate, generating oxaloacetate. This is then reduced to malate by malate dehydrogenase in an NADH-dependent fashion allowing the reaction catalyzed by ODC to be monitored by measuring the decrease in the absorbance of NADH. The optimization of this technique for high throughput applications will be reported elsewhere. Herein, we report the use of this method to identify active compounds from a library of 316,114 unique compounds. This effort led to the discovery of four novel inhibitory chemotypes possessing previously uncharacterized modes of inhibition. A subset of the inhibitors appeared to bind to a novel site that was characterized by molecular docking and mutagenesis techniques.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were used as purchased from their vendors. Deionized water was filtered with a MilliQ Synthesis Ultra-Pure water system (Millipore, Billerica, MA) immediately before use. InfinityTM carbon dioxide liquid stable reagent was purchased from Thermo Fisher Scientific (Waltham, MA). l-Ornithine, PLP, and dithiothreitol (DTT) were purchased from Sigma. DFMO was purchased from Chem-Impex International (Wood Dale, IL). All plate-based enzymatic assays were performed in 384-well black-sided, clear-bottomed polystyrene microtiter plates (catalog no. 3702) from Corning Life Sciences (Acton, MA).
Screening Library
The compound library at St. Jude Children's Research Hospital was assembled from commercially available collections, including the following: Prestwick Chemical Library (Prestwick Chemical, Illkirch, France); the LOPAC Collection (Sigma); the Spectrum Collection, NINDS Collection (National Institutes of Health); Natural Product Collection and Killer Plate Collection (Microsource Discovery Systems, Gaylordsville, CT); Chemical Diversity (San Diego); ChemBridge (San Diego); Life Chemicals (Burlington, Ontario, Canada); and Tripos (St. Louis, MO). The library was constructed by filtering available compounds using a combination of physiochemical metrics chosen to improve bioavailability and functional group metrics chosen to reduce the likelihood of nonspecific or artifactual hits (32, 33). The filtered compound list was used to generate maximally diverse clusters by reducing the compounds to core fragments using the method of Bemis and Murcko (34). The clusters were prioritized based on the diversity of the existing library. Five to 20 compounds are required per cluster. Clusters of more than 20 available compounds were preferred, with a maximum of 20 compounds being purchased from within each cluster.
Purification of T. brucei ODC (TbODC) and Human ODC (hODC)
TbODC and hODC were expressed as N-terminal His6 tag fusion proteins in Escherichia coli BL21(DE3) cells as described previously (31). Protein was purified by Ni2+-nitrilotriacetic acid-agarose column followed by Superdex 200 gel filtration column chromatography. Fractions containing the desired protein were identified by SDS-PAGE. Those containing ODC were combined and concentrated using an Amicon ultracentrifugal filter device (10-kDa cutoff, Millipore, UFC901024) to concentrations of ∼40 mg/ml. Yields of purified TbODC were generally 7–13 mg/liter of cultured cells. Protein concentration was determined by Bradford assay. Yields of purified hODC were ∼2–5 mg/liter of cultured cells.
Site-directed Mutagenesis of TbODC
The S367A and S420A TbODC mutants were produced using the pODC29 plasmid that encodes the wild-type TbODC with the QuikChangeTM mutagenesis kit (Stratagene, La Jolla, CA using the following forward primers: 5′-GTCGTAGGAACTTCTGCCTTTAATGGATTCCAG-3′ and 5′-CCTTTAATGGATTCCAGGCTCCGACTATTTACTATG-3′ for S396A and S402A, respectively (desired mutations in boldface type). The TbODC D364E mutant was generated using the standard Kunkel technique in the Bluescript vector (Stratagene, La Jolla, CA) using the M13 helper phage (Stratagene, La Jolla, CA) and the Kunkel strain BO265 (35). The primer used for this mutant was 5′-ATGTGATGGGCTCGAGCAGATAG.
Assay Automation
All screening data were generated on a High Resolution Engineering (Woburn, MA) integrated screening system using Liconic plate incubators (Woburn, MA) and a Stabuli T60 robotic arm (Stabuli, SC). All automated screening was performed under a nitrogen atmosphere as described previously (36). Assay solutions were dispensed using Matrix Wellmates (Matrix Technologies, NH) equipped with 1-μl rated tubes. Plates were centrifuged after all bulk liquid additions using a Vspin plate centrifuge (Velocity11, Menlo Park, CA). Compound transfers were performed using a 384-well pin tool equipped with 10-nl slotted hydrophobic surface-coated pins (V&P Scientific, San Diego, CA). This allowed delivery of ∼25 nl of DMSO stock solution with coefficients of variation of less than 10%. All absorbance data were measured using an EnVision Multilabel Plate Reader equipped with a 340 nm narrow bandwidth filter (PerkinElmer Life Sciences, 2100-5740).
ODC-Phosphoenolpyruvate Carboxylase-Malate Dehydrogenase-linked Assay
This assay was performed under nitrogen atmosphere as described previously (36). Assay buffer (66 mm Tris, 25 mm NaCl, 8 mm MgSO4, 0.01% Triton X-100, pH 8.05) was prepared daily in water. Plates and compounds were allowed to equilibrate in the presence of ODC for 20 min before l-Orn was added to start the reaction. Final primary screening assay conditions were 2.3 mm DTT, 60 μm PLP, 625 μm l-Orn, 150 nm TbODC, 10 μm test compound, and 60% InfinityTM CO2 (v/v) in assay buffer with a final volume of 25 μl. Reaction progress was monitored by decrease in absorbance at 340 nm using an Envision plate reader (PerkinElmer Life Sciences) equipped with a narrow bandwidth 340 nm filter (PerkinElmer Life Sciences, 2100-5740).
Compounds for screening were placed in 384-well polypropylene plates (Corning Life Sciences, Acton, MA) at 10 mm concentrations in DMSO. Sixteen positive controls (DFMO, 1 m in DMSO) and 16 negative controls (DMSO) were placed in a single separate 384-well polypropylene plate and pin-transferred to test plates after the addition of variable compounds.
Cuvette assays for low throughput re-testing were performed as described above with the following minor modifications: the final assay volume in cuvettes was 500 μl at 40% InfinityTM carbon dioxide liquid stable reagent, 50 μm PLP, 50 μm DTT, 1% DMSO, and varied ornithine concentrations from 10 mm to 100 μm. As with microplate assays, assay buffer (66 mm Tris, 25 mm NaCl, 8 mm MgSO4, 0.01% Triton-X, pH 8.05) was prepared fresh daily. Cuvette assays were performed under normal atmosphere at 37 °C.
Primary Screening Data Analysis and Reaction Rate Calculation
Primary screening data analysis was performed using custom protocols (RISE 3.0) written in Pipeline Pilot (version 7.0, Accelrys) and the R program (version 2.5.0) (37). Kinetic data from the full 6-min observation (6 points in total) were fit to a linear model using a robust, iteratively re-weighted least squares algorithm (“lmrob” function in robustbase R package, version 0.2-7) that reduces the influence of outliers compared with classical least squares (38). The slope values from kinetic data fits were taken as end points that were then used for the calculations of all other plate statistics. Only plates passing the minimum Z-prime and Z-factor thresholds of 0.4 and 0.4, respectively, were accepted. Initial screening hits were determined on a plate-by-plate basis by identifying compounds with activities that were simultaneously outliers from the negative control and variable compound populations. The outlier cutoffs were calculated as the upper fourth plus 1.5 times the fourth spread (the upper fourth and fourth spread are similar to the third quartile and interquartile range, respectively), which corresponds to a p value ∼0.005 for normal distributions. However, such cutoff criteria are more robust to population deviations from normality (39). For calculations of Z-scores, 16 positive and 16 negative controls were used unless otherwise stated. Z-scores were calculated as described previously (40). Reaction rate was changed from absorbance units/min to millimolar NADH/min using an extinction coefficient of 6.349 absorbance units mm−1 cm−1 for NADH and an approximate path length of 0.4 cm for assays performed at 25 μl of final volume in a 384-well plate. Dose-response data were fit using a nonlinear regression to a four-parameter sigmoidal curve model, resulting in fit values for maximum and minimum responses, Hill slope, and EC50.
Phosphoenolpyruvate Carboxylase-Malate Dehydrogenase-linked Assay
For assay of the linking enzymes, assay buffer (66 mm Tris, 25 mm NaCl, 8 mm MgSO4, 0.01% Triton X-100, pH 8.05) was prepared daily using water. Compounds were allowed to equilibrate in the presence of enzymes for 20 min before substrate was added.
Final assay concentrations were 2.3 mm DTT, 60 μm PLP, 0.75 mm sodium bicarbonate, 60% InfinityTM carbon dioxide liquid stable reagent, and 0.01% Triton X-100. Reaction progress was monitored by decrease in absorbance at 340 nm using an Envision plate reader (PerkinElmer Life Sciences) equipped with a narrow bandwidth 340 nm filter (PerkinElmer Life Sciences, 2100-5740) for 10 min with time points taken every minute. Data from minutes 2–7 were fit to a linear model using statistical methods as described below.
Radiolabeled Ornithine ODC Assay
14CO2 released from l-[1-14C]ornithine by the decarboxylation activity of TbODC was directly measured as described previously (41, 42) at pH 7.5 at 37 °C in the absence or presence of inhibitors. IC50 values were determined in an 8-point dose-response curve performed in singlicate. Data were fit to a simple IC50 model as described previously (43).
Reductive Assay
Reductive activity of all hits was determined using a high throughput assay based on the detection of molecules capable of reducing resazurin (catalog no. R7017, Sigma) to resorufin as described previously (44). Briefly, 25 μl of assay solution (5 μm resazurin, 50 mm HEPES, 50 mm NaCl, pH 7.5, 50 μm DTT, prepared immediately prior to usage) was added to a black 384-well polystyrene plate (catalog no. 3573, Corning, Lowell, MA) using a Matrix WellMate (Thermo Fisher, Hudson, NH). Test compounds were added by pin transfer using 10-nl hydrophobic coated pins (FP1CS10H, V&P Scientific, Inc., San Diego) to a final concentration of 10 μm. Test plates were incubated in the dark at room temperature for 60 min, and fluorescence intensity was read on an Envision (PerkinElmer Life Sciences) with excitation = 560 nm and emission = 590 nm. Each compound was read in quadruplicate, and the signals were averaged to generate relative activity levels. Compounds that differed significantly from DMSO levels as determined by use of in-house statistical software (RISE 3.0) were scored as being potentially redox-active.
Reversibility Assay
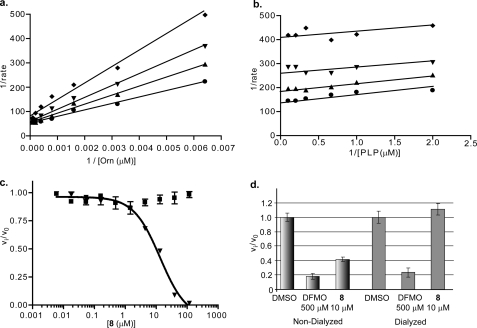
Reversibility assays were performed by dialysis. ODC (1500 nm) was incubated with inhibitors at 10× IC50 concentrations for 40 min in assay buffer. Samples were then dialyzed in assay buffer overnight using Mr 3000 cutoff Slide-A-Lyzer Mini dialysis units (Pierce) and assayed at 600 μm Orn, 60 μm PLP, and 150 nm ODC. Percent activity recovered was defined as the difference between the ratio of the dialyzed rate to the uninhibited rate and the ratio of the inhibited (10× IC50) rate to the uninhibited rate. Compounds were labeled reversible if >90% activity was recovered.
Kinetic Analysis and Dose-response Analysis
Mode of inhibition for ornithine was determined by monitoring the reaction rate in the presence of increasing substrate concentration (0–10 mm Orn in the presence of 60 μm PLP) with varying concentrations of inhibitors (0, IC50, 3× IC50, and 4× IC50). For PLP mode of inhibition studies, ornithine concentration was held constant at 600 μm, whereas PLP was varied from 0 to 600 μm. All kinetic data were gathered from experiments performed in 384-well format, as described above. The data were used in Lineweaver-Burk analysis followed by fitting the data to the appropriate Michaelis-Menten inhibition equation (competitive, mixed competition with varied α values, noncompetitive, or uncompetitive) for determination of Ki values (45). In specific cases reported in this study, the mode of inhibition and the Ki values were confirmed by analysis using the cuvette-based assay. Km value for ornithine was calculated by the fitting data to the Michaelis-Menten equation with variable Km and Vmax values. Data were fit using GraphPad Prism 4.03 (GraphPad Software, La Jolla, CA).
Reaction rates for dose-response data were calculated as described below. Rates were then normalized to DMSO and DFMO (1 mm) controls, and sigmoidal curves with variable slopes (four-parameter fit) were fit using GraphPad Prism 4.03 or custom protocols in Pipeline Pilot (version 7.0, Accelrys).
Computational Chemistry and Molecular Modeling
Structural data used in modeling experiments were taken from the Protein Data bank (1QU4, apo-TbODC; 1D7K, apo-hODC) (13, 46). All molecular modeling was performed using molecular operating environment (version 2007.09, Chemical Computing Group, Inc.). Site Finder, a geometric method for finding potential binding pockets similar to LigSite (47), was used to identify potential binding sites. The analysis was run with 1.4 and 1.8 Å probe radii for hydrophobic and hydrophilic probes, respectively. A minimum site size of 25 α-spheres was used, and a minimum bounding sphere radius of 2.5 Å was used to eliminate smaller pockets unlikely to bind with drug-like molecules. Dummy atoms were placed at the center of the α-spheres generated by Site Finder and used as superposition targets for docking calculations.
Docking studies were also performed using molecular operating environment (version 2007.09). Docking was performed using rigid ligand and receptors, using the Alpha Triangle placement method with 80,000 maximum generations for each ligand conformation. Scoring was done using the London dG scoring function. This is a five-parameter function taking into account rotational and translational entropy, ligand flexibility, hydrogen bonding, metal ligations, and desolvation energies. The predefined ligand conformer library was generated using the systematic conformational search function in molecular operating environment with a 10 kcal/mol cutoff. Results were visualized in PyMOL (version 0.99, Delano Scientific LLC), and receiver-operator curve scores were generated in Pipeline Pilot (version 7.0, Accelrys).
RESULTS
Primary Screen Results
For the purposes of identifying novel inhibitors of TbODC, a collection of 316,114 unique molecules was tested at St. Jude Children's Research Hospital. The compounds were screened at 10 μm against the TbODC-phosphoenolpyruvate carboxylase-malate dehydrogenase-linked enzyme system. Plates with Z′ values of less than 0.4 were rejected for the purposes of identifying hits. This somewhat liberal data quality cutoff was chosen to maximize the number of hits chosen for further examination, because preliminary examination of the data showed that the hit rate was quite low. After filtering for poor Z′, 625 plates, with an average Z′ of 0.52 and an average Z-factor of 0.47, containing a total of 240,000 unique compounds remained. These plates were used for selecting hits. Hits were picked using robust methods corresponding roughly to using a p value cutoff of 0.005, resulting in 883 primary hits (0.3% hit rate) (39). No minimum activity was required for characterization as a “hit.” This technique is more completely described under “Experimental Procedures.”
To further characterize the hits, samples of each unique compound were cherry-picked and subjected to full dose-response studies using a dilution series of 10 points in a 1:3 dilution steps (top = 100 μm). Of the initial 883 compounds picked, 189 displayed a saturating dose response in the ODC-phosphoenolpyruvate carboxylase-malate dehydrogenase system, whereas 310 displayed partial dose responses. Upon retesting, 384 of the initially identified hits were inactive, giving a validated hit rate of 43.4%. None of the compounds detectably affected the phosphoenolpyruvate carboxylase-malate dehydrogenase system at concentrations up to 100 μm concentrations. Comparison of inhibited reaction rates at both 150 and 300 nm ODC showed that all compounds were acting via inhibition of ODC and that ODC inhibition was the rate-limiting step in the coupled assay system. The remaining validated hits were filtered to remove those containing potentially undesirable chemical moieties, such as highly reactive groups or metal-containing compounds, and checked for commercial availability, leaving 179 commercially available validated hits. These validated hits were re-ordered from their suppliers. At the same time, expanded sets were designed, centered on promising compound series, and sourced from the same vendors. This produced a total of 260 compounds that were subjected to the secondary assay panel.
Secondary Testing
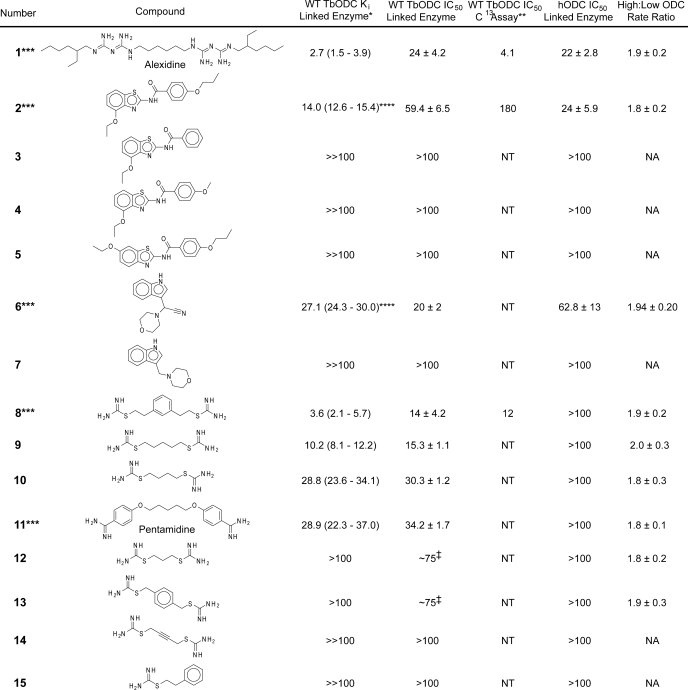
The identity and purity of all 260 re-ordered compounds were validated using ultra high pressure liquid chromatography-mass spectrometry (48). The average purity was 79%, and any compound less than 90% pure was rejected from further analysis. Seventy five of the compounds failed purity or identity checks. Stock solutions were prepared at a putative concentration of 10 mm in DMSO, and the concentrations were confirmed using chemiluminescent nitrogen detection (49). The remaining compounds were re-tested in the primary assay to validate their activities. Of the 185 pure compounds tested, 76 showed activity in the primary assay. At this point, the compounds were also screened versus hODC and at high (300 nm) concentrations of TbODC to determine selectivity and verify that ODC inhibition was the rate-limiting step in the system. Inhibition of TbODC was determined to be rate-limiting in all cases. The compounds were also screened for redox activity to eliminate nonspecific radical based inhibitors (44). A representative compound from each scaffold series was also tested using the radiolabeled ornithine assay to confirm inhibition of TbODC in an orthogonal assay system. The representative compound was also tested for reversibility by dialysis. Of the 76 compounds active in the primary assay, only 7 were acceptably active in all assays. The results of these are summarized in Table 1. The active compounds represented four scaffold classes. Following confirmation of activity in this fashion, the compounds were screened at varying l-Orn concentrations (0.31, 1.3, 2.5, 5.0, and 10 mm) as well as varying PLP concentrations (0.5, 1.5, 1, 3, 5, and 10 μm) to determine the mode of inhibition and Ki values. These compounds, spanning four scaffold series, along with small structure-activity groups are discussed below.
TABLE 1.
Inhibitors of TbODC
All values are in micromolar unless otherwise noted.
* Ki values were determined by global fitting of raw rate data to appropriate Michaelis-Menten equations and are expressed as fit value followed by 95% confidence limits. Ki values were determined using data from three separate triplicate experiments.
** IC50 values were determined at 37 °C at 400 μm l-Orn as described under “Experimental Procedures.” Data were fit to simple IC50 models.
*** Primary HTS hit compound is shown. All other compounds were part of reordered analog series.
**** αKi value from uncompetitive inhibition model is shown.
‡ Single point is active at 100 μm. All IC50 values were determined at isokinetic conditions (1.5× Km l-Orn, 60 μm PLP) as described under “Experimental Procedures.” IC50 values are expressed as the mean of three measurements taken in triplicate ± S.D. ODC concentrations were 150 nm for all experiments. High and low ODC rates were determined at 150 and 300 nm ODC, respectively, in the presence of 625 μm l-Orn. NT indicates not tested; NA indicates not active.
Novel Inhibitors of ODC
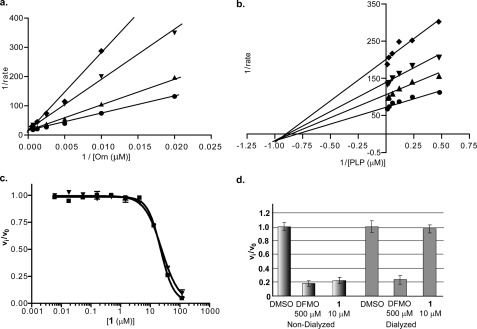
The first well behaved inhibitor discovered during the course of this effort (compound 1, Table 1, and Fig. 2) was a bisbiguanide compound. This compound was quite potent, with a Ki value of 2.7 μm, and displayed competitive inhibition with respect to ornithine but noncompetitive inhibition with respect to PLP. However, despite its resemblance to the dithioamidines, it was not a selective inhibitor of TbODC, inhibiting hODC with a similar potency. This, as well as the difference in the mode of inhibition with respect to PLP, suggests that it has a different binding mode than the dithioamidines. This compound was reversible and inactive in the redox assay. No close analogs were available for structure-activity relationship studies.
FIGURE 2.
Bisbiguanide inhibitor data. a and b, Lineweaver-Burk plots for ornithine versus compound 1 and PLP versus 1. ♦ = 10 μm, ▾ = 6 μm, ▴ = 4 μm, and ● = uninhibited. c, species selectivity analysis for compound 1. Data for TbODC (▾) and hODC (■) enzyme-linked assays were collected at 22 °C in 384-well plates. Data were fitted to a four-parameter sigmoidal dose response for determination of IC50 values. All data were collected under isokinetic conditions at 1.5× Km for ornithine. d, reversibility of compound 1. TbODC was incubated with inhibitor for 1 h followed by overnight dialysis into assay buffer. Data were collected as described under “Experimental Procedures.”
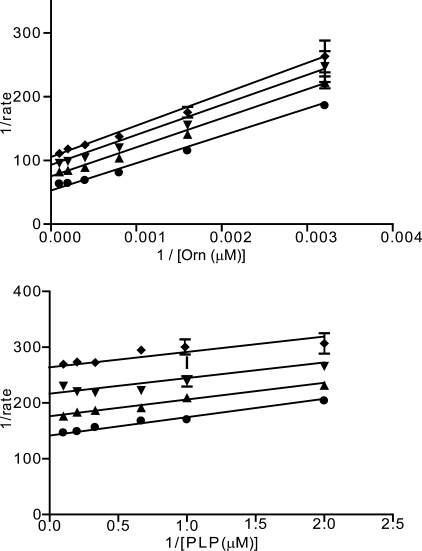
The second and third chemotypes were the benzthiazoles (compounds 2 to 5, Table 1, and Fig. 3) and the indoles (compounds 5 and 6, Table 1, and Fig. 4) that were moderately potent, with Ki values of 14.0 and 27.1 μm, respectively. Both chemotypes were nonselective inhibitors, inhibiting both hODC and TbODC. Both chemotypes displayed uncompetitive inhibition with respect to PLP. The benzthiazoles were uncompetitive with respect to ornithine, whereas the indoles were noncompetitive. Both chemotypes were also reversible inhibitors of TbODC. Although there was only a single commercially available active member in each of these re-ordered series, the preliminary structure-activity relationships from the screening collection suggest a specific binding interaction is responsible for the inhibition. This is particularly true for the benzthiazoles, which were heavily represented in the compound library. After compound 2 was verified as a promising compound, all compounds containing the benzthiazole core were cherry-picked from our screening collection and subjected to full dose-response analysis. Although the small amounts of available compound prevented determination of Ki values for these compounds, the IC50 values at 625 μm Orn, 60 μm PLP, and 150 nm TbODC suggest that very little modification to the scaffold is acceptable. The structures and IC50 values for these compounds can be found in the supplemental material. To maintain activity, the 4-position ethoxy group must be present. Moving this group to the 6-position on the benzthiazole ring or substitution with a methoxy group eliminates activity. Furthermore, the p-propoxy group on the phenyl amide moiety must also be preserved. This group can be substituted with a p-isopropoxyl moiety with a similar potency being maintained, but all other substitutions on this ring were inactive. In the case of the indole compounds, only one close analog could be obtained for testing. This compound, SJ000360927, showed that the nitrile group was necessary for activity.
FIGURE 3.
Benzthiazole inhibitor data. Selectivity curves and reversibility data for these compounds can be found in the supplemental material. Lineweaver-Burk plot for ornithine versus compound 2 and PLP versus 2. ♦ = 28 μm, ▾ = 9.5 μm, ▴ = 3.2 μm, and ● = uninhibited.
FIGURE 4.
Indole inhibitor data. Selectivity curves and reversibility data for these compounds can be found in the supplemental material. Lineweaver-Burk plot for ornithine versus compound 6 and PLP versus 6. ♦ = 40 μm, ▾ = 13 μm, ▴ = 4.4 μm, and ● = uninhibited.
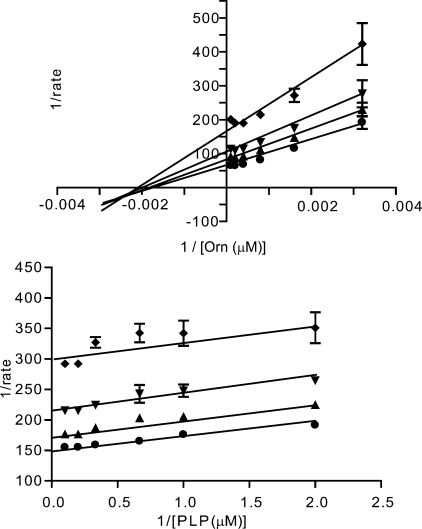
The final novel chemotype discovered was the dithioamidines (compounds 8 to 15, Table 1, and Fig. 5). These compounds were both selective and potent inhibitors of TbODC, with the most potent showing a Ki of 3.6 μm. Importantly, this compound has no detectable effect on hODC up to 100 μm. The compounds exhibit competitive inhibition with respect to ornithine and uncompetitive inhibition with respect to PLP. They also displayed reversible inhibition. A reasonable preliminary structure-activity relationship exists in the series. To be active, these compounds must contain two thioamidine moieties connected by a flexible linker of at least four atoms. Longer linkers were tolerated, with no upper limit on linker length being detected within the test set. These compounds were not active in the redox assay and are among the most potent reversible inhibitors of TbODC known. Interestingly, the anti-trypanosomal drug pentamidine (11) was also part of this inhibitor group, but it does not appear to act by this mechanism in whole cells.
FIGURE 5.
Dithioamidine inhibitor data. a and b, Lineweaver-Burk plots for ornithine versus compound 8 and PLP versus 8. ♦ = 40 μm, ▾ = 13 μm, ▴ = 4.4 μm, and ● = uninhibited. c, species selectivity analysis for compound 8. Data for TbODC (▾) and hODC (■) enzyme-linked assays were collected at 22 °C in 384-well plates. Data were fitted to a four-parameter sigmoidal dose response for determination of IC50 values. All data were collected under isokinetic conditions at 1.5× Km for ornithine. d, reversibility of compound 8. TbODC was incubated with inhibitor for 1 h followed by overnight dialysis into assay buffer. Data were collected as described under “Experimental Procedures.”
Identification of Potential Inhibitor Binding Sites on TbODC
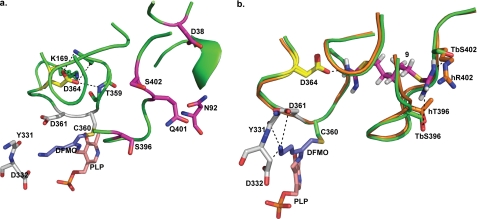
Because the active sites of TbODC and hODC are highly homologous, it seemed unlikely that the TbODC selective dithioamidines were binding at the active site, despite the fact that they were competitive with ornithine (21, 50). Therefore, an effort was made to identify other potential binding sites on the surface of the TbODC homodimer. Site Finder was used to identify three major possible binding sites. The first of these (site 1) is a pocket just below the active site bounded by Arg-337, His-333, Gly-201, and Pro-245. The second (site 2) is a groove at the dimer interface just above the active site bounded by Lys-173, Val-168, Pro-297, and Phe-179. The final site (site 3) is a small, relatively deep pocket above and behind the active site bound by Asp-364, Ser-396, and Ser-402, with a possible entrance to the protein hydrophobic core defined by Asn-92, Asp-38, Gln-401, and Glu-36. After computational identification, each site was utilized for DOCKing using a pre-generated conformer library containing all active dithioamidines as well as inactive analogs. Sites 1 and 2 were both predicted to be poor binding sites in comparison with the active site for all active compounds in the test set. However, site 3 was predicted to be a better binding site for the dithioamidines than the active site. Furthermore, site 3 yielded better discrimination between active and inactive dithioamidines than did any other site, with a receiver-operator characteristic area under curve score of 0.99 versus scores of ∼0.91 at the three other sites. The receiver-operator characteristic area under curve score is the probability that the assay will rank a randomly chosen true positive ahead of a randomly chosen true negative. A score of >0.5 in this metric represents discriminatory ability. Binding at site 3 would also explain the selectivity seen with this scaffold series. Residues at both possible entrances to the site are unconserved between TbODC and hODC. The apo structure of this site and the top docked pose of the dithioamidine 9 at this site are pictured in Fig. 6, a and b, respectively. Enrichment plots (ROC plots) of the docking results for the three alternate sites, as well as the active site, can be found in supplemental material.
FIGURE 6.
Proposed dithioamidine-binding site. a, TbODC bound to DFMO. DFMO is in purple; PLP is salmon; active site residues are white, and residues that form the proposed binding site are indicated as follows: Asp-364 is colored yellow and forms the bottom of the binding site, and magenta residues form the two possible entrances to the proposed dithioamidine-binding site. b, compound 9 docked into the proposed binding site on TbODC and superimposed with the apo human ODC structure. TbODC residues are orange, and hODC residues are light blue. Asp-364 is yellow, and active site residues are white, DFMO is purple, and PLP is salmon. Note that Ser-402 from TbODC is not conserved in hODC, and that the opening to the proposed site is completely occluded in the human enzyme. This could help account for the remarkable selectivity seen with the dithioamidine series of inhibitors.
As the DOCKing studies suggested that site 3 might represent the bone fide site for the dithioamidenes, this site was examined in greater detail. The majority of the high scoring dock poses involved favorable interactions of one thioamidine moiety with Asp-364, a negatively charged residue at the bottom binding site. The second thioamidine moiety assumed placements allowing a wide variety of interactions with charged or polar groups in and around the binding pocket. Previous studies have shown that Asp-364 is critical to enzyme function and that substitution with alanine renders the enzyme inactive (51). To test our binding hypothesis, a more conservative mutant (D364E) was used to evaluate the role of the residue in the binding of the dithioamidine compounds. Although this mutant did have dramatically increased Km (60 versus 0.37 mm) and decreased kcat values (0.02 versus 7 s−1), some activity was still measurable. This mutant was then used to test compounds 1 and 8, two competitive inhibitors with similar Ki values emerging from our studies that possess theoretically different binding modes. The non-TbODC selective inhibitor 1 was able to inhibit 20 μm D364E TbODC with a Ki value of 10.5 ± 0.7 μm, whereas compound 8 had no detectable effect up to 100 μm, indicating that Asp-364 does play a role in the inhibitory activity of the dithioamidine compounds. The results of these assays, along with steady state kinetic data for all enzymes used in the comparison, are included in Table 2.
TABLE 2.
Steady state kinetic analysis of TbODC and hODC enzymes with inhibition data
All data were collected at 37 °C using the linked enzyme assay as described under “Experimental Procedures.” All IC50 values were determined at isokinetic conditions (400 μm l-Orn for WT-TbODC and S396A-TbODC, 60 mm l-Orn for D364E TbODC, 150 μm l-Orn for WT-hODC, and 60 μm PLP was used for all experiments). IC50 values were determined with the cuvette-based enzyme-linked assay using the same protocol as for the 14CO2 assay. ODC concentrations were 150 nm for all experiments except for D364E, in which 20 μm ODC was used to compensate for the low kcat of the mutant. Values in parentheses are Ki values in micromolars. Ki values were obtained as described under “Experimental Procedures.” A value of ≫100 indicates no detectable effect at the maximum concentration of 100 μm.
| ODC protein | Km (l-Orn) | kcat | kcat/Km (l-Orn) | Compound 1 IC50 | Compound 8 IC50 |
|---|---|---|---|---|---|
| mm | s−1 | μm | μm | ||
| Wild type TbODC | 0.37 ± 0.03 | 7.0 ± 0.2 | 18.9 | 5.1 ± 0.6 (2.7 ± 1.2) | 6.2 ± 0.8 (3.6 ± 1.5) |
| D364E TbODC | 55.2 ± 4.6 | 0.015 ± 0.001 | 0.00027 | 22.6 ± 2.6 (10.5 ± 0.7) | ≫100 (≫100) |
| S396A TbODC | 0.27 ± 0.03 | 10.0 ± 0.4 | 37.0 | 6.9 ± 1.1 | 12.9 ± 0.5 |
| Wild type hODC | 0.170 ± 0.015 | 5.3 ± 0.2 | 31.2 | 15.9 ± 3.8 | ≫100 |
Two other mutants, S402A and S396A, at one possible entrance to the binding site were also tested. The S402A mutant expressed as an insoluble aggregate, and S396A had no detectable effect on binding of compound 1 or 8. However, in light of the fact that many energetically close docked poses of compound 8 placed the second thioamidine moiety at the entrance defined by Asp-38, this is not unexpected.
To further evaluate the effect of the D364E mutation on the geometry of site 3, the residue was virtually modified using the apo structure 1QU4 as a template. After performing a rotomer search and minimizing all residues within 8 Å of the mutation using the AMBER99 force field, no large scale structural changes were seen. However, Glu-364 is able to move closer to the backbone amine of Thr-359 and assume a more optimal hydrogen bonding position (2.10 versus 2.44 Å). Furthermore, Glu-364 is able to move within hydrogen bonding distance of Lys-169, a residue located in a loop whose flexibility is thought to be important for proper functioning of the catalytic cycle (52). This movement also orients the carboxylate of the residue away from the thioamidine groups of the inhibitors, making the binding interaction less favorable.
DISCUSSION
This study was the first large scale screening effort to discover new chemotypes for ODC inhibition. Screening a large chemical library has led to the discovery of several potent and selective inhibitors, including the first known inhibitors that are selective for TbODC over the highly homologous hODC. We also report the first nonsubstrate, nonproduct-based inhibitors of ODC. The identification of pentamidine, a known weakly binding inhibitor of TbODC (Ki > 30 μm), as a hit compound in the primary screen shows that our assay was sensitive (53). This suggests that most reversible ODC inhibitors present in our screening library with Ki values below 30 μm are likely to have been identified.
The four classes of inhibitors identified in this screening effort represent novel chemotypes for ODC inhibition. They also possess novel modes of inhibition. The benzthiazoles, typified by compound 2, are the most potent ornithine uncompetitive inhibitors known for ODC and are also uncompetitive with respect to PLP. Although the exact binding modes of these compounds are unknown, it is unlikely to be at the active site, because addition of substrate increases the potency of the inhibitor. It is known that binding of ornithine stabilizes the dimerization of ODC (54). This suggests that the benzthiazoles bind to the ODC homodimer. Although other uncompetitive inhibitors of ODC have been characterized previously, their Ki values have been in the millimolar range (55), significantly weaker than compound 2, which possesses a Ki of 12.6 to 15.4 μm. Identification of the binding site for this inhibitor would allow further optimization with the potential for an improvement in binding affinity. Because the compound is nonselective for TbODC over hODC, it is also likely that the binding site is conserved between the human and trypanosomal enzymes.
The indole compound 6 is noncompetitive with respect to ornithine and uncompetitive with respect to PLP, suggesting a binding mode that differs from the benzthiazoles and is not at the active site. Because this compound is nonselective for TbODC versus hODC, the binding site must be conserved between the two enzymes. Although the presence of an electrophilic nitrile that is required for activity suggests the possibility of a covalent mechanism of action, the fact that inhibition is completely reversible indicates any modification is reversible. As with compound 2, the identification of this binding site would prove useful in the further development of novel ODC inhibitors for both TbODC and hODC.
Two classes of potent competitive inhibitors were also discovered during the screen. The first of these, exemplified by compound 1 (alexidine), is bisbiguanides. Reports of the antimicrobial activity of alexidine date back to the 1950s (56), and it has antifungal activity and has been assayed as a potential chemotherapeutic compound (57, 58). Alexidine represents an interesting molecule for use in characterizing ODC more fully. Although it is a competitive inhibitor, alexidine is too large to fit completely within the active site, suggesting that the binding mode is more than a simple interaction with a single active site. Prior studies have suggested that ODC is susceptible to allosteric modulation by Gly-418, another large, basic antimicrobial compound (59). However, Gly-418 binds very weakly (Ki of 3–8 mm), severely hampering mechanistic studies. Alexidine, with the much lower Ki of 3.8 μm, represents a much better opportunity to investigate the modulation of ODC through allosteric binding sites.
The final class of inhibitors discovered in this screening effort is the most interesting. The dithioamidine series and compound 8 in particular represent the first selective inhibitors for TbODC. Compound 8 is a known inhibitor of nitric-oxide synthase and is one of the most potent reversible inhibitors of ODC known, with a Ki of 3.6 μm. This compound is structurally significantly different from the simple putrescine and ornithine analogs previously reported as reversible inhibitors of ODC (28, 60), suggesting that either the active site of ODC tolerates larger compounds or that these compounds are not binding at the active site. Because compound 8 is not highly functionalized, it may be possible for medicinal chemistry efforts to improve its potency. This proposition is supported by the work of Tidwell and co-workers (61, 62), who have previously reported similar diamidine scaffolds as anti-trypanosomal compounds with unknown mechanism and have shown clear structure-activity relationships within these compound series. In addition, other diamidine compounds such as berenil have been shown to be effective inhibitors of S-adenosylmethionine decarboxylase, another enzyme in the polyamine biosynthetic pathway (53). This raises the possibility that diamidine or dithioamidine compounds could be developed that are able to inhibit both TbODC and S-adenosylmethionine decarboxylase, effectively shutting off both rate-limiting steps in the polyamine biosynthetic pathway.
The remarkable selectivity for TbODC versus hODC displayed by the dithioamidines is likely due to the binding of these compounds to a secondary site, located behind the active site and bounded by TbAsp-364, TbSer-396, and TbSer-402, with a second possible entrance defined by TbAsn-92, TbAsp-38, TbGln-401, and TbGlu-36. Binding at this site may explain selectivity, because the residues at the entrance to the site are not conserved between the trypanosomal and the human enzymes. In hODC, TbSer-402 is replaced by hArg-402, and TbAsn-92 is substituted by hLys-92. Examination of the crystal structure of the human enzyme (1D7K) shows these changes place greater positive charge density at the binding site entrances and, in the case of the TbSer-402 entrance, completely occlude access to the binding pocket. Both molecular modeling experiments and mutagenesis data support the idea that TbAsp-364, a residue known to be important for ODC catalytic activity, is important for the inhibitory properties of the dithioamidines. However, crystallographic data will be necessary to confirm this hypothesis and establish the exact binding modes of the inhibitors.
Asp-364 is conserved across a wide range of eukaryotic ODC enzymes, indicating an important role for maintaining enzymatic functionality (51). The residue is uniquely positioned at the center of interaction between two important loops. The first loop, in which Asp-364 resides, contains both Asp-361 and Cys-360. Asp-361 is an active site residue involved in the stabilization of the terminal amine of ornithine during substrate binding. The precise positioning of the substrate by Asp-361, along with Asp-332 and the backbone carbonyl of Tyr-331, has been hypothesized to be necessary for the substrate binding. Cysteine 360, the residue thought to be responsible for protonating the anion generated by decarboxylation of ornithine, would also likely be highly sensitive to small movements (63). The second loop, which interacts with Asp-364, contains Lys-169 (directly hydrogen bonded to Asp-364) and Leu-166, a residue that has been predicted to interact with the carboxylate of l-ornithine (52). This loop is known to be flexible and that flexibility is thought to be important in the ODC catalytic cycle. Perturbations in the positioning of Asp-364 would affect the populations of conformers available to this loop. In short, Asp-364 is at the center of what is likely to be a highly dynamic set of hydrogen bond interactions involving many residues known to be vital to enzymatic function. The importance of its positioning is supported by mutagenic data for this residue, where substitution with an alanine renders the enzyme inactive, and even the relatively conservative substitution with glutamic acid increases Km by more than 175-fold and decreases kcat by 280-fold.
The relatively small number of inhibitors discovered in this screen and the similarity of these inhibitors to polyamines underscore the difficulty of developing inhibitors for this enzyme. The bulk of available evidence, including the results of this screen, suggests that the small and charged active site of ODC is unable to bind conventional drug-like molecules. However, this study has shown that TbODC is susceptible to inhibition by compounds likely to be acting at allosteric sites. Furthermore, allosteric inhibition of this enzyme may allow it to escape the stabilization typically seen in the presence of other reversible inhibitors in mammalian systems.
In conclusion, we report the use of a high throughput assay to screen over 300,000 molecules and identify potent and selective inhibitors of TbODC. These inhibitors are likely to bind at novel nonactive site locations on the enzyme and represent valuable tools for further development of more drug-like inhibitors of this clinically relevant drug target.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI34432 (to M. A. P.). This work was also supported by the American Lebanese Syrian Associated Charities, St. Jude Children's Research Hospital, and Welch Foundation Grant I-1257 (to M. A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental material.
- ODC
- ornithine decarboxylase
- PLP
- pyridoxal 5′-phosphate
- DFMO
- α-difluoromethylornithine
- DTT
- dithiothreitol
- Orn
- ornithine
- TbODC
- T. brucei ODC
- hODC
- human ODC.
REFERENCES
- 1.World Health Organization (2006) African Trypanosomiasis (Sleeping Sickness), Fact Sheet, 259 Ed., World Health Organization Media Centre, Geneva [Google Scholar]
- 2.Pays E., Nolan D. P. (1998) Mol. Biochem. Parasitol. 91, 3–36 [DOI] [PubMed] [Google Scholar]
- 3.Barrett M. P., Boykin D. W., Brun R., Tidwell R. R. (2007) Br. J. Pharmacol. 152, 1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell D., Snyder S. H. (1968) Proc. Natl. Acad. Sci. U.S.A. 60, 1420–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Monem M. M., Newton N. E., Weeks C. E. (1974) J. Med. Chem. 17, 447–451 [DOI] [PubMed] [Google Scholar]
- 6.Casero R. A., Jr., Marton L. J. (2007) Nat. Rev. Drug Discov. 6, 373–390 [DOI] [PubMed] [Google Scholar]
- 7.Iwami K., Wang J. Y., Jain R., McCormack S., Johnson L. R. (1990) Am. J. Physiol. 258, G308–G315 [DOI] [PubMed] [Google Scholar]
- 8.Childs A. C., Mehta D. J., Gerner E. W. (2003) Cell. Mol. Life Sci. 60, 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amadasi A., Bertoldi M., Contestabile R., Bettati S., Cellini B., di Salvo M. L., Borri-Voltattorni C., Bossa F., Mozzarelli A. (2007) Curr. Med. Chem. 14, 1291–1324 [DOI] [PubMed] [Google Scholar]
- 10.Coffino P. (2001) Nat. Rev. Mol. Cell Biol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 11.Wallace H. M., Fraser A. V., Hughes A. (2003) Biochem. J. 376, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalf B. W., Bey P., Danzin C., Jung M. J., Casara P., Vevert J. P. (1978) J. Am. Chem. Soc. 100, 2551–2553 [Google Scholar]
- 13.Grishin N. V., Osterman A. L., Brooks H. B., Phillips M. A., Goldsmith E. J. (1999) Biochemistry 38, 15174–15184 [DOI] [PubMed] [Google Scholar]
- 14.Carbone P. P., Douglas J. A., Thomas J., Tutsch K., Pomplun M., Hamielec M., Pauk D. (2000) Clin. Cancer Res. 6, 3850–3854 [PubMed] [Google Scholar]
- 15.Griffin C. A., Slavik M., Chien S. C., Hermann J., Thompson G., Blanc O., Luk G. D., Baylin S. B., Abeloff M. D. (1987) Invest. New Drugs 5, 177–186 [DOI] [PubMed] [Google Scholar]
- 16.Meyskens F. L., Jr., McLaren C. E., Pelot D., Fujikawa-Brooks S., Carpenter P. M., Hawk E., Kelloff G., Lawson M. J., Kidao J., McCracken J., Albers C. G., Ahnen D. J., Turgeon D. K., Goldschmid S., Lance P., Hagedorn C. H., Gillen D. L., Gerner E. W. (2008) Cancer Prev. Res. 1, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simoneau A. R., Gerner E. W., Nagle R., Ziogas A., Fujikawa-Brooks S., Yerushalmi H., Ahlering T. E., Lieberman R., McLaren C. E., Anton-Culver H., Meyskens F. L., Jr. (2008) Cancer Epidem. Biomark. Prev. 17, 292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shantz L. M., Levin V. A. (2007) Amino Acids 33, 213–223 [DOI] [PubMed] [Google Scholar]
- 19.Xiao Y., McCloskey D. E., Phillips M. A. (2009) Eukaryot. Cell 8, 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoda L., Phillips M. A., Bass K. E., Wang C. C., Coffino P. (1990) J. Biol. Chem. 265, 11823–11826 [PubMed] [Google Scholar]
- 21.Phillips M. A., Coffino P., Wang C. C. (1987) J. Biol. Chem. 262, 8721–8727 [PubMed] [Google Scholar]
- 22.Willert E. K., Fitzpatrick R., Phillips M. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8275–8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willert E. K., Phillips M. A. (2008) PLoS Pathog. 4, e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairlamb A. H., Henderson G. B., Bacchi C. J., Cerami A. (1987) Mol. Biochem. Parasitol. 24, 185–191 [DOI] [PubMed] [Google Scholar]
- 25.Priotto G., Kasparian S., Mutombo W., Ngouama D., Ghorashian S., Arnold U., Ghabri S., Baudin E., Buard V., Kazadi-Kyanza S., Ilunga M., Mutangala W., Pohlig G., Schmid C., Karunakara U., Torreele E., Kande V. (2009) Lancet 374, 56–64 [DOI] [PubMed] [Google Scholar]
- 26.Milord F., Pépin J., Loko L., Ethier L., Mpia B. (1992) Lancet 340, 652–655 [DOI] [PubMed] [Google Scholar]
- 27.Iten M., Matovu E., Brun R., Kaminsky R. (1995) Trop. Med. Parasitol. 46, 190–194 [PubMed] [Google Scholar]
- 28.Bey P., Danzin C., Jung M. (1987) in Inhibition of Polyamine Metabolism, Biological Significance and Basis for New Therapies (McCann P. P., Pegg A. E., Sjoerdsma A. eds) p. 32, Academic Press, San Diego [Google Scholar]
- 29.Bitonti A. J., McCann P. P., Sjoerdsma A. (1982) Biochem. J. 208, 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thyssen S. M., Libertun C. (2002) Neurosci. Lett. 323, 65–69 [DOI] [PubMed] [Google Scholar]
- 31.Osterman A., Grishin N. V., Kinch L. N., Phillips M. A. (1994) Biochemistry 33, 13662–13667 [DOI] [PubMed] [Google Scholar]
- 32.Shelat A. A., Guy R. K. (2007) Curr. Opin. Chem. Biol. 11, 244–251 [DOI] [PubMed] [Google Scholar]
- 33.Hann M., Hudson B., Lewell X., Lifely R., Miller L., Ramsden N. (1999) J. Chem. Inf. Comp. Sci. 39, 897–902 [DOI] [PubMed] [Google Scholar]
- 34.Bemis G. W., Murcko M. A. (1996) J. Med. Chem. 39, 2887–2893 [DOI] [PubMed] [Google Scholar]
- 35.Kunkel T. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smithson D. C., Shelat A., Baldwin J., Phillips M. A., Guy K. R. (2010) Assay Drug Dev. Technol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team R. D. (2007) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna [Google Scholar]
- 38.Rousseeuw P., Croux C., Todorov V., Ruckstuhl A., Salibian-Barrera M., Maechler M. (2006) robustbase: Basic Robust Statistics, R package 0.2–7 Ed [Google Scholar]
- 39.Hoaglin D. C., Mosteller F., Tukey J. W. (eds) (1983) Understanding Robust and Exploratory Data Analysis, pp. 57–64, John Wiley & Sons, Inc., New York [Google Scholar]
- 40.Zhang J. H., Chung T. D., Oldenburg K. R. (1999) J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 41.Pegg A. E., McGill S. (1979) Biochim. Biophys. Acta 568, 416–427 [DOI] [PubMed] [Google Scholar]
- 42.Lee J., Michael A. J., Martynowski D., Goldsmith E. J., Phillips M. A. (2007) J. Biol. Chem. 282, 27115–27125 [DOI] [PubMed] [Google Scholar]
- 43.Baldwin J., Michnoff C. H., Malmquist N. A., White J., Roth M. G., Rathod P. K., Phillips M. A. (2005) J. Biol. Chem. 280, 21847–21853 [DOI] [PubMed] [Google Scholar]
- 44.Lor L. A., Schneck J., McNulty D. E., Diaz E., Brandt M., Thrall S. H., Schwartz B. (2007) J. Biomol. Screen. 12, 881–890 [DOI] [PubMed] [Google Scholar]
- 45.Segel I. H. (1975) Enzyme Kinetics, Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 249–250, John Wiley & Sons, Inc., New York [Google Scholar]
- 46.Almrud J. J., Oliveira M. A., Kern A. D., Grishin N. V., Phillips M. A., Hackert M. L. (2000) J. Mol. Biol. 295, 7–16 [DOI] [PubMed] [Google Scholar]
- 47.Hendlich M., Rippman F., Barnickel G. (1997) J. Mol. Graphics 15, 359–363 [DOI] [PubMed] [Google Scholar]
- 48.Lemoff A., Yan B. (2008) J. Comb. Chem. 10, 746–751 [DOI] [PubMed] [Google Scholar]
- 49.Yan B., Zhao J., Leopold K., Zhang B., Jiang G. (2007) Anal. Chem. 79, 718–726 [DOI] [PubMed] [Google Scholar]
- 50.Steglich C., Schaeffer S. W. (2006) Infect. Genet. Evol. 6, 205–219 [DOI] [PubMed] [Google Scholar]
- 51.Osterman A. L., Kinch L. N., Grishin N. V., Phillips M. A. (1995) J. Biol. Chem. 270, 11797–11802 [DOI] [PubMed] [Google Scholar]
- 52.Jackson L. K., Baldwin J., Akella R., Goldsmith E. J., Phillips M. A. (2004) Biochemistry 43, 12990–12999 [DOI] [PubMed] [Google Scholar]
- 53.Bitonti A. J., Dumont J. A., McCann P. P. (1986) Biochem. J. 237, 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solano F., Peñafiel R., Solano M. E., Lozano J. A. (1985) FEBS Lett. 190, 324–328 [DOI] [PubMed] [Google Scholar]
- 55.Henley C. M., 3rd, Mahran L. G., Schacht J. (1988) Biochem. Pharmacol. 37, 1679–1682 [DOI] [PubMed] [Google Scholar]
- 56.Rose F. L., Swain G. (1956) J. Chem. Soc. 4422–4425 [Google Scholar]
- 57.Ganendren R., Widmer F., Singhal V., Wilson C., Sorrell T., Wright L. (2004) Antimicrob. Agents Chemother. 48, 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yip K. W., Ito E., Mao X., Au P. Y., Hedley D. W., Mocanu J. D., Bastianutto C., Schimmer A., Liu F. F. (2006) Mol. Cancer Ther. 5, 2234–2240 [DOI] [PubMed] [Google Scholar]
- 59.Jackson L. K., Goldsmith E. J., Phillips M. A. (2003) J. Biol. Chem. 278, 22037–22043 [DOI] [PubMed] [Google Scholar]
- 60.Garvey E. P., Oplinger J. A., Tanoury G. J., Sherman P. A., Fowler M., Marshall S., Harmon M. F., Paith J. E., Furfine E. S. (1994) J. Biol. Chem. 269, 26669–26676 [PubMed] [Google Scholar]
- 61.Athri P., Wenzler T., Ruiz P., Brun R., Boykin D. W., Tidwell R., Wilson W. D. (2006) Bioorg. Med. Chem. 14, 3144–3152 [DOI] [PubMed] [Google Scholar]
- 62.Mathis A. M., Holman J. L., Sturk L. M., Ismail M. A., Boykin D. W., Tidwell R. R., Hall J. E. (2006) Antimicrob. Agents Chemother. 50, 2185–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson L. K., Brooks H. B., Osterman A. L., Goldsmith E. J., Phillips M. A. (2000) Biochemistry 39, 11247–11257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.