Abstract
Purpose
Variants at chromosomal loci 8q24 and 17q are established risk factors for prostate cancer. Many studies have confirmed the findings for risk, but few have examined aggressiveness and other clinical variables in detail. Additionally, Gleason score is typically used as a surrogate for the primary endpoint of prostate cancer mortality. We investigated whether the 8q24 and 17q risk variants are associated with clinical variables as well as prostate cancer mortality.
Experimental Design
In the Physicians’ Health Study (1347 cases, 1462 controls), the Dana-Farber Harvard Cancer Center SPORE (Gelb Center) (3714 cases), and the Fred Hutchinson Cancer Research Center King County Case-Control Studies (1308 cases, 1266 controls), we examined eight previously identified 8q24 and 17q risk variants for association with prostate cancer mortality in men of European ancestry. We considered associations with other surrogate markers of prostate cancer aggressiveness such as Gleason score, pathological stage, PSA at diagnosis, and age at diagnosis.
Results
Six of the eight variants were confirmed as prostate cancer risk factors. Several variants were nominally associated with age at diagnosis; when totaling all alleles for SNPs significantly associated with risk, each additional allele decreased age at diagnosis by an average of 6 months in the Physicians’ Health Study (p=0.0005) and 4 months in the Dana-Farber Harvard Cancer Center SPORE (Gelb Center) cohort (p=0.0016). However, there were no statistically significant associations with prostate cancer mortality.
Conclusions
Our results suggest that the 8q24 and 17q prostate cancer risk variants may influence age at diagnosis but not disease aggressiveness.
Keywords: genetic variation, prostate cancer, mortality
Introduction
Chromosomal loci 8q24 and 17q have emerged recently as bona fide prostate cancer risk loci(1–17). Fine mapping (18) and additional genome scans (4, 5) have identified three 8q24 regions containing variants independently associated with risk (hg17 build 35; region 1: 128.54–128.62 Mb; region 2: 128.12–128.28; region 3: 128.47–128.54) (19). Notably, none of the variants in regions 1–3 lie in or near known coding regions. A single nucleotide polymorphism (SNP) outside these regions was recently identified as a novel risk factor in African Americans (20). This SNP lies in an intron of the non-SMC element 2 (NSMCE2) gene. The risk SNPs identified on chromosome 17q lie in two regions, one in a non-coding region at 17q24.3, the other in the second intron of TCF2/HNF1β at 17q12 (3).
While variants in these regions unambiguously confer risk of developing disease, their association with mortality or with other important prostate cancer traits is unknown. Most men with clinically localized prostate cancer are treated as if they have lethal disease because of the limited ability to predict outcome. However, most men with localized prostate cancer have indolent disease that would not be lethal even in the absence of early aggressive therapy (21, 22). Since aggressive therapy is not without associated morbidity and costs, it is imperative to find markers that can distinguish indolent from fatal disease at diagnosis.
In previous studies, some of the 8q24 and 17q alleles have demonstrated modest associations with prostate cancer clinical parameters, such as Gleason score (8, 16) or age at diagnosis (1, 3, 4, 6, 17, 18), while other studies failed to show an association with aggressive characteristics (1, 2, 5–7, 9, 23) or age at diagnosis (5, 7, 11, 23). Additionally, the majority of work to date has focused on one of the first 8q24 SNPs detected, rs1447295; no single study has yet examined multiple 8q24 and 17q risk variants in relation to prostate cancer mortality.
Mortality is inarguably the most important prostate cancer endpoint. As substantial longitudinal follow-up is required to capture this information, many studies define “aggressive” cancers as those with high Gleason score. Gleason score is a strong predictor of mortality; however, the positive predictive value (PPV) for mortality of a high Gleason score (≥7) is only 29% (24) and therefore far from optimal. Both prostate cancer mortality and Gleason score likely have genetic determinants. A recent study showing concordance of survival among family members with prostate cancer suggests that prostate cancer prognosis may have a hereditary component (25). Several linkage scans have identified regions of the genome associated with Gleason score (26–28). However, there is a trade-off when choosing which of these outcomes to investigate – studies of Gleason score are likely to have more statistical power because of the larger sample size, but the endpoint of mortality will provide more useful information. In the largest study of its kind, the present investigation evaluated the association of genetic risk variants with mortality (n=491 prostate cancer deaths) across three cohorts.
We evaluated six 8q24 and two 17q risk variants in participants from three large, well-annotated studies: the Physicians’ Health Study, the Dana-Farber Harvard Cancer Center SPORE (Gelb Center) cohort, and the Fred Hutchinson Cancer Research Center King County Case-Control Studies. We tested for associations between these eight SNPs and several clinico-pathologic traits (Gleason score, pathologic stage at radical prostatectomy, age at diagnosis, and PSA at diagnosis) and prostate cancer mortality.
Materials and Methods
Study Populations
This study received Institutional Review Board approval at each participating institution.
Physicians’ Health Study (PHS)
The PHS began as a randomized, double-blind trial of aspirin and β-carotene in the prevention of cardiovascular disease and cancer among 22,071 healthy US physicians. Written consent was obtained from each participant and the investigation was approved by the Human Subjects Committee at Brigham and Women’s Hospital. Men were excluded at baseline if they had any serious medical conditions including all cancers (except non-melanoma skin cancer). Blood samples were collected from 68% of the physicians in 1982–1984, as described previously (29). Participants are followed through annual questionnaires to collect data on diet, health and lifestyle behaviors, and medical history, and biannually through postcards to ascertain health endpoints, including prostate cancer. All self-reported prostate cancer cases are verified through medical record and pathology review. Through this systematic medical record review, we also abstract data on PSA at diagnosis, tumor stage, and Gleason score. Cause of death is determined by review of death certificates, medical records, and information from the family by a panel of three physicians. There is high follow-up for both cancer incidence (96%) and mortality (98%). Metastases are reported on follow-up questionnaires sent to all men living with prostate cancer. We utilized a nested case-control study, with controls selected by risk-set sampling and matched on age at baseline (± 1 year and, for older cases, within 5 years), smoking status (never, former, current), and follow-up time. For the current study, we restricted participants to self-reported Caucasians. 1347 cases and 1462 controls were included from PHS for the matched case-control analysis; controls that were later diagnosed with prostate cancer were included in the case-only analyses. During follow-up through March 1, 2008, 187 men died of prostate cancer. In this cohort, follow-up for vital status was over 99% complete.
Dana-Farber Harvard Cancer Center SPORE (Gelb Center)
Dana-Farber Harvard Cancer Center SPORE (Gelb Center) is a case series of prostate cancer patients diagnosed between 1976 and 2007. A detailed description of this study has been published previously (30). The study includes detailed clinical information from multiple sources, including medical records and patient registration, and a blood sample collected after diagnosis. Follow-up of the participants occurs at clinic visits to the Dana-Farber Cancer Institute and by searching the National Death Index. Because cause of death is not always available or known, if an individual was known to have metastases before death they were assumed to have died from prostate cancer. Long-term survivors were defined as men who survived at least 10 years beyond their prostate cancer diagnosis; these cases were additionally restricted to those who had a follow-up visit at Dana-Farber at least 10 years after diagnosis and were free of metastases at that time. Individuals who reported their ethnicity as non-Caucasian or were missing information on ethnicity or age at diagnosis were excluded.
Fred Hutchinson Cancer Research Center King County Case-Control Studies (FHCRC)
The study population consists of participants from two population-based case-control studies in residents of King County, Washington (Study I and Study II), which have been described previously (31, 32). Incident cases diagnosed between 1993 and (Study I) and 2002 and 2005 (Study II) with histologically confirmed prostate cancer were ascertained from the Seattle-Puget Sound SEER cancer registry. Of the 2244 eligible prostate cancer patients identified, 1754 (78.2%) were interviewed. Controls were male residents of King County, Washington without a self-reported history of prostate cancer. They were identified using one-step random digit dialing and recruited evenly throughout both ascertainment periods. Controls were frequency matched to case patients by five-year age groups. Of the 2448 men identified who met the eligibility criteria, 1645 (67.2%) completed a study interview. Subjects in both studies completed in-person interviews using a questionnaire that collected information on social and demographic factors, family structure, cancer history and medical history. Clinical information on cases, including Gleason score, stage of disease, serum PSA level at diagnosis, and primary treatment was obtained from the cancer registry. For this analysis, only Caucasian cases (n=1308) and controls (n=1266) were included. Cases from both studies are under long-term surveillance with ascertainment of vital status and underlying cause of death. The patient file is linked to the SEER cancer registry and for each deceased case a death certificate is requested to confirm the underlying cause of death. Long-term survivors had not progressed according to self-administered survey and medical record review. The date of last follow-up for vital status for this analysis was November 1, 2007, with an average length of follow-up of 7.6 years (11.9 years for Study I and 4.0 years for Study II). All study subjects signed informed consent prior to participation, and the studies were approved by the Fred Hutchinson Cancer Research Center’s Institutional Review Board.
DNA, SNPs, and Genotyping
In all cohorts, DNA was extracted from whole blood. SNPs in the 8q24 and 17q regions associated with prostate cancer incidence were genotyped. These SNPs include rs13254738, rs1447295, rs6983267, rs6983561, rs7000448, and rs7008482 (not in FHCRC) at 8q24 and rs4430796 and rs1859962 at 17q. On the PHS and Gelb Center specimens, genotyping was performed with Sequenom iPLEX matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry technology; see (33) for reaction details. For quality control, 6 SNPs were genotyped on a subset of PHS (n=598) and Gelb Center (n=772) specimens twice; concordance was >99%. In the FHCRC studies, SNPs were genotyped using the Applied Biosystems (ABI) SNPlex™ Genotyping System. Identification of the specific SNP allele was carried out with the ABI 3730xl DNA Analyzer with GeneMapper® software used for allele assignment. Quality control included genotyping of 140 blind duplicate samples distributed across all genotyping batches, with 100% agreement between duplicates for the SNPs. For PHS and FHCRC, batches genotyped incorporated similar numbers of case and control samples and laboratory personnel were blinded to the case-control status of samples. All SNPs had >94% genotype passing rates.
Statistical Analysis
We tested for Hardy-Weinberg proportions in the PHS and FHCRC controls using Pearson’s goodness-of-fit test. No SNPs violated Hardy-Weinberg equilibrium in either the PHS or FHCRC controls (p>0.01). SNPs that had a minor allele frequency of >10% were analyzed under a co-dominant model, while the less common SNPs were analyzed assuming a dominant inheritance model. P-values reported were for trend (1 degree of freedom test). We created a variable for total number of risk alleles, using only the SNPs that were significantly associated with prostate cancer risk in the PHS and FHCRC combined results.
Prostate cancer incidence
Prostate cancer incidence was investigated only in the PHS and the FHCRC case-control studies as there are no controls in the Gelb Center. These data were analyzed by unconditional logistic regression, adjusting for the matching factors, to estimate odds ratio as an approximation of the rate ratio, and 95% confidence intervals. The resulting odds ratios were combined into a summary estimate across the two cohorts using a random effects model, with cohort as the random effect.
Prostate cancer mortality
To study the extremes of prostate cancer, we selected cases who died of prostate cancer and cases who survived for at least ten years of follow-up post-diagnosis without developing metastases (to represent indolent cases). An analysis comparing these two groups was carried out using unconditional logistic regression, adjusting for age at diagnosis. The resulting odds ratios were combined across the three cohorts into a summary estimate using a random effects model.
A survival analysis using the Cox proportional hazards model, adjusting for age at diagnosis, was performed in only the PHS and FHCRC studies, where date and cause of death was known for all participants who died. Individuals who died of other causes or were still alive at end of follow-up (2008 in PHS, 2007 in FHCRC) were censored.
Gleason score
We examined the association between SNPs and Gleason score in a case-only analysis using a three category variable (<7 (referent), 7, >7) with polytomous logistic regression, adjusting for age at diagnosis. For the men who underwent prostatectomy, Gleason score was preferentially from prostatectomy; for all others, Gleason score from biopsy was used. The resulting odds ratios were combined into a summary estimate using a random effects model.
Pathologic stage
Among the cases, we classified men as either having evidence of extraprostatic disease (T3/T4) or localized disease (T2) at prostatectomy. We examined the relationship between each of the SNPs and this binary variable with unconditional logistic regression, adjusted for age at diagnosis. The resulting odds ratios were combined into a summary estimate using a random effects model.
Age and PSA at diagnosis
The relationships between the 8q24 and 17q SNPs and age at diagnosis and PSA at diagnosis were analyzed with ANOVA; the analysis of total number of risk alleles was performed with linear regression. The FHCRC studies over-sampled younger patients and cases in older age groups were randomly selected at diagnosis; thus, the age distribution is a result of the sampling scheme. Individuals who were known to be clinical stage T4 at diagnosis were excluded from the PSA analysis. PSA values were log-transformed and this analysis was adjusted for age at diagnosis.
Statistical significance was declared when the 2-sided p-value was <0.05. Analyses were performed using SAS v.9.1 statistical software.
Results
Selected clinical characteristics of the PHS, the Gelb Center, and the FHCRC studies are described in Table 1. These studies and the participants differ, PHS being a prospective cohort study, the Gelb Center a referral hospital-based case-series, and FHCRC two population-based case-control studies. The mean age of diagnosis is 70.5 in the PHS compared with 62 in the Gelb Center and 59.9 in FHCRC. While most tumors are Gleason 7 or less in all three studies, a large proportion of the Gelb Center patients is Gleason score 8–10 (21.5%). Many PHS cases were diagnosed before 1992 in the pre-PSA era (28%). Genotype frequencies for all eight SNPs are similar to those reported in previous studies for both cases and controls (Supplementary Table 1).
Table 1.
Clinical characteristics of study participants
| PHS | Gelb Center | FHCRC | ||
|---|---|---|---|---|
| Cases/controls (n) | 1438 (1347)*/1462 | 3714/-- | 1308/1266 | |
| Deaths/long-term survivors (n) | 187/584 | 250/158 | 56/526 | |
| Age at diagnosis (n) | 1438 | 3714 | 1308 | |
| mean (st. dev.) | 70.5 (7.7) | 62.0 (8.2) | 59.9 (7.0) | |
| Gleason (n) | 1270 | 3458 | 1308 | |
| <7 (%) | 51.3 | 41.1 | 57.3 | |
| 7 (%) | 33.5 | 37.4 | 33.1 | |
| >7 (%) | 15.2 | 21.5 | 9.7 | |
| Pathologic stage (n) | 469 | 1452 | 740 | |
| T2 | 72.7 | 67.5 | 71.3 | |
| T3/T4 | 27.3 | 32.5 | 28.7 | |
| PSA at diagnosis (n) | 687 | 3278 | 1177 | |
| median (Q1, Q3) | 7.6 (5.4, 13.0) | 6.5 (4.7, 11.0) | 6.2 (4.6, 9.6) | |
1347 cases were part of the PHS matched case-control study and included in the analysis for prostate cancer risk
Prostate cancer incidence
Results for the association of 8q24 variants with prostate cancer incidence in the FHCRC studies were previously published (14). In the PHS and FHCRC combined case-control analyses, 4 of the variants in the 8q24 region were significantly associated with risk of prostate cancer (rs13254738, rs6983561, rs6983267, and rs1447295) (Supplementary Table 2). Of note, the two variants that were not significantly associated with risk in our study have been reported as significant risk factors only in non-white populations. We found that both 17q risk SNPs, rs4430796 and rs1859962, were also significantly associated with prostate cancer (Supplementary Table 2). The odds ratios for the homozygote risk alleles in the combined analysis ranged from 1.28 (95% confidence interval (CI): 1.06–1.54) for rs13254738 to 1.60 (95% CI: 1.37–1.88) for rs4430796. We created a variable for the total number of alleles by adding up the alleles for all SNPs significantly associated with risk (rs13254738, rs6983561, rs6983267, rs1447295, rs4430796, and rs1859962; range 0–9); with each additional risk allele, the risk of prostate cancer increased 19% (p<0.0001) in PHS and 23% (p<0.0001) in the FHCRC studies.
Prostate cancer mortality
We examined the association of these variants with prostate cancer mortality by comparing prostate cancer deaths to long-term survivors (alive ≥10 years after diagnosis). rs1859962 was associated with prostate cancer mortality in the Gelb Center cohort only; having the risk allele was associated with an increased risk of mortality of ~70% (Table 2). In all cohorts, the total number of risk alleles was not associated with mortality, with p-values for trend ranging from 0.23–0.99 (results not shown). When results from the three studies were combined, none of the SNPs were significantly associated with prostate cancer mortality (Table 2). We also performed a survival analysis among all the cases in the PHS and FHCRC studies; again, no significant associations with mortality were observed (data not shown).
Table 2.
Associations between prostate cancer mortality and 8q24/17q variants, comparing prostate cancer deaths to long-term survivors
| SNP | PHS OR (95% CI)† |
Gelb Center OR (95% CI)† |
FHCRC OR (95% CI)† |
Combined OR (95% CI) |
|---|---|---|---|---|
| rs13254738 | p*=0.54 | p=0.38 | p=0.75 | p=0.35 |
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC | 1.23 (0.86,1.77) | 1.21 (0.80,1.85) | 0.88 (0.44,1.78) | 1.17 (0.90,1.51) |
| CC | 1.12 (0.65,1.91) | 1.24 (0.60,2.56) | 0.90 (0.29,2.76) | 1.12 (0.75,1.68) |
| rs6983561 | p=0.81 | p=0.54 | p=0.66 | p=0.49 |
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC/CC | 0.93 (0.52,1.68) | 0.80 (0.38,1.65) | 0.79 (0.27,2.36) | 0.86 (0.56,1.32) |
| rs6983267 | p=0.97 | p=0.57 | p=0.31 | p=1.00 |
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 0.82 (0.52,1.27) | 1.51 (0.89,2.54) | 0.85 (0.36,2.00) | 1.04 (0.68,1.58) |
| GG | 0.96 (0.59,1.54) | 1.24 (0.70,2.21) | 0.59 (0.21,1.66) | 1.00 (0.70,1.41) |
| rs7000448 | p=0.06 | p=0.39 | p=0.57 | p=0.10 |
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CT | 0.80 (0.56,1.15) | 1.17 (0.75,1.83) | 1.21 (0.58,2.52) | 0.97 (0.74,1.27) |
| TT | 0.60 (0.34,1.06) | 0.70 (0.40,1.24) | 1.30 (0.44,3.83) | 0.70 (0.48,1.03) |
| rs1447295 | p=0.88 | p=0.64 | p=0.84 | p=0.65 |
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CA/AA | 1.03 (0.69,1.55) | 1.12 (0.70,1.80) | 1.08 (0.51,2.30) | 1.07 (0.80,1.42) |
| rs7008482 | p=0.47 | p=0.50 | - | p=0.87 |
| TT | 1.00 (ref) | 1.00 (ref) | - | 1.00 (ref) |
| GT | 1.04 (0.72,1.49) | 1.05 (0.69,1.61) | - | 1.04 (0.79,1.38) |
| GG | 1.26 (0.73,2.17) | 0.66 (0.32,1.35) | - | 0.95 (0.51,1.79) |
| rs4430796 | p=0.70 | p=0.57 | p=0.91 | p=0.43 |
| GG | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AG | 1.13 (0.72,1.77) | 0.96 (0.57,1.64) | 0.44 (0.18,1.07) | 0.89 (0.57,1.39) |
| AA | 0.93 (0.56,1.56) | 0.86 (0.48,1.51) | 0.80 (0.34,1.86) | 0.88 (0.62,1.25) |
| rs1859962 | p=0.82 | p=0.08 | p=0.90 | p=0.47 |
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 1.02 (0.67,1.56) | 1.68 (1.01,2.81)‡ | 1.03 (0.44,2.42) | 1.22 (0.87,1.71) |
| GG | 0.95 (0.60,1.52) | 1.73 (0.97,3.06) | 0.95 (0.36,2.47) | 1.18 (0.78,1.79) |
All results from unconditional logistic regression, adjusted for age at diagnosis
All p-values for 1 d.f. test
p<0.05
Gleason score and pathologic stage
Using polytomous logistic regression, cases with Gleason score 7 (total n=2151) (Supplementary Table 3) and Gleason score 8–10 (total n=1063) (Table 3) were compared to the referent of Gleason score 2–6 (total n=2822). In the combined summary estimate, the rs6983267 heterozygotes had a lower risk of Gleason 8–10 cancer (OR=0.82, 95% CI: 0.67–0.99); however no association was observed for the GG homozygote (Table 3).
Table 3.
Associations between Gleason score and 8q24/17q variants, comparing men with Gleason score 8–10 to Gleason score 2–6
| SNP | PHS OR (95% CI)† |
Gelb Center OR (95% CI)† |
FHCRC OR (95% CI)† |
Combined OR (95% CI) |
|---|---|---|---|---|
| rs13254738 | ||||
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC | 1.10 (0.77,1.57) | 0.90 (0.74,1.09) | 0.85 (0.56,1.29) | 0.93 (0.79,1.09) |
| CC | 1.09 (0.64,1.87) | 0.90 (0.66,1.23) | 1.35 (0.75,2.40) | 1.01 (0.79,1.29) |
| rs6983561 | ||||
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC/CC | 1.10 (0.62,1.95) | 1.01 (0.72,1.41) | 0.98 (0.53,1.83) | 1.02 (0.79,1.33) |
| rs6983267 | ||||
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 0.84 (0.54,1.30) | 0.80 (0.63,1.00) | 0.86 (0.52,1.42) | 0.82 (0.67,0.99)* |
| GG | 1.15 (0.72,1.82) | 0.68 (0.53,0.87)* | 0.93 (0.54,1.61) | 0.85 (0.61,1.20) |
| rs7000448 | ||||
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CT | 0.97 (0.68,1.39) | 0.88 (0.72,1.08) | 1.00 (0.66,1.52) | 0.91 (0.78,1.07) |
| TT | 0.84 (0.49,1.41) | 0.89 (0.68,1.17) | 0.82 (0.42,1.58) | 0.87 (0.70,1.09) |
| rs1447295 | ||||
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CA/AA | 0.88 (0.59,1.31) | 1.10 (0.89,1.36) | 0.88 (0.56,1,39) | 1.02 (0.86,1.22) |
| rs7008482 | ||||
| TT | 1.00 (ref) | 1.00 (ref) | - | 1.00 (ref) |
| GT | 0.82 (0.58,1.17) | 0.87 (0.72,1.06) | - | 0.86 (0.73,1.02) |
| GG | 1.08 (0.63,1.85) | 0.76 (0.56,1.03) | - | 0.84 (0.62,1.15) |
| rs4430796 | ||||
| GG | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AG | 1.08 (0.69,1.69) | 0.97 (0.77,1.23) | 1.03 (0.62,1.71) | 1.00 (0.82,1.21) |
| AA | 1.04 (0.63,1.71) | 0.96 (0.74,1.25) | 0.92 (0.53,1.59) | 0.97 (0.78,1.20) |
| rs1859962 | ||||
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 1.28 (0.84,1.93) | 1.05 (0.84,1.32) | 0.88 (0.55,1.42) | 1.06 (0.88,1.28) |
| GG | 0.98 (0.61,1.59) | 1.00 (0.77,1.30) | 0.82 (0.48,1.42) | 0.97 (0.78,1.20) |
All results from unconditional logistic regression, adjusted for age at diagnosis
p<0.05
Carriers of the rs6983267 risk allele had a decreased risk of pathologic stage T3/T4 cancer in the Gelb Center (Table 4), but this did not persist when the results from the three studies were combined. In the combined results, the rs7008482 TT homozygote was associated with a decreased risk of stage T3/T4 cancer (p=0.01), while the GT heterozygote of rs1859962, but not the homozygote, was associated with an increased risk of T3/T4 cancer (p =0.03).
Table 4.
Associations between stage and 8q24/17q variants, comparing men with pathologic stage T3/T4 to T2
| SNP | PHS OR (95% CI)† |
Gelb Center OR (95% CI)† |
FHCRC OR (95% CI)† |
Combined OR (95% CI) |
|---|---|---|---|---|
| rs13254738 | p*=0.61 | p=0.78 | p=0.43 | p=0.68 |
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC | 1.23 (0.77,1.97) | 1.15 (0.91,1.47) | 1.11 (0.79,1.57) | 1.15 (0.96,1.38) |
| CC | 0.63 (0.29,1.38) | 0.94 (0.64,1.37) | 1.19 (0.72,1.97) | 0.96 (0.73,1.28) |
| rs6983561 | p=0.44 | p=0.24 | p=0.23 | p=0.20 |
| AA | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AC/CC | 0.73 (0.33,1.63) | 1.28 (0.85,1.93) | 1.36 (0.83,2.25) | 1.21 (0.90,1.63) |
| rs6983267 | p=0.94 | p=0.007 | p=0.89 | p=0.27 |
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 0.56 (0.31,0.99)† | 0.73 (0.55,0.97)‡ | 0.92 (0.59,1.43) | 0.74 (0.60,0.93) |
| GG | 0.91 (0.50,1.68) | 0.65 (0.47,0.88)‡ | 1.01 (0.62,1.63) | 0.79 (0.59,1.05) |
| rs7000448 | p=0.38 | p=0.14 | p=0.70 | p=0.09 |
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CT | 0.64 (0.39,1.03) | 0.87 (0.68,1.12) | 1.03 (0.73,1.45) | 0.87 (0.70,1.08) |
| TT | 0.91 (0.48,1.73) | 0.78 (0.55,1.11) | 0.86 (0.51,1.45) | 0.82 (0.63,1.07) |
| rs1447295 | p=0.49 | p=0.75 | p=0.37 | p=0.67 |
| CC | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| CA/AA | 1.21 (0.71,2.05) | 0.96 (0.73,1.25) | 0.84 (0.57,1.23) | 0.96 (0.78,1.17) |
| rs7008482 | p=0.83 | p=0.02 | p=0.03 | |
| TT | 1.00 (ref) | 1.00 (ref) | - | 1.00 (ref) |
| GT | 1.12 (0.70,1.78) | 0.89 (0.70,1.13) | - | 0.93 (0.76,1.15) |
| GG | 0.76 (0.33,1.75) | 0.57 (0.37,0.87)‡ | - | 0.61 (0.41,0.89)‡ |
| rs4430796 | p=0.63 | p=0.77 | p=0.95 | p=0.97 |
| GG | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| AG | 1.06 (0.56,1.97) | 0.84 (0.62,1.13) | 0.82 (0.54,1.25) | 0.86 (0.69,1.07) |
| AA | 0.89 (0.45,1.74) | 1.01 (0.73,1.39) | 0.95 (0.60,1.49) | 0.98 (0.76,1.25) |
| rs1859962 | p=0.21 | p=0.07 | p=0.36 | p=0.54 |
| TT | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) | 1.00 (ref) |
| GT | 0.99 (0.57,1.72) | 1.36 (1.01,1.83)‡ | 1.32 (0.87,1.99) | 1.28 (1.03,1.60)‡ |
| GG | 0.68 (0.36,1.28) | 1.39 (1.00,1.92) | 1.26 (0.80,2.00) | 1.14 (0.79,1.65) |
All results from unconditional logistic regression, adjusted for age at diagnosis
All p-values for 1 d.f. test
p<0.05
Age and PSA at diagnosis
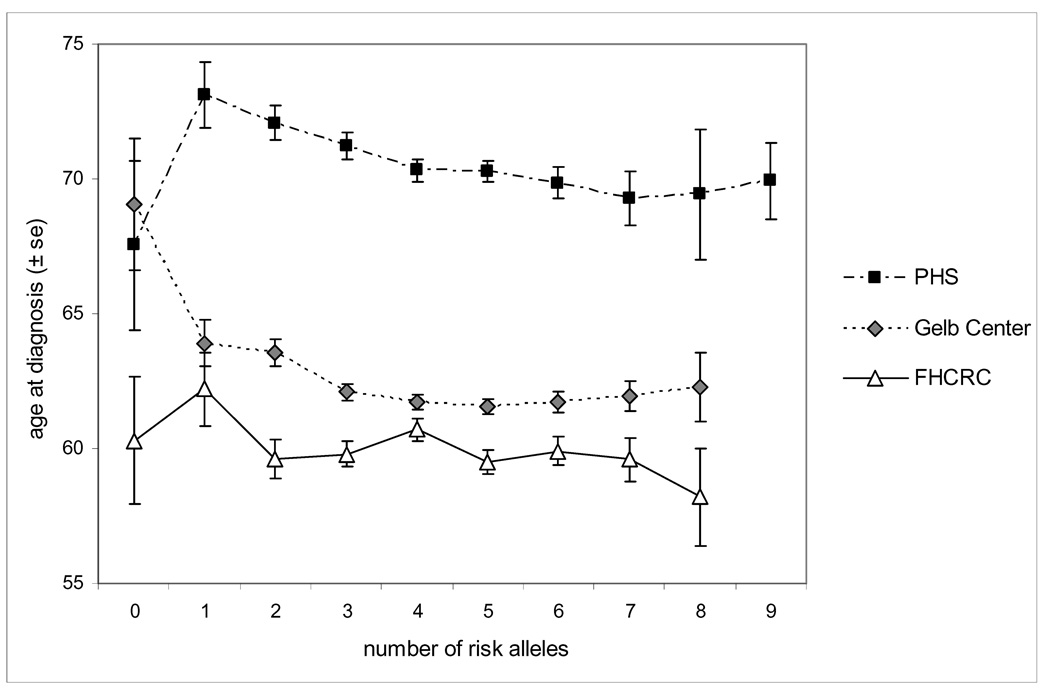
Several variants were significantly associated with age at diagnosis in the Gelb Center study (rs13254738, rs6983267, rs4430796, and rs1859962) (Table 5). The total number of risk alleles was strongly associated with a younger age of diagnosis in both the PHS and Gelb Center; with each additional risk allele, the age at diagnosis decreased by 6 months (p=0.0005) or 4 months (p=0.0016), respectively (Figure 1). However, number of risk alleles was not associated with age at diagnosis in the FHCRC studies (p=0.26).
Table 5.
Mean values of age at diagnosis by 8q24/17q genotypes and cumulative number of risk alleles
| SNP | PHS Mean (s.e.)† |
Gelb Center Mean (s.e.)† |
FHCRC Mean (s.e.)† |
|---|---|---|---|
| rs13254738 | p=0.94 | p=0.0004 | p=0.26 |
| AA | 70.6 (0.3) | 61.4 (0.2) | 59.7 (0.3) |
| AC | 70.4 (0.3) | 62.4 (0.2) | 60.3 (0.3) |
| CC | 70.5 (0.6) | 62.8 (0.4) | 59.6 (0.6) |
| rs6983561 | p=0.43 | p=0.25 | p=0.80 |
| AA | 70.5 (0.2) | 62.0 (0.1) | 60.0 (0.2) |
| AC/CC | 70.0 (0.7) | 61.4 (0.5) | 59.8 (0.6) |
| rs6983267 | p=0.20 | p=0.003 | p=0.59 |
| TT | 71.0 (0.5) | 62.7 (0.3) | 60.3 (0.5) |
| GT | 70.6 (0.3) | 62.1 (0.2) | 59.9 (0.3) |
| GG | 70.0 (0.4) | 61.4 (0.2) | 59.7 (0.4) |
| rs7000448 | p=0.88 | p=0.10 | p=0.81 |
| CC | 70.6 (0.3) | 62.1 (0.2) | 60.1 (0.3) |
| CT | 70.4 (0.3) | 61.8 (0.2) | 59.9 (0.3) |
| TT | 70.7 (0.5) | 62.6 (0.3) | 59.7 (0.6) |
| rs1447295 | p=0.33 | p=0.06 | p=0.21 |
| CC | 70.6 (0.2) | 62.2 (0.2) | 59.8 (0.2) |
| CA/AA | 70.1 (0.4) | 61.5 (0.3) | 60.4 (0.4) |
| rs7008482 | p=0.70 | p=0.22 | |
| TT | 70.7 (0.30 | 61.8 (0.2) | - |
| GT | 70.4 (0.3) | 62.1 (0.2) | - |
| GG | 70.3 (0.6) | 62.4 (0.4) | - |
| rs4430796 | p=0.08 | p=0.02 | p=0.30 |
| GG | 71.0 (0.5) | 62.5 (0.3) | 60.3 (0.5) |
| AG | 70.7 (0.3) | 62.1 (0.2) | 60.1 (0.3) |
| AA | 69.8 (0.4) | 61.5 (0.3) | 59.5 (0.3) |
| rs1859962 | p=0.25 | p=0.03 | p=0.67 |
| TT | 70.6 (0.4) | 62.4 (0.3) | 60.2 (0.4) |
| GT | 70.8 (0.3) | 62.1 (0.2) | 60.0 (0.3) |
| GG | 70.0 (0.4) | 61.5 (0.3) | 59.7 (0.4) |
| # of risk alleles | p=0.008 | p=0.0004 | p=0.40 |
| 0–2 | 72.1 (0.6) | 63.7 (0.4) | 60.2 (0.6) |
| 3 | 71.2 (0.5) | 62.1 (0.3) | 59.8 (0.5) |
| 4 | 70.3 (0.4) | 61.7 (0.3) | 60.7 (0.4) |
| 5 | 70.0 (0.4) | 61.6 (0.3) | 59.5 (0.4) |
| 6–9 | 69.7 (0.5) | 61.8 (0.3) | 59.7 (0.4) |
All results from ANOVA
Figure 1.
Age at diagnosis by number of risk alleles
*Note: FHCRC was limited to patients with age at diagnosis <65 years
Though rs6983267 was significantly associated with PSA level at diagnosis in the FHCRC studies (p=0.04), no other associations with PSA were observed (Supplementary Table 4).
Discussion
This study comprehensively examined the association of known prostate cancer risk variants at 8q24 and 17q with prostate cancer mortality and other clinical variables. Additionally, this study replicated the findings that polymorphisms in these regions are associated with prostate cancer risk. Six of the eight previously reported risk variants that we tested were significantly associated with prostate cancer incidence in a large case-control study nested in the PHS and in the FHCRC Case-Control Studies. The SNPs that were not significantly associated with risk were previously associated only in non-white populations; we did not attempt to replicate these findings as our analyses were only powered for populations of European ancestry.
While prostate cancer is very common, a minority of those diagnosed will die from their disease. The ability to predict a patient’s course of disease at diagnosis would be of tremendous clinical utility. In our study populations, we examined the association of the 8q24 and 17q variants with the primary endpoint of prostate cancer mortality, the most important clinical outcome. None of the SNPs were significantly associated with mortality in our combined analysis. This combined analysis is the largest of its kind (493 prostate cancer deaths, 1268 long-term survivors).
It takes years of follow-up to identify lethal prostate cancer cases, so many studies use Gleason score as a surrogate for prostate cancer mortality. Gleason score is frequently used as a marker of aggressive disease because it readily obtainable. However, there are significant limitations in using Gleason score as a predictor of lethal disease, such as inter-observer variability, grading changes over time (34, 35), and the relatively low positive predictive value for death at higher Gleason scores (24). There may be some misclassification when using long-term survivors to represent “indolent” cancers, i.e., some will still die of prostate cancer or some may have only survived due to aggressive therapy. However, we attempted to limit the first concern by selecting only patients who survived a minimum of 10 years after diagnosis. The second issue is present in any study of survival in a treated population; if we assume that this misclassification is non-differential, we could be missing small effects. We believe, however, that the majority of the long-term survivors would have survived even in the absence of treatment. As the results of the Swedish randomized trial of prostatectomy versus watchful waiting suggest, the number needed to treat to save the life of one man is 19 (36).
In this study, one marker, rs6983267, was significantly associated with Gleason score and pathologic stage (in the Gelb Center only); this variant was previously shown to be more strongly associated with aggressive cancer (15). Two other markers, rs7008482 and rs1859962, were associated with pathologic stage but not with Gleason score. Since these variants were not associated with mortality, it is possible that they are truly associated with these clinical traits but due to treatment, and the resulting misclassification, we did not observe associations with mortality. However, given the large number of tests performed and the moderate level of significance, one must consider that these results could be due to chance.
Several markers were associated with age at diagnosis in the Gelb Center patients. When we examined total number of risk alleles (adding together all alleles significant for risk), we observed a strong association with age at diagnosis in both the PHS and Gelb Center studies. It is of note that, as one might expect for a genetic disease, men with more risk alleles were more likely to develop cancer at a younger age. Several studies have suggested an association of these alleles with younger age at diagnosis (3, 4, 12, 15, 18); however, most dichotomize age and few have looked at the number of risk alleles combined. There was no significant association with age at diagnosis in the FHCRC studies; this may be due to the sampling scheme which selected participants with a younger age of diagnosis (<65) and therefore there is less variability making it more difficult to detect a significant trend. Additionally, these cases were all diagnosed in the PSA era, which may affect age at diagnosis. When we stratified PHS cases by PSA era, the association between number of risk alleles and age at diagnosis was stronger in the pre-PSA era (diagnosed before 1992) than the PSA era (decrease of 9 months per risk allele (p=0.0017) vs. decrease of 4 months (p=0.034), respectively).
Although markers in the 8q24 and 17q regions are definitively associated with prostate cancer risk, their public health and clinical utility remains uncertain if they do not affect clinically relevant outcomes, particularly prostate cancer mortality. Given the high proportion of men who develop prostate cancer in their lifetime, it is crucial to determine whose life expectancy will be shortened by this disease. Understanding the genetic basis of lethal disease could enhance clinical decision making; based on our results, the 8q24 and 17q markers do not appear to contribute information to a predictive model for lethal disease in cases of European ancestry.
Statement of Translational Relevance
The majority of prostate cancer cases diagnosed by PSA screening will have an indolent course of disease, even in the absence of therapy. However, one of the key questions in prostate cancer research is how to predict at diagnosis which patients have lethal cancer and which patients have indolent cancer. Identifying genetic determinants of prostate cancer aggressiveness would have clinical utility. Genetic studies primarily have focused on surrogates of prostate cancer aggressiveness, such as Gleason grade. In this study, we investigated the association between genetic markers and prostate cancer mortality, the primary endpoint of interest. The genetic markers examined are known prostate cancer risk polymorphisms at chromosomes 8q24 and 17q; determining whether these markers predict prostate cancer mortality or other clinical covariates is required for evaluating if and how these markers will inform clinical decision making.
Supplementary Material
Acknowledgments
Funding Source: This work was supported by a grant from the Dana-Farber Harvard Cancer Center SPORE in Prostate Cancer (P50CA090381-06). The Fred Hutchinson Cancer Research studies were supported by grants CA56678, CA092579, CA097186 from the National Cancer Institute. The PHS was supported by grants CA-34944, CA-40360, and CA-097193 from the National Cancer Institute and grants HL-26490 and HL-34595 from the National Heart, Lung, and Blood Institute, Bethesda, Maryland. KLP was supported by National Research Service Awards (T32 CA009001-32; R25 CA098566); FRS was supported by National Research Service Award (T32 CA009001-32). We gratefully acknowledge the Intramural Program of the National Human Genome Research Institute.
References
- 1.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 5.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 7.Severi G, Hayes VM, Padilla EJ, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 8.Suuriniemi M, Agalliu I, Schaid DJ, et al. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–814. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Lange EM, Isaacs SD, et al. Chromosome 8q24 risk variants in hereditary and non-hereditary prostate cancer patients. Prostate. 2008;68:489–497. doi: 10.1002/pros.20695. [DOI] [PubMed] [Google Scholar]
- 11.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 13.Zheng SL, Sun J, Wiklund F, et al. Cumulative Association of Five Genetic Variants with Prostate Cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 14.Salinas CA, Kwon E, Carlson CS, et al. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1203–1213. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 15.Beebe-Dimmer JL, Levin AM, Ray AM, et al. Chromosome 8q24 markers: risk of early-onset and familial prostate cancer. Int J Cancer. 2008;122:2876–2879. doi: 10.1002/ijc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helfand BT, Loeb S, Cashy J, et al. Tumor characteristics of carriers and noncarriers of the deCODE 8q24 prostate cancer susceptibility alleles. J Urol. 2008;179:2197–2201. doi: 10.1016/j.juro.2008.01.110. [DOI] [PubMed] [Google Scholar]
- 17.Levin AM, Machiela MJ, Zuhlke KA, Ray AM, Cooney KA, Douglas JA. Chromosome 17q12 variants contribute to risk of early-onset prostate cancer. Cancer Res. 2008;68:6492–6495. doi: 10.1158/0008-5472.CAN-08-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witte JS. Multiple prostate cancer risk variants on 8q24. Nat Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 20.Robbins C, Torres JB, Hooker S, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. Jama. 2005;293:2095–2101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 22.Bill-Axelson A, Holmberg L, Ruutu M, et al. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med. 2005;352:1977–1984. doi: 10.1056/NEJMoa043739. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Isaacs SD, Sun J, et al. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clin Cancer Res. 2008;14:5819–5824. doi: 10.1158/1078-0432.CCR-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andren O, Fall K, Franzen L, Andersson SO, Johansson JE, Rubin MA. How well does the Gleason score predict prostate cancer death? A 20-year followup of a population based cohort in Sweden. J Urol. 2006;175:1337–1340. doi: 10.1016/S0022-5347(05)00734-2. [DOI] [PubMed] [Google Scholar]
- 25.Hemminki K, Ji J, Forsti A, Sundquist J, Lenner P. Concordance of survival in family members with prostate cancer. J Clin Oncol. 2008;26:1705–1709. doi: 10.1200/JCO.2007.13.3355. [DOI] [PubMed] [Google Scholar]
- 26.Witte JS, Goddard KA, Conti DV, et al. Genomewide scan for prostate cancer-aggressiveness loci. Am J Hum Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slager SL, Zarfas KE, Brown WM, et al. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–179. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- 28.Schaid DJ, Stanford JL, McDonnell SK, et al. Genome-wide linkage scan of prostate cancer Gleason score and confirmation of chromosome 19q. Hum Genet. 2007;121:729–735. doi: 10.1007/s00439-007-0368-5. [DOI] [PubMed] [Google Scholar]
- 29.Final report on the aspirin component of the ongoing Physicians' Health Study. Steering Committee of the Physicians' Health Study Research Group. N Engl J Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 30.Oh WK, Hayes J, Evan C, et al. Development of an integrated prostate cancer research information system. Clin Genitourin Cancer. 2006;5:61–66. doi: 10.3816/CGC.2006.n.019. [DOI] [PubMed] [Google Scholar]
- 31.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:881–886. [PubMed] [Google Scholar]
- 32.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin Use and Risk of Prostate Cancer: Results from a Population-based Epidemiologic Study. Am J Epidemiol. 2008;168:250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross RW, Oh WK, Xie W, et al. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J Clin Oncol. 2008;26:842–847. doi: 10.1200/JCO.2007.13.6804. [DOI] [PubMed] [Google Scholar]
- 34.Albertsen PC, Hanley JA, Barrows GH, et al. Prostate cancer and the Will Rogers phenomenon. J Natl Cancer Inst. 2005;97:1248–1253. doi: 10.1093/jnci/dji248. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell RE, Shah JB, Desai M, et al. Changes in prognostic significance and predictive accuracy of Gleason grading system throughout PSA era: impact of grade migration in prostate cancer. Urology. 2007;70:706–710. doi: 10.1016/j.urology.2007.06.1084. [DOI] [PubMed] [Google Scholar]
- 36.Bill-Axelson A, Holmberg L, Filen F, et al. Radical prostatectomy versus watchful waiting in localized prostate cancer: the Scandinavian prostate cancer group-4 randomized trial. J Natl Cancer Inst. 2008;100:1144–1154. doi: 10.1093/jnci/djn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.