Abstract
In eukaryotic nuclei the majority of genomic DNA is believed to exist in higher order chromatin structures. Nonetheless, the nature of direct, long range nucleosome interactions that contribute to these structures is poorly understood. To determine whether these interactions are directly mediated by contacts between the histone H4 amino-terminal tail and the acidic patch of the H2A/H2B interface, as previously demonstrated for short range nucleosomal interactions, we have characterized the extent and effect of disulfide cross-linking between residues in histones contained in different strands of nucleosomal arrays. We show that in 208-12 5 S rDNA and 601-177-12 nucleosomal array systems, direct interactions between histones H4-V21C and H2A-E64C can be captured. This interaction depends on the extent of initial cross-strand association but does not require these specific residues, because interactions with residues flanking H4-V21C can also be captured. Additionally, we find that trapping H2A-H4 intra-array interactions antagonizes the ability of these arrays to undergo intermolecular self-association.
Keywords: Chromatin Structure, Disulfide, Histones, Nuclear Organization, Nucleosome
Introduction
In eukaryotic nuclei, DNA is packaged into chromatin to facilitate and regulate the storage, segregation, organization, and utilization of the genome. Chromatin is a complex DNA-protein assembly that exhibits multiple levels of structures. Its most basic structural unit, the nucleosome, is composed of an octamer (two copies each of histone proteins H2A, H2B, H3, and H4), wrapped by 147 base pairs of DNA (1, 2). In general, the majority of genomic DNA is sequestered in nucleosomes (3), and in their most extended form these nucleosomal arrays form a 10-nm fiber. However, even for cells in interphase, it is widely believed that most of the chromatin adopts higher order structures. In these cells a variety of chromatin fibers have been observed, including fibers that are more than 100 nm thick (4).
The understanding of higher order chromatin structures has been significantly aided by in vitro studies of isolated and reconstituted nucleosomal array systems, where reversible short range intra-array and long range interarray nucleosome associations can be induced even in the absence of additional chromatin-associated proteins (5, 6). From these studies a number of factors important for higher order chromatin structure have been identified. Within the nucleosome, the amino-terminal portions of the histones that extend past the nucleosomal DNA, the histone tails, have been shown to affect both intra- and interarray associations (7–9). Among these tails, the histone H4 tail has the largest effect on both types of association (9–11), in a manner dependent on the charge (12), modification state (13, 14), and position of the H4 tail (12). Another important region within the nucleosome is the acidic patch interface of histones H2A and H2B (1), where mutations to this region can change both intra- and interarray interactions (10, 15, 16).
How these nucleosome components contribute to internucleosomal interactions is not completely clear. However, based on crystal contacts observed in the first high resolution structure of a mononucleosome, i.e. a nucleosome not linked to other nucleosomes through intervening linker DNA, it was proposed that one way in which nucleosome interactions occur in nucleosomal arrays is through direct contact between the H4 tail of one nucleosome and the H2A/H2B interface of another nucleosome (1, 17). Indeed, for intra-array nucleosomal interactions, this contact has been captured by disulfide and photo-affinity cross-linking (18, 19). Whether such interactions also occur for interarray associations is not as clear, because support for such a model has been indirect and potentially conflicting. In some cases, changes to the H4 tail and the H2A surface result in similar changes in intra- and interarray associations (9–11, 13, 15), suggesting either that both types of interactions share a common mechanism or that intra-array associations facilitate interarray associations. In contrast, there are a number of examples where changes to the H4 tail-H2A/H2B interface interaction result in different intra- and interarray association effects (10, 16). This is seen, perhaps most dramatically, in the case of studies with tetramer arrays that lack the H2A and H2B subunits entirely. Studies of these systems have shown that they are highly defective in intra-array association (20). However, despite the absence of any H2A or H2B histone, these arrays are just as capable of forming interarray associations as arrays with a complete complement of histones (8). These results may indicate that the major mechanism of interarray association is different from than that for intra-array associations or that these two types of interactions are interrelated in a way such that changes in one type of nucleosome to nucleosome interaction can affect the nature of the other.
To provide a foundation for interpreting these observations, we sought to determine the extent and nature of direct cross-strand interactions between the H4 tail and the H2A/H2B surface. Because of the complexity and large size of the self-associated array species, standard structural techniques are not readily applicable. Moreover, because the H4 interaction with the H2A/H2B surface is already known to be directly involved in intra-array interactions, any technique used must be able to separate this contribution from those involved in interarray interactions. By adapting a nucleosomal array system previously developed to trap intra-array nucleosome interactions (18), we have been able to isolate and better understand the interactions of the H4 tail and H2A/H2B surface in interarray self-association.
EXPERIMENTAL PROCEDURES
Template and Carrier DNA Preparation
208-12 5 S rDNA and 601-177-12 DNA templates were excised from plasmids and purified by gel filtration chromatography according to standard methods (9, 21). The 174-bp carrier DNA was prepared by PCR amplification of the purified 196-bp fragment that results from complete EcoRI digestion of the 208-12 template. Carrier DNA was purified by phenol chloroform extraction and gel electroelution. The purity and quantity of the template and carrier DNA were determined by gel electrophoresis and absorbance spectroscopy.
Histone Octamer Preparation
Xenopus histones were recombinantly expressed, purified, and characterized using standard methods (22). Cysteine-containing histones H2A-E64C, H4-K20C, H4-V21C, H4-L22C, and H4-R23C were generated by QuikChange mutagenesis (Stratagene) of the bacterial expression vectors containing histone genes (22). Octamers were assembled and purified in the presence of 0.1 mm Tris (2-carboxyethyl)phosphine hydrochloride (TCEP)2 reductant using standard methods (22). All of the octamers were prepared using H3-C110A. The octamers were characterized by SDS-PAGE gel electrophoresis and absorbance spectroscopy (22).
Nucleosomal Array Assembly
Nucleosomal arrays were assembled by mixing histone octamers and DNA components followed by the stepwise salt dialysis method as previously described (13, 21). 0.1 mm TCEP reductant was added to the DNA and octamer mixture and to the dialysis solutions. For 208-12 arrays, molar ratios of octamer to template varied from 0.9 to 1.1. For 601-177-12 arrays, octamer to template to carrier DNA molar ratios were 1.15–1.3:1.0:0.3. Carrier DNA and mononucleosomes were removed by differential centrifugation as previously described (9). The final composition of the array solutions included array buffer (2.5 mm NaCl, 10 mm Tris-HCl, pH 8.0, 0.25 mm EDTA for 208-12; 2.5 mm NaCl, 10 mm HEPES, pH 8.0, for 601-177-12) and 0.1 mm TCEP, and the arrays were stored at 0 °C. The arrays were quantified based on amount of DNA by measuring absorbance at 260 nm. Array saturation was analyzed by restriction endonuclease analysis, sedimentation velocity analysis, and differential centrifugation assay as described below. Arrays with similar nucleosome saturation were used for studies.
Disulfide Cross-linking of Nucleosomal Arrays
2× cross-linking solutions were prepared by adding 1 m Tris-HCl, pH 9.0 (2× final concentration of 100 mm), 100 mm glutathione (2× final concentration of 2 mm, molar ratios of oxidized (Sigma) to reduced (Acros Organics) glutathione of 1:15, 1:7, 1:3, 1:1, 3:1, 7:1, and 15:1), and 500 mm MgCl2 (2× final concentrations from 0 to 12.0 mm for interarray cross-linking, 2.0 mm for intra-array cross-linking) to the appropriate array buffer. To initiate cross-linking, this solution was mixed in an equal volume with nucleosomal arrays (concentration of 50 ng/μl of DNA template). For inter-molecular cross-linking, the arrays were incubated at room temperature for 16 h followed by the addition of an equal volume of EDTA solution (final concentration, 20 mm) and further incubation at room temperature for 2 h. For intra-molecularly cross-linked arrays, the samples were incubated at 37 °C for 16 h. These samples were then dialyzed three times at 4 °C in array buffer. After dialysis, the absorbance values of samples at 260 nm were measured before and after centrifugation at 14,000 × g for 10 min at room temperature to quantify and remove any highly cross-linked species. Array concentrations were determined by subtracting the 260-nm absorbance of a mock reaction consisting of all of the components except the array.
Preparation of Cross-linked Histone Standards
Ideal solution conditions for generating cross-linked histone standards are those in which the histone is fully reduced and denatured, and where the solution is at a high ionic strength to allow close approach of the highly basic histones. 7 m guanidine hydrochloride fulfills these criteria and was used for the reduction and cross-linking steps. Urea was used in the dialysis steps used to remove the reductant because of its lower cost. Specifically, lyophilized histones H2A-E64C and H4-V21C were resuspended in unfolding buffer (7 m guanidine HCl, 20 mm Tris-HCl, pH 7.4, 10 mm dithiothreitol), dialyzed two times in 7 m urea and 20 mm Tris-HCl, pH 7.4, and finally dialyzed in 7 m guanidine-HCl and 20 mm Tris-HCl, pH 7.4. Dialyzed histones were then quantified by absorbance. H2A-E64C alone, H4-V21C alone, or a one-to-one mixture of H2A-E64C and H4-V21C were mixed with 4 mm 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) prepared in 0.1 m phosphate buffer, pH 7.6, to achieve a final concentration of 100:50 μm histone to DTNB for single histones and 200:100 μm histones to DTNB for mixed histones. This mix was incubated at room temperature for 12–16 h and then dialyzed in 0.1% trifluoroacetic acid at 4 °C. The histones were then dried, resuspended in protein loading dye with no reducing agents, and stored at −20 °C.
Restriction Endonuclease Analysis
As previously described, EcoRI and ScaI digestion was used to characterize the extent of nucleosome saturation of the 208-12 and 601-177-12 arrays, respectively (9, 21). The resulting mononucleosomes and free DNA were analyzed on 4% native PAGE gel followed by ethidium bromide staining.
Sedimentation Velocity
Sedimentation velocity experiments were performed with a Beckman XLA ultracentrifuge. Nucleosomal arrays were analyzed at concentrations ranging from 18 to 25 ng/μl of DNA template and at speeds from 12,000 to 16,000 rpm. TCEP to a final concentration of 0.1 mm and 0.1 mm additional EDTA were added to non-cross-linked nucleosomal arrays prior to analysis. For intramolecularly cross-linked arrays, the mock sample was used as reference, and no TCEP or EDTA was added to the samples. The data were analyzed using the method of van Holde and Weischet on Ultrascan data analysis software (Dr. B. Demeler, University of Texas Health Science Center, San Antonio, TX) as described previously (23).
Differential Centrifugation
Differential centrifugation analysis of nucleosomal arrays prior to cross-linking was performed largely as previously described (8). In short, nucleosomal arrays (∼30 ng/μl of DNA template) were mixed with an equal volume of array buffer containing both 0.1 mm TCEP reductant and MgCl2 at twice the desired final concentration. Following 15 min of incubation at room temperature, the arrays were centrifuged at 14,000 × g for 10 min at room temperature. The absorbance of array in the supernatant was then determined at 260 nm. To calculate the fraction of nucleosomal array remaining in solution, this absorbance was divided by the absorbance of array that remains in solution when treated similarly, but with a 0 mm final MgCl2 concentration. The differential centrifugation analysis of cross-linked arrays was performed in a similar manner, but with the following differences. No TCEP reductant was present and the absorbance of array in the supernatant was determined relative to a mock cross-linked sample, which lacked nucleosomal arrays, but was otherwise treated identically. For the inter-molecularly cross-linked arrays, no additional array buffer or MgCl2 was added prior to measuring the absorbance. For arrays containing no cysteine residues, ∼75% recovery of initial signal was observed, potentially because of incomplete magnesium ion sequestration or a different array subpopulation, and the absolute recovered absorbance has been reported. For intra-molecularly cross-linked array, 10× MgCl2 in array buffer was added in a ratio of 1:9 instead of the 2× MgCl2 solution prior to measuring the absorbance. All of the trials were repeated at least three times mostly with the same array preparation and presented either as the representative data or the mean values, with error bars representing the standard deviation.
Nonreducing SDS-PAGE
Histones from cross-linked arrays were trichloroacetic acid-precipitated (final concentration, 20% trichloroacetate) and resuspended in protein loading dye with no reducing agent. The histones were separated on an 18% SDS-PAGE gel and visualized using Coomassie Blue.
RESULTS
Interarray Cross-linking in 208-12 Arrays via H4-V21C and H2A-E64C
Direct interactions between the histone H4 tail of one nucleosome and the H2A/H2B acidic patch of another have been previously demonstrated within the same nucleosomal array by Richmond and co-workers (18) using oxidative cross-linking. In this system, substitution of cysteines for histone residues H4-Val21 and H2A-Glu64 allowed them to trap interactions that occurred under conditions where nucleosomal arrays exhibit intramolecular but not intermolecular compaction. To adapt this technique to study the extent of interarray contacts between the H4 tail and H2A/H2B acidic patch, our strategy was to generate two different sets of arrays, where each array consisted of nucleosomes that included either H4-V21C histones or H2A-E64C histones. With a mixture of these arrays, if this interaction is involved in interarray association, then disulfide cross-linking would be induced under conditions that cause interarray self-association. The products of this cross-linking can then be characterized to assess how much of the array association persists under conditions that do not normally promote array self-association. Additionally, the nature of the cross-linked histone species can be determined, where any observed H2A-H4 cross-linking is only possible via interarray contacts.
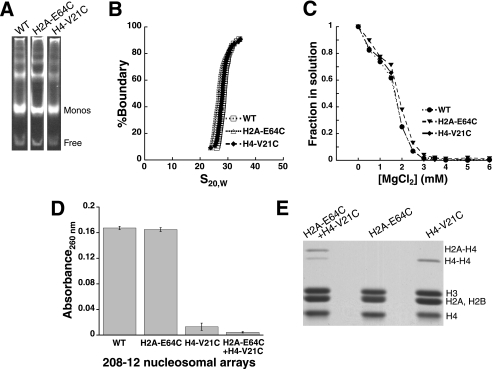
To generate well defined nucleosomal arrays, wild-type and cysteine-containing Xenopus laevis histones were recombinantly expressed, purified, and assembled into histone octamers. These octamers were then deposited onto recombinantly expressed and purified DNA templates by stepwise salt dialysis. In our initial experiments 208-12 DNA templates containing 12 head to tail repeats of the naturally occurring Lytechinus variegatus 5 S rDNA sequence were used (24). Arrays were assembled to be significantly saturated, but not oversaturated, and to be closely matched in saturation. To confirm this, the arrays were characterized by EcoRI digestion (Fig. 1A), sedimentation velocity analysis (Fig. 1B), and cation-dependent differential centrifugation assays (Fig. 1C). Comparison of the results for the three arrays indicate that the arrays are well matched in saturation, that the arrays are nearly saturated (21, 25), and that the presence of the cysteine residues does not change their properties under nonoxidizing conditions. In the differential centrifugation analysis (Fig. 1C), in contrast to arrays assembled from isolated, endogenous histones (26), no plateau is observed for lower magnesium ion concentration. Nonetheless, this behavior is consistent with previous studies of arrays reconstituted from recombinantly expressed core histones (10, 25) and occurs regardless of whether or not the histones in the arrays contain cysteine residues (Fig. 1C).
FIGURE 1.
For self-associated nucleosomal arrays, disulfide cross-linking captures direct interarray interactions between histones H4-V21C and H2A-E64C. A, EcoRI digestion of arrays prior to cross-linking treatment. Mononucleosomes (Monos) and free DNA liberated from the 208-12 arrays via cleavage of the linker DNA between nucleosomes were separated by electrophoresis on a native 4% PAGE gel and stained with ethidium bromide. B, sedimentation velocity characterization of arrays prior to cross-linking treatment. Shown are representative integrated sedimentation coefficient distribution plots for 208-12 5 S rDNA nucleosomal arrays in the absence of divalent cation. Each array contains either wild-type (WT), H4-V21C, or H2A-E64C histones. S20,W is the sedimentation coefficient corrected for water at 20 °C. C, cation-dependent self-association of arrays prior to cross-linking treatment. Shown is a representative differential centrifugation plot for non-cross-linked arrays, where the fraction of array remaining in the supernatant is plotted as a function of MgCl2 concentration. D, differential centrifugation of individual arrays and array mixtures after treatment of the self-associated arrays with 1:1 ox/red glutathione and MgCl2 removal. Shown is the amount of array absorbance remaining in the supernatant, where the averages and standard deviations were derived from three independent trials. E, histone composition of the arrays in D. The histones were separated by SDS-PAGE analysis under nonreducing conditions and stained with Coomassie Blue. The identities of the cross-linked histones were assigned by comparison with H4-V21C and H2A-E64C histones cross-linked individually and as a mixture.
To capture interarray interactions, equal amounts of H2A-E64C and H4-V21C arrays were mixed with a solution containing magnesium chloride and glutathione. With a final concentration of divalent magnesium ion of either 4.0 or 6.0 mm, the arrays were expected to be fully self-associated and form species that exhibit a large sedimentation coefficient (Fig. 1C). The glutathione in solution contained equal molar amounts of its oxidized and reduced forms, creating a redox buffer in which histone disulfide formation could reach equilibrium with respect to the redox potential established by the ratio of glutathione species. Prior to analysis of the cross-linked arrays, EDTA was added to chelate the divalent magnesium and thereby limit interarray association caused by noncovalent interactions.
Differential centrifugation analysis of these arrays, in which the amount of nonsedimented arrays is assessed after centrifugation, revealed that the glutathione-treated array mixture generated one or multiple species with very large sedimentation coefficients relative to wild-type arrays, consistent with extensive interarray cross-linking (Fig. 1D). Significant sedimentation was not observed with similar treatment of arrays containing H2A-E64C histones (Fig. 1D), indicating that H2A-H2A cross-linking was not responsible for the interarray association. However, arrays containing only H4-V21C did show significant sedimentation, suggesting that H4-H4 cross-linking could be responsible for the interarray association in the mixed array experiment. To directly assess the nature of the histone cross-linking, the histone composition of these reactions was analyzed by nonreducing denaturing gel electrophoresis (Fig. 1E). Consistent with the differential centrifugation, this analysis shows that H2A-E64C arrays do not generate cross-linked histones, whereas H4-V21C arrays, as well as mixtures of H2A-E64C and H4-V21C arrays, do. The H4-V21C arrays generate H4-H4 cross-linked histones, whereas the H2A-E64C and H4-V21C array mixture predominantly captures direct interarray interactions between histones H4-V21C and H2A-E64C while also producing some of the H4-H4 cross-linked species.
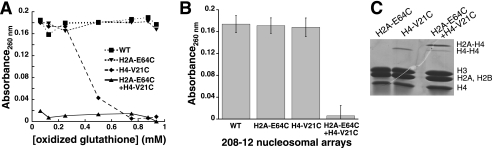
To determine the relative stability and role of the H2A-H4 cross-link in interarray association, we investigated the effects of decreasing the oxidizing potential in the cross-linking reaction. For the H2A-E64C and H4-V21C array mixture, the formation of interarray cross-linked species that sediments remains constant over decreasing ratios of oxidized to reduced glutathione (Fig. 2A). In contrast, formation of such species for the H4-V21C arrays alone decreases with decreasing oxidizing potentials. At an oxidized to reduced glutathione ratio of 1:15, the array mixture shows complete sedimentation, whereas all of the individual array species show no discernable sedimentation (Fig. 2B). Analysis of the histones under these conditions shows that for the mixed array experiments, cross-linked H2A-H4 histones are the predominant cross-linked species, although some H4-H4 cross-linking is still observed (Fig. 2C). For the H4 arrays alone, H4-H4 cross-linking is still present, although slightly reduced relative to the higher oxidation potentials (data not shown). This indicates that the nature of the remaining H4-H4 cross-linked histones is insufficient to facilitate interarray sedimentation. In the mixed arrays, the amount of H4-H4 cross-linking is even less and suggests that, because the larger amounts of H4-H4 cross-linking observed in the H4-V21C arrays alone were not sufficient to promote interarray sedimentation, the smaller amounts present in the mixed arrays are not likely to be the species responsible for the interarray sedimentation, i.e. the H2A-H4 cross-linked species are the predominant species responsible for the interarray sedimentation.
FIGURE 2.
Stable interarray interactions via H4-V21C to H2A-E64C cross-linking persist at decreased oxidizing potentials. A, differential centrifugation characterization of individual arrays and array mixtures following glutathione treatment of the self-associated arrays at varying ratios of oxidized to reduced glutathione, where total glutathione remained constant at 1.0 mm. Shown is the amount of array absorbance remaining in the supernatant for a representative trial. B, differential centrifugation characterization of individual and mixed arrays following 1:15 ox/red glutathione treatment. The array remaining in the supernatant was analyzed as described for Fig. 1D. C, histone composition of individual and mixed arrays following 1:15 ox/red glutathione treatment. Histone components were analyzed as described for Fig. 1E. WT, wild type.
Dependence of Interarray Cross-linking on Array Self-association
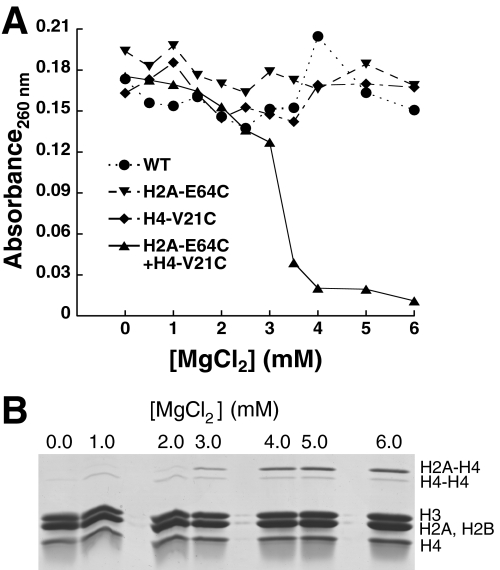
In self-associated arrays, interarray cross-linking between H4-V21C and H2A-E64C is favored over the other potential modes of cross-linking. We expected that this cross-linking was because these sites are brought into close spatial proximity with one another in self-associated arrays. To directly test this idea, we determined the relationship between array self-association and interarray cross-linking.
As has been previously shown (8) and is apparent in Fig. 1C, self-association of arrays is facilitated by divalent cations. Thus, if the observed interarray cross-linking requires array self-association, the degree of interarray cross-linking should decrease with decreasing amounts of divalent cation. Indeed, formation of species sufficiently cross-linked to sediment in the absence of divalent cations occurs only when initial cross-linking is performed at higher concentrations of divalent cation (Fig. 3A). Formation of this stably associated species appears to require H2A-H4 interactions, because arrays with only one of these components do not show significant differential centrifugation. Further, analysis of the histones from the H2A-E64C and H4V21C array mixture after cross-linking treatment shows the H2A-H4 cross-linked pair to be the predominant species, with its presence increasing with increasing concentrations of divalent cation (Fig. 3B). Thus, these data suggest that the observed interarray cross-linking requires array self-association.
FIGURE 3.
Array self-association is required for H4-V21C to H2A-E64C cross-linking. A, differential centrifugation characterization of individual and mixed arrays following 1:15 ox/red glutathione treatment at varying MgCl2 concentrations. Shown is a representative plot of the amount of array absorbance remaining in the supernatant after the removal of MgCl2. B, histone composition of mixed arrays containing both H2A-E64C and H4-V21C arrays following 1:15 ox/red glutathione treatment at varying MgCl2 concentrations. The histone components were analyzed as described for Fig. 1E. WT, wild type.
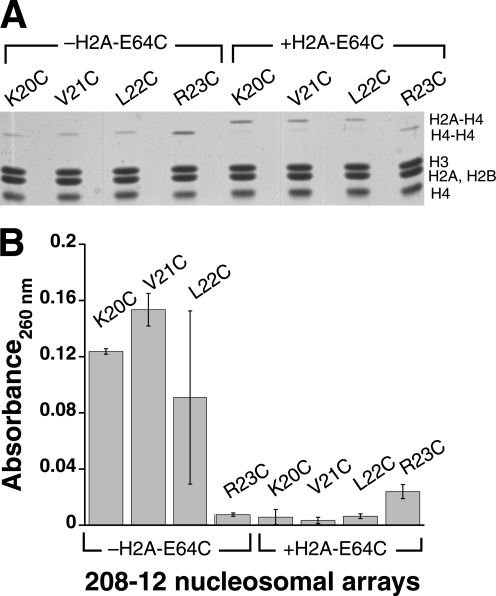
Specificity of Interarray H4 to H2A Interactions
Because interarray interactions between H4-V21C and H2A-E64C are captured in self-associated arrays, we wondered how specific this interaction was. To test the specificity, we investigated cross-linking of other sites in the H4 tail to histone H2A-E64C. Like H4-V21, substitution of residues directly adjacent to this site with cysteine, i.e. H4-K20C and H4-L22C, resulted in arrays with an ability to cross-link to themselves in the absence of H2A-E64C but that preferentially cross-link to H2A-E64C when it is present (Fig. 4A). Also like H4-V21C, the products generated by H4-H4 cross-links resulted in less differential centrifugation than products generated by the H4-H2A cross-linking (Fig. 4B). The similarity in behavior suggests that the interarray interaction between the H4 tail and the H2A/H2B acidic patch does not have to involve specific residue contacts, because multiple H4 tail residue contacts with H2A-E64C can be captured. However, the preference for H2A-E64C contacts appears to be constrained to a specific region of the H4 tail, because substitution of H4-Arg23 with cysteine does not favor cross-linking to H2A-E64C but rather predominantly cross-links to itself (Fig. 4A).
FIGURE 4.
Multiple, but not all, positions on the H4 tail preferentially cross-link to histone H2A-E64C. A, histone composition of cross-linked arrays (1:15 ox/red glutathione treatment, 6.0 mm MgCl2) containing a cysteine residue at various histone H4 tail locations. Histone components were analyzed as described for Fig. 1E. B, differential centrifugation characterization of arrays analyzed in A. Shown is the amount of array absorbance remaining in the supernatant in the absence of MgCl2 for various H4 arrays alone (left bracket) or combined with arrays containing H2A-E64C histone (right bracket). Analysis was performed as described for Fig. 1D.
Intra-array Cross-linking Affects Interarray Associations
Prior studies indicate that the H4 histone tail contributes to intra-array interaction through direct contacts with the surface of histone H2A, whereas our results suggest that the same interactions contribute to interarray nucleosome contacts. The dual role for this interaction raises the question as to how intra- and interarray nucleosome associations are related. To address this, we wanted to see to what extent interarray associations would change when intra-array H4-H2A associations were favored. Because these intra-array associations might not persist to a significant extent under the usual conditions necessary to observe interarray associations, we sought to trap the intra-array association irreversibly through a covalent interaction.
To generate arrays with intramolecular nucleosomal cross-links, we turned to the strategy devised by Richmond and co-workers (18), where nucleosomal arrays containing both H4-V21C and H2A-E64C are oxidized under conditions that form only intra-array disulfide cross-links. Although 208-12 arrays containing both cysteine-containing histones were readily generated and were similar to our wild-type arrays (supplemental Fig. S1), attempts to apply this strategy to 208-12 arrays proved problematic, because sedimentation velocity analysis of the oxidation products showed them to be highly heterogeneous (data not shown). Because a different nucleosomal array DNA template, 601-177-12, was utilized for the majority of the previous intra-array cross-linking studies (18), this template was utilized for our subsequent experiments.
The 601-177-12 DNA template is composed of 12 head-to-tail repeats of a SELEX-selected 177-bp octamer-binding sequence (9, 27). This template has a shorter linker length and a stronger positioning sequence than the 208-12 template, and arrays assembled on this template show greater homogeneity of octamer positioning and saturation than 208-12 arrays (9, 21). Additionally, non-H4 histone tails appear to have a less significant role in interarray associations than in 208-12 arrays (9, 11). Nonetheless, in both types of arrays, the H4 tail is the most important mediator of interarray association, suggesting that the H4 tail in 601-177-12 arrays might also mediate direct interarray contacts with histone H2A.
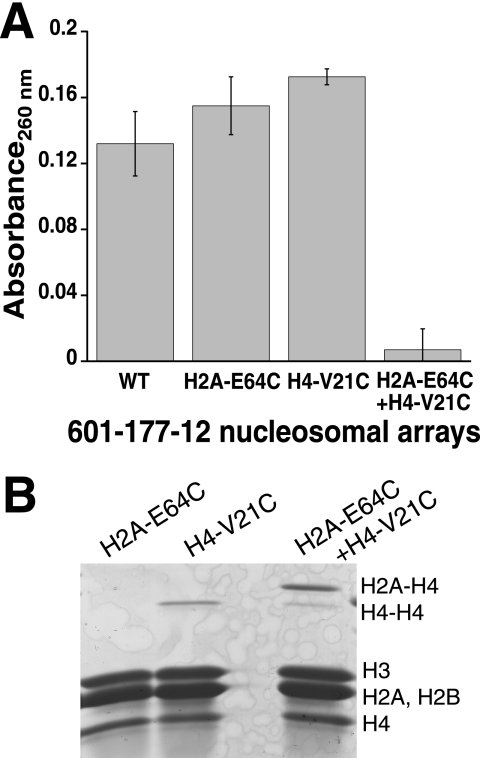
To confirm that direct H4-H2A contacts mediate 601-177-12 array self-association, interarray cross-linking studies were performed with these arrays. Arrays that were well matched and nearly saturated (supplemental Fig. S2) (9) exhibited cross-linking properties very similar to the analogous 208-12 arrays (Fig. 5 and supplemental Fig. S3). In particular, under 1:15 ox/red glutathione oxidation conditions, solutions with a mix of H4-V21C and H2A-E64C arrays show a preference for heterotypic cross-linking and generate a cross-linked species that is readily sedimented (Fig. 5). Thus, the 601-177-12 array system also appears to exhibit direct interarray H4-H2A interactions, making it a suitable system for investigating the relationship between intra- and interarray interactions.
FIGURE 5.
601-177-12 arrays also undergo direct interarray cross-linking. A, differential centrifugation characterization of individual and mixed arrays following 1:15 ox/red glutathione treatment in 4.0 mm MgCl2. The arrays remaining in the supernatant were analyzed as described for Fig. 1D. B, histone composition of individual and mixed arrays following 1:15 ox/red glutathione treatment. Histone components were analyzed as described for Fig. 1E. WT, wild type.
To assess the effect of intramolecular cross-linking on interarray self-association, 601-177-12 arrays were assembled with either octamers containing wild-type histones or with octamers containing both H4-V21C and H2A-E64C histones. Hydrodynamic and self-association characterization of these arrays confirmed that the arrays were nearly saturated and similar in both composition and gross structure (supplemental Fig. S4) (9), making them appropriate for assessing the effects of intra-array cysteine cross-linking.
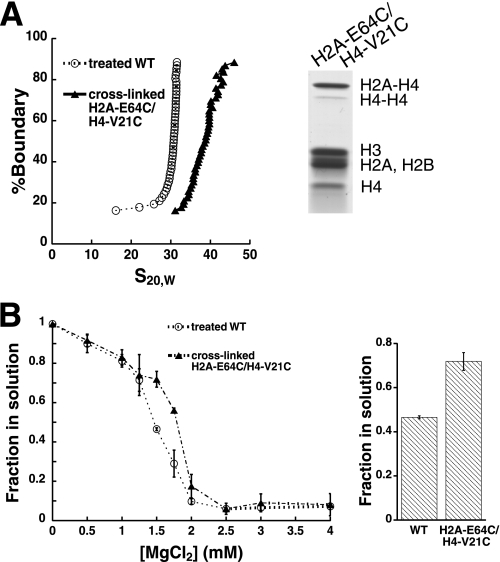
The arrays were subjected to oxidation in 1.0 mm MgCl2, a divalent cation concentration in which intramolecular nucleosome interactions predominate. Following removal of the divalent magnesium cation, sedimentation velocity analysis showed that the cysteine-containing array had a sedimentation coefficient of ∼10 S greater than the wild-type array (Fig. 6A, left panel). This cross-linked species was the predominant form of the array, because relatively little cross-linked species with a large sedimentation coefficient was observed to pellet during initial sedimentation (supplemental Fig. S5). Coupled with the relative uniformity of the distribution of the cross-linked species, these results suggest that array oxidation under these conditions results in reasonably homogeneous arrays with a similar extent of cross-linking. The magnitude of the sedimentation coefficient for the internally cross-linked array is consistent with previous reports, where intra-array cross-linking traps individual arrays in a more compacted state that has an increased propensity to undergo intramolecular compaction to the fully compacted 30-nm fiber (18). Further, this change in array compaction appears to be due to H2A-H4 cross-linking, because it is the predominant cross-linking species (Fig. 6A, right panel).
FIGURE 6.
Intra-array cross-linking disrupts interarray self-association. A, glutathione treatment (1:1 ox/red glutathione with 1.0 mm MgCl2) results in intra-array cross-linking. Shown is the characterization of the arrays described in A following cross-linking treatment. On the left are representative integrated sedimentation coefficient distribution plots obtained in the absence of divalent magnesium ion. On the right are the histone components of these arrays. B, intra-array cross-linking decreases cation-dependent self-association. Shown on the left are differential centrifugation plots for the arrays described in A following cross-linking treatment. The average and standard deviations of the fraction in solution are calculated from three independent cross-linking experiments. The differential centrifugation of these arrays at 1.5 mm MgCl2 is highlighted on the right. The p value is 0.00021 and is calculated from the single tail Student's t test for unpaired data with equal variance. WT, wild type.
The arrays subjected to cross-linking conditions were then assessed for their ability to undergo interarray self-association. Comparison of the wild-type and intramolecularly cross-linked arrays shows that the cross-linked species requires a greater amount of divalent cation to induce comparable differential centrifugation (Fig. 6B). This difference is observable throughout the range of magnesium ion concentrations in which the greatest differential sedimentation occurs (Fig. 6B, left panel) and is highly statistically significant, because the p value for the observed differential centrifugation at 1.5 mm MgCl2 is 0.00021 (Fig. 6B, right panel). Thus, even though the 601-177-12 intra-array cross-linked species exhibits greater compaction that the analogous untreated array, this cross-linking has an opposing effect on interarray association.
DISCUSSION
Role of Direct H4-H2A Interarray Interactions
In chromatin, the acid patch of the histone H2A/H2B dimer serves as a key protein interaction site. Chromatin-associated proteins, such as the Kaposi's sarcoma herpesvirus latency-associated nuclear antigen protein, directly interact with this surface (15, 28). Additionally, direct interaction of the histone H4 tail with this site occurs in forming short range intra-array nucleosome contacts involved in 30-nm chromatin fiber formation (18). Our interarray cross-linking results demonstrate that this site is also directly contacted in long range nucleosome interactions that can mediate higher order chromatin structures beyond the 30-nm fiber. Importantly, this cross-linking increases with increasing interarray interaction, suggesting that the two components are brought into closer proximity during the association process. Such an increase in direct contact is not a general effect of increasing the proximity of cysteine-containing residues during array associations, because cross-linking of H2A-E64C with itself is not observed, and cross-linking of H4-V21C with itself does not increase with increasing array self-association.
Interestingly, the initial divalent magnesium concentration required for half-sedimentation of the cross-linked arrays occurs at 3.0–3.5 mm (Fig. 3A), a midpoint concentration greater than that observed for interarray association of non-cross-linked arrays (Fig. 1C). This suggests that the cross-linked species generated at a given divalent magnesium concentration is not equivalent to the interarray associated species induced by divalent magnesium at that same concentration. This observation may be due to several nonmutually exclusive reasons. One possibility is that divalent magnesium can induce multiple types of species that cannot be distinguished by differential centrifugation. Some of these species may not generate H2A-H4 cross-linking, but the amount of the species that can undergo H2A-H4 cross-linking increases with increasing divalent cation concentration. Another possibility is that because the glutathione system is only weakly oxidizing, at lower magnesium ion concentrations there might not be enough of a driving force to achieve extensive cross-linking. However, at a higher magnesium concentration, an increase in the stability of interarray association could drive cross-linking under the relatively weak oxidizing conditions.
The presence of interarray contact between the H4 tail and the H2A core is further supported by prior observations. In a recent photo-cross-linking study, the amount of cross-linking between these histones was shown to increase with the formation of more extensive higher order chromatin structure, although the extent to which these contacts occurred within versus between arrays was not distinguishable (19). Additionally, the importance of this direct contact is consistent with the wealth of experimental data in which changes in either the H4 tail or H2A core result in changes in array self-association (9–11, 15, 16, 20).
Flexibility in H4-H2A Interarray Interactions
The pattern of interarray cross-linking as a function of the sites of cysteine substitutions indicates some conformational flexibility in the mode of H4-H2A interactions. In the crystal contacts of the first high resolution mononucleosome structure, the H4 tail residue H4 valine 21 is oriented toward histone residue H2A glutamate 64, whereas the H4 tail residues directly flanking this valine residue, lysine 20 and leucine 22, do not (1). Nonetheless, both of these flanking residues show preferential H2A-H4 cross-linking with a magnitude similar to cross-linking between H4-V21C and H2A-E64C. This shows that some slippage in the mode of interaction between the H4 tail and the H2A/H2B acidic patch can be accommodated without significantly disrupting the extent of nucleosomal array self-association. This flexibility in interaction may be a feature specific to interarray associations, because only a minor degree of cross-linking was observed between H4-K20C and H2A-E64C when intra-array cross-linking was studied (18).
When cross-linking of residue H2A-Glu64 with residues flanking H4-Val21 occurs, the changes in the overall H4 tail-H2A interaction may be confined to residues near this site of interaction. Alternatively, this change might result in movement of the entire H4 tail relative to the H2A/H2B acidic patch. Although the latter possibility presents a more dramatic change in structure, recent experimental data suggests that such a structural change is possible. Specifically, Hansen and co-workers (12) have shown that in nucleosomal arrays, the H4 tail can be replaced with a number of other histone tail sequences with similar charge densities with maintenance or improvement in the degrees of array self-association. This suggests that the H4 tail interactions with the H2A/H2B acidic patch may not require a single, defined mode of interaction. Nonetheless, there does appear to be limits as to how much reorientation of the tail can occur. Replacement of H4-Arg23 with a cysteine does not result in cross-linking to H2A-E64C. This suggests that the proximity of these two sites is reduced under the physical constraints of the orientation of the residues imposed by array self-association,.
Role of the H4 Tail and H2A Core Outside of Their Direct Contacts
The less than full extent of interarray cross-linking suggests that the H4 tail and H2A-H2B acid patch have other potential roles in facilitating array self-association. In our experiments, H2A-H4 cross-linking is not quantitative, because significant non-cross-linked H4 histone is observed. This is presumably true of H2A, as well. However, the recombinant H2A histones are not readily resolved from H2B histones under our experimental conditions. This lack of total H2A-H4 cross-linking could be accounted for in a variety of ways. One, up to half of the actual H2A-H4 interaction may not result in disulfide cross-links because not every interaction in the H2A-E64C/H4-V21C self-associated arrays would necessarily have two cysteine residues present. For example, a nucleosome in an H2A-E64C array could be equally likely to be next to an array containing H2A-E64C nucleosomes (and wild-type H4) as to be next to one containing H4-V21C nucleosomes (and a wild-type H2A). Two, not every H2A-H4 interaction would result in a disulfide linkage if the reaction did not go to completion. This could be due to glutathione oxidation not reaching a steady state. However, in our system we do not believe that this is the case, because longer reaction times do not appear to significantly change the extent of cross-linking (data not shown). Alternatively, such incomplete reaction might be due to cysteines being inaccessible within densely packed self-associated arrays. However, it is likely that such inaccessibility does not explain incomplete cross-linking in its entirety. Similar nonquantitative disulfide cross-linking is observed for intra-array cross-linking, where accessibility is not expected to be an issue (Fig. 6 and Ref. 18). As a third possibility, the lack of quantitative H2A-H4 cross-linking may reflect that not all H4 tails and H2A histones are involved in direct contacts and are available to play alternative roles in facilitating array self-association. Prior data support several alternative roles. The H4 histone alone can interact with DNA, H2A/H2B histones, and H3/H4 histones (15), and in the context of nucleosomal arrays, the H4 tail has been shown to interact with array DNA in trans, especially at residues near the amino terminus (19). It is also important to note that the H4 tail and H2A/H2B patch are not the only mediators of interarray association, because neither the H4 tail nor the H2A/H2B histones are absolutely required (9, 20).
In addition to H2A-H4 cross-linking, H4-H4 cross-linking can also be observed. In mixed arrays with the H4 cysteine at position 21, some of this H4-H4 cross-linking occurs between arrays, because these arrays can be differentially sedimented (Fig. 1D). However, it appears that stable H4 interactions are not predominantly interarray in nature. H4-H4 cross-linking persists under less forcing oxidation conditions in which differential sedimentation does not occur (Fig. 2) but does not increase at the higher MgCl2 concentrations that increase interarray association (Figs. 3 and supplemental Fig. S3). The extent and potential nature of these H4-H4 interactions also appear to vary with H4 cysteine position (Fig. 4), suggesting that certain locations on the H4 tail might be better positioned to engage in H4-H4 interactions.
Interplay between Intra- and Interarray Associations
In nucleosomal array systems the interplay between intra- and interarray interactions is poorly understood. When arrays containing wild-type histones are subjected to lower concentrations of divalent magnesium cation, intra-array interactions occur preferentially over interarray interactions (Figs. 1C and supplemental Fig. S2) (9, 21). This suggests that at the higher divalent magnesium concentrations required for stable interarray interactions (Fig. 1C and supplemental Fig. S2) and H2A-H4 cross-linking (Figs. 3 and supplemental Fig. S3), intra-array interactions precede interarray interactions. However, how existing intra-array interactions influence interarray interactions is not clear. Formation of the intra-array compacted species could present interaction sites in a way that facilitates or hinders nucleosome interactions between strands (or potentially not affect them at all). Indeed, existing data with arrays containing mutated or truncated wild-type histones and histone variants can be used to support each of these models (8–11, 13, 15, 16, 20). Further, the necessity that intra-array interactions precede interarray ones in wild-type array is not certain, because the relative stability of intra-array versus interarray associations may be different at higher divalent magnesium concentration.
Our results indicate that stabilizing internucleosomal associations through intra-array H2A-H4 cross-linking antagonizes interarray self-association. The magnitude of the shift in divalent magnesium sensitivity is statistically significant and is greater than one-third the effect of the complete loss of the H4 tail (13), which of all of the histone tails is the only one whose loss affects interarray association of 601-177-12 arrays (9). Moreover, we expect that what we observe is less than the full magnitude of the antagonism, because complete antagonism is difficult to reproduce experimentally. In our experiments the intra-array cross-linked species does not have a sedimentation coefficient of 50–60 S, characteristic of fully compacted 30-nm fiber (9). This result is similar to those obtained by other groups (18) and might be due to incomplete intra-array H2A-H4 cross-linking (Fig. 6B). With some of these interaction sites still available to facilitate interarray association, the full degree of potential antagonism between intra- and interarray associations is likely to be masked.
The antagonism of interarray association through intra-array cross-linking can be interpreted in at least two different ways. The observed effect may be a specific result of using 601-177-12 arrays. These arrays were originally designed to facilitate intra-array association by virtue of their well positioned nucleosomes and relatively short linker lengths. This results in quantitative differences between these arrays and less homogeneous array systems, such as 208-12 arrays. Specifically, 601-177-12 arrays form inter- and intra-array interactions and interarray cross-links at lower concentrations of divalent magnesium than the analogous 208-12 arrays (8, 9, 21) (Figs. 1C and 3A and supplemental Figs. S2 and S3). However, in the interarray association experiments with 601-177-12 arrays that have been cross-linked within the array (Fig. 6B), this difference is minimized, because the midpoint association MgCl2 concentration (1.8 mm) is nearly the same as that for non-cross-linked 208-12 arrays (Fig. 1C). This could mean that intra-array cross-linking of the 601-177-12 array makes this array function more like the non-cross-linked 208-12 array. However, this resemblance seems limited to interarray association. Prior studies with the 601-177-12 intra-array cross-linked species showed that this cross-linking increased its propensity to undergo intra-array compaction (18), making its behavior resemble a 208-12 array even less so than when the 601-177-12 array is not cross-linked.
An alternative interpretation of the antagonism of interarray association by intra-array cross-linking is that these forms of interactions are antagonistic for all chromatin. Qualitatively, 601-177-12 and 208-12 arrays behave similarly with respect to interarray and intra-array interactions (8, 9, 21) (Fig. 1C and supplemental Fig. S2). Similarly, the H4-H4 interarray cross-linking in the 601 array is largely independent on the MgCl2 concentration, whereas the H4-H2A interarray cross-linking is highly dependent (Figs. 3A and supplemental Fig. S3). The quantitative differences seen in 601-177-12 arrays could be attributed to the propensity of the well positioned nucleosomes and relatively short linker lengths to facilitate interactions that already occur in 208-12 systems. With respect to linker length differences, experimental data support this structural similarity. Although 601 arrays with a repeat length less than 177 bp, such as those used for the first tetranucleosome structure (29), do show structural differences from 601 arrays with a longer repeat length (30), arrays with nucleosome repeat lengths from 177 to 207 bp can form similar structures (31).
In a general model of intra- and interarray antagonism, the direct H4-H2A interaction between nucleosomes in a strand would prevent such interactions between strands. This could occur by direct competition, where tying up H2A-H4 contacts in one type of interaction would prevent them from being used for other interactions. Additionally, the structure formed by one type of interaction, such as the intra-array compacted species, could be less compatible with interarray associations. For example, if interarray associations required interdigitation between nucleosomes within an individual strand, intra-array compaction would disfavor this interaction. For such models, some histone mutations or variants would stabilize one type of interaction at the cost of the other (10, 16), whereas others, such as the loss of the H4 tail, as well as other tails, could detrimentally affect the stability of both interactions (9–11).
Altogether, our data shed new light on the nature and role of the interaction between the histone H4 tail and the H2A/H2B acidic patch in intra-array compaction and interarray self-association. Further studies to place this interaction in the context of other factors in intra- and interarray association will provide a fuller understanding of higher order chromatin structure.
Supplementary Material
Acknowledgment
We thank Melissa Blacketer for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant GM79663 (to M. A. S.-K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- ox/red
- oxidized to reduced.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. (2002) J. Mol. Biol. 319, 1097–1113 [DOI] [PubMed] [Google Scholar]
- 3.van Holde K. E. (1988) Chromatin, Springer-Verlag, New York [Google Scholar]
- 4.Belmont A. S., Bruce K. (1994) J. Cell Biol. 127, 287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Holde K., Zlatanova J. (2007) Semin. Cell Dev. Biol. 18, 651–658 [DOI] [PubMed] [Google Scholar]
- 6.Lu X., Klonoski J. M., Resch M. G., Hansen J. C. (2006) Biochem. Cell Biol. 84, 411–417 [DOI] [PubMed] [Google Scholar]
- 7.Fletcher T. M., Hansen J. C. (1995) J. Biol. Chem. 270, 25359–25362 [DOI] [PubMed] [Google Scholar]
- 8.Schwarz P. M., Felthauser A., Fletcher T. M., Hansen J. C. (1996) Biochemistry 35, 4009–4015 [DOI] [PubMed] [Google Scholar]
- 9.Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 10.Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 11.Gordon F., Luger K., Hansen J. C. (2005) J. Biol. Chem. 280, 33701–33706 [DOI] [PubMed] [Google Scholar]
- 12.McBryant S. J., Klonoski J., Sorensen T. C., Norskog S. S., Williams S., Resch M. G., Toombs J. A., 3rd, Hobdey S. E., Hansen J. C. (2009) J. Biol. Chem. 284, 16716–16722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Hayes J. J. (2008) Mol. Cell Biol. 28, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chodaparambil J. V., Barbera A. J., Lu X., Kaye K. M., Hansen J. C., Luger K. (2007) Nat. Struct. Mol. Biol. 14, 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. (2007) Nat. Struct. Mol. Biol. 14, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 17.Luger K., Richmond T. J. (1998) Curr. Opin. Gen. Dev. 8, 140–146 [DOI] [PubMed] [Google Scholar]
- 18.Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 19.Kan P. Y., Caterino T. L., Hayes J. J. (2009) Mol. Cell Biol. 29, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen J. C., Wolffe A. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2339–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carruthers L. M., Tse C., Walker K. P., 3rd, Hansen J. C. (1999) Methods Enzymol. 304, 19–35 [DOI] [PubMed] [Google Scholar]
- 22.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 23.Demeler B., van Holde K. E. (2004) Anal. Biochem. 335, 279–288 [DOI] [PubMed] [Google Scholar]
- 24.Simpson R. T., Thoma F., Brubaker J. M. (1985) Cell 42, 799–808 [DOI] [PubMed] [Google Scholar]
- 25.Horn P. J., Crowley K. A., Carruthers L. M., Hansen J. C., Peterson C. L. (2002) Nat. Struct. Biol. 9, 167–171 [DOI] [PubMed] [Google Scholar]
- 26.Schwarz P. M., Hansen J. C. (1994) J. Biol. Chem. 269, 16284–16289 [PubMed] [Google Scholar]
- 27.Lowary P. T., Widom J. (1998) J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 28.Barbera A. J., Chodaparambil J. V., Kelley-Clarke B., Joukov V., Walter J. C., Luger K., Kaye K. M. (2006) Science 311, 856–861 [DOI] [PubMed] [Google Scholar]
- 29.Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 30.Routh A., Sandin S., Rhodes D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson P. J., Fairall L., Huynh V. A., Rhodes D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.