During the progression of metastasis, cancer cells detach from the solid primary tumor, enter the blood stream, and travel to different tissues of the body. These breakaway cancer cells in the peripheral blood are known as circulating tumor cells (CTCs).[1] In addition to conventional diagnostic imaging and serum marker detection, quantification of CTCs in patient blood provides new and valuable information about managing cancer.[2–5] Over the past decade, CTC counting has been used for examining cancer metastasis, predicting patient prognosis, and monitoring the therapeutic outcomes of cancer.[6] However, isolation of CTCs has been technically challenging due to the extremely low abundance (a few to hundreds per milliliter) of CTCs among a large number of hematologic cells in the blood (109 mL−1).[4, 7, 8] Several technology platforms for isolating/counting CTCs have been developed with strategies that involve immunomagnetic beads or microfluidic devices.[3, 4,9, 10] The former utilizes capture-agent-coated magnetic beads to immunologically recognize CTCs in the blood, followed by magnetic isolation. However, these bead-based approaches are limited by their low CTC-capture yield and purity. Recently, a number of microfluidic technologies[9, 10] has been established for capturing viable CTCs from whole-blood samples with improved efficiency and selectivity compared to the bead-based approach.[3, 7] While different device architectures were applied in these CTC-sorting microchips, the improved CTC-capture efficiencies were achieved by increasing CTC/substrate contact frequency and duration.
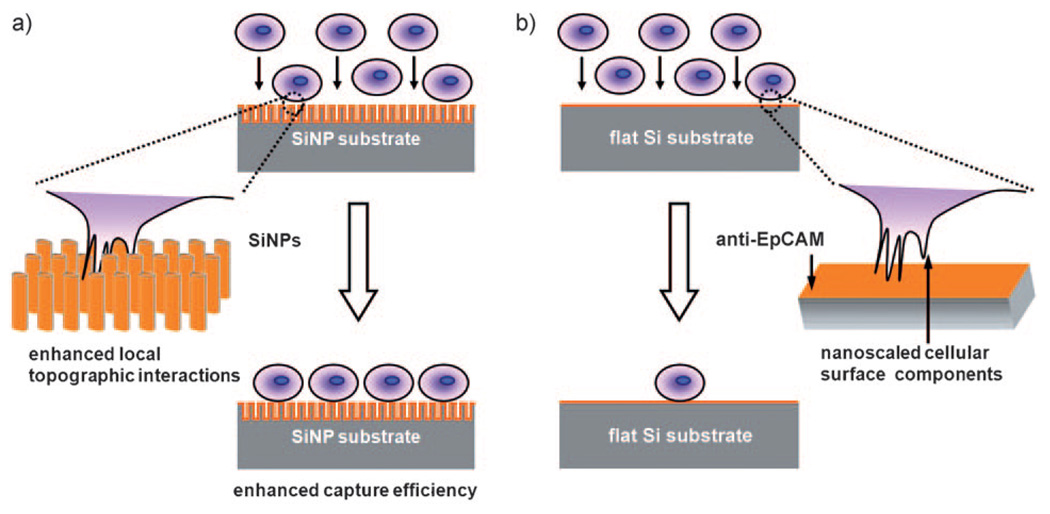
Herein we demonstrate that a three-dimensionally (3D) nanostructured substrate coated with epithelial-cell adhesion-molecule antibody (anti-EpCAM) exhibits outstanding cell-capture efficiency when employed to isolate viable cancer cells from whole-blood samples. We foresaw that this new cell capture platform could provide a convenient and cost-efficient alternative for isolating/counting CTCs. EpCAM is a transmembrane glycoprotein that is frequently overexpressed in a variety of solid-tumor cells and is absent from hematologic cells.[11] The uniqueness of this new approach (Figure 1a) lies in the use of 3D nanostructured substrates—specifically, a silicon-nanopillar (SiNP) array—which allow for enhanced local topographic interactions[12–14] between the SiNP substrates and nanoscale components of the cellular surface (e.g., microvilli and filopodia) and result in vastly improved cell-capture affinity compared to unstructured (i.e., flat Si) substrates (Figure 1 b). The rationale of our approach is indirectly supported by a recent study in which the enhanced adhesive force between a SiNP-coated bead and mucosal epithelial cells was attributed to local topographic interactions between SiNPs bound to the bead and nanoscale microvilli on the cell surfaces.[12] Enormous research efforts have been devoted to studying local topographic interactions between cells and a diversity of nanostructured substrates,[13–22] which share nanoscale feature dimensions similar to those of cellular surface components and extracellular matrix (ECM) structures. However, most of this research has focused on achieving a better understanding of how nanostructures affect cellular behavior,[16–21, 23–25] for example, adhesion,[17, 26–28] viability,[16, 23] migration,[25, 29, 30] differentiation,[21, 22, 31] and morphology.[27, 31, 32]
Figure 1.
Conceptual illustration of how an anti-EpCAM-coated 3D nanostructured (i.e., SiNP) substrate can be employed to achieve significantly enhanced capture of EpCAM-positive cells (i.e., CTCs) from cell suspension in contrast to an anti-EpCAM-coated unstructured (i.e., flat Si) substrate. a) Interdigitation of nanoscale cellular surface components and SiNPs enhances local topographic interactions, resulting in vastly improved cell-capture efficiency. b) Lack of local topographic interactions between cells and flat Si substrate compromises the respective cell-capture efficiency.
The 3D nanostructured cell-capture substrates were prepared as illustrated in Scheme 1. First, we fabricated densely packed nanopillars with diameters of 100–200 nm on silicon wafers using a wet chemical etching method (Scheme 1a).[33] The lengths of these chemically etched SiNPs can be controlled by applying different etching times. Thus, we were able to obtain a series of SiNP substrates with SiNP lengths varying from 1 to 20 µm. After preparing the SiNP substrates, we employed N-hydroxysuccinimide (NHS)/maleimide chemistry[9] to introduce streptavidin onto the surfaces of the SiNP substrates (Scheme 1b and Supporting Information). Biotinylated anti-EpCAM (R&D Systems) was introduced onto the streptavidin-coated substrates prior to the cell-capture experiments.
Scheme 1.
A) Chemical etching by Ag+ and HF was employed to produce a silicon nanopillar (SiNP) array on a silicon wafer. The SEM images reveal that well-defined SiNPs with diameters ranging from 100 to 200 nm and lengths around 10 µm were produced. B) Grafting of biotinylated epithelial-cell adhesion-molecule antibody (anti-EpCAM) onto silicon substrates.
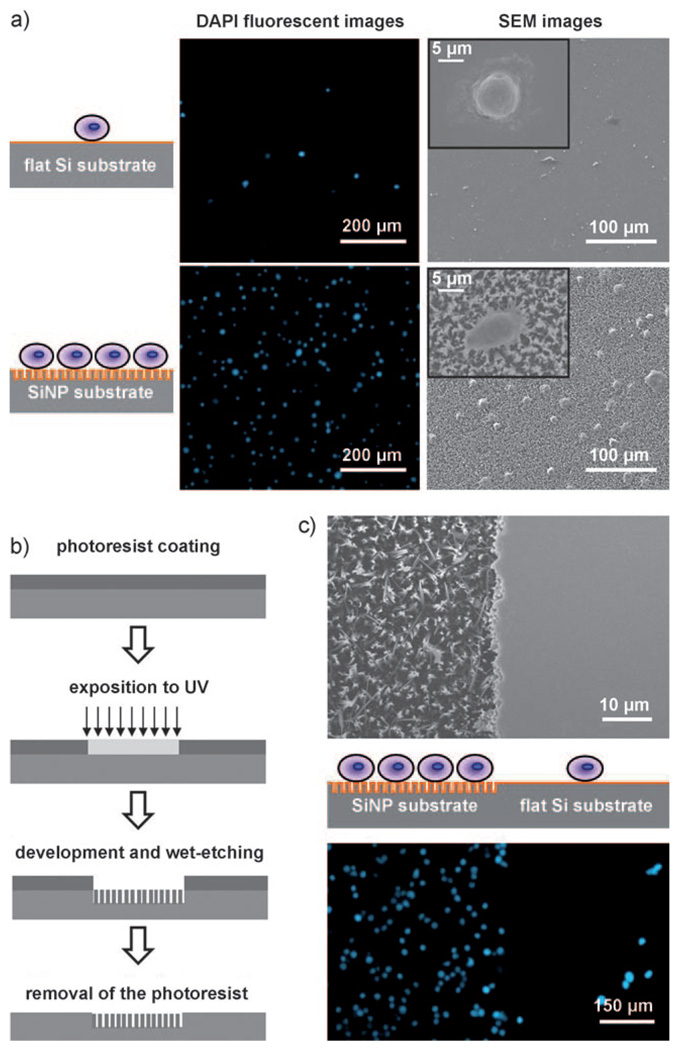
To test the cell-capture performance of the SiNP substrates, we prepared a cell suspension (105 cellsmL−1) of an EpCAM-positive breast-cancer cell line (i.e., MCF7)[9, 10, 34] in cell culture medium (DMEM). The MCF7 cell suspension (1 mL) was introduced onto a 10 µm-long SiNP substrate (1 × 2 cm), which was placed into a commercial cell chamber slide and kept in an incubator (5% CO2, 37°C) for 1 h. As a control, a flat Si substrate modified with anti-EpCAM was also examined in parallel. After rinsing, fixing, and nuclear staining, the substrate-immobilized cells were imaged and counted by using a fluorescence microscope (Nikon, TE2000). As shown in Figure 2, the SiNP substrates can capture up to ten times more cells than the flat Si substrates. The cell-capture yield was only 4–14%on flat Si substrates as opposed to 45–65% on SiNP substrates, and this suggests that 3D nanostructures are possibly responsible for the enhanced cell-capture yields. As shown on the right-hand side of Figure 2a, the cells captured on the flat Si substrates and on the SiNP substrates exhibited marked morphology differences, in addition to the vastly different capture yields. In order to maintain the morphologies of the substrate-immobilized cells for scanning electron microscopy (SEM) observation, the samples were processed by glutaraldehyde fixation, treatment with osmium tetroxide, and dehydration.[35] The insets in Figure 2a show typical morphologies of cells captured on flat Si and SiNP substrates. The cells on the flat substrate appear more rounded, and no nanoscale cellular protrusions are observed. In contrast, observation of many interdigitated cellular protrusions (with diameters of about 100–200 nm) on the SiNP substrates validates[12] the working mechanism of our 3D nanostructure-based cell-capture approach.
Figure 2.
a) Fluorescence micrographs and SEM images of SiNP substrates and flat Si substrates on which MCF7 cells were captured. The SiNP substrates exhibited significantly better cell-capture efficiency than the flat ones. DAPI: 4’,6-Diamidino-2-phenylindol. b) Photolithography process for patterning alternate SiNP and flat substrates on the silicon wafer for comparing their cell-capture efficiencies in a close experimental setup. c) SEM images of patterned and flat substrates before cell capture (top) and fluorescence micrographs of cells captured on patterned and flat substrates (bottom).
To compare SiNP and flat Si substrates in a spatially close experimental setting, a lithographic method (Figure 2b) and chemical etching process were combined to make a patterned substrate that alternates between flat Si and SiNP surfaces (upper part of Figure 2c). As shown at the bottom of Figure 2c, the flat Si surface captured few target cells, while the SiNP-covered area captured more cells. This is consistent with the results obtained individually under the separate experimental conditions. Moreover, we were able to confirm minimal nonspecific interactions between the MCF7 cells and the SiNP substrates by performing similar cell-capture experiments on three different substrates: 1) a SiNP substrate without any surface modification, 2) a SiNP substrate with only streptavidin coating, and 3) a SiNP substrate modified with anti-EpCAM (see Supporting Information).
During optimization, we noticed that the number of substrate-immobilized cells increases with increasing incubation time. To determine the minimum time required to achieve cell capture, we examined the cell-capture performances of both the modified 10 µm SiNPs and flat Si substrates (with anti-EpCAM coating) at different incubation times. Two EpCAM-positive cancer-cell lines (i.e., MCF7 and PC3 human prostate adenocarcinoma cells[9, 34]) were examined. For comparison, two EpCAM-negative cancer-cell lines (i.e., HeLa cervical cancer cell line and Daudi lymphoma cell line,[36] representing both adherent and suspension cell types, respectively) were employed as controls. Prior to our cell-capture studies, we confirmed EpCAM expression levels on the four cell lines using flow cytometry (see Figure S4 in Supporting Information). Figure 3a summarizes the correlation between incubation time and the number of substrate-immobilized cells on SiNP substrates. For the EpCAM-positive cells (i.e., MCF7 and PC3), the maximal cell-capture numbers were achieved at an incubation time of 45 min. In general, the 10 mm SiNPs exhibit up to ten times better cell-capture yield than the flat Si substrates (Figure S5 in Supporting Information). On the contrary, relatively low cell numbers were observed for EpCAM-negative cells (i.e., HeLa[37] and Daudi), that is, the 3D nanostructured substrates are specific for capturing EpCAM-positive cells.
Figure 3.
Quantitative evaluations of cell-capture yields a) at different capture times and b) with different SiNP lengths ranging from 0 to 20 mm. Each plot and error bar represents a mean standard deviation from three repeats.
To unveil how SiNP length affects the cell-capture yields of the 3D SiNP substrates, we performed cell-capture experiments with the four cancer cell lines (i.e., MCF7, PC3, HeLa, and Daudi) on a series of SiNP substrates with SiNP lengths of 4, 6, 8, 10, and 20 µm (see cross-sectional SEM images in Supporting Information). As shown in Figure 3b, the numbers of captured EpCAM-positive cells increased with increasing SiNP lengths, but relatively minor changes were observed for EpCAM-negative cells. When the SiNPs were longer than 6 mm, maximum cell-capture yields were achieved. This minimum SiNP length required for optimal capture yield is believed to be compatible with cell-protrusion lengths of tumor cells.[38] This observation led us to believe that the 3D local topographic interactions are correlated with characteristics of the SiNPs, including but not limited to length, and that SiNPs lead to improved cell-capture affinity.[12]
From the aforementioned studies, optimal cell-capture conditions (i.e., 10 µm anti-EpCAM-coated SiNP substrates and 45 min cell incubation time) were obtained for performing static cell-capture studies on whole blood samples. A series of artificial CTC blood samples was prepared by spiking rabbit blood with Red-Dye-stained MCF7 cells at densities of 1000–1250, 80–100, and 5–20 cellsmL−1 of blood. The results are summarized in Table S1 of the Supporting Information. Our approach shows vastly improved capture yields (>40%), compared to those observed for the commercialized technology.[3]
In conclusion, a cell-capture platform based on 3D nanostructured substrates was demonstrated to exhibit efficient cell capture. This likely originated from enhanced local topographic interactions between the 3D nanostructured substrates and extracellular extensions, in addition to anti-EpCAM/EpCAM biological recognition. Using these nanostructured substrates we can reliably capture cancer cells from artificial CTC blood samples. It is conceivable that our platform can provide a convenient and cost-efficient alternative for CTC sorting in clinics. The impact of this cell-capture platform beyond CTCs will potentially benefit the diagnosis of early diseases that are currently detected by means of cell-capture technologies.[3, 4, 9, 10, 39] Importantly, cell viability with this platform is as high as 84–91% (see Supporting Information), which is conducive to subsequently releasing the cells, culturing them, and performing molecular biological diagnosis.
Supplementary Material
Footnotes
This research was supported by NIH-NCI NanoSystems Biology Cancer Center (U54A119347).
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200901668.
References
- 1.Steeg PS. Nat. Med. 2006;12:895. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 2.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, Matera J, Repollet M, Doyle GV, Terstappen LWMM, Hayes DF. Clin. Cancer Res. 2006;12:6403. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 3.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AGJ, Uhr JW, Terstappen LWMM. Clin. Cancer Res. 2004;10:6897. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 4.Zieglschmid V, Hollmann C, Böcher O. Crit. Rev. Clin. Lab. Sci. 2005;42:155. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. N. Engl. J. Med. 2004;351:781. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LWMM, Uhr JW. Proc. Natl. Acad. Sci. USA. 1998;95:4589. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. Proc. Natl. Acad. Sci. USA. 2004;101:10501. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams A, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Hum. Pathol. 2004;35:122. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Fischer KE, Aleman BJ, Tao SL, Daniels RH, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Lett. 2009;9:716. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis ASG, Varde M. J. Natl. Cancer Inst. 1964;33:15. [PubMed] [Google Scholar]
- 14.Liu WF, Chen CS. Adv. Drug Delivery Rev. 2007;59:1319. doi: 10.1016/j.addr.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard PP, Cavalcanti-Adam EA, Kemkemer R, Spatz JP. Soft Matter. 2007;3:307. doi: 10.1039/b614008d. [DOI] [PubMed] [Google Scholar]
- 16.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 17.Sun TL, Han D, Rhemann K, Chi LF, Fuchs H. J. Am. Chem. Soc. 2007;129:1496. doi: 10.1021/ja0686155. [DOI] [PubMed] [Google Scholar]
- 18.Yang MT, Sniadecki NJ, Chen CS. Adv. Mater. 2007;19:3119. [Google Scholar]
- 19.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Science. 2002;295:1702. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 20.Kim W, Ng JK, Kunitake ME, Conklin BR, Yang PD. J. Am. Chem. Soc. 2007;129:7228. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- 21.Dang JM, Leong KW. Adv. Mater. 2007;19:2775. doi: 10.1002/adma.200602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan E, Kotov NA. Nano Lett. 2007;7:1123. doi: 10.1021/nl0620132. [DOI] [PubMed] [Google Scholar]
- 23.Park J, Bauer S, von der Mark K, Schmuki P. Nano Lett. 2007;7:1686. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 24.Sun TL, Tan H, Han D, Fu Q, Jiang L. Small. 2005;1:959. doi: 10.1002/smll.200500095. [DOI] [PubMed] [Google Scholar]
- 25.Yim EKF, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Biomaterials. 2005;26:5405. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang WC, Lee LP, Liepmann D. Lab Chip. 2005;5:64. doi: 10.1039/b400455h. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Park SY, Namgung S, Kim B, Im J, Kim Y, Sun K, Lee KB, Nam JM, Park Y, Hong S. Adv. Mater. 2007;19:2530. [Google Scholar]
- 28.Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. J. Cell Sci. 2004;117:3153. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang XY, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Proc. Natl. Acad. Sci. USA. 2005;102:975. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yousaf MN, Houseman BT, Mrksich M. Angew. Chem. 2001;113:1127. [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2001;40:1093. [Google Scholar]
- 31.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat. Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 32.Kandere-Grzybowska K, Campbell C, Komarova Y, Grzybowski BA, Borisy GG. Nat. Methods. 2005;2:739. doi: 10.1038/nmeth796. [DOI] [PubMed] [Google Scholar]
- 33.Peng KQ, Yan YJ, Gao SP, Zhu J. Adv. Mater. 2002;14:1164. [Google Scholar]
- 34.The EpCAM expression levels of MCF7 and PC3 cell lines are >500000 and ca. 50000 antigens per cell, respectively: Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Int. J. Oncol. 2005;27:49.
- 35.de La Fuente JM, Andar A, Gadegaard N, Berry CC, Kingshott P, Riehle MO. Langmuir. 2006;22:5528. doi: 10.1021/la053045s. [DOI] [PubMed] [Google Scholar]
- 36.a) http://www.abcam.com/EpCAM-antibody-E144-ab32392.html.; b) Gillies SD, Lan Y, Williams S, Carr F, Forman S, Raubitschek A, Lo K-M. Blood. 2005;105:3972. doi: 10.1182/blood-2004-09-3533. [DOI] [PubMed] [Google Scholar]
- 37.We observed that HeLa cells seem to exhibit higher nonspecific adhesion than Daudi cells on both SiNP and flat substrates, likely due to their adherent nature.
- 38.Domagala W, Koss LG. Virchows Arch. 1977;26:27. doi: 10.1007/BF02889533. [DOI] [PubMed] [Google Scholar]; Iyer S, Gaikwad RM, Subba-Rao V, Woodworth CD, Sokolov I. Nat. Nanotechnol. 2009;4:389. doi: 10.1038/nnano.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J. Am. Chem. Soc. 2007;129:1959. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.