Abstract
Glutathione (GSH) transport is vital for maintenance of intracellular and extracellular redox balance. Only a few human proteins have been identified as transporters of GSH, glutathione disulfide (GSSG) and/or GSH conjugates (GS-X). Human epithelial MDA1586, A549, H1975, H460, HN4, and H157 cell lines were exposed to 2′,5′-dihydroxychalcone, which induces a GSH efflux response. A real-time gene superarray for 84 proteins found in families that have a known role in GSH, GSSG, and/or GS-X transport was employed to help identify potential GSH transporters. ABCG2 was identified as the only gene in the array that closely corresponded with the magnitude of 2′,5′-dihydroxychalcone (2′,5′-DHC)-induced GSH efflux. The role of human ABCG2 as a novel GSH transporter was verified in a Saccharomyces cerevisiae galactose-inducible gene expression system. Yeast expressing human ABCG2 had 2.5-fold more extracellular GSH compared with those not expressing ABCG2. GSH efflux in ABCG2-expressing yeast was abolished by the ABCG2 substrate methotrexate (10 μm), indicating competitive inhibition. In contrast, 2′,5′-DHC treatment of ABCG2-expressing yeast increased extracellular GSH levels in a dose-dependent manner with a maximum 3.5-fold increase in GSH after 24 h. In addition, suppression of ABCG2 with short hairpin RNA or ABCG2 overexpression in human epithelial cells decreased or increased extracellular GSH levels, respectively. Our data indicate that ABCG2 is a novel GSH transporter.
Keywords: ABC Transporter, Antioxidant, Membrane Proteins, Transport Amino Acids, Yeast, Glutathione
Introduction
Glutathione (GSH) is a tripeptide (γ-glutamyl-cysteinyl-glycine) that is maintained at millimolar concentrations within the cell. It is important in many signaling processes, including cell proliferation, post-translational modification of proteins, immune responses, and apoptosis (1, 2). The importance of GSH in normal cell function as well as its role in disease states such as cancer, cystic fibrosis, Parkinson disease, AIDS, liver dysfunction, and a host of other maladies has been well documented (3–8). For the purpose of detoxification and to respond to extracellular oxidative stress, cells express membrane proteins capable of transporting GSH, glutathione disulfide (GSSG), and GSH conjugates (GS-X).2 Currently, there are only a handful of proteins identified as capable of transporting GSH across membranes.
The first human protein identified as a GS-X and/or GSSG transporter was the ATP-binding cassette (ABC) protein ABCC1, first called the GSH S-conjugate pump and later identified as the multidrug resistance protein MRP1 (9–11). Since the discovery of ABCC1 as a GSH transporter, five other ABC family members have been shown to transport GSH and/or GS-X, including ABCC2, ABCC3, ABCC4, ABCC5, and ABCC7 (12–17). GSH transport has also been linked to the organic anion transporting polypeptide protein family, a subfamily of the solute carrying (SLC) protein superfamily. The SLC superfamily also contains mitochondria-specific membrane proteins (SLC25) capable of transporting GSH (18).
Numerous compounds stimulate GSH, GSSG, and GS-X efflux through the ABC proteins. Flavonoids are an example of naturally occurring compounds that have been shown to trigger MRP1-mediated GSH efflux (19). Hydroxychalcones also induce GSH efflux, but the MRP(s) involved remain unidentified (20). Cells expressing higher levels of ABCG2 had a high capacity to efflux reduced GSH, and an inducible yeast model was used to test further the hypothesis that ABCG2 transports GSH. In addition, standard molecular biology approaches were employed to show that changes in ABCG2 expression in human epithelial cells correlate with GSH efflux. Our findings indicate that ABCG2 is a novel GSH transporter.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
All cell lines were cultured at 37 °C with 5% CO2. Human epithelial lung cancer cell lines A549, H157, H460, and H1975 were grown in RPMI 1640 medium supplemented with l-glutamine (Mediatech, Manassas, VA) and 5% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA). Human epithelial head and neck cancer cell lines HN4 and MDA1586 were grown in Dulbecco's modified Eagle's medium and RPMI 1640 medium supplemented with l-glutamine and 10% fetal bovine serum. All compounds and reagents were purchased from Sigma-Aldrich unless otherwise stated. Cells were plated into 24-well plates at a 70–80% confluence at least 18 h prior to treatments. Fresh medium containing 2′,5′-DHC and 100 μm acivicin (γ-glutamyl transpeptidase inhibitor to prevent the breakdown of extracellular GSH) per well was added to the cells for the indicated treatment periods (21). At the time of cell harvest, media were removed and centrifuged at 2,000× g for 5 min at 4 °C. The cells were lysed with a sonicating probe (Cole-Parmer Instruments, Vernon Hill, IL), and lactate dehydrogenase activity was determined in both media and lysate samples. To measure toxicity, the percent of total lactate dehydrogenase release was utilized as described previously (22). Cell lysate protein values were used to normalize GSH values and were determined spectrophotometrically with the Coomassie Protein Assay kit according to the manufacturer's protocol (Pierce). Proteins were precipitated using 10% (w/v) meta-phosphoric acid added to both media and lysates. The acidified samples were vortexed and centrifuged at 23,000× g for 5 min at 4 °C. All samples awaiting analysis were stored at −20 °C until ready for processing.
ABCG2 Transient Overexpression and Short Hairpin (shRNA) Transfection in Cell Culture
HN4 cells were transiently transfected with TrueClone® transfection ready plasmid pCMV6-XL5 (OriGene, Rockville, MD). ABCG2 silencing in MDA1586 cells was achieved by inserting the ABCG2 shRNA target sequence (NM_004827.2) into pSilencer 4.1-CMV hygro vector (Applied Biosystems/Ambion, Austin, TX). Both plasmids were transfected into the appropriate cell model with Effectene Transfection Reagent according to the manufacturer's protocol (Qiagen, Valencia, CA).
GSH Measurement
GSH was measured in cell culture experiments using HPLC. The mobile phase was composed of 0.125 m sodium phosphate solution with 0.7% HPLC-grade methanol (Fisher Scientific); concentrated phosphoric acid was used to bring the solution to pH 3 (Fisher Scientific), and it was sterile filtered. Reverse phase HPLC was performed using a CoulArray model 5600 system (ESA Laboratories, Chelmsford, MA) coupled with an electrochemical detector as described previously (4). GSH was separated using an isocratic method with a flow rate of 0.5 ml/min through a Synergi 4u Hydro-RP 80A, 150- × 4.6-mm column with a guard column (Phenomenex, Torrance, CA), and detected on a 4-channel electrochemical cell (ESA) with electrode voltages set to 250, 525, 575, and 800 mV on channels 1–4, respectively. After a minimum electrode equilibration time of 24 h, 10 μl of sample and GSH standard solutions, prepared in either medium or phosphate-buffered saline, was injected. Samples were held at 4 °C in the cooled sample tray. Under these conditions, GSH had an approximate retention time of 7.5 min, which was quantified using the dominant signal on channel 3. Total GSH was measured in yeast experiments by the enzymatic recycling method according to Rahman et al. (23) and detected using a SpectraMax 340PC (Molecular Devices, Sunnyvale, CA).
Real-time Reverse Transcriptase (SYBR Green) SuperArray Assay
Isolation of mRNA from cells grown in a T-75 was completed using the Qiagen RNeasy Mini Spin Column-QIAshredder kit (Qiagen), according to the manufacturer's protocol. Purity and quantity of total RNA were determined using the Nanodrop UV-visible spectrophotometer (NanoDrop Technologies, Wilmington, DE). First strand DNA was created using RT2 First Strand kit (SABiosciences Corporation, Frederick, MD), and the protocol and cycling times as recommended by the manufacturer. The reverse transcriptase step utilized a 9800 FAST 96-well block PCR (Applied Biosystems). Two-stage real-time reverse transcriptase PCR of ABC family members and several GSH and SLC family members was performed on a custom designed 96-well format RT2 SuperArray (SABiosciences). The super arrays were designed for the Opticon 2 thermal cycler (Bio-Rad) with sample preparation and cycle times performed according to the RT2 SuperArray protocol. All solutions, including the SYBR Green reverse transcriptase PCR mix, were purchased from SABiosciences Corporation. Relative mRNA abundance for each target was determined by comparing the cycle time of the target against a 6-point (1:1 to 1:100,000) glyceraldehyde-3-phosphate dehydrogenase standard curve created by serial dilutions of loaded cDNA template within each plate.
Yeast Strains and Growth
Wild-type Saccharomyces cerevisiae express the plasma membrane proton-coupled oligopeptide transporter Opt 1p (YJL212C) which is a high affinity GSH importer (24). A S. cerevisiae strain with a null allele for Opt 1p to prevent interference from GSH import was utilized. The genotype of our strain is Sc opt1Δ leu lys his ura (uracil is used as a transformation selection gene). Yeast were grown in minimal essential medium containing 50 mg/ml galactose or glucose, 1 mg/ml leucine, 0.3 mg/ml lysine, 0.2 mg/ml histidine, 0.2 mg/ml methionine (for GSH synthesis supplement), and 67 mg/ml yeast nitrogen base. A human ABCG2 plasmid construct was created (see supplemental Experimental Procedures), and yeast were transformed according to the detailed methods described in supplemental Experimental Procedures. Fresh single colonies from agar plates containing either glucose or galactose were routinely used at the start of each experiment. All experiments were completed as strains were in exponential growth phase.
Statistical Analysis
Data are expressed as the means ± S.E., and significance was set at a p value < 0.05. All experiments included at least triplicate treatment groups, and each experiment was repeated at least three times. One-way analysis of variance (ANOVA) and Tukey post comparison test or Student's t test were performed using Prizm version 5 (GraphPad, San Diego, CA).
RESULTS
Increased ABCG2 Expression Resulted in Higher Basal and 2′,5′-DHC Stimulated Extracellular GSH Levels
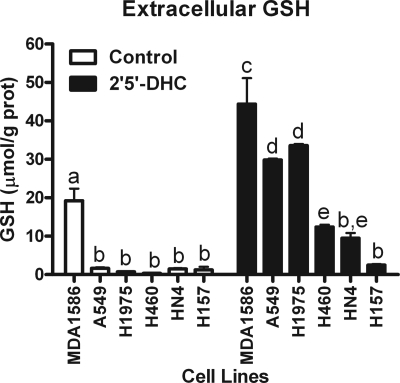
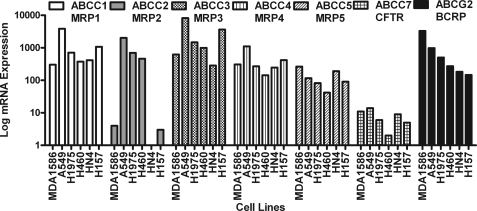
Previously, the chalcone 2′,5′-DHC was reported as an effective stimulant promoting cellular GSH efflux (20). Several human cancer epithelial cell lines were treated for 6 h with 25 μm 2′,5′-DHC, and extracellular GSH levels were determined. The GSH efflux response to 2′,5′-DHC was cell line-dependent with the highest response observed in MDA1586 followed by A549 ∼ H1975 > H460 > HN4 ≈ H157 (Fig. 1). The mRNA expression for all ABC family members and RLIP76 was profiled to understand better which membrane proteins might account for the difference in the extracellular GSH responses observed between cell types (supplemental Table 1). For illustration purposes, the real-time reverse transcriptase PCR signals of the known MRP family members that have been shown to transport GSH with that of ABCG2 were compared (Fig. 2). This comparison illustrates that only ABCG2 mRNA expression profile corresponded with GSH efflux in our human epithelial cell lines. See supplemental Fig. 1 for a side-by-side comparison of each cell type for their ABCG2 expression and the GSH efflux response after 2′,5′-DHC treatment. In MDA1586 cells, the apparent rate of GSH transport through ABCG2 at steady-state conditions was 0.83 μmol of GSH/g of protein/h over an 18-h time period. After treatment with 25 μm 2′,5′-DHC, this rate increased in MDA1586 cells to 4.87 μmol of GSH/g of protein/h.
FIGURE 1.
2′,5′-DHC increases extracellular GSH levels in human epithelial cells. Extracellular GSH levels were determined after 6-h exposure of 2′,5′-DHC (25 μm) in human epithelial MDA1586, A549, H1975, H460, HN4, and H157 cancer cell lines. 2′,5′-DHC-induced GSH efflux was most robust in MDA1586 cells followed in order by A549, H1975, H460, HN4, and H157 cells. Data are presented as the mean ± S.E. (error bars), and columns with nonidentical superscripts are significantly different (p < 0.05) as determined using ANOVA with the Tukey post comparison test.
FIGURE 2.
Comparison of GSH transport-related MRPs and ABCG2 mRNA expression in epithelial MDA1586, A549, H1975, H460, HN4, and H157 cancer cells. Relative basal mRNA expression of the five known MRPs shown to transport GSH are compared with ABCG2 in epithelial cancer cells. The expression levels of ABCG2 best correspond to 2′5′-DHC-induced GSH efflux and follow the order of 1586 > A549 > H1975 > H460 > HN4 ≈ H157. Values were normalized to a glyceraldehyde-3-phosphate dehydrogenase standard curve. The pattern of GSH efflux positively corresponds with ABCG2 expression in the cells. BCRP, breast cancer resistance protein; CFTR, cystic fibrosis transmembrane conductance regulator.
The observation of the relationship between ABCG2 expression and 2′,5′-DHC-induced GSH efflux led us to explore further the role of ABCG2 as a GSH transporter. A yeast-inducible expression system for human ABCG2 was created because human epithelial cells express multiple ABC transporters, which could confound the 2′,5′-DHC-induced GSH efflux results. The yeast model allows one to focus specifically on ABCG2-mediated GSH transport activity.
Human ABCG2 Expression in the Inducible Sc opt1Δ Yeast Model Results in Increased GSH Efflux
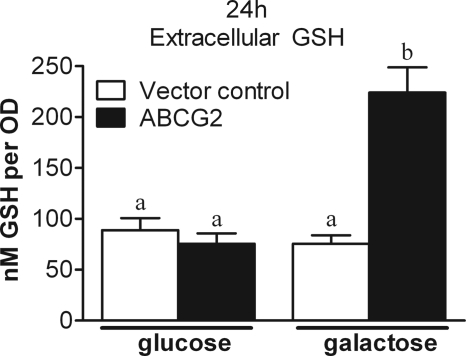
Human ABCG2 was expressed in the yeast strain S. cerevisiae mutant for the yeast GSH importer protein Opt 1p, designated here as Sc opt1Δ. The inducible yeast model was grown in galactose for expression of plasma membrane-bound ABCG2. Yeast grown in glucose do not express the protein (supplemental Figs. 2 and 3). Extracellular GSH was measured in glucose- (without ABCG2) and galactose- (expressing ABCG2) grown yeast to determine ABCG2-mediated GSH efflux directly. After a 24-h incubation in galactose, extracellular GSH reached 2.5-fold greater concentration than all other control groups, demonstrating ABCG2-dependent GSH transport (Fig. 3). At the 48-h time point GSH efflux in the galactose-grown control strain increases, but ABCG2-expressing yeast still maintain a 2-fold greater ability to efflux GSH than the galactose control and a 10-fold greater ability to efflux GSH compared with glucose controls. Intracellular GSH concentration is not significantly different between cultures at the 48-h time point (supplemental Fig. 4).
FIGURE 3.
Elevated levels of extracellular GSH in ABCG2-expressing yeast. Yeast expressing human ABCG2 protein (filled bars) for 24 h had 2.5-fold higher extracellular GSH compared with yeast not expressing the ABCG2 protein (open bars). Data are presented as the mean ± S.E. (error bars), and columns with nonidentical superscripts are significantly different (p < 0.05) as determined using ANOVA with the Tukey post comparison test.
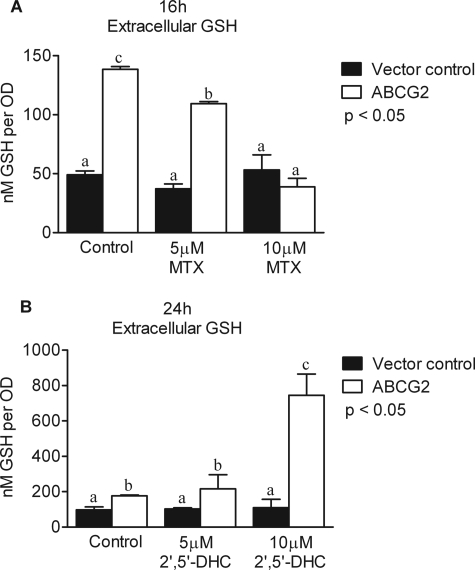
Methotrexate (MTX) Inhibits and 2′,5′-DHC Increases GSH Efflux of ABCG2-expressing Yeast
One of the early names for ABCG2 was the MTR protein, named for its ability to confer MTX resistance to cancer cells expressing the protein (25). The ability of MTX to alter GSH efflux through ABCG2 in our yeast expression system was tested. Yeast were cultured for 16 h with or without MTX (5 μm and 10 μm) under glucose or galactose conditions. MTX treatment did not alter yeast growth. Complete inhibition of GSH efflux through ABCG2 was observed with 10 μm MTX at the 16-h time point (Fig. 4A). These data suggest that MTX competes with GSH for export through ABCG2. GSH efflux response to 2′,5′-DHC was also tested in ABCG2-expressing yeast. Yeast expressing ABCG2 demonstrated an increase in GSH efflux when treated with 10 μm 2′,5′-DHC with ∼3.5-fold more extracellular GSH than controls (Fig. 4B). These results corroborate our cell culture data which indicated that cells with high ABCG2 expression efflux more GSH than cells expressing less ABGC2 (Fig. 1) and suggest that 2′,5′-DHC increases ABCG2-mediated GSH transport.
FIGURE 4.
MTX and 2′,5′-DHC alter extracellular GSH levels in ABCG2-expressing yeast. A, the ABCG2 substrate MTX decreased extracellular GSH levels in human ABCG2-expressing yeast (open bars) but not in the vector controls (filled bars). B, 2′,5′-DHC treatments (5 μm and 10 μm) significantly increased extracellular GSH levels in ABCG2-expressing yeast (open bars) but not in the vector controls (filled bars). Data are presented as the mean ± S.E. (error bars), and columns with nonidentical superscripts are significantly different (p < 0.05) as determined using ANOVA with the Tukey post comparison test.
Altering ABCG2 Protein Expression in Human HN4 or MDA1586 Cells Changes Basal Extracellular GSH Levels
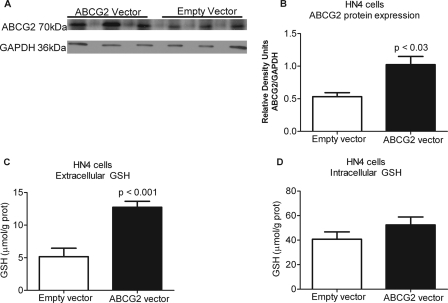
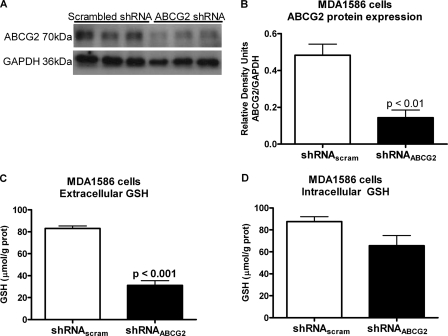
Finally, to establish a role for ABCG2 in human cells as a regulator of extracellular GSH levels, ABCG2 was overexpressed in human HN4 cells and suppressed in MDA1586 cells. Both MDA1586 and HN4 cells are derived from human head and neck cancer. ABCG2 overexpression in HN4 cells resulted in a 2.0-fold increase in ABCG2 protein levels (Fig. 5, A and B) and a 2.5-fold increase in basal extracellular GSH levels (Fig. 5C). Intracellular GSH levels were not significantly altered (Fig. 5D). Finally, transfection of MDA1586 cells with an ABCG2 shRNA plasmid resulted in a 2.6-fold decrease in ABCG2 protein expression (Fig. 6, A and B), correlating to a 2.7-fold decrease in basal extracellular GSH levels (Fig. 6C). Intracellular GSH levels were not significantly altered (Fig. 6D). Both systems support our results from the ABCG2-inducible yeast system.
FIGURE 5.
Overexpressing human ABCG2 in epithelial head and neck cancer HN4 cells increases extracellular GSH levels. A and B, HN4 cells were transfected with the OriGene ABCG2 transfection-ready plasmid, which resulted in a 2-fold increase in ABCG2 protein expression. C and D, the increase in ABCG2 protein expression resulted in 2.5-fold increase in extracellular GSH levels over vector control (C) and no significant changes in intracellular GSH levels over 48 h (D). Data are presented as the mean ± S.E. (error bars), and statistical significance was determined using Student's t test.
FIGURE 6.
Silencing ABCG2 in epithelial head and neck cancer MDA1586 cells decreased extracellular GSH levels. MDA1586 cells were transfected with a shRNA plasmid targeting ABCG2. A and B, silencing ABCG2 resulted in a 2.6-fold decrease in ABCG2 protein expression compared with the vector control. C and D, the loss of ABCG2 protein expression correlated with a 2.7-fold decrease in extracellular GSH levels compared with vector control (C) and no significant changes in intracellular GSH levels (D) over 48 h. Data are presented as the mean ± S.E. (error bars), and statistical significance was determined using Student's t test.
DISCUSSION
GSH plays important and diverse roles in cell biology. Major advances have been made in the areas of GSH metabolism and catabolism, but less is known about GSH transport. Our results demonstrate a novel GSH transport function for ABCG2. This was shown pharmacologically with the ability of 2′,5′-DHC to increase extracellular GSH dramatically in an ABCG2-dependent manner and inhibition of extracellular GSH release with a known ABCG2 substrate, MTX. In addition, the overexpression of ABCG2 in both yeast and human epithelial cells dramatically increased basal extracellular GSH levels, and silencing ABCG2 mRNA corresponded with decreased extracellular GSH levels.
ABCG2 has recently been investigated for its role in drug metabolism in the kidney, intestines, placenta, at the blood–brain barrier, and for conferring drug resistance to cancer cells (26–30). In addition to its role in drug disposition, ABCG2 has become a popular marker for identifying progenitor cells in a wide array of tissues, although its function in these cells is unclear (31–35). As an ABC superfamily member, ABCG2 is similar to other MRPs in its ability to protect cells against chemical insults; but it is unlike other MRPs in that ABCG2 is a member of the half-transporter G (WHITE) subfamily. Half-transporter ABC members require dimerization to function. ABCG2 is a 72-kDa protein comprising six transmembrane-spanning domains and one ATP-binding domain. The protein is believed to function minimally as a homodimer, although recent three-dimensional structural analysis indicates it may function as a tetrameric complex made up of homodimers (36). Several cysteine residues appear to be important for the dimerization of the functional protein (37, 38). Functional ABCG2 acts on a wide range of substrates, which is a common feature of the MRP class of transporters (30). The protein also has a number of polymorphisms that affect its substrate specificity and alter drug metabolism (39).
Transcriptional regulation of ABCG2 is complex. Evidence suggests that several signaling molecules alter ABCG2 expression, including the peroxisome proliferator-activated receptor γ, protein kinase B, the hypoxia-inducible factor 2α, and the NF-E2-related factor-2 (40–45). A common theme among these molecules is their broad control over redox regulation and their ability to alter GSH metabolism and catabolism. Alterations in intracellular and extracellular GSH involve a host of enzymes and transporters, and these proteins must work in concert for the cell to maintain an appropriate redox balance within the intracellular compartment and its surrounding environment.
This is the first study to demonstrate ABCG2-mediated GSH transport, a function that may be important in maintenance of redox balance for cells found in redox-sensitive areas such as the stem cell niche. The redox environment in progenitor cells has an important role in altering the balance between maintaining stem cell self-renewal properties or driving the cell in the direction of differentiation (46, 47). Because the extracellular environment can directly affect intracellular redox, progenitor cells may use ABCG2 to efflux GSH to help maintain a more reducing niche. More research into the role of GSH transport through ABCG2 is required to understand the potential importance this protein may have in altering redox balance of progenitor cells and their ability to self-renew or differentiate.
Identification of GSH transport by ABCG2 may open avenues to target diseases involving an extracellular oxidative stress component. Our data demonstrate that compounds like MTX decrease GSH efflux through ABCG2, whereas others such as 2′,5′-DHC increase GSH efflux. Recent evidence from Han et al. (48) indicates that some chalcones are potent inhibitors of MTX efflux through ABCG2; however, our data regarding 2′,5′-DHC induced GSH efflux suggest that ABCG2 may be altered by some chalcones to favor GSH efflux over MTX. Such results suggest that ABCG2 is a potential target for modulation of cellular GSH levels. Identifying compounds that increase the ABCG2-mediated GSH efflux may provide a pharmaceutical approach for increasing GSH concentrations in diseases associated with decreased extracellular GSH. A number of lung diseases such as cystic fibrosis (4, 49) and idiopathic pulmonary fibrosis (50) are associated with low extracellular GSH levels.
These studies show that ABCG2 has novel GSH transporter functions. The presented findings provide additional insight into the mechanisms used by a cell to move GSH from one compartment to another and highlight the multiple functions of proteins in cellular biology. Further examination of the role of ABCG2 in maintenance of cellular redox balance is warranted given its many and diverse functions in drug disposition, chemotherapy resistance, and stem cell modulation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL084469 (to B. J. D.), R01 HL075523 (to B. J. D), R01 ES0175825, R37-GM32453 (to D. R. V), and 1F32-GM076798 (to W. R. R.). This work was also supported by American Cancer Society Great-West Division Postdoctoral Fellowship Award PF-06-288-01-CSM (to W. R. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and additional references, Figs. 1–5, and Table 1.
- GS-X
- glutathione conjugate
- ABC
- ATP-binding cassette
- MRP
- multidrug-resistant protein
- SLC
- solute-carrying
- 2′,5′-DHC
- 2′,5′-dihydroxychalcone
- shRNA
- short hairpin RNA
- HPLC
- high performance liquid chromatography
- ANOVA
- analysis of variance
- MTX
- methotrexate.
REFERENCES
- 1.Day R. M., Suzuki Y. J. (2005) Dose Response 3, 425–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 3.Franco R., Schoneveld O. J., Pappa A., Panayiotidis M. I. (2007) Arch. Physiol. Biochem. 113, 234–258 [DOI] [PubMed] [Google Scholar]
- 4.Day B. J., van Heeckeren A. M., Min E., Velsor L. W. (2004) Infect. Immun. 72, 2045–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantin A. M., Hubbard R. C., Crystal R. G. (1989) Am. Rev. Respir. Dis. 139, 370–372 [DOI] [PubMed] [Google Scholar]
- 6.Pacht E. R., Diaz P., Clanton T., Hart J., Gadek J. E. (1997) Chest 112, 785–788 [DOI] [PubMed] [Google Scholar]
- 7.Pacht E. R., Timerman A. P., Lykens M. G., Merola A. J. (1991) Chest 100, 1397–1403 [DOI] [PubMed] [Google Scholar]
- 8.Roum J. H., Buhl R., McElvaney N. G., Borok Z., Crystal R. G. (1993) J. Appl. Physiol. 75, 2419–2424 [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa T. (1989) J. Biol. Chem. 264, 17343–17348 [PubMed] [Google Scholar]
- 10.Jedlitschky G., Leier I., Buchholz U., Center M., Keppler D. (1994) Cancer Res. 54, 4833–4836 [PubMed] [Google Scholar]
- 11.Müller M., Meijer C., Zaman G. J., Borst P., Scheper R. J., Mulder N. H., de Vries E. G., Jansen P. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 13033–13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Q., Deeley R. G., Cole S. P. (2000) J. Biol. Chem. 275, 34166–34172 [DOI] [PubMed] [Google Scholar]
- 13.Hagmann W., Nies A. T., König J., Frey M., Zentgraf H., Keppler D. (1999) Eur. J. Biochem. 265, 281–289 [DOI] [PubMed] [Google Scholar]
- 14.Zelcer N., Saeki T., Reid G., Beijnen J. H., Borst P. (2001) J. Biol. Chem. 276, 46400–46407 [DOI] [PubMed] [Google Scholar]
- 15.Rius M., Nies A. T., Hummel-Eisenbeiss J., Jedlitschky G., Keppler D. (2003) Hepatology 38, 374–384 [DOI] [PubMed] [Google Scholar]
- 16.Sadzuka Y., Sugiyama T., Suzuki T., Sonobe T. (2001) Toxicol. Lett. 123, 159–167 [DOI] [PubMed] [Google Scholar]
- 17.Kogan I., Ramjeesingh M., Li C., Kidd J. F., Wang Y., Leslie E. M., Cole S. P., Bear C. E. (2003) EMBO J. 22, 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lash L. H. (2006) Chem. Biol. Interact. 163, 54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothnie A., Conseil G., Lau A. Y., Deeley R. G., Cole S. P. (2008) Mol. Pharmacol. 74, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 20.Kachadourian R., Day B. J. (2006) Free Radic. Biol. Med. 41, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith T. K., Ikeda Y., Fujii J., Taniguchi N., Meister A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velsor L. W., Kariya C., Kachadourian R., Day B. J. (2006) Am. J. Respir. Cell Mol. Biol. 35, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman I., Kode A., Biswas S. K. (2006) Nat. Protoc. 1, 3159–3165 [DOI] [PubMed] [Google Scholar]
- 24.Bourbouloux A., Shahi P., Chakladar A., Delrot S., Bachhawat A. K. (2000) J. Biol. Chem. 275, 13259–13265 [DOI] [PubMed] [Google Scholar]
- 25.Miyake K., Mickley L., Litman T., Zhan Z., Robey R., Cristensen B., Brangi M., Greenberger L., Dean M., Fojo T., Bates S. E. (1999) Cancer Res. 59, 8–13 [PubMed] [Google Scholar]
- 26.Huls M., Brown C. D., Windass A. S., Sayer R., van den Heuvel J. J., Heemskerk S., Russel F. G., Masereeuw R. (2008) Kidney Int. 73, 220–225 [DOI] [PubMed] [Google Scholar]
- 27.Urquhart B. L., Ware J. A., Tirona R. G., Ho R. H., Leake B. F., Schwarz U. I., Zaher H., Palandra J., Gregor J. C., Dresser G. K., Kim R. B. (2008) Pharmacogen. Genomics 18, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeboah D., Sun M., Kingdom J., Baczyk D., Lye S. J., Matthews S. G., Gibb W. (2006) Can. J. Physiol. Pharmacol. 84, 1251–1258 [DOI] [PubMed] [Google Scholar]
- 29.Eisenblätter T., Hüwel S., Galla H. J. (2003) Brain Res. 971, 221–231 [DOI] [PubMed] [Google Scholar]
- 30.Hardwick L. J., Velamakanni S., van Veen H. W. (2007) Br. J. Pharmacol. 151, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho M. M., Ng A. V., Lam S., Hung J. Y. (2007) Cancer Res. 67, 4827–4833 [DOI] [PubMed] [Google Scholar]
- 32.Majka S. M., Beutz M. A., Hagen M., Izzo A. A., Voelkel N., Helm K. M. (2005) Stem Cells 23, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 33.Naylor C. S., Jaworska E., Branson K., Embleton M. J., Chopra R. (2005) Bone Marrow Transplant. 35, 353–360 [DOI] [PubMed] [Google Scholar]
- 34.Pfister O., Oikonomopoulos A., Sereti K. I., Sohn R. L., Cullen D., Fine G. C., Mouquet F., Westerman K., Liao R. (2008) Circ. Res. 103, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda T., Brenner S., Malech H. L., Langemeijer S. M., Perl S., Kirby M., Phang O. A., Krouse A. E., Donahue R. E., Kang E. M., Tisdale J. F. (2005) J. Biol. Chem. 280, 991–998 [DOI] [PubMed] [Google Scholar]
- 36.McDevitt C. A., Collins R. F., Conway M., Modok S., Storm J., Kerr I. D., Ford R. C., Callaghan R. (2006) Structure 14, 1623–1632 [DOI] [PubMed] [Google Scholar]
- 37.Wakabayashi K., Nakagawa H., Tamura A., Koshiba S., Hoshijima K., Komada M., Ishikawa T. (2007) J. Biol. Chem. 282, 27841–27846 [DOI] [PubMed] [Google Scholar]
- 38.Xu J., Peng H., Chen Q., Liu Y., Dong Z., Zhang J. T. (2007) Cancer Res. 67, 4373–4381 [DOI] [PubMed] [Google Scholar]
- 39.Tamura A., Watanabe M., Saito H., Nakagawa H., Kamachi T., Okura I., Ishikawa T. (2006) Mol. Pharmacol. 70, 287–296 [DOI] [PubMed] [Google Scholar]
- 40.Mogi M., Yang J., Lambert J. F., Colvin G. A., Shiojima I., Skurk C., Summer R., Fine A., Quesenberry P. J., Walsh K. (2003) J. Biol. Chem. 278, 39068–39075 [DOI] [PubMed] [Google Scholar]
- 41.Takada T., Suzuki H., Gotoh Y., Sugiyama Y. (2005) Drug Metab. Dispos. 33, 905–909 [DOI] [PubMed] [Google Scholar]
- 42.Szatmari I., Vámosi G., Brazda P., Balint B. L., Benko S., Széles L., Jeney V., Ozvegy-Laczka C., Szántó A., Barta E., Balla J., Sarkadi B., Nagy L. (2006) J. Biol. Chem. 281, 23812–23823 [DOI] [PubMed] [Google Scholar]
- 43.Adachi T., Nakagawa H., Chung I., Hagiya Y., Hoshijima K., Noguchi N., Kuo M. T., Ishikawa T. (2007) J. Exp. Ther. Oncol. 6, 335–348 [PubMed] [Google Scholar]
- 44.Hagiya Y., Adachi T., Ogura S., An R., Tamura A., Nakagawa H., Okura I., Mochizuki T., Ishikawa T. (2008) J. Exp. Ther. Oncol. 7, 153–167 [PubMed] [Google Scholar]
- 45.Martin C. M., Ferdous A., Gallardo T., Humphries C., Sadek H., Caprioli A., Garcia J. A., Szweda L. I., Garry M. G., Garry D. J. (2008) Circ. Res. 102, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 46.Noble M., Smith J., Power J., Mayer-Pröschel M. (2003) Ann. N. Y. Acad. Sci. 991, 251–271 [DOI] [PubMed] [Google Scholar]
- 47.Smith J., Ladi E., Mayer-Pröschel M., Noble M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10032–10037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han Y., Riwanto M., Go M. L., Ee P. L. (2008) Eur. J. Pharm. Sci. 35, 30–41 [DOI] [PubMed] [Google Scholar]
- 49.Gao L., Kim K. J., Yankaskas J. R., Forman H. J. (1999) Am. J. Physiol. Lung Cell. Mol. Physiol. 277, L113–L118 [DOI] [PubMed] [Google Scholar]
- 50.Rahman I., MacNee W. (2000) Eur. Respir. J. 16, 534–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.