Abstract
MnmE is a GTP-binding protein conserved between bacteria and eukarya. It is a dimeric three-domain protein where the two G domains have to approach each other for activation of the potassium-stimulated GTPase reaction. Together with GidA, in a heterotetrameric α2β2 complex, it is involved in the modification of the wobble uridine base U34 of the first anticodon position of particular tRNAs. Here we show, using various spin-labeled MnmE mutants and EPR spectroscopy, that GidA binding induces large conformational and dynamic changes in MnmE. It stimulates the GTPase reaction by stabilizing the GTP-bound conformation in a potassium-independent manner. Surprisingly, GidA binding influences not only the GTP- but also the GDP-bound conformation. Thus GidA is a new type of regulator for a G protein activated by dimerization.
Keywords: Biophysics, Electron Paramagnetic Resonance (EPR), G Proteins, Protein-Protein Interactions, Transfer RNA (tRNA), G Protein Activated by Dimerization, GidA, MnmE, tRNA Modification
Introduction
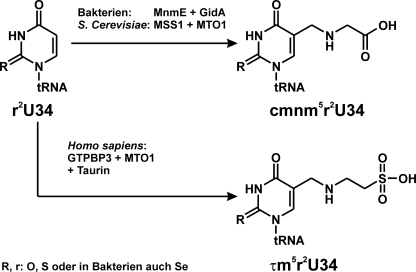
MnmE belongs to the class of guanine nucleotide-binding proteins (G proteins) that are involved in the regulation of many cellular processes such as protein biosynthesis, vesicular and nucleo-cytoplasmic transport, and signal transduction. It is a multidomain GTP-binding protein conserved between bacteria and eukarya and, together with the also conserved FAD-binding protein GidA (also named MnmG), accomplishes the first step in the modification of the wobble uridine base at the first anticodon position of particular transfer RNAs (1). Specifically, MnmE together with MnmE/GidA catalyzes the formation of a carboxymethylaminomethyl (cmnm)4 group at the 5 position of the wobble uridine (U34) of tRNAs reading 2-fold degenerated codons ending with A or G in a number of bacteria (Fig. 1) (2–4). It has been shown that the presence of these two enzymes is sufficient for the formation of cmnm5U34 in vitro with albeit very slow kinetics and that the GTPase reaction is required (4, 5). This modification (cmnm5U34) combined with thiolation at the 2 position favors the interaction with A and G but suppresses base pairing with C and U (6–10). Moreover, hypermodified U34 plays a regulatory role in gene expression (11). The eukaryotic homologues of MnmE and MnmE/GidA (termed MSS1 and Mto1, respectively, in yeast) are targeted to mitochondria (12, 13). The human homologues, hGTPBP3 and Mto1 have been implicated in the development of mitochondrial myopathies such as MERRF (myoclenic epilepsy ragged red fibers), MELAS (mitochondrial encephalomyopathy lactic acidosis stroke), and nonsyndromic deafness (14–18). Schemes depicting the tRNA modification reaction of MnmE-GidA and its eukaryotic homologues are shown in Fig. 1.
FIGURE 1.
Modification of tRNA by MnmE and GidA. Modifications at the C5 atom of uridine 34 by MnmE-GidA in bacteria (cmnm), MSS1-MTO1 in yeast (cmnm), and GTPBP3-MTO1 in Homo sapiens (taurinomethyl, τm).
MnmE has been shown to be dimeric with each monomer consisting of three domains: an N-terminal domain responsible for constitutive dimerization, a central helical domain, and the G domain (1). In contrast to Ras-like small G proteins, which require a guanine nucleotide exchange factor to drive the nucleotide exchange and a GTPase-activating protein to stimulate hydrolysis (19, 20), MnmE is a G protein activated by dimerization (21). G proteins activated by nucleotide-dependent dimerization display lower affinities toward nucleotides and have a higher intrinsic GTP hydrolysis activity, which in the case of MnmE is stimulated by the presence of K+ (1, 22, 23). Although in the crystal structures of full-length protein, the G domains are more or less apart from each other, the structures of the isolated G domain in the presence of the transition state analogue GDP-AlFx (24) and potassium indicated that the G domains come close to each other and that the close juxtaposition rearranges the active site to a catalytic competent conformation. Here, K+ is responsible for dimerization and for stabilization of the transition state. This stabilizing effect is mediated by providing a positive charge into the catalytic site in an arginine finger-like manner (23), as deduced from its localization found in the crystal structure of the isolated G domains with GDP-AlF4− of Escherichia coli MnmE, where it is coordinated by the K-loop of switch I, the phosphates, and AlF4− (23). The exact role of GTP hydrolysis in the modification reaction is still not clear, but it has been shown that the G domains have to cycle between an open, dissociated conformation and a closed, dimerized conformation to drive the modification reaction, suggesting that conformational changes coupled to G domain dimerization are required to orchestrate the complex modification reaction (4). However, the role of these structural dynamics for the activity of the other cofactor active sites (NADH and FAD sites on GidA; 5-formyl-THF-site on MnmE) is not known.
Conformational changes in proteins can be addressed by measuring the distances between sites appropriately located in the protein-protein complex under investigation. Site-directed spin labeling in combination with EPR techniques provide the means to measure distances between paramagnetic centers by quantifying dipole-dipole coupling. Double-electron electron resonance (DEER) spectroscopy (25, 26) is a pulse EPR technique with an accessible distance range of ∼2–7 nm. This technique measures the dipole-dipole coupling frequency, which is inversely proportional to the cube of the distance, by monitoring the influence of a spin population excited by a 180° microwave pulse on the amplitude of a refocused echo of another spin population at a different microwave frequency.
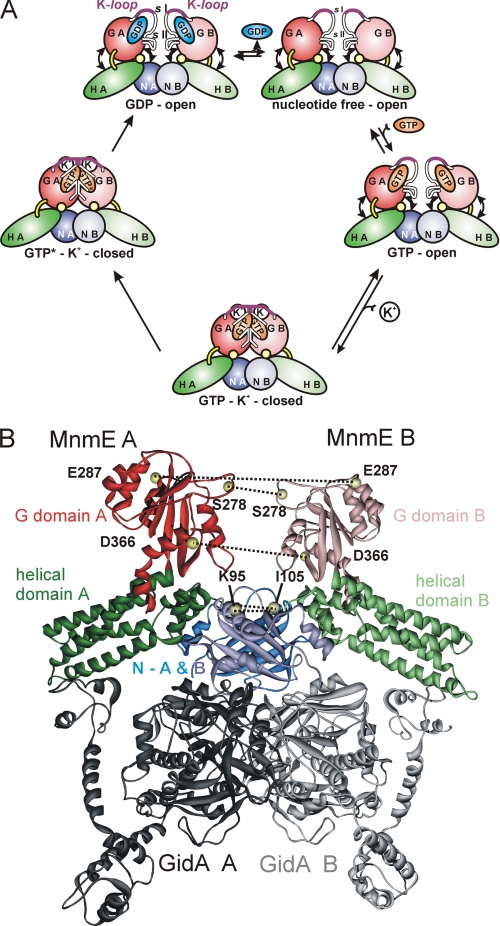
Using EPR, we previously showed that the G domains adopt an open conformation in the nucleotide-free and the GDP-bound states, an open-closed two-state equilibrium in the GppNHp-bound state (a stable triphosphate analogon; Fig. 2A), and a closed conformation in the presence of GDP-AlFx (27). In addition we corroborated the dependence of G domain dimerization on the presence of potassium, or more precisely of monovalent cations of a specific size, as suggested before by biochemical and fluorescence data (23). Recapitulation of these data suggests a model for the G domain conformational states of the GTPase cycle as shown in Fig. 3A.
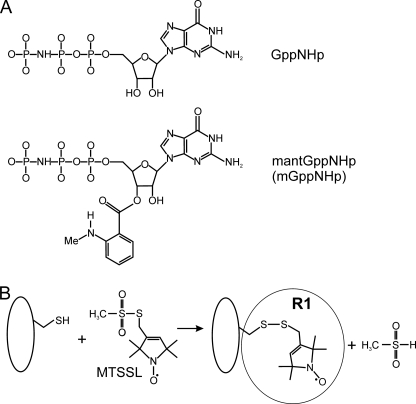
FIGURE 2.
Structures of the nucleotide analogues GppNHp und mantGppNHp (A) and reaction of the MTSSL with the sulfhydryl group of a cysteine side chain, generating the spin label side chain R1 (B).
FIGURE 3.
A, schematic model for the G domain conformational states during the GTPase cycle. The G domains are colored in red, the α-helical domains are in green, and the α/β-domains are in blue. The key structural elements of the G domain (K-loop, Switches I and II (sI and sII, respectively) are indicated. In the GDP state the G domains adopt an open conformation, independent of the presence of K+. Switches I and II and the K-loop are disordered. After GTP binding, which because of the low nucleotide affinity takes places via a nucleotide-free intermediate, the G domains exhibit an equilibrium between the open and closed conformations. In the presence of K+, this equilibrium is shifted toward the closed state, in which K+ binds to the K-loop, which becomes structured, and the switch regions form the dimer interface. Thereby the catalytic machinery for GTP hydrolysis is assembled, and the transition state of GTP hydrolysis (indicated by GTP*) is stabilized by G domain dimerization. Details of the steps following hydrolysis, dissociation of the γ-phosphate group and GDP, have not been elucidated so far. B, structural model of the MnmE-GidA complex based on interaction studies with truncated and mutated protein constructs (29). MnmE (Protein Data Bank entry 1XZP) and GidA (Protein Data Bank entry 3CP8) are shown in ribbon representation. Positions mutated to cysteine and subsequently labeled with MTSSL are indicated by yellow spheres at the positions of the respective Cα atoms. The MnmE domains are colored as in A. The GidA dimer is shown in gray.
Based on recent studies with the E. coli proteins, it was further suggested that GidA and MnmE form a functional complex in which they directly cooperate instead of independently catalyze sequential steps in the modification reaction as previously stated (28, 29). Site-directed mutagenesis data led to the conclusion that complex formation between MnmE (α) and GidA (β) most likely takes place by generating a 2-fold symmetrical α2β2 heterotetramer in which the α-helical domain of MnmE and the last three C-terminal helices of GidA represent the main interprotein contact sites (29).
GidA comprises three domains: (i) an FAD-binding domain and (ii) an NADH-binding domain, which are involved in the homodimerization of GidA, as well as (iii) a C-terminal domain that is in ideal geometry to interact with the helical domain of MnmE (29). In a recent publication Meyer et al. (4) showed by fluorescence and biochemical techniques that GidA influences G domain dimerization and the GTPase reaction of MnmE and partly abolishes the potassium dependence described for the GTPase activity. The current model for the MnmE-GidA complex suggests that GidA is not in contact with the G domains but bound to the opposite side of the MnmE dimer (Fig. 3B), indicating that GidA binding is communicated to the G domains by conformational changes in other parts of MnmE.
In the present study, applying site-directed spin labeling in combination with DEER spectroscopy to sites in the MnmE G domains and the α/β domains (Fig. 3B), we provide direct evidence that GidA directly influences the switch cycle of MnmE by stabilizing both the GDP- and GTP-bound conformations. Furthermore, we demonstrate that the potassium dependence of G domain dimerization is largely abolished in the presence of GidA.
MATERIALS AND METHODS
Mutant Construction, Protein Preparation, and Spin Labeling
Cloning, expression, and purification of E. coli MnmE and GidA proteins (EcMnmE and EcGidA, respectively) and preparation of nucleotide-free MnmE was carried out as described elsewhere (4, 27). GidA preparations, after the final purification step via gel filtration, do not contain FAD and NADH, as confirmed by UV absorption spectroscopy. Purified, nucleotide-free Cys mutants of E. coli MnmE-C451S were pretreated with dithioerythritol (4 °C). After removal of dithioerythritol, the protein solutions were incubated with 1–5 mm 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethio-sulfonate spin label (MTSSL; Toronto Research, Alexis Biochemicals) for 16 h (4 °C). The unbound free MTSSL was removed by gel filtration. The labeling efficiencies were determined to be >50% in all cases.
Fluorescence Stopped Flow Kinetics of mGppNHp Binding
In mGppNHp binding experiments, 60 μm MnmE or preformed MnmE-GidA complex was mixed in a stopped flow apparatus (SM-17; Applied Photophysics) in a 1:1 ratio with 10 μm mGppNHp, a fluorescently labeled stable GTP analogue. The mant fluorophor was excited at 360 nm, and the fluorescence time course was monitored through a 408-nm cut-off filter. The experiments were carried out at 20 °C in 50 mm Tris, pH 7.5, 5 mm MgCl2, 2 mm dithioerythritol, and 100 mm of either NaCl or KCl.
Fluorescence Equilibrium Titration
Fluorescence equilibrium titrations to determine the dissociation constants (KD) of spin-labeled mutant MnmE proteins and GidA was performed as described in Ref. 4: GidA at a fixed concentration (ranging between 2 and 3 μm) was titrated with spin-labeled mutant MnmE protein at 20 °C in 50 mm Tris, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 1 mm GDP using the quench of the GidA-bound FAD fluorescence (excitation, 450 nm; emission, 526 nm), and titration data points were fitted to a quadratic function.
MnmE-GidA Complex Preparation for EPR Measurements
MnmE and GidA were mixed in a 1:1 molar ratio at protein concentrations between 200 and 500 μm. According to the dissociation constants of 0.07 (with GppCp and KCl) to 3.0 μm (in the presence of GDP and NaCl) determined for the MnmE-GidA complex (4), the complex is always saturated at the protein concentrations used. Buffer conditions for the EPR experiments were 100 mm KCl or NaCl, 50 mm Tris-HCl, 5 mm MgCl2, pH 7.4, with 5% (v/v) ethylene glycol (for H2O buffer) or 12.5% (v/v) glycerol-d8 (for D2O buffer), and 1 mm GDP, 1 mm GppNHp, or 1 mm GDP, 1 mm AlCl3, 10 mm NaF, respectively.
Continuous Wave EPR Measurements
Room temperature continuous wave (cw) EPR spectra were recorded using a Miniscope X-band benchtop EPR spectrometer (MS200; Magnettech GmbH, Berlin, Germany) equipped with a rectangular TE102 resonator fluxed with gaseous nitrogen to keep the temperature stable. The microwave power was set to 10 milliwatts, and the B-field modulation was set to 0.15 mT. 10 μl of sample volume containing protein concentrations of 200–500 μm were filled in EPR glass capillaries (0.9-mm inner diameter).
Pulse EPR Measurements
Pulse EPR experiments (DEER) were accomplished at X-band frequencies (9.3–9.4 GHz) with a Bruker Elexsys 580 spectrometer equipped with a Bruker Flexline split-ring resonator ER 4118X-MS3 and a continuous flow helium cryostat (ESR900; Oxford Instruments) controlled by an Oxford Intelligent Temperature Controller ITC 503S.
All of the measurements were performed using the four-pulse DEER sequence: π/2(υobs) − τ1 − π(υobs) − t′ − π(υpump) − (τ1 + τ2 − t′) − π(υobs) − τ2 − echo (25, 26). A two-step phase cycling (+ <x , − <x
, − <x ) was performed on π/2(υobs). Time t′ is varied, whereas τ1 and τ2 are kept constant, and the dipolar evolution time is given by t = t′ − τ1. The data were analyzed only for t > 0. The resonator was overcoupled to Q = ∼100; the pump frequency υpump was set to the center of the resonator dip and coincided with the maximum of the nitroxide EPR spectrum, whereas the observer frequency υobs was 65 MHz higher, coinciding with the low field local maximum of the spectrum. All of the measurements were performed at a temperature of 50 K with observer pulse lengths of 16 ns for π/2 and 32 ns for π pulses and a pump pulse length of 12 ns. Proton modulation was averaged by adding traces at eight different τ1 values, starting at τ1,0 = 200 ns and incrementing by Δτ1 = 8 ns. For proteins in D2O buffer with deuterated glycerol used to prolong the phase relaxation, the corresponding values were τ1,0 = 400 ns and Δτ1 = 56 ns. The data points were collected in 8-ns time steps or, if contributions to the distance distribution below an appropriate threshold were absent, in 16- or 32-ns time steps. The total measurement time for each sample was 4–24 h. Analysis of the data were performed with DeerAnalysis 2006.1/2008 (30). A detailed description of the analysis exemplified on the data set for MnmE-E287R1 + GidA + GppNHp + K+ is given in the supplementary material.
) was performed on π/2(υobs). Time t′ is varied, whereas τ1 and τ2 are kept constant, and the dipolar evolution time is given by t = t′ − τ1. The data were analyzed only for t > 0. The resonator was overcoupled to Q = ∼100; the pump frequency υpump was set to the center of the resonator dip and coincided with the maximum of the nitroxide EPR spectrum, whereas the observer frequency υobs was 65 MHz higher, coinciding with the low field local maximum of the spectrum. All of the measurements were performed at a temperature of 50 K with observer pulse lengths of 16 ns for π/2 and 32 ns for π pulses and a pump pulse length of 12 ns. Proton modulation was averaged by adding traces at eight different τ1 values, starting at τ1,0 = 200 ns and incrementing by Δτ1 = 8 ns. For proteins in D2O buffer with deuterated glycerol used to prolong the phase relaxation, the corresponding values were τ1,0 = 400 ns and Δτ1 = 56 ns. The data points were collected in 8-ns time steps or, if contributions to the distance distribution below an appropriate threshold were absent, in 16- or 32-ns time steps. The total measurement time for each sample was 4–24 h. Analysis of the data were performed with DeerAnalysis 2006.1/2008 (30). A detailed description of the analysis exemplified on the data set for MnmE-E287R1 + GidA + GppNHp + K+ is given in the supplementary material.
RESULTS
Stopped Flow Analysis of mGppNHp Binding Kinetics
Previously, real time fluorescence measurements indicated that GidA stabilizes the dimerized, closed state of the MnmE G domains, which can be trapped by binding of the transition state analogue GDP-AlFx to the nucleotide-binding pocket of MnmE (4). Incorporation of AlFx into the γ-phosphate binding site of MnmE and concomitant G domain dimerization strictly requires K+ or ions with similar ionic radii such as Rb+ or NH4+ (23, 27). However, in the presence of GidA, the potassium dependence is relieved, and AlFx-induced G domain dimerization is possible in the absence of K+ (4). In contrast to the transition state analogue GDP-AlFx, the stable triphosphate analogue GppNHp is not capable of fully stabilizing the dimerized state of the MnmE G domains. Although in the presence of Na+ only the open state is observed, an equilibrium between an open and a closed conformation is observable in the presence of K+, as revealed by distance distributions obtained from four-pulse DEER experiments (27).
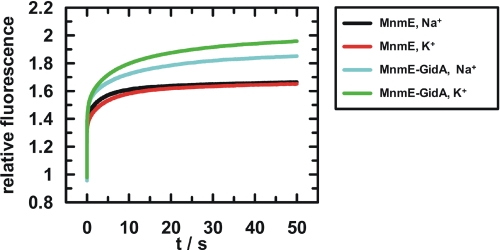
To further investigate whether GidA has a stabilizing effect on G domain dimerization, we followed the stopped flow kinetics of binding of the fluorescence-labeled stable GTP analogue mGppNHp (Fig. 2A) to MnmE or to the MnmE-GidA complex in the presence of Na+ or K+ (Fig. 4). For the isolated MnmE protein, an increase in the fluorescence amplitude indicates binding of mGppNHp (Fig. 4, black and red curves). In contrast to experiments in the presence of GidA, the kinetics and the equilibrium fluorescence amplitude levels are almost equal in the presence of Na+ and K+. For the MnmE-GidA complex a higher equilibrium fluorescence amplitude is observed (Fig. 4, cyan and green curves) in comparison with the MnmE protein alone, which we attribute to G domain dimerization stabilized by GidA. The even higher fluorescence amplitude in the presence of K+ indicates that the effect of GidA and K+ on the closed state of the G domains is additive and that both components are required for full stabilization. This is well in line with the previously reported real time fluorescence analysis of transition state analogue binding by the MnmE-GidA complex (4).
FIGURE 4.
Stopped flow analysis of mGppNHp binding to MnmE or the MnmE-GidA complex. 60 μm MnmE with or without 60 μm GidA was mixed in a stopped flow apparatus with 10 μm mGppNHp in the presence of either 100 mm NaCl or KCl as indicated in the legend. Mant fluorescence was excited at 360 nm and monitored through a 408-nm cut-off filter.
DEER Spectroscopic Analysis of the Conformation States of the G Domains in the MnmE-GidA Complex
To gain direct structural information on the effect of GidA on the conformational states of the MnmE G domains and to specify and characterize the influence of GidA on the GTPase reaction of MnmE, we applied four-pulse DEER spectroscopy (25, 26, 31–33) to measure distances between nitroxide spin labels in the G domains of full-length EcMnmE together with EcGidA in different steps of the GTPase cycle. Residues mutated to cysteines for spin labeling with MTSSL are Glu287, close to the top of the G domain in Gα2; Ser278 in switch II; and Asp366, located in Gα5, as shown in Fig. 3B. Introduction of a spin label results in two symmetry-related labels in the functional MnmE dimer. Furthermore, we labeled positions 95 and 105 in the N-terminal domain, for which no distance changes are expected. The calculated distances between these sites derived from the structures of the open state and the model of the closed state of MnmE are listed in Table 1.
TABLE 1.
Cα -Cα distances
The residue numbering is according to the E. coli MnmE sequence.
| Residue mutated to Cys | Nucleotide state | Cα-Cα distance from x-ray structures |
|---|---|---|
| Å | ||
| S278R1, switch II | apo | 23a |
| GDP-AlFx | 14b | |
| E287R1, Gα2 | apo | 54a |
| GDP-AlFx | 30b | |
| D366R1, Gα5 | apo | 66a |
| GDP-AlFx | 48b | |
| K95R1, N-terminal domain | apo | 38 |
| GDP-AlFx | NDc | |
| I105R1, N-terminal domain | apo | 35a |
| GDP-AlFx | ND |
a Thermotoga maritima homodimer model (generated with Protein Data Base entry 1XZP).
b From E. coli G domain dimer (Protein Data Base entry 2GJ8).
c ND, not determined as no crystal structure of full-length MnmE in the GDP-AlFx state is available.
Previously it was established that no impairment of the GTPase activity in comparison with the wild type protein could be observed for any of the mutations with and without the bound spin label (27). Intact complex formation between spin-labeled MnmE proteins and GidA was verified by determining the dissociation constants (KD) for the spin-labeled, GDP-bound complexes with a previously established fluorescence titration assay (4) (supplemental Table S1). As previously shown the interaction between MnmE and GidA is drastically strengthened in the presence of triphosphate or transition state analogues, whereas K+ or Na+ ions only have a minor effect on the affinity (4). The protein concentrations in the EPR experiments (200–500 μm) well above the KD values of the spin-labeled complexes (between 1.3 and 4.2 μm in the GDP-bound state) thus ensure that ∼90% of complex is present in the GDP-bound state, as calculated from the bimolecular mass law equation (supplemental Table S1). The fraction of complex is even higher in the nucleotide-free or GDP-AlFx/GppNHp-bound state, because the affinity is higher in these states (4). The experimental distances given in the following represent interspin distances. Resulting from the conformational distribution of the spin label side chain (Fig. 2B), this distance can deviate from the Cα-Cα distance by 0.4–1.2 nm (32), and this possible deviation has to be taken into account when the DEER distances are compared with the distances shown in Table 1.
Conformational Changes in MnmE upon Association with GidA
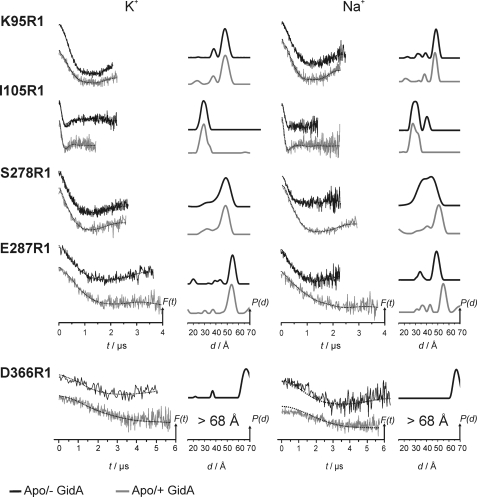
To reveal possible conformational changes in the N-terminal dimerization domains as well as in the G domains of MnmE upon binding of GidA, distance distributions for the nucleotide-free state were determined for the sites K95R1, I105R1, S278R1, E287R1, and D366R1 (R1 denotes the MTSSL side chain) in the presence of two different cations, i.e. K+ and Na+. A comparison of the dipolar time traces (left column) and the corresponding distance distributions (right column, obtained by Tikhonov regularization; see “Materials and Methods”) for MnmE in the absence and in the presence of GidA is shown in Fig. 5. Positions 95 and 105 show just minor influences of GidA on the distance distributions. First, the major distance for K95R1 with Na+ is slightly shifted to shorter distances (∼1 Å). Second, the distance distributions for position 105, both with Na+ and with K+, become slightly narrower and more defined, as shoulders indicating the presence of two distinct spin label rotamers become visible. Third, for I105R1, the fraction at ∼40 Å disappears upon binding of GidA, although we cannot exclude that the 40 Å distance peak for position 105 in the absence of GidA might be caused by insufficient background subtraction because of the short dipolar evolution time in this experiment. Nevertheless, changes caused by the presence of GidA are clearly visible from the dipolar evolution data. These results indicate that binding of GidA to MnmE also affects the structure of the N-terminal dimerization domain, but major structural rearrangements are not observed.
FIGURE 5.
Conformational changes in MnmE upon association with GidA in the apo state in the presence of K+ (left panel) and Na+ (right panel). In each panel, the left column shows background corrected dipolar evolution data, F(t), and the right column shows distance distributions obtained by Tikhonov regularization, P(t). All of the plots are normalized by amplitude. The modulation depths in all cases coincided with the labeling efficiencies obtained from the cw EPR measurements. The broken lines in the left columns are fits to the data obtained by Tikhonov regularization.
In contrast, the G domains of MnmE are strongly affected by the presence of GidA, and these effects are more pronounced in the presence of Na+. Complex formation with GidA in the presence of Na+ leads to a significantly narrower and more defined distance distribution for the spin label attached at position 278, indicating that switch II becomes significantly more ordered in the presence of GidA. In addition, the mean distance is shifted to longer distances by ∼10 Å, leading in combination with peak narrowing to a distance distribution almost identical to that found for K+.
The distance distributions for position 287 in Gα2 are almost not affected by the presence of GidA with K+ but are significantly shifted to longer distances by ∼5 Å in the presence of Na+. As a result, similar to the observation for S278R1, the distance becomes almost identical to that found for K+. GidA in the presence of Na+ thus induces or stabilizes a “more open” conformation of the G domains, as it is normally observed for K+.
Interestingly, for position 366, located in Gα5, analysis of the DEER data obtained in the presence of GidA and K+ reveals a distance larger than 68 Å (because of the limited dipolar evolution time, it cannot be reliably quantified). Therefore, as can be seen also directly from the dipolar evolution traces, the distance between positions 366 in the MnmE dimer increases significantly in the presence of GidA, indicating structural changes in the Gα5 region.
GidA Stabilization of the On-Off Conformational Switch in the Presence of Potassium Ions
Considering the significant changes in the distance distributions in nucleotide-free MnmE upon binding of GidA, we wondered whether and how GidA would influence specific intermediate steps of the GTPase cycle, i.e. the GDP-, GppNHp- (a nonhydrolyzable GTP analogue), and the GDP-AlFx-bound (transition mimic) states.
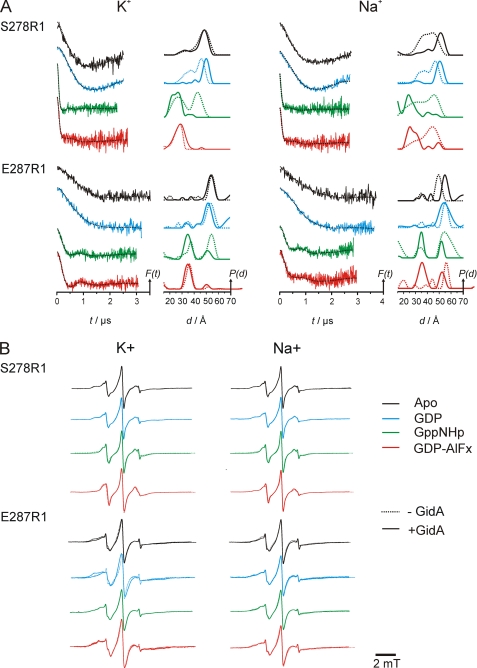
The results of the DEER measurements in the presence of 100 mm KCl (left columns) or NaCl (right column) are shown in Fig. 6A. Comparison of the data in the absence (dashed lines) and presence (solid lines) of GidA reveals a clear influence of the latter on the relative positions of the MnmE G domains in all steps of the GTPase cycle. For mutant S278R1 in the presence of K+, the data show that binding of GidA in the GDP-bound state induces a shift of the major distance peak from 46 to 50 Å and a strong reduction of the broad shoulder at smaller distances. The major effect of GidA is thus to stabilize the open conformation in the nucleotide-free and GDP-bound states.
FIGURE 6.
Characterization of the influence of GidA on the MnmE GTPase cycle in the presence of K+ (left panel) and Na+ (right panel). A, DEER measurements. Data analysis was performed by Tikhonov regularization. In each panel, the left column shows background corrected dipolar evolution data for the apo-, GDP-, GppNHp-, and GDP-AlFx-bound state of the respective MnmE mutant, and the right column shows distance distributions obtained by Tikhonov regularization. All of the plots are normalized by amplitude. The modulation depths in all cases coincided with the labeling efficiencies obtained from the cw EPR measurements. The broken lines in the left columns are fits to the data obtained by Tikhonov regularization. The broken lines in the right columns represent the distance distribution for MnmE without GidA (data taken from Ref. 27). For each mutant, the dipolar evolution data and the respective distance distributions are colored according to the nucleotide-bound state (Apo, black; GDP, blue; GppNHp, green; GDP-AlFx, red). B, overlay of room temperature cw EPR spectra of MnmE-C451S/S278R1 and -C451S/E287R1 both with (continuous line) and without (dotted line) GidA. Color coding is the same as in A.
The effect of GidA on the GppNHp state is even more pronounced but in the opposite direction. Here, MnmE-S278R1 shows a broad distance distribution ranging from 22 to 27 Å representing the G domains in the closed state, whereas another fraction exhibits an average distance of ∼43 Å representing the open conformation. The latter distance peak disappears by the addition of GidA, showing that the equilibrium between the two states is shifted toward the closed state. Previously we showed that the G domains in the absence of GidA showed a dynamic equilibrium in the presence of GppNHp between the open (43 Å interspin distance) and the closed (27 Å) conformation (27). In the GDP-AlFx-bound state the equilibrium between open and closed state is no longer observed. In both cases the distance distribution is characterized by a single but asymmetric peak at ∼28 Å, but the distribution width becomes broader and more symmetric in the presence of GidA.
Although the details differ, similar observations were made for mutant E287R1. The distance distributions for the apo and the GDP-bound state remain largely unchanged on GidA addition but slightly shift to shorter distances by ∼1 (apo) or 2 Å (GDP). For the GppNHp conformation, the equilibrium between the two states with interspin distances of 35 and 51 Å is shifted toward the closed state, again demonstrating the effect of GidA on the MnmE G domains already in the GppNHp-bound state. Complete association and juxtaposition of the G domains occurs only in the presence of GDP-AlFx, with no significant influence of GidA on the state of MnmE. There is a major population maximum with a distance of 35 Å, which is being shifted by ∼1 Å to shorter distances by GidA. A minor fraction of proteins with a peak at 50 Å indicates a small population in the open state in both the presence and the absence of GidA. The differences observed for the GppNHp and GDP-AlFx conformations of the two mutants is possibly due to a weak inhibitory effect of the E287R1 mutation on G domain dimerization, although both mutants exhibit a GTPase activity close to the wild type protein (27).
GidA Abolishes the Potassium Dependence of G Domain Association in MnmE
Previously we demonstrated that full G domain closure can only be reached in the presence of GDP, AlFx, and K+ (27). The extent of AlFx-induced dimerization of the G domains correlates with the ionic radii of the cation present and its ability to stimulate GTP hydrolysis (23, 27). Full dimerization is observed with K+ (r = 1.38 Å), whereas similar size cations (Rb+, 1.52 Å; NH4+, 1.44 Å) lead to partial dimerization, and no dimerization is observed for significantly smaller (Na+, 0.99 Å) or larger (Cs+, 1.67 Å) ions.
Recent biochemical data indicated that this dependence of the GTPase reaction on the presence of a specific cation of appropriate radius is partly abolished in the presence of GidA (4). To further characterize the cation dependence of G domain dimerization in the MnmE-GidA complex, we measured the mutants S278R1 and E287R1 in the apo-, GDP-, GppNHp-, and GDP-AlFx-bound states in the presence of Na+ (Fig. 6A, right panel) and compared them with the K+ data (left panel) described above.
For S278R1 in the GDP-bound state, the broad distance distribution from ∼25–55 Å observed with Na+ converts to a defined distribution upon addition of GidA, with one major distance peak shifted by 4 Å to a more open conformation. This peak overlays well with the distance peak of the nucleotide-free state, which has also been shifted considerably by GidA. This behavior is almost identical to that observed with K+, indicating that the structure of switch II in the GDP bound and nucleotide-free state is affected by GidA but does not depend on the presence of a specific cation. The distance distributions for the GppNHp and GDP-AlFx conformations again show remarkable shifts upon complex formation with GidA. Without GidA a broad distance distribution from 15 to 55 Å with a maximum at ∼45 Å is observed in both states. Binding of GidA induces a shift of the major distance distribution to ∼25 Å, indicating conversion to and/or stabilization of the closed conformation. Small peaks in the distance range from 35 to 55 Å indicate the presence of a small fraction of proteins in the open state.
The results for mutant E287R1 with Na+ are well in line with those obtained for S278R1. In the apo state the major distance peak is shifted by 7 Å to larger distances. In the presence of GDP, the mean distance remains basically unchanged with a significant increase in the distribution width. Comparison with the K+ data reveals that E287R1 and therefore Gα2 is influenced by GidA only if K+ is replaced by Na+.
In the GppNHp state, the equilibrium between the open and closed conformations is shifted toward the closed state, similar to the observation in the presence of K+. Here the broad distance peak observed without GidA converts to a small narrow peak at 51 Å, the distribution width of which is smaller by a factor of 3. This indicates that, besides the shift of the equilibrium, the open conformation becomes more structured and that GidA in the presence of sodium stabilizes Gα2 in a similar way as K+ in the absence of GidA. In the GDP-AlFx-bound state after the addition of GidA, an equilibrium between the closed and open conformations can be observed, with a major distance peak at 35 Å and a smaller one at 52 Å. Compared with the results for S278R1, the fraction of MnmE dimers with G domains in the open state appears to be somewhat larger, being well in line with the observations in the presence of K+.
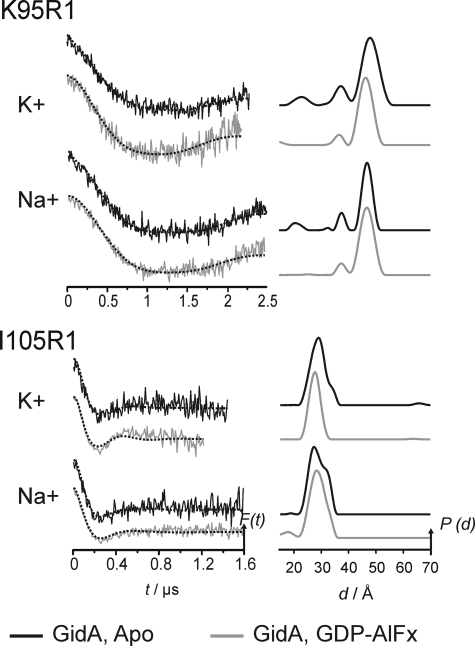
Comparisons for the two spin label positions in the α/β domain (K95R1, I105R1) of the apo state with the active, GDP-AlFx-bound state in the presence of K+ and Na+ are shown in Fig. 7. In general, the differences between the two states are minor, judged by comparison of the distance distributions as well as the dipolar evolution data, although a closer examination of the data reveals small differences for each mutant in the width of the distributions. Thus binding of GidA to MnmE induces changes in the structure and dynamics in MnmE, which are much more pronounced in the G domains as compared with the central α/β domain.
FIGURE 7.
Influence of GidA on MnmE mutants K95 and I105 during GTP hydrolysis in the presence of K+ and Na+. Left column, background corrected dipolar evolution data for the apo (black) and GDP-AlFx-bound (red) state of the respective MnmE mutant; right column, distance distributions obtained by Tikhonov regularization. The broken lines in the left column are fits to the data obtained by Tikhonov regularization.
These results indicate that in the heterotetrameric protein complex formed by MnmE and GidA, the latter at least partially abolishes the requirement of K+ for dimerization of the G domains. In the presence of the nonhydrolyzable GTP analogue GppNHp, the distance distributions are characterized by a significant shift of the equilibrium toward the closed conformation of the G domains also in the absence of K+. Moreover, in the presence of the transition state mimic GDP-AlFx and Na+, where in MnmE alone the G domains are found solely in the open conformation, GidA induces formation of the closed G domain conformation. Compared with the distances characterizing the closed conformation in the presence of GppNHp, the population maxima are slightly shifted, and the distance distributions for the GDP-AlFx-bound state are marginally broader. As observed in the presence of K+, the distance distribution of E287R1 comprises a second minor fraction with significantly larger distances, corresponding to the open conformation. This second fraction is much more pronounced in the presence of Na+, implying the necessity of K+ to be present for a full stabilization of the closed conformation. Binding of K+ to the nucleotide-binding pocket structures a highly conserved segment in switch I, involved in the coordination of K+, the so-called K-loop (23), thereby enabling the K-loop to form a part of the G domain interface. Hence, the whole dimer interface is stabilized upon K+ coordination (23). Nevertheless, within experimental error, the data for both spin label positions indicate that with GDP-AlFx in the presence of Na+, the conformational equilibrium is shifted toward the closed state.
Association with GidA Alters Backbone Dynamics in MnmE
To investigate whether the changes in the distribution widths especially in the presence of Na+ could be assigned to changes in protein dynamics or an altered motional freedom of the spin label side chain caused by structural changes in the protein, we compared the room temperature cw EPR spectra of the samples subjected to DEER. The spectra for S278R1 and E287R1 in the apo, GDP, GppNHp, and GDP-AlFx state for K+ and Na+, respectively, are shown in Fig. 6B. Although in some of the cases slight changes of the spin label mobility upon binding of GidA to MnmE are visible (for E287R1 in the apo state, the spin label mobility is slightly decreased in the presence of GidA, indicated by an increased width of the central resonance line and larger amplitude of the leftmost spectral features; for E287R1 in the GDP-bound state, the contrary effects, corresponding to an increased spin label side chain mobility, are observed upon complex formation with GidA), there is no clear correlation between the changes in the DEER distance distribution widths and the mobility of the spin label in the cw spectra. In fact, in most of the cases, especially that where significant changes in the distance distribution widths are observed (e.g. position 278 with Na+ in the apo and GDP-bound states; Fig. 6A), the cw spectra of MnmE with and without GidA are almost identical. This leads us to the conclusion that the observed changes in the distance distributions should be ascribed to changes in the mobility of the protein backbone, being on a time scale at or above the EPR rigid limit (>50 ns) rather than changes of the spin label side chain mobility and rotamer occupancy.
DISCUSSION
In bacteria, the modification of the wobble uridine of certain tRNAs is catalyzed by a heterotetrameric complex comprising the proteins MnmE and GidA. hGTPBP3 and Mto1, the human orthologues of the two enzymes, are responsible for the corresponding 5-taurinomethyl tRNA modification. In the absence of this modification, which can be due to mutation of one of the two enzymes, neurological disorders such as myoclenic epilepsy ragged red fibers, mitochondrial encephalomyopathy lactic acidosis stroke, and nonsyndromic deafness occur (14–18). The communication between MnmE and GidA is mandatory for both in vitro and in vivo activity of the complex (4, 28, 33, 34), and although structural information is available for MnmE from different bacterial organisms and EcGidA, the exact role of the two proteins and specifically their interplay in the modification reaction remain largely unknown (1, 29, 33, 35).
Here we have investigated the influence of GidA on the G domain dimerization taking place in the GTP-bound and the transition state mimic of the MnmE GTPase reaction cycle. The results demonstrate that interaction with GidA on a site remote from the G domains (Fig. 3B), according to the present model (29), stabilizes their activated GTPase competent dimer. Binding of GidA induces conformational changes in MnmE that in turn mediate dimerization of the G domains even in the absence of potassium ions, indicating that GidA binding significantly alters the relative localization of the G domains and thus acts as a co-stimulator of the GTPase reaction.
The assumption that GidA binds to a site opposite to the MnmE G domain dimerization interface implies that the presence of GidA is communicated to the G domains through the central α/β domains and/or the α-helical domains. Because we did not observe significant changes in the distance distributions for the two positions located in the α/β domain, communication most likely takes place via conformational changes of the α-helical domains, either by means of a structural rearrangement or a wing-like rigid body motion of the entire domain. The exact nature of these conformational changes is the subject of a current investigation.
We have shown by structural analysis using anomalous scattering from Rb, an efficient substitute of K, that a potassium ion is coordinated by the K-loop of the switch I region, the GTP phosphate chain, and Asn266 in the P-loop. There it acts as the GTPase-activating factor in MnmE. Sodium was shown to be unable to replace potassium (23, 27). Because the G domains of MnmE after dimerization and juxtaposition contain all of the necessary elements for catalysis, GidA can be considered to be a GTPase co-stimulator as recently defined by Gasper et al. (21). The stimulation mechanism is different from that of GTPase-activating proteins for Ras-like small G proteins, which supply a catalytic residue into the active site. The fact that G domain dimerization and GTPase activity can be induced by GidA even in the absence of potassium indicates significant structural rearrangements in this region. In the case of the E. coli proteins, GTP hydrolysis is 5-fold accelerated by GidA but 24-fold by potassium, whereas in presence of both components a 28-fold stimulation is observed (4). This indicates that K+ is much more potent to stimulate GTP hydrolysis than GidA. Hence, whereas GidA stabilizes the closed state of the G domains, potassium is still required to fully accelerate GTP hydrolysis.
Although the tRNA modifying reaction has been reconstituted in vitro (4, 17), the reaction rates measured were still very slow, and the role and nature of the tetrahydrofolate substrate are not completely understood. The physiological nature and relative importance of potassium and GidA to the overall modification reaction and the role of the GTPase reaction are still unknown. The GTPase co-stimulator activity of GidA might be of larger importance in the human homologues, because hGTPBP3 was recently shown to have a very low intrinsic GTP activity compared with MnmE and is not activated in the presence of K+, in contrast to the bacterial systems (17). Our data reveal that GidA performs its stimulatory function by stabilizing the closed, GTPase competent conformation and/or by shifting the equilibrium between open and closed very strongly toward the latter.
For the signal recognition particle and its receptor, another system of G proteins activated by nucleotide-dependent dimerization, similar effects caused by signal recognition particle cargo, the ribosome-nascent chain complex (RNC), have been described recently. The G domain cycle of signal recognition particle and its receptor has been reported to initially oscillate between the open and closed GTP-bound states via an early intermediate. The closed state converts then, accompanied by conformational changes, to a hydrolysis competent (active) closed state, to finally revert back to an open, GDP-bound state (36). Zhang et al. (37) showed that RNC stabilizes the early intermediate but disfavors the closed and active conformations, creating a time window in which the RNC can dissociate from the signal recognition particle-RNC complex. Although RNC effects only the open conformation rather than both the open and closed as found for the effect of GidA on MnmE, RNC and GidA nevertheless use the same basic principle, stabilization/destabilization of G domain conformations to modulate the timing of the GTPase cycle for optimal function.
We have previously shown that MnmE and GidA form a heterotetrameric complex in the presence of both GDP and GTP, although the affinity is higher in the presence of nucleoside triphosphate (4). As shown by the EPR experiments, GidA also influences the apo and particularly the GDP-bound conformation of MnmE. In the presence of sodium or potassium the G domains preferentially adopt a single, stable, and more open conformation and are far apart from each other. This shows that GidA is not a classical effector defined as a protein interacting with the GTP-bound form of the G proteins. It can thus be considered a new type of regulatory protein that acts on both ends of the switch cycle, presumably for a better coordination between the GTPase cycle and the enzymatic modification of tRNA.
Supplementary Material
This work was supported by funds from the Fond der Chemischen Industrie (to S. M.) and the International Max-Planck Research School (to S. M.), Deutsche Forschungsgemeinschaft Grant STE640/7-2 (to S. B., J. P. K., and H.-J. S.), and Graduate College Grant 612 (to S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- cmnm
- carboxymethylaminomethyl
- GppNHp
- guanosine-5′-(β,γ-imino)triphosphate
- DEER
- double electron electron resonance
- m
- mant (2′-/3′-O-(N′-methylanthraniloyl moiety)
- mGppNHp
- P2′-/3′-O-(N′-methylanthraniloyl)-guanosine-5′-(β,γ-imino)triphosphate
- MTSSL
- (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate spin label
- cw
- continuous wave
- RNC
- ribosome-nascent chain complex.
REFERENCES
- 1.Scrima A., Vetter I. R., Armengod M. E., Wittinghofer A. (2005) EMBO J. 24, 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elseviers D., Petrullo L. A., Gallagher P. J. (1984) Nucleic Acids Res. 12, 3521–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brégeon D., Colot V., Radman M., Taddei F. (2001) Genes Dev. 15, 2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer S., Wittinghofer A., Versées W. (2009) J. Mol. Biol. 392, 910–922 [DOI] [PubMed] [Google Scholar]
- 5.Moukadiri I., Prado S., Piera J., Velázquez-Campoy A., Björk G. R., Armengod M. E. (2009) Nucleic Acids Res. 37, 7177–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agris P. F., Vendeix F. A., Graham W. D. (2007) J. Mol. Biol. 366, 1–13 [DOI] [PubMed] [Google Scholar]
- 7.Yarian C., Townsend H., Czestkowski W., Sochacka E., Malkiewicz A. J., Guenther R., Miskiewicz A., Agris P. F. (2002) J. Biol. Chem. 277, 16391–16395 [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto K., Kawai G., Watanabe S., Niimi T., Hayashi N., Muto Y., Watanabe K., Satoh T., Sekine M., Yokoyama S. (1996) Biochemistry 35, 6533–6538 [DOI] [PubMed] [Google Scholar]
- 9.Agris P. F., Sierzputowska-Gracz H., Smith W., Malkiewicz A., Sochacka E., Nawrot B. (1992) J. Am. Chem. Soc. 114, 2652–2656 [Google Scholar]
- 10.Yokoyama S., Nishimura S. (1995) in tRNA: Structure, Biosynthesis and Function (Söll D., RajBhandary U. L. eds) pp. 207–223, American Society for Microbiology, Washington, D.C. [Google Scholar]
- 11.Gustilo E. M., Vendeix F. A., Agris P. F. (2008) Curr. Opin. Microbiol. 11, 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decoster E., Vassal A., Faye G. (1993) J. Mol. Biol. 232, 79–88 [DOI] [PubMed] [Google Scholar]
- 13.Colby G., Wu M., Tzagoloff A. (1998) J. Biol. Chem. 273, 27945–27952 [DOI] [PubMed] [Google Scholar]
- 14.Li X., Guan M. X. (2002) Mol. Cell. Biol. 22, 7701–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Li R., Lin X., Guan M. X. (2002) J. Biol. Chem. 277, 27256–27264 [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. (2002) EMBO J. 21, 6581–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villarroya M., Prado S., Esteve J. M., Soriano M. A., Aguado C., Pérez-Martínez D., Martínez-Ferrandis J. I., Yim L., Victor V. M., Cebolla E., Montaner A., Knecht E., Armengod M. E. (2008) Mol. Cell. Biol. 28, 7514–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bykhovskaya Y., Mengesha E., Wang D., Yang H., Estivill X., Shohat M., Fischel-Ghodsian N. (2004) Mol. Genet. Metab. 83, 199–206 [DOI] [PubMed] [Google Scholar]
- 19.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 20.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 21.Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. (2009) Nat. Rev. Mol. Cell Biol. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 22.Cabedo H., Macián F., Villarroya M., Escudero J. C., Martínez-Vicente M., Knecht E., Armengod M. E. (1999) EMBO J. 18, 7063–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scrima A., Wittinghofer A. (2006) EMBO J. 25, 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittinghofer A. (1997) Curr. Biol. 7, R682–R685 [DOI] [PubMed] [Google Scholar]
- 25.Martin R. E., Pannier M., Diederich F., Gramlich V., Hubrich M., Spiess H. W. (1998) Angew. Chem. Int. Ed. 37, 2833–2837 [DOI] [PubMed] [Google Scholar]
- 26.Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W. (2000) J. Magn. Res. 142, 331–340 [DOI] [PubMed] [Google Scholar]
- 27.Meyer S., Böhme S., Krüger A., Steinhoff H. J., Klare J. P., Wittinghofer A. (2009) PLoS Biology 7, e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yim L., Moukadiri I., Björk G. R., Armengod M. E. (2006) Nucleic Acids Res. 34, 5892–5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer S., Scrima A., Versées W., Wittinghofer A. (2008) J. Mol. Biol. 380, 532–547 [DOI] [PubMed] [Google Scholar]
- 30.Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J. E., Timmel C. R., Hilger D., Jung H. (2006) Appl. Magn. Reson. 30, 473–498 [Google Scholar]
- 31.Steinhoff H. J. (2004) Biol. Chem. 385, 913–920 [DOI] [PubMed] [Google Scholar]
- 32.Jeschke G., Polyhach Y. (2007) Phys. Chem. Chem. Phys. 9, 1895–1910 [DOI] [PubMed] [Google Scholar]
- 33.Schiemann O., Prisner T. F. (2007) Q. Rev. Biophys. 40, 1–53 [DOI] [PubMed] [Google Scholar]
- 34.Shi R., Villarroya M., Ruiz-Partida R., Li Y., Proteau A., Prado S., Moukadiri I., Benítez-Páez A., Lomas R., Wagner J., Matte A., Velázquez-Campoy A., Armengod M. E., Cygler M. (2009) J. Bacteriol. 191, 7614–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osawa T., Ito K., Inanaga H., Nureki O., Tomita K., Numata T. (2009) Structure 17, 713–724 [DOI] [PubMed] [Google Scholar]
- 36.Shan S. O., Stroud R. M., Walter P. (2004) PLoS Biol. 2, e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Schaffitzel C., Ban N., Shan S. O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.