Abstract
Binding of epidermal growth factor (EGF) to the EGF receptor (EGFR) initiates signal transduction, ultimately leading to altered gene expression. Ligand-activated EGFR is also rapidly internalized and then targeted to lysosomes for degradation or recycled back to the plasma membrane. Endocytosis is a major regulator of EGFR signaling. Therefore, elucidation of the mechanisms of EGFR endocytosis is essential for a better understanding of EGFR biology. In order to achieve a comprehensive analysis of these mechanisms, reliable methods for measuring the rates of EGFR protein turnover and the rate parameters for individual steps of EGFR endocytic trafficking must be employed. The protocols in this unit describe methodologies to measure the rates of EGFR synthesis and degradation, to monitor EGF-induced down-regulation of surface EGFR, to measure the kinetic rate parameters of internalization, recycling, and degradation of radiolabeled EGF, and to perform radioiodination of EGF by the chloramine T method.
Keywords: EGF receptor, endocytosis, recycling, degradation, synthesis
INTRODUCTION
At the molecular level, binding of a growth factor to EGFR triggers several signal-transduction cascades, ultimately leading to altered gene expression. There are at least six ligands of EGFR, with EGF being the best characterized. Upon ligand binding, activated EGFR is rapidly internalized through clathrin-dependent and -independent pathways of endocytosis. After internalization to early endosomes, ligand-receptor complexes are either retained in endosomes during their maturation into late endosomes or recycled back to the plasma membrane. Acceleration of internalization and degradation of EGFR by EGF results in dramatic decreases in the amount of EGFR at the cell surface (termed ligand-induced receptor down-regulation) and an eventual decrease of total cellular EGFR protein. The efficiency of receptor targeting to late endosomes and subsequent degradation by lysosomal enzymes determines the overall rate of ligand-induced receptor down-regulation.
CAUTION: When working with radioactivity, take appropriate precautions to avoid contamination of the experimenter and the surroundings. Carry out the experiment and dispose of wastes in appropriately designated area, following the guidelines provided by the local radiation safety officer (also see APPENDIX 1D).
NOTE: All solutions and equipment coming into contact with cells must be sterile, and proper aseptic technique should be used accordingly.
NOTE: All culture incubations should be performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified.
MEASURING EGFR PROTEIN SYNTHESIS
The turnover rate of a protein is a function of its rates of biosynthesis and degradation. Transmembrane proteins like EGFR are synthesized by ribosomal complexes that are associated with the endoplasmic reticulum (ER), allowing for the simultaneous translocation of the nascent transmembrane protein through the ER membrane. EGFR is N-glycosylated in the Golgi complex and then delivered to the plasma membrane (Todderud and Carpenter, 1989). Fully glycosylated EGFR protein runs on SDS-PAGE as a ~170-kDa band.
Measurement of the rate of EGFR biosynthesis relies on determining the rate of incorporation of radiolabeled amino acids (methionine and cysteine) into EGFR (see UNIT 7.1). These two amino acids are the only ones that contain sulfur and, therefore, can be labeled with the 35S isotope. 35S offers a stronger radioactivity and other advantages as compared to 14C, 3H, and other isotopes used for radiolabeling of proteins in the research laboratory. Methionine and cysteine are rapidly transported into the cell by a concentration gradient–dependent mechanism. The cellular concentration of labeled amino acids is, therefore, maintained constant during the assay if the concentration of these amino acids is constant in the medium. Hence, the amount of the label incorporated into EGFR per unit of time can be used as a measure of the rate of EGFR biosynthesis. Since all newly synthesized proteins incorporate labeled amino acids, EGFR has to be isolated from cell lysates by immunoprecipitation (UNIT 7.2) followed by SDS-PAGE (UNIT 6.1). The amount of radioactivity associated with EGFR bands is then measured at each time point.
Materials
EGFR-expressing cells (most cultured cells of epithelial and fibroblast origin express EGFR at levels from 5,000 to 2,000,000 per cell)
Complete DMEM (UNIT 1.1; use 5% to 10% FBS) or other appropriate cell culture medium
DMEM or other appropriate culture medium free of l-methionine, l-cysteine, and l-glutamine (Invitrogen)
EasyTag 35S-labeled amino acids (PerkinElmer)
Phosphate-buffered saline (DPBS; Invitrogen
TGH lysis buffer (see recipe) containing 150 mM NaCl
EGFR antibody mAb528 (mouse hybridoma, ATCC #HB8509)
20% (v/v) protein A–Sepharose resin (Sigma) in 50 mM HEPES
TGH lysis buffer (see recipe) containing no NaCl and 500 mM NaCl
2× sample buffer for SDS-PAGE (see recipe for 1×)
7.5% acrylamide gel (UNIT 6.1)
EGFR antibody 1005 (Santa Cruz Biotechnology)
60-mm plastic culture dishes
Rubber policemen
Platform rotator (e.g., Nutator from Clay Adams)
Image analysis software: e.g., ImageJ (NIH freeware)
Additional reagents and equipment for cell culture (UNIT 1.1), determining protein concentration (APPENDIX 3H), SDS-PAGE (UNIT 6.1), immunoblotting and immunodetection (UNIT 6.2), and phosphor imaging (UNIT 6.3)
Label cells
-
1.Seed EGFR-expressing cells at ~0.5–1.0 × 106 per 60-mm dish in 3 ml of complete DMEM (with 5% to 10% FBS depending on the cell line). Grow the cells for 2 days or until the cells are confluent.A basic experiment will require a minimum of four time points for each experimental condition, with two dishes per time point. Repeat the entire experiment a minimum of two times.The SCC2 cells used in Figure 15.14.1 are grown in 5% FBS, whereas many other cell types require 10% FBS.The amount of cells to use depends on the abundance of EGFR in the particular cell line. This protocol is geared towards cell lines that express high levels of EGFR (>100,000 receptors per cell).
-
2.
Wash cells once with 3 ml DMEM lacking FBS, l-methionine, l-cysteine, and l-glutamine.
-
3.
Add 3 ml of freshly prepared 35S protein labeling medium to each dish and incubate the cells at 37°C for appropriate periods of time (e.g., 0.75, 1.5, 3.0, and 4.5 hr, as in Fig. 15.14.1A).
-
4.After each desired labeling time, place the cells on ice, collect radiolabeled medium, and dispose of it according to proper institutional procedures for radioactive waste.Properly dispose of all radiolabeled medium, subsequent washes, extracts and disposable labware.
-
5.Wash the cells three times with 3 ml ice-cold PBS. Aspirate as much as possible of the PBS after the last wash. Store dishes with washed monolayers at −70°C until further analysis.All dishes from the different time points of the pulse-chase are eventually frozen. Subsequent thawing and solubilization, and all remaining steps of the protocol, are performed simultaneously to minimize sample variability.
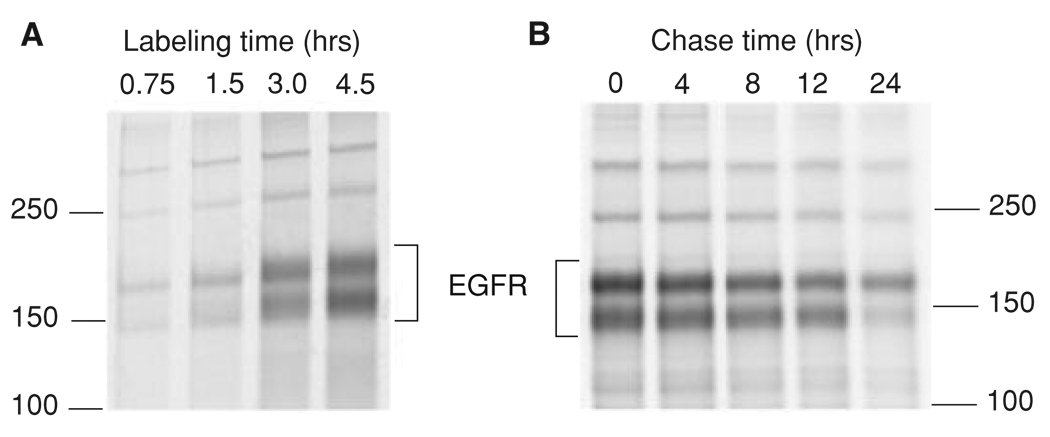
Figure 15.14.1.
Examples of EGFR biosynthesis and degradation experiments. (A) Squamous cell carcinoma (SCC2) cells (~106 EGFR/cell) were incubated with the [35S]amino acid labeling mix in the methionine-free medium containing 5% dialyzed FBS for 45 min to 4.5 hr. The cells were solubilized and EGFR-immunoprecipitated as described in Basic Protocol 1. (B) SCC2 cells were incubated with the [35S]amino acid labeling mix in the methionine-free medium containing 5% dialyzed FBS for 4 hr. After washing,the cells were incubated in complete medium containing 5% FBS for 1 hr before initiating chase times of 0 to 24 hr. The cells were solubilized and EGFR immunoprecipitated as described in Basic Protocol 1. Note that EGFR immunoprecipitated from SCC2 cells runs as a doublet of 170- and 150-kDa (probably degradation-product) bands.
Lyse the cells
-
6.
Place the frozen dishes on ice to thaw.
-
7.
Add 300 µl of freshly prepared ice-cold TGH lysis buffer (with 150 mM NaCl) to each dish. Scrape the cells using a rubber policeman and transfer the lysates to 1.5-ml tubes.
-
8.
Carry out the lysis for 10 min by gently rotating tubes on a platform rotator at 4°C.
-
9.
Clear the lysates by microcentrifugation for 10 min at 15,000 × g, 4°C, and transfer the supernatants to new tubes.
-
10.
Determine protein concentration (BradFord or Lowry assay; APPENDIX 3H) for each sample to assure that an equal amount of protein is added to each immunoprecipitation reaction.
-
11.
Save an 10-µl aliquot of each cell extract for future analysis of immunoprecipitation efficiency.
Immunoprecipitate EGFR
-
12.Add 20 µg of affinity-purified mouse monoclonal EGFR antibody 528 (mAb528) to each tube containing an equal amount of protein and mix the tubes gently on a platform rotator for 2 hr or longer at 4°C (this amount of mAb528 is sufficient for cells with >100,000 receptors per cell, e.g., human epidermal carcinoma A-431 and human head-and-neck squamous cell carcinoma SCC2 cells).The amount of mAb528 needed will vary depending upon the abundance of EGFR in a particular cell line. Other antibodies that efficiently precipitate EGFR can be used. The dose of antibodies necessary for efficient immunoprecipitation should be determined in preliminary experiments using unlabeled cell lysates and immunoblotting.
-
13.
Add 200 µl of protein A–Sepharose (20% v/v suspension in 50 mM HEPES) and continue the incubation for 1 hr at 4°C.
-
14.
Pellet the protein A–Sepharose and bound EGFR immunoprecipitates by microcentrifugation for 2 min at 2000 × g, 4°C.
-
15.
Collect supernatants and save 10-µl aliquots for future analysis of immunoprecipitation efficiency.
-
16.
Wash the immunoprecipitates (pellets) three times with 1 ml of TGH lysis buffer containing decreasing amounts of NaCl (500 mM, 150 mM, and finally 0 mM NaCl).
-
17.
Mix washed immunoprecipitates with 2× sample buffer for SDS-PAGE at a ratio of 1:1. Mix aliquots of cell extracts and supernatants from immunoprecipitation reactions at a ratio of 1:4 with 4× sample buffer for SDS-PAGE. Heat all samples at 95°C for 5 min, and subject to SDS-PAGE in a 7.5% acrylamide gel (UNIT 6.1).
Analyze labeling
-
18.
Cut out the very bottom part of the gel (2 cm) containing most of the unincorporated low-molecular-weight radioactivity, and dispose of it in a designated 35S waste container.
-
19.
Transfer the portion of the gel that contains aliquots of cell extract input and output to nitrocellulose for subsequent analysis by immunoblotting with EGFR antibody 1005 and an appropriate loading control (see UNIT 6.2).
-
20.
Carefully place the portion of the gel that contains the immunoprecipitation reactions onto chromatography paper and vacuum dry (UNIT 6.3).
-
21.
For quantification of radioactivity using a phosphor imager, place the dried portion of the gel containing the immunoprecipitation reactions against an appropriate phosphor imaging screen in a phosphor imaging cassette. Expose for the necessary time (typically 12 to 48 hr), being careful not to generate a saturating signal.
-
22.
Perform analysis of the phosphor imaging screen on an imager such as a Typhoon8600.
-
23.
Quantify signal intensities using software such as ImageJ.
MEASURING EGFR PROTEIN DEGRADATION
After synthesis and delivery to the cell surface, EGFR is turned over, typically with half-lives ranging from 8 to 24 hr, or longer, depending on the cell type and the level of EGFR expression. These turnover rates can be dramatically accelerated if EGFR is activated by EGF or other ligands. Measurement of the degradation rate involves pre-labeling of cells with [35S]methionine and cysteine, followed by a chase incubation in the presence of an excess of the corresponding unlabeled amino acids. Since it takes several minutes for the newly synthesized receptor to acquire post-translational modifications in the Golgi complex and reach the plasma membrane, the pre-labeled cells are incubated without the label for ~30 min prior to the beginning of the chase incubation. This allows most of the newly labeled synthesized receptors to complete glycosylation and move to the plasma membrane. As in the biosynthesis assay, EGFR is immunoprecipitated before and during the chase incubation, electrophoresis is performed, and the amount of radioactivity in each EGFR band is measured. In contrast to the biosynthesis assay, the amount of labeled EGFR decreases during the chase incubation due to degradation. An example of a degradation experiment is shown in Figure 15.14.1B.
Additional Materials (also see Basic Protocol 1)
Experimental compounds of interest (e.g., EGF)
-
1.
Prepare the cell cultures as in Basic Protocol 1, steps 1 and 2.
-
2.Add 3 ml of freshly prepared [35S]protein labeling medium to each dish and incubate the cells at 37°C for 4 hr or longer.Incubation of cells with labeling medium for less than 4 hr may result in labeling of only a small pool of EGFR, since the half-life of EGFR is typically 8 to 24 hr. However, long incubations of some cells (in particular, cancer cells with enhanced metabolism) in methionine-cysteine-free medium and dialyzed serum may cause a substantial toxicity.
-
3.
After an appropriate incubation time, dispose of the radioactive medium properly and rinse the cells with 3 ml of DMEM lacking FBS, l-methionine, l-cysteine, and l-glutamine.
-
4.
Add 4 ml of fresh, complete medium with 10% (v/v) FBS to each dish. Incubate the cells at 37°C for 30 min to allow newly synthesized EGFR to reach the plasma membrane.
-
5.Following the 30-min incubation time, add any desired experimental compounds (for instance, EGF at 1 to 200 ng/ml dosage) and chase the cells for appropriate times at 37°C (e.g., 0, 4, 8, 12, and 24 hr as in Fig.15.14.1B).The range of chase incubations is discussed below in Commentary.
-
6.
At the end of each chase incubation period, wash the appropriate dishes three times with 3 ml ice-cold PBS and store at −70°C for later analysis.
-
7.
Thaw and lyse the labeled cultures, immunoprecipitate, and analyze the samples by following steps 6 through 23 of Basic Protocol 1.
MEASURING EGFR DOWN-REGULATION
Calculation of the number of EGFR molecules at the cell surface is based on measuring the number of EGF binding sites per intact cell. Because the stoichiometry of the EGF-EGFR interaction is 1:1, radiolabeled [125I]EGF with known specific activity can be used to determine the number of EGF binding sites or surface EGFR per cell. This [125I]EGF binding assay is performed at 4°C to avoid the uptake of [125I]EGF inside the cell due to endocytosis. For an example of results of a down-regulation experiment, see Figure 15.14.2.
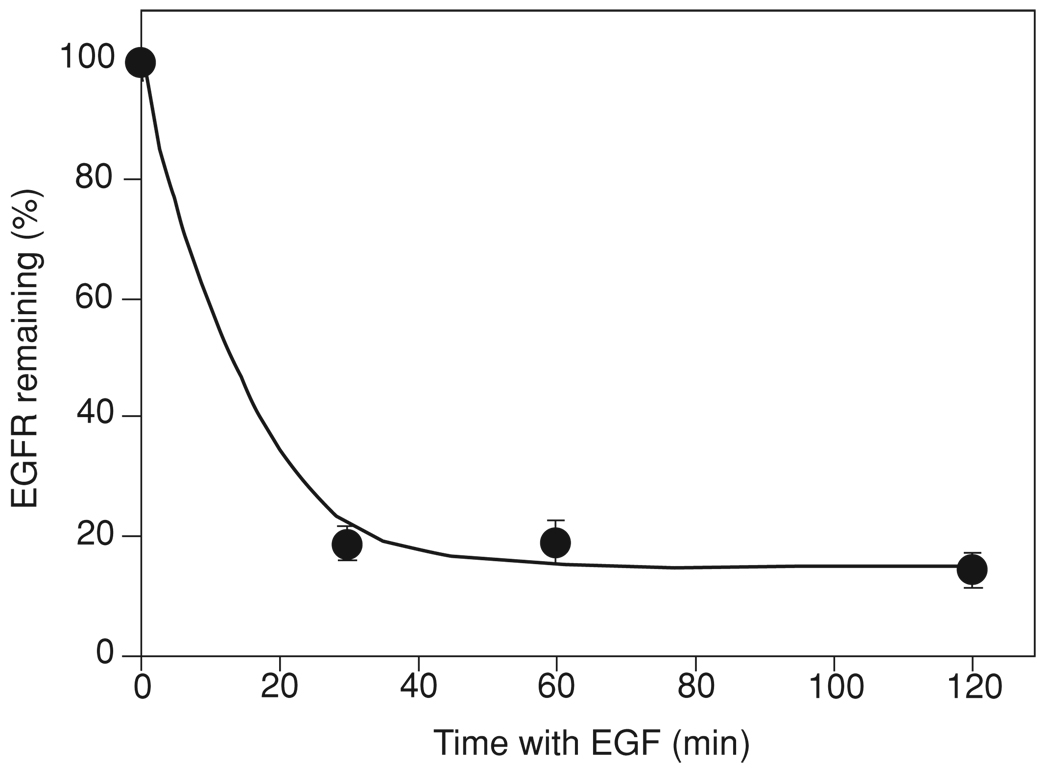
Figure 15.14.2.
Example of EGFR down-regulation experiment. Porcine aortic endothelial cells stably expressing EGFR were incubated with 100 ng/ml EGF for 30 to 120 min at 37°C, acid washed, and further incubated with 50 ng/ml [125I]EGF for 1 hr at 4°C. The cells were solubilized in NaOH. The specific radioactivity of cell lysates was determined and expressed as percent of the specific radioactivity associated with cells that were not incubated with unlabeled EGF (mean ± SE).
Materials
EGFR-expressing cells (most cultured cells of epithelial and fibroblast origin express EGFR at levels from 5,000 to 2,000,000 per cell)
Complete DMEM (UNIT 1.1; use 5% to 10% FBS) or other appropriate cell culture medium
Recombinant human culture-grade EGF (BD Biosciences, cat. no. 354052)
Binding medium; DMEM or other medium containing 0.1% (w/v) BSA
Sodium acetate buffer, pH 4.5: 0.2 M sodium acetate, pH 4.5 (APPENDIX 2A)/0.5 M NaCl
1 to 200 ng/ml [125I]EGF (sp. act. ~200,000 cpm/ng; Support Protocol)
1 N NaOH
12-well plastic culture plates
Additional reagents and equipment for cell culture (UNIT 1.1)
Prepare cells
-
1.Seed EGFR-expressing cells at ~0.125–0.250 × 106 cells per well of a 12-well dish in 2 ml of complete DMEM (with 5% to 10% FBS depending on the cell line). Grow the cells for 2 days or until the cells are confluent.A basic experiment will require a minimum of four time points for each experimental condition, with four wells of a 12-well dish per time point. Repeat the entire experiment a minimum of two times. Three additional wells are used for cell counting.A confluent monolayer helps to maximize specific binding of [125I]EGF to cells and minimize nonspecific binding of [125I]EGF to plastic surfaces of culture dishes.
Incubate cells with and without EGF
-
2.Incubate the cells with or without unlabeled EGF in binding medium (1 ml/well of 1 to 200 ng/ml) for various lengths of time at 37°C starting from the longest incubation time.The start of shorter incubations is delayed to make it possible harvest all cells in the 12-well plate in the same time.Possible lengths of incubations with EGF and EGF concentrations are discussed in Commentary.
-
3.
At the end of the longest incubation time, place the cells on ice and rinse once with 1 ml ice-cold medium to remove unbound EGF.
-
4.Strip surface-bound, unlabeled EGF by incubating the cells with sodium acetate buffer, pH 4.5, for 2.5 min (1 ml/well) at 4°C, then washing the cells twice, each time with 1 ml ice-cold culture medium, to neutralize the acid.This is an important step that has been omitted in many studies. If cold EGF is not stripped from receptors, binding of [125I]EGF at step 5 will be underestimated. The mild acidic wash does not affect EGFR properties.
Incubate the cells with labeled EGF
-
5.
Incubate the cells in 1 ml of binding medium containing saturating concentrations of [125I]EGF (up to 200 ng/ml) alone (specific binding) or together with a 50-fold molar excess of unlabeled EGF (controls for nonspecific binding) at 4°C for 1 hr.
-
6.Rinse the wells three times with 1 ml ice-cold DMEM or other medium to remove unbound [125I]EGF. Aspirate the medium as much as possible after the last wash.There are high- and low-affinity EGF binding sites in most studied cells. The mechanisms determining the EGF binding affinity are unknown. If [125I]EGF is used at low concentrations it may predominantly bind to high-affinity receptors that would yield misleading results. The number of high-affinity sites may not be proportional to the total receptor number under various experimental conditions.
Measure radioactivity
-
7.
Lyse the cells in 1 ml of 1 N NaOH for 1 hr or longer at 37°C to determine cell-bound radioactivity.
-
8.Measure nonspecific binding for each time point in the presence of unlabeled EGF (see step 5).Typically nonspecific binding is not more than 10% to 20% of the total [125I]EGF binding.
Make calculations
-
9.
Calculate the specific binding by subtracting the nonspecific radioactivity from total bound radioactivity for each time point of the time course.
-
10.Translate cpm values into the number of EGFR molecules per well (number of surface EGFRs) based on the specific activity of each [125I]EGF preparation.No. of EGFRs per well = [(cpm/well)/(spec.act. – cpm/ng)] × 1011.
-
11.Divide the number of surface EGFR per well by the number of cells in the well to obtain a number of EGFRs per cell.No. of EGFRs per cell = (EGFR/well)/(no. of cells/well).
MEASURING INTERNALIZATION OF [125I]EGF
Binding of EGF to EGFR at the cell surface results in acceleration of internalization of receptors. After internalization into early endosomes, EGF-receptor complexes are either recycled back to the plasma membrane or sorted to late endosomes and lysosomes where both EGF and EGFR are degraded. Availability of EGF, its high level of stability, and the ease in preparing fully active labeled forms of EGF has led to wide use of various EGF conjugates for the quantitative analysis of EGFR endocytosis. Endocytosis assays that use radiolabeled EGF ([125I]EGF) remain the gold standard of quantitative endocytosis assays because 125I-radioactivity detection is highly sensitive and linear within a large range of radioactivity concentrations.
The internalization assay is based on short incubations of cells with [125I]EGF followed by separation of surface bound [125I]EGF from intracellular (internalized) [125I]EGF. Internalization is highly temperature-dependent and, therefore, this assay is performed at 37°C. The internalization is stopped by placing cells on ice followed by the stripping of surface [125I]EGF. The most reliable method to strip [125I]EGF bound to surface receptors without affecting intracellular [125I]EGF is a low-pH treatment (pH 2.5 to 2.8; Haigler et al., 1980). The stripped [125I]EGF is then quantitated as the surface-bound [125I]EGF, whereas acid-wash-resistant [125I]EGF corresponds to internalized [125I]EGF. The ratio of internalized to surface [125I]EGF (I/S) plotted against incubation time is considered to be a measure of the apparent internalization rates. Internalization is considered to be a first-order kinetics process. Therefore, the specific rate of internalization depends on the concentration of EGF-receptor complexes at the cell surface. The precise and easy calculation of the internalization rate constant ke can be performed if the [125I]EGF concentration at the cell surface remains constant during the time-course of [125I]EGF uptake. Under these conditions, the I/S ratio displays a linear dependence on time, and, therefore, ke corresponds to the linear regression coefficient of I/S dependence on time. See Wiley and Cunningham (1982) for detailed explanations.
NOTE: Rapid handling of all steps of this procedure, particularly washes, is critical because of the short incubations of cells with [125I]EGF.
Materials
EGFR-expressing cells (most cultured cells of epithelial and fibroblast origin express EGFR at levels from 5,000 to 2,000,000 per cell)]
Complete DMEM (UNIT 1.1; use 5% to 10% FBS) or other appropriate cell culture medium
Binding medium; DMEM or other medium containing 0.1% (w/v) BSA
0.1 to 200 ng/ml [125I]EGF (sp. act., ~200,000 cpm/ng; Support Protocol)
Recombinant human culture-grade EGF (BD Biosciences, cat. no. 354052)
Sodium acetate buffer, pH 2.8: 0.2 M acetic acid/0.5 M NaCl, pH 2.5 to 2.8
1 N NaOH
12-well plastic cell culture plates
γ-counter vials
γ counter
Additional reagents and equipment for cell culture (UNIT 1.1)
Prepare cells and binding medium
-
1.Seed EGFR-expressing cells at 0.125 to 0.25 × 106 cells/well in 12-well plates in 2 ml of DMEM containing 5% to 10% FBS and grow for 2 days or until confluent.Typically, each experiment includes at least five time points, each represented by two to four wells of a 12-well dish. The time-course experiment needs to be repeated at least twice to obtain statistically significant data.The use of confluent monolayers helps to minimize nonspecific binding of [125I]EGF to plastic surfaces of a culture dish.UNIT 1.1 describes basic cell culture techniques.
-
2.Prepare [125I]EGF-containing binding medium (see Commentary for consideration of [125I]EGF concentrations) and the same [125I]EGF-containing medium supplemented with 50× to 100× molar excess of unlabeled human recombinant EGF (“nonspecific” medium).For each 12-well plate, 3 ml each of “specific” (not containing unlabeled EGF) and “nonspecific” medium are needed and should be warmed to 37°C prior to the assay.IMPORTANT NOTE: Prepare 6.5 ml of [125I]EGF-containing medium and then add unlabeled EGF to 3 ml of this medium. After the experiment, the remaining ~0.5 ml is used to measure the specific activity of [125I]EGF.
-
3.
Place the plate in a water bath at 37°C.
Bind [125I]EGF to the cells
-
4.Quickly wash two wells with 1 ml of warm (37°C) binding medium without labeled or unlabeled EGF. To these wells, add 0.5 ml of specific or nonspecific [125I]EGF-containing medium (one well each).These cells correspond to a 6-min time point.It is advisable to add specific and nonspecific medium to the two wells simultaneously using a pipettor in each hand.
-
5.Repeat the same procedure (step 4) with the other five pairs of wells at 1-min intervals while the dish is kept at 37°C.In this way each pair of wells (specific and nonspecific) receives the radioactive medium at precisely 1-min intervals.
Remove unbound [125I]EGF
-
6.
After the last pair of wells has incubated for 1 min, place the dish on ice and rapidly aspirate the radioactive medium and discard as radioactive waste.
-
7.Rapidly wash the monolayers three times with the ice-cold medium by adding ~1ml per well, followed by vacuum aspiration to remove as much unbound [125I]EGF as possible after the last wash.This step is performed as quickly as possible to minimize the dissociation of [125I]EGF from surface receptors during washes.
Strip the surface-bound [125I]EGF
-
8.
Strip surface-bound [125I]EGF from all wells by incubating cells for 5 min with sodium acetate buffer, pH 2.8, at 1 ml/well, 4°C. Collect this acid wash in γ-counter vials, perform another short rinse with 0.5 ml/well of acetate buffer, pH 2.8, and combine with the original wash in the vial to determine the amount of surface-bound [125I]EGF by gamma counting.
Measure internalized [125I]EGF
-
9.
Lyse the cells in 1 ml of 1 N NaOH for 1 hr at 37°C to determine the amount of internalized [125I]EGF by gamma counting.
Perform calculations
-
10.Calculate the specific radioactivity of the acid-sensitive (surface) and acid-insensitive (internalized) fractions by subtracting nonspecific counts from the corresponding counts of specific wells (Figure 15.14.3A).Nonspecific radioactivity is typically not more than 3% to 10% of the total counts.Internalization is considered to be a first-order kinetics process. Therefore, the specific rate of internalization depends on the concentration of EGF-receptor complexes at the cell surface.
-
11.To estimate the rate of internalization, plot the ratio of specific internalized to specific surface radioactivity (I/S) against time (Figure 15.14.3B).The slope of this plot corresponds to the apparent rate of internalization. If this slope is linear, it can be used to calculate the rate constant of internalization (ke). ke is the linear regression coefficient of the dependence of I/S ratio on time. Such a simple calculation of the values is, however, possible only if the amount of occupied surface receptors stays relatively constant during the time course of [125I]EGF uptake.Figure 15.14.3 shows an example of internalization data from one 12-well plate. In Figure 15.14.3A the amount of surface [125I]EGF is relatively constant (does not decrease due to EGF-induced down-regulation of surface EGFR). This results in a near linear dependence of I/S [125I]EGF ratio on time and, therefore, allows calculation of the ke value (Fig. 15.14.3B).
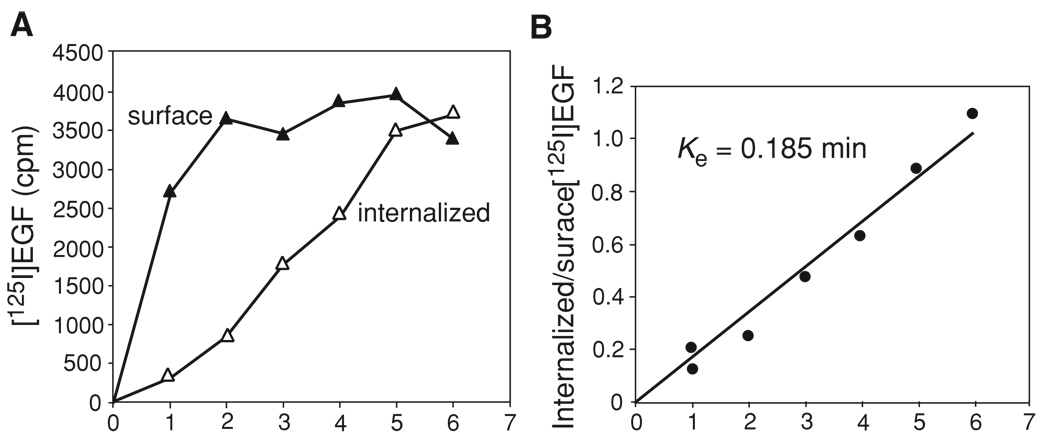
Figure 15.14.3.
Example of the [125I]EGF internalization assay. NIH 3T3 cells stably expressing EGFR tagged with GFP were incubated with 1 ng/ml [125I]EGF for 1 to 6 min according to Basic Protocol 3. (A)The specific [125I]EGF radioactivity of acid-wash (surface) and acid-resistant [125I]EGF (internalized) are plotted against time. (B)The ratio of internalized to surface [125I]EGF is plotted against time. The linear regression coefficient was calculated to obtain the ke value.
MEASURING [125I]EGF RECYCLING AND DEGRADATION
The rationale to use [125I]EGF for estimating both the recycling and degradation rates of EGFR is underscored by two important facts. First, EGF, which is sorted at the early endosome for degradation in the lysosome, does not significantly dissociate from the internalized EGFR until it reaches the lysosome (Sorkin et al., 1988). Second, degradation of [125I]EGF results in a product that can be biochemically separated from intact [125I]EGF which has been recycled or remains internalized. The recycling/degradation assay is based on loading early endosomes with [125I]EGF-receptor complexes by allowing cells to endocytose [125I]EGF. The endocytosis is then stopped at 4°C, and [125I]EGF that has not been internalized is stripped from the cell surface by the mild acidic wash treatment. When cells are returned to 37°C, a pool of [125I]EGF-receptor complexes are recycled back to the cell surface, where [125I]EGF is released from the receptor or re-internalized. Another pool of internalized [125I]EGF-receptor complexes are sorted from early endosomes to late endosomes and lysosomes where [125I]EGF-receptor complexes are degraded by proteolytic enzymes. This degradation results in the release of mono- and di-[125I]iodotyrosines, which readily penetrate membranes and accumulate in the extracellular medium. This form of 125I radioactivity is not precipitable by trichloroacetic acid (TCA) and is used as a measure of the rate of [125I]EGF degradation. Only negligible amounts of TCA-soluble 125I can be detected in the cells. On the other hand, intact [125I]EGF (TCA-precipitable) detected in the medium and at the cell surface (acid-wash stripped) after the chase incubation of [125I]EGF-loaded cells corresponds to [125I]EGF-receptor complexes that have recycled from endosomes to the plasma membrane.
Recycling and degradation can also be considered first-order processes. This means that their rates are proportional to the concentration of [125I]EGF in early/sorting endosomes. However, the calculation of specific rate constants is difficult due to heterogeneity of endosomal compartments and the decrease in the concentration of [125I]EGF in early/sorting endosomes during the chase time. Also, the presence of unlabeled EGF (Basic Protocol 4) and re-internalization of [125I]EGF-receptor complexes (Alternate Protocol 2) make calculations of specific rate constants challenging. Therefore, it is important to interpret the results of the recycling/degradation assay described in these protocols only as apparent rates. In theory, kinetic rate parameters can be calculated by using computational modeling that takes into account all EGF-receptor association-dissociation parameters and apparent trafficking rates (see, for example, Resat et al., 2003). For a schematic of the assay and example of results, see Figure 15.14.4.
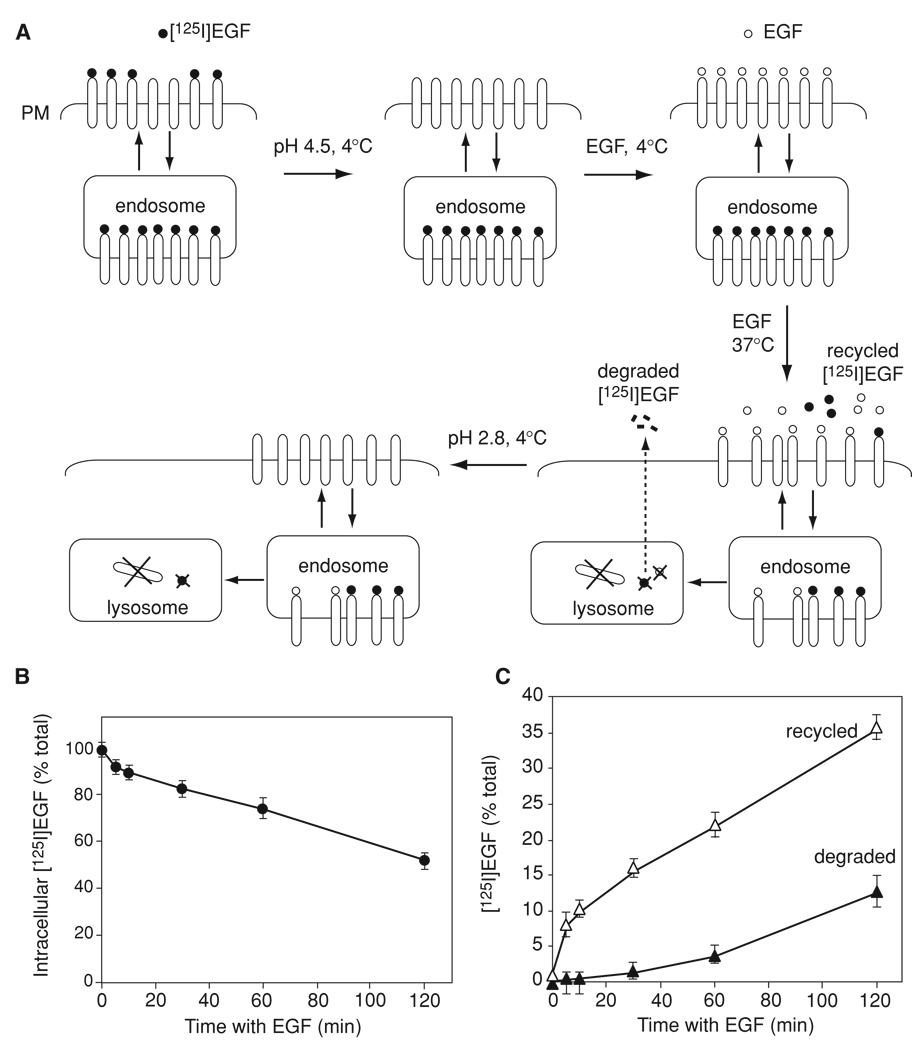
Figure 15.14.4.
[125I]EGF recycling/degradation assay (Basic Protocol 4). (A) Schematic representation of the main steps of the assay. PM, plasma membrane. (B) and (C) Representative example of the [125I]EGF recycling/degradation experiment. Mouse embryonic fibroblasts were incubated with 5 ng/ml [125I]EGF for 5 min at 37°C, and[125I]EGF that has not been internalized was stripped by the acid wash. [125I]EGF-loaded cells were incubated with unlabeled EGF at 4°C and then chased with unlabeled EGF for 0 to 120 min according to the protocol. The specific radioactivities of all fractions were determined and expressed as percent of the total radioactivity associated with cells and present in the medium (mean ±SE). The amount of intracellular [125I]EGF (NaOH lysates) plotted against the chase time is shown in (B), whereas the time course of [125I]EGF recycling (surface plus medium trichloroacetic acid/phosphotungstic acid-insoluble [125I]EGF) and degradation (trichloroacetic acid/phosphotungstic acid 125I-radioactivity) is shown in (C).
Materials
EGFR-expressing cells (most cultured cells of epithelial and fibroblast origin express EGFR at levels from 5,000 to 2,000,000 per cell)
Complete DMEM (UNIT 1.1; use 5% to 10% FBS) or other appropriate cell culture medium
Binding medium: DMEM or other medium containing 0.1% (w/v) BSA
1 to 200 ng/ml [125I]EGF (sp. act., ~200,000 cpm/ng; Support Protocol)
Recombinant human culture-grade EGF (BD Biosciences, cat. no. 354052)
Sodium acetate buffer, pH 4.5: 0.2 M sodium acetate buffer, pH 4.3 to 4.5 (APPENDIX 2A)/0.5 M NaCl
Sodium acetate buffer, pH 2.8: 0.2 M acetic acid/0.5 M NaCl, pH 2.5 to 2.8
1 N NaOH
10% (w/v) trichloroacetic acid (TCA)/2% (w/v) phosphotungstic acid (PTA) in H2O
35-mm plastic cell culture dishes
γ-counter vials
γ counter
Refrigerated centrifuge
Additional reagents and equipment for cell culture (UNIT 1.1)
Prepare cells
-
1.Seed 0.25 to 0.50 × 106 cells in 35-mm culture dishes in 2 ml of DMEM containing 5% to 10% FBS and grow for 2 days or until confluent.UNIT 1.1 describes basic cell culture techniques.Typically, at least five time points are required, each represented by two to three dishes. The entire time-course experiment should be repeated at least twice.
-
2.Incubate the cells at 37°C with 1 ml [125I]EGF in binding medium for 5 to 10 min (see Commentary for consideration of [125I]EGF concentration and incubation times) to allow for accumulation of [125I]EGF-receptor complexes in early endosomes. Incubate control dishes with the same [125I]EGF-containing medium supplemented with a 50-fold molar excess of unlabeled EGF to determine nonspecific radioactivity.Because handling of multiple dishes in this type of experiments is difficult, it is recommended to use two “specific” dishes and one “nonspecific” dish for each time point of the chase incubation (step 7 below). The entire time-course experiment can be repeated to ensure the statistical significance of the data.
-
3.
After 37°C incubation, place the dishes on ice and then rapidly wash three times with 1 ml of ice-cold DMEM. Briefly rinse cells with DMEM (2 ml/dish) to neutralize the acid.
-
4.
Incubate the cells with 1 ml/dish of sodium acetate buffer, pH 4.5, for 2.5 min, remove supernatants, and then rinse briefly with 1 ml of the same buffer to remove [125I]EGF that had not been internalized during the 37°C incubation.
-
5.Combine both of the acidic washes from step 4 to determine the amount of [125I]EGF at the cell surface by gamma counting.At this point the cells contain[125I]EGF only in endosomes and are referred to as “[125I]EGF-loaded cells.”
-
6.Incubate the [125I]EGF-loaded cells in binding medium containing 200 ng/ml unlabeled EGF for 40 min at 4°C to occupy surface EGFRs, and aspirate the medium.This EGF concentration is sufficient to occupy >90% of surface receptors in all types of cells, including cells expressing the highest levels of EGFR.
-
7.Add 1 ml fresh pre-warmed (37°C) medium containing 200 ng/ml EGF. Rapidly place the cells in the water bath or incubator (for incubations longer than 15 min) at 37°C to allow trafficking of [125I]EGF-receptor complexes in loaded cells. Keep three dishes (two “specific” medium and one nonspecific medium) on ice for the zero chase time point.The lengths of chase incubations are discussed in the Commentary, below.An excess of unlabeled EGF bound to surface receptors and in the medium minimizes re-internalization of recycled [125I]EGF-receptor complexes and rebinding of dissociated [125I]EGF from the medium.
-
8.While other dishes are incubating at 37°C, add 1 ml prewarmed medium to zero-time dishes and immediately collect the medium in γ-counter vials.This step is necessary to have the zero-time dishes exposed to medium that is the same as that used for the chase incubation of other dishes in step 7.
-
9.
At the end of each of the chase incubations, place the dishes on ice, add 1 ml medium, and collect the medium into γ-counter vials.
-
10.
Strip surface-bound [125I]EGF from all wells by incubating cells for 5 min with sodium acetate buffer, pH 2.8, at 1 ml/dish, 4°C. Collect this acid wash in γ-counter vials, perform another short rinse at 0.5 ml/dish, and combine this rinse with the original wash to determine the amount of surface-bound (recycled) [125I]EGF by gamma counting.
-
11.
Solubilize the cells in 1 N NaOH for 1 hr to measure the amount of intracellular [125I]EGF.
-
12.Mix the medium collected at step 8 with 0.3 ml of 10% (w/v) TCA/2% (w/v) PTA and incubate 1 hr or longer at 4°C. Centrifuge 10 min at 5000 × g, 4°C.Because of its small size, EGF polypeptide is not efficiently precipitated by TCA alone. Therefore, addition of PTA is important for complete precipitation of [125I]EGF.
-
13.
Transfer supernatants to new γ-counter tubes to determine the amount of degraded [125I]EGF by gamma counting.
-
14.
Dissolve the pellets in 1 ml of 1 N NaOH to determine the amount of [125I]EGF by gamma counting.
Make calculations
-
15.
First, obtain specific counts by subtracting nonspecific radioactivity of all fractions from the counts of the same fractions of corresponding specific dishes.
-
16.
Calculate the total amount of [125I]EGF specifically associated with cells for each dish as the sum of the second (pH 2.8) acid wash (surface [125I]EGF, step 10), intracellular [125I]EGF (step 11), intact-medium [125I]EGF (TCA-precipitated radioactivity, step 12), and degraded-medium [125I]EGF (step 13).
-
17.
Calculate the amount of recycled [125I]EGF by summing the surface [125I]EGF and intact medium [125I]EGF.
-
18.Calculate the percent of each [125I]EGF pool (intracellular, recycled, and degraded) relative to the total cell-associated [125I]EGF for each time point.The example of the dynamics of recycled, degraded and intracellular [125I]EGF is presented in Figure 15.14.4B and C.
-
19.Calculate the ratios of the percent of recycled and degraded [125I]EGF to percent of intracellular [125I]EGF and plot against time.The slopes of these graphs correspond to the apparent recycling and degradation rates, respectively.
MEASURING [125I]EGF RECYCLING/DEGRADATION IN THE ABSENCE OF EXCESS UNLABELED EGF
This assay for recycling/degradation differs from Basic Protocol 4 at the step of incubation of [125I]EGF-loaded cells at 37°C, during which no unlabeled EGF is added.
For materials, see Basic Protocol 4.
-
1.
Follow steps 1 through 4 of Basic Protocol 4 to prepare cells and load the cells with labeled EGF.
-
2.Initiate trafficking of [125I]EGF-receptor complexes in loaded cells by incubating the cells in fresh binding medium with neither labeled nor unlabeled EGF at 37°C for 0 to 60 min or longer.See Commentary for considerations in the choice of the chase incubation time.
-
3.
Follow steps 7 to 19 in Basic Protocol 4 to measure the various EGF pools in the cells.
PREPARATION OF [125I]EGF
For the binding and trafficking of EGF studies, it is necessary to have [125I]labeled EGF. This protocol describes the chloramine T method for labeling proteins.
Materials
0.05 M sodium phosphate buffer, pH 7.5 (APPENDIX 2A) containing 0.075 M NaCl
20 mg/ml BSA in 0.05 M sodium phosphate buffer, pH 7.5 (APPENDIX 2A) containing 0.075 M NaCl
Receptor-grade mouse EGF (BD Biosciences, cat. no. 354010)
0.5 M sodium phosphate buffer, pH 7.5 (APPENDIX 2A)
0.1 mCi/µl Na[125I] (PerkinElmer; cat. no. NEZ33A)
2 mg/ml chloramine T (Sigma) in 0.05 M sodium phosphate buffer, pH 7.5 (see APPENDIX 2A for buffer); prepare fresh
4 mg/ml sodium metabisulfite in 0.05 M sodium phosphate buffer, pH 7.5 (see APPENDIX 2A for buffer); prepare fresh
PD-10 (G-25) Sepharose disposable column (GE Healthcare)
γ counter
NOTE: The entire procedure is carried out at room temperature.
Prepare the column
-
1.
Wash a PD-10 (G-25) Sepharose disposable column with 50 ml Milli-Q water and equilibrate with 50 ml 0.05 M sodium phosphate buffer, pH 7.5. containing 0.075 M NaCl.
-
2.
Load 1 ml of 20 mg/ml BSA in 0.05 M sodium phosphate buffer, pH 7.5, containing 0.075 M NaCl onto the column.
Label the EGF
-
3.In a 1.5-ml microcentrifuge tube, mix 24 µl of 0.5 M sodium phosphate buffer, pH 7.5, 28 µl of 0.05 sodium phosphate buffer, pH 7.5, and 5 µg of receptor-grade mouse EGF (5 µl of 1 mg/ml EGF in water).This gives a precisely 100 mM final sodium phosphate concentration in the mixture.The following steps are performed in a designated fume hood.
-
4.
Add 10 µl of 0.1 mCi/µl Na[125I] (1 mCi) to the EGF mixture from step 3 and initiate the reaction by adding 10 µl of 2 mg/ml chloramine T in the dark.
-
5.After a 40-sec incubation in the dark, stop the reaction by adding 20 µl of 4 mg/ml sodium metabisulfite.Both the chloramine T and sodium metabisulfite solutions should be freshly prepared.
Separate the 125I-labeled EGF
-
6.
Mix 100 µl of 20 mg/ml BSA in 0.05 M sodium phosphate buffer, pH 7.5, containing 0.075 M NaCl into the reaction mixture (final volume ~200 µl) and load onto the PD-10 column. Allow the column to equilibrate for 3 min.
-
7.
After 3 min of equilibration, start gel filtration by eluting with 0.05 M sodium phosphate buffer, pH 7.5, containing 0.075 M NaCl.
-
8.
Allow the first 1.8 ml to flow through.
-
9.Collect subsequent fractions at the rate of 10 drops per tube into 10 tubes, each containing 50 µl of 20 mg/ml BSA.BSA serves as a protein carrier to minimize binding of [125IEGF] to the tube walls.
-
10.
Measure the amount of radioactivity in 2 µl from each tube in the γ counter and plot the number of counts against the tube number.
-
11.
Based on the curve, combine the first peak (usually tubes 2 to 5 or 2 to 6) and mix in a larger plastic tube.
-
12.
Measure the specific activity of the final [125I]EGF preparation in a 2-µl sample of the pool.
-
13.
Divide the final [125I]EGF preparation into 0.1-ml aliquots and store at −20°C.
Make calculations
-
14.Calculate the specific activity of the preparation.Considering that the approximate yield of EGF in this procedure is 90%, then the total amount of [125I]EGF is ~4.5 µg. Typically, the volume of the peak is ~2 ml, which results in the final concentration of [125I]EGF being ~2.25 ng/µl. The amount of radioactivity is usually ~450,000 cpm per µl in our experiments. Therefore, the specific activity of [125I]EGF is 450,000 cpm/2.25 ng = 200,000 cpm/ng.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX.
Sample buffer for SDS-PAGE, 1 × (prepare at 2× or 4× concentration as indicated for protocol)
50 mM Tris·Cl, pH 6.8 (APPENDIX 2A)
10% (v/v) glycerol
2% (w/v) SDS
5% (v/v) 2-mercaptoethanol
Bromphenol blue (a few small crystals per 15 ml)
Store up to 1 year at room temperature
[35S]Protein labeling medium
DMEM free of l-methionine, l-cysteine, and l-glutamine (Invitrogen) supplemented with:
5% to 10% (v/v) dialyzed FBS (typically)
300 µCi Easy-Tag 35S-labeled amino acids
Prepare fresh
TGH lysis buffer
1% (v/v) Triton X-100
10% (v/v) glycerol
50 mM HEPES, pH 7.4
150 mM NaCl (or 0 mM or 500 mM as specified in protocol)
2 mM EGTA
2 mM EDTA
1 mM phenylmethylsulfonyl fluoride
10 ng/µl aprotinin
10 ng/µl leupeptin
Store up to 2 to 3 months at 4°C
COMMENTARY
Background Information
EGFR is a member of the ErbB tyrosine kinase family. EGFR is an important regulator of eukaryotic development and is involved in regulation of proliferation, survival, and differentiation of many types of cells in adult organisms. EGFR and other ErbBs are frequently overexpressed in various types of human carcinoma, glioblastoma, and glioma. In recent years, EGFR became an important diagnostic and prognostic marker in several carcinomas. Several EGFR inhibitors have been developed and approved for clinical use as anti-cancer targeted therapies.
Endocytosis and lysosomal degradation of active EGFR is thought to serve as the negative feedback loop in the regulation of EGFR signaling. However, endocytic trafficking regulates more than just receptor down-regulation. Several lines of evidence suggest that EGF-receptor complexes continue to signal after endocytosis and that signaling from endosomes may generate outcomes different from those triggered from the plasma membrane. Thus, mechanisms of EGFR endocytosis and its role in signaling has been the subject of more than 1000 publications. However, the large amount of data in the field is impossible to reconcile into a single reasonable model. One of the major reasons for such a slow pace towards the complete understanding of EGFR trafficking is the use of incomparable and often inadequate methodologies by different laboratories. This underscores the importance of standardizing the methodological approaches to monitoring EGFR endocytosis, specifically the methodologies that allow for reliable quantification of kinetics rates. Therefore, we focus this article on describing standard quantitative methods used to measure the rates of EGFR turnover (synthesis and degradation), EGFR down-regulation, and EGF internalization, both recycling and degradation.
Methods to study EGF and EGFR endocytosis using [125I]EGF were the first to be introduced and remain the most quantitative and sensitive. Most of these methodologies are designed to study active, EGF-occupied receptors. However, similar methods using 125I-labeled antibodies have been developed to study the kinetics of endocytosed EGFR which is not occupied by ligand (Wiley et al., 1991). It is important to be aware that cross-linking of EGFR by antibodies can alter trafficking parameters. Therefore, Fab fragments are the most appropriate in this type of experiment. Also, methods should use only those antibodies that do not interfere with any functions of EGFR. Other conventional methods of measuring surface receptor density and receptor endocytosis include those using surface biotinylation and reversible biotinylation (Shtiegman et al., 2007). These methods can also be used to study the endocytic trafficking of EGFR. However, they involve multiple steps, including pull-downs and SDS-PAGE. Each of these steps increases the chances of experimental error while not being as sensitive and quantitative as the methods using [125I]EGF.
Critical Parameters and Troubleshooting
Consideration of EGFR expression levels
The design and interpretation of the quantitative analysis of EGFR turnover and trafficking must take into consideration the expression levels of EGFR in the cell line to be studied. Normal mammalian cells express from 5,000 (most epithelial and fibroblast cells) to 200,000 (hepatocytes) EGFR per cell. Cancer cells express up to 2–3 × 106 EGFR/cell. Recombinant EGFR can be stably expressed in cells at different levels to directly analyze the dependency of the parameters of the EGFR biosynthesis, endocytosis, recycling, and degradation on the EGFR concentration in the cell.
Synthesis of EGFR
In Basic Protocol 1 it is important to use the shortest time course possible because 35S-labeled amino acids can be rapidly depleted from the medium. A significant decrease in available free [35S]amino acids will result in nonlinear incorporation, and thus an inaccurate determination of the rate of EGFR biosynthesis. Different cell lines have different rates of metabolism, and, therefore, different rates of methionine/cysteine incorporation. Control experiments using variable concentrations of labeled amino acids and incubation times should be performed to confirm linear incorporation of radiolabeled methionine/cysteine for the desired concentration and pulse time. For most cell lines, this will be in the 1- to 4-hr range. Alternatively, unlabeled methionine/cysteine may be simultaneously added to extend the half-life of labeled methionine/cysteine in the medium.
EGFR degradation
Two types of methods have been used to determine the rates of EGFR degradation. In the first method, the amount of EGFR is determined by immunoblotting (UNIT 6.2) of lysates from cells incubated without or with EGF and in the presence of an inhibitor of protein synthesis, such as cycloheximide. This assay relies on the assumption that cycloheximide completely inhibits protein synthesis and does not have other, unrelated effects.
The second method, which is described in the Alternate Protocol 1, involves radiolabeling of cellular proteins followed by chasing the cells in the absence of label and subsequent immunoprecipitation of EGFR at various time points. In this method, inhibition of synthesis is not necessary. The amount of time cells are incubated with labeled amino acids in these experiments should be sufficient to label a significant pool of EGFR. This is especially important in cells with high levels of EGFR expression, in which the turnover of the receptor is very slow.
Down-regulation assay
Typically, the effect that low amounts of EGF have on EGFR surface expression is difficult to detect in cells expressing moderate and high levels of EGFR. Under these conditions, there is not a significant number of EGFR being internalized in these cells. However, low EGF concentrations can be used in experiments with cells expressing low EGFR levels. When measuring the number of [125I]EGF binding sites at the cell surface, the time of incubation with [125I]EGF at 4°Cmustbe1 hr or longer. This time is necessary to reach a binding equilibrium for [125I]EGF at 4°C in various types of cells.
Internalization
It has been demonstrated that EGF-induced internalization of EGFR via the clathrin-dependent pathway is saturated if too many EGF-receptor complexes are present at the cell surface. This is typically observed with [125I]EGF concentrations higher than 1 to 2 ng/ml when [125I]EGF occupies more than 10,000 to 15,000 receptors per cell. Therefore, these low concentrations of [125I]EGF are recommended for experiments comparing clathrin-dependent endocytosis of EGFR mutants or the effects of inhibitors and modulators of this process.
One caveat of this type of internalization experiment is the possible underestimation of the internalization rate because of rapid recycling of internalized [125I]EGF. Thus, the internalization assay must be performed using a short time course so that the bulk of internalized [125I]EGF does not have time to return to the cell surface.
It is also important to note that Basic Protocol 3 uses a low-pH wash with acetate to strip [125I]EGF from the cell surface. This treatment removes ~95% of surface-bound [125I]EGF. In fact, a 1-min treatment with the pH 2.5 to 2.8 acetate buffer is sufficient to strip 90% of surface [125I]EGF. However, other low-pH buffers can also be used. For instance, glycine-Cl buffer has been used in other studies (Wiley et al., 1991). Also, addition of 2 M urea further improves the efficiency of stripping (Wiley et al., 1991).
Recycling and degradation
In the recycling/degradation assay, it is important to load the cells with sufficient concentrations of [125I]EGF for a sufficient amount of time to obtain a substantial concentration of [125I]EGF in early endosomes. These parameters should be chosen based on both EGFR cellular levels and internalization rates for each particular cell type. The initial incubation time should be 5 to 10 min in most cell types and not longer than 15 min in cells with the slowest endocytic trafficking. This helps ensure that most endosomal [125I]EGF is still in early endosomes and has not been sorted to late compartments or back to the plasma membrane. Similarly, it is critical to keep the loading time consistent between all samples in the experiment to obtain comparable pools of endosomal [125I]EGF. Thus, it is advisable to limit the number of variants in each experiment, depending upon the experience level of the investigator with this type of experiments.
This report offers two versions of the recycling/degradation assay (Basic Protocol 4 and Alternate Protocol 2). Basic Protocol 4 is designed to prevent any rebinding and re-internalization of [125I]EGF after one round of recycling because of the presence of an excess of unlabeled EGF during the chase incubation. However, the caveat of this protocol is that unlabeled EGF-receptor complexes compete with [125I]EGF-receptor complexes during sorting to recycling and degradation pathways. This is especially important for estimating the degradation rates of [125I]EGF, because the degradation pathway has been shown to be saturable (Herbst et al., 1994). On the other hand, Alternate Protocol 2 is designed to avoid interference of unlabeled EGF, although recycled [125I]EGF can dissociate and rebind as well as re-internalize before dissociation from the receptor at the cell surface. Typically, degradation products of [125I]EGF (TCA-soluble) can be detected after about 30 min of chase incubation in [125I]EGF-loaded cells. However, it is necessary to extend the time of chase incubation to 2 to 4 hr to enable detection of significant amounts of degraded [125I]EGF. Thus, in experiments aiming to measure recycling rates, short chase incubations are recommended (1 to 15 min). In experiments focusing on degradation rates, a time course of 30 to 240 min is recommended. Finally, it should be noted that as an alternative to trichloroacetic acid/phosphotungstic acid precipitation, a G-25 gel filtration procedure can be used to separate intact [125I]EGF and low-molecular-weight products of [125I]EGF degradation (Wiley et al., 1991).
Iodination
The protocol for EGF iodination using chloramine T was developed and optimized in the laboratories of Stanley Cohen and Graham Carpenter (Carpenter and Cohen, 1976). This protocol is designed to carry out iodination under mild conditions, avoid excessive exposure to chloramine T, and avoid formation of highly reactive [125I]EGF products that can covalently cross-link to the plasma membrane. It is critical to limit the time of iodination to 40 sec. Alternative protocols of EGF iodination can also be used. For instance, EGF can be iodinated using Iodobeads (Pierce). In our experience, the specific activity of [125I]EGF made using Iodobeads is lower as compared with preparations obtained using the chloramine T protocol.
Anticipated Results
Biosynthesis of EGFR
In the biosynthesis assay, the amount of labeled EGFR should correlate in a linear fashion with the time of label incorporation. The apparent rate of the label incorporation into newly synthesized EGFR varies depending on the cell type.
EGFR degradation
The half-life of EGFR ranges from 8 to 24 hr (low to high levels of receptor respectively) and even longer in other cells. The half-life of EGFR is dramatically shortened following EGF stimulation, to as low a value as t1/2 = 1 hr in cells expressing low levels of the receptor. Examples of EGFR biosynthesis and degradation analysis performed in our laboratory have recently been published (Duex and Sorkin, 2009).
EGFR down-regulation
Significant EGFR down-regulation in cells treated with high EGF concentrations (20 to 100 ng/ml) is observed beginning at 15 to 20 min after EGF stimulation. The amount of surface EGFR usually stabilizes after 30 to 60 min of EGF stimulation. Therefore, a complete dynamics of the down-regulation process can be observed within the range of incubation times ranging from 15 to 120 min.
EGF internalization
The values of ke measured in various types of cells using low [125I]EGF concentrations (clathrin-mediated pathway) are typically within the range of 0.15 to 0.40 min−1. This range in values is quite similar to the rate constants measured for other clathrin-coated pit-internalized cargo, such as the transferrin receptor. ke values measured using high EGF concentrations (resulting in a significant contribution from clathrin-independent pathways) are typically below 0.1 min−1 and can be as low as 0.03 min−1. Such values correspond to the rates of constitutive endocytosis of membrane proteins.
EGF recycling and degradation
In both Basic Protocol 4 and Alternate Protocol 2, recycling of [125I]EGF is evident immediately when the [125I]EGF-loaded cells are chased at 37°C. The protocols can produce similar kinetics of [125I]EGF recycling if a short time course is used for the chase (5 to 10 min). However, in Alternate Protocol 2, re-internalization of [125I]EGF results in a slight underestimation of the recycling rates, especially, at longer chase times. Typically, as much as 50% of endosomal [125I]EGF in [125I]EGF-loaded cells is recycled during a 2-hr chase if re-internalization is prevented (Alternate Protocol 2). In contrast, under conditions of this protocol, only about 20% to 25% of endosomal [125I]EGF is recycled. In both protocols, degradation products of [125I]EGF can be detected after about 30 min of chase incubation of [125I]EGF-loaded cells. At maximum, 10% to 20% of endosomal [125I]EGF is degraded after 2 to 4 hr of chase incubation in the presence of excess cold EGF (Basic Protocol 4), whereas up to 40% to 50% of internalized [125I]EGF is degraded in the absence of unlabeled EGF during the same time (Alternate Protocol 2).
Iodination
Typically, the specific activity of [125I]EGF is within the range of 120,000 to 200,000 cpm/ng.
Time Considerations
Synthesis of EGFR
The incubation of cells with radiolabeled methionine/cysteine is up to 6 hr. Since the cells are frozen at −70°C at the end of the incubation, the experiment can be interrupted at this step and continued later. Preparation of cell lysates takes ~1 hr. A minimal time of incubation of lysates with primary antibodies is 2 hr, although the incubation can be run overnight. Subsequent incubation with protein A–Sepharose, washes and loading the samples for SDS-PAGE take ~2 hr. Thus, the minimal length of the entire assay until starting electrophoresis is ~11 hr.
EGFR degradation
The cells are labeled with the [35S]amino acid mix for 4 to 16 hr. Subsequent washes and a 30 to 60 min preincubation take ~1 to 1.5 hr. The longest chase incubation of cells in unlabeled medium should extend to at least 4 hr for cells with low levels of EGFR and up to 30 hr for cells with high EGFR levels. If the goal of the experiment is to measure the degradation rates of EGF-activated EGFR, the length of the longest chase incubation can be reduced to 2 and 10 hr in cells with low and high EGFR levels, respectively. The time required for cell lysis and EGFR immunoprecipitation is the same as described above for the EGFR biosynthesis assay. Therefore, the length of the entire experiment until SDS-PAGE can be from 12 hr at minimum and up to 2 days.
EGFR down-regulation
A typical experiment includes a 2-hr incubation with unlabeled EGF, 15-min cell washing, 1-hr incubation with [125I]EGF, 5-min washing, and about 1-hr solubilization with NaOH. Therefore, the entire experiment with two to three 12-well plates, including radioactivity counting, can be completed in 5 hr.
EGF internalization
One internalization experiment (one 12-well plate) is performed in about 20 min followed by a 1-hr cell solubilization in NaOH. More internalization experiments can be carried out during solubilization of the first set of cells. Practically, internalization measurements in four 12-well plates can be accomplished within 3 hr, including radioactivity counting on a γ-counter.
EGF recycling and degradation
Loading of cells with [125I]EGF including the first acid wash in Basic Protocol 4 and Alternate Protocol 2 takes ~20 min. In Basic Protocol 4, pre-incubation of loaded cells with unlabeled EGF and the following chase can take together up to 5 hr. In Alternate Protocol 2, the manipulations with loaded cells are ~1 hr shorter. The acid wash treatment of the last time point takes only 10 min, but the trichloroacetic acid/phosphotungstic acid precipitation procedure and separation of the soluble from the insoluble radioactivity takes at least 2 hr. Therefore, a degradation/recycling experiment can last up to 7.5 hr, although it can be done in a significantly shorter period of time if the main goal is to measure the recycling rates.
Iodination
The total length of the iodination procedure is ~3 hr.
Acknowledgments
This work was supported by NCI grant CA089151(A.S.) and NCI/NRSA grant F32CA126344 (J.E.D.).
Literature Cited
- Carpenter G, Cohen S. 125I-Labeled human epidermal growth factor: Binding internalization, and degradation in human fibroblasts. J. Cell Biol. 1976;71:159–171. doi: 10.1083/jcb.71.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of EGF receptor synthesis. Mol. Biol. Cell. 2009;20:1833–1844. doi: 10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler HT, Maxfield FR, Willingham MC, Pastan I. Dansylcadaverine inhibits internalization of 125I-epidermal growth factor in BALB 3T3 cells. J. Biol. Chem. 1980;255:1239–1241. [PubMed] [Google Scholar]
- Herbst JJ, Opresko LK, Walsh BJ, Lauffenberger DA, Wiley HS. Regulation of postendocytic trafficking of the epidermal growth factor receptor through endosomal retention. J. Biol. Chem. 1994;269:12865–12873. [PubMed] [Google Scholar]
- Resat H, Ewald JA, Dixon DA, Wiley HS. An integrated model of epidermal growth factor receptor trafficking and signal transduction. Biophys. J. 2003;85:730–743. doi: 10.1016/s0006-3495(03)74516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Teslenko L, Nikolsky N. The endocytosis of epidermal growth factor in A431 cells: A pH of microenvironment and the dynamics of receptor complexes dissociation. Exp. Cell Res. 1988;175:192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben-Basat Y, Benjamin S, Corso S, Gan J, Yosef RB, Giordano S, Yarden Y. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26:6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- Todderud G, Carpenter G. Epidermal growth factor: The receptor and its function. BioFactors. 1989;2:11–15. [PubMed] [Google Scholar]
- Wiley HS, Cunningham DD. The endocytotic rate constant: A cellular parameter for quantitating receptor-mediated endocytosis. J. Biol. Chem. 1982;257:4222–4229. [PubMed] [Google Scholar]
- Wiley HS, Herbst JJ, Walsh BJ, Lauffenberger DA, Rosenfeld MG, Gill GN. The role of tyrosine kinase activity in endocytosis, compartmentalization and down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 1991;266:11083–11094. [PubMed] [Google Scholar]