Abstract
LovF is a highly reducing polyketide synthase (HR-PKS) from the filamentous fungus Aspergillus terreus. LovF synthesizes the α-S-methylbutyrate side chain that is subsequently transferred to monacolin J to yield the cholesterol-lowering natural product lovastatin. In the report, we expressed the full length LovF and reconstituted the megasynthase activities in vitro. We confirmed the diketide product of LovF is offloaded from the LovF ACP domain by the dissociated acyltransferase LovD. This represents the first example of acyltransferase-mediated release of polyketide products from fungal PKSs. We determined LovD primarily interacts with the ACP domain of LovF and the protein-protein interactions leads to highly efficient transfer of the diketide product. The catalytic efficiency is enhanced nearly one million-fold when LovF was used as the acyl carrier instead of N-acetylcysteamine. Reconstitution and characterization of the LovF offloading mechanism provide new insights into the functions of fungal HR-PKS.
The highly-reducing polyketide synthases (HR-PKSs)1 from filamentous fungi are a large family of megasynthases that produce important natural products such as lovastatin 1,2 squalestatin,3 and fumonisin.4 Unlike other families of PKSs, the biosynthetic programming rules of fungal HR-PKSs remain largely unexplored. One particularly unique feature of HR-PKSs is the lack of a built-in offloading domain that facilitates the release of completed products. This is in sharp contrast to bacterial type I or fungal nonreducing PKSs, in which a dedicated thioesterase (TE) domain is appended at the end of the megasynthase and catalyzes the release of polyketides via macrocyclization,5 Claisen cyclization6 or hydrolysis.7 Understanding the mechanisms of product release is therefore an important goal in demystifying the functions of HR-PKSs.
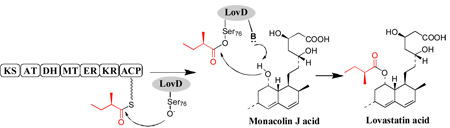
The product release mechanisms of LovB and LovF, the two HR-PKS from the lovastatin biosynthetic pathway,2 have not been investigated to date. The diketide synthase LovF consists of seven linearly arranged domains and has been proposed to biosynthesize the α-S-methylbutyrate side chain of 1 (Figure 1A).2 Transfer of the diketide side chain from LovF to monacolin J 2 is hypothesized to be catalyzed by a dissociated acyltransferase LovD.2 The proposed acyltransferase-mediated product release mechanism is unprecedented among known PKSs. In this report, we demonstrate LovF-LovD protein-protein interactions play an important role in LovF product release and lovastatin biosynthesis.
Figure 1.
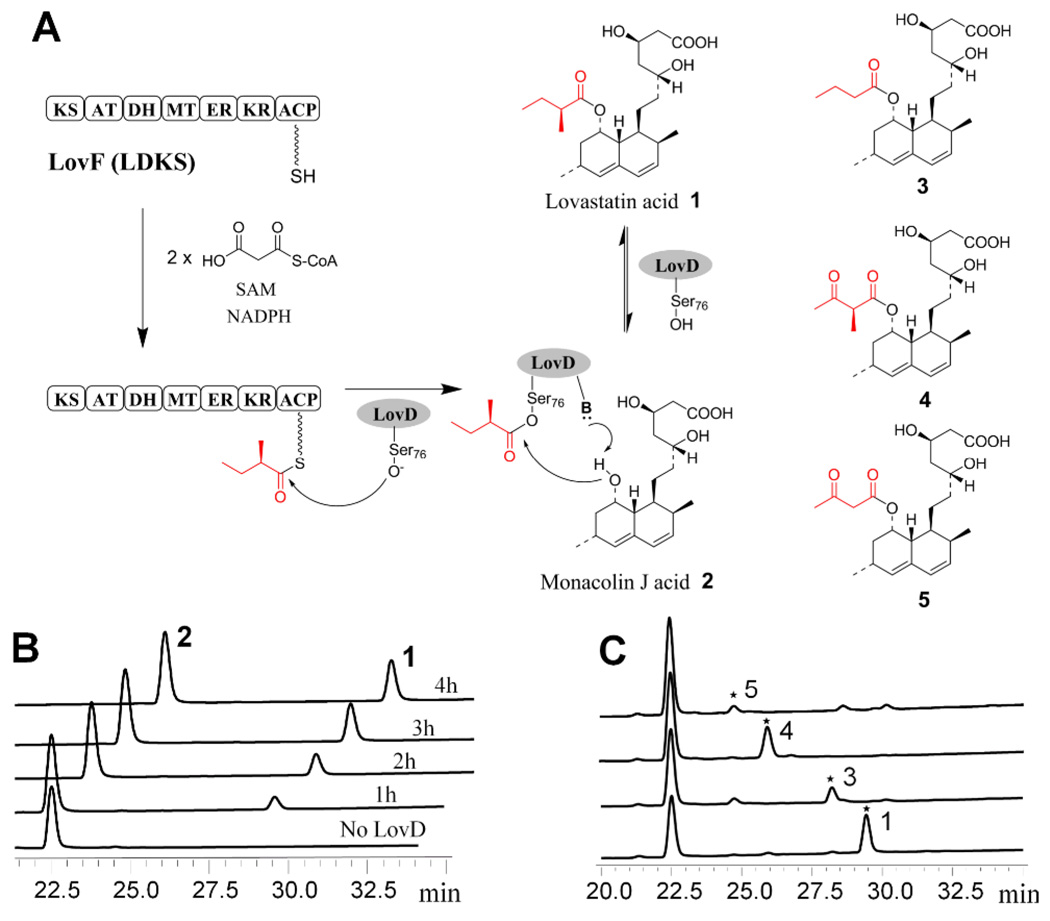
Reconstitution of LovF and LovD activity in vitro. (A) LovF is a megasynthase consists of ketosynthase (KS), malonyl-CoA:ACP acyltransferase (MAT), dehydratase (DH), methyltransferase (MT), enoylreductase (ER), ketoreductase (KR), and acyl-carrier protein (ACP). LovF is proposed to synthesize the α-S-methylbutyryl diketide, which is hypothesized to be offloaded by LovD. Other lovastatin analogues 3, 4, and 5 are shown. (B) The formation of lovastatin catalyzed by both LovF and LovD was confirmed by LC/MS. (C) Lovastatin analogues were formed when cofactors were excluded from the assay.
To obtain soluble LovF megasynthase for in vitro studies, the lovF gene (7.6 kB) from Aspergillus terreus was inserted into a yeast 2µ expression vector under the control of the ADH2 promoter (Figure S1A).8 The resulting vector was transformed into Saccharomyces cerevisiae BJ5464-NpgA, which contains a chromosomal copy of the phosphopantetheinyl (Ppant) transferase NpgA.9 Recombinant LovF (277 kDa) was purified to homogeneity by affinity and anion exchange chromatography steps (Figure S1B). The ACP domain of LovF was determined to be phosphopantetheinylated after tryptic digestion, offline HPLC purification, and Ppant ejection analysis by FTMS (Figure S2).10 To test the activity of LovF, we incubated LovF with malonyl-CoA, SAM and NADPH. When malonyl-CoA was supplied at substoicheometric concentrations of LovF, a new species with a molecular weight of 84.0576 Da attached to the Ppant arm of the ACP was detected using FTMS (Figure S3). The Ppant ejection ion that resulted from source fragmentation of the fraction containing the ACP active site peptide was determined to have a mass of 345.1843 Da. This mass is consistent with that of Ppant covalently bound to the expected diketide product α-methylbutyrate. Furthermore, an additional round of fragmentation confirmed that this ion was indeed the α-methylbutyrate loaded phosphopantetheinyl ejected ion10b (Figure S3B). As expected, no released diketide products were detected in the reaction mixture, confirming the inability of LovF alone to offload the product.
In order to verify the role of LovD, we incubated LovF with LovD and the acyl acceptor 2. LovD was supplied at low concentrations to minimize the reverse hydrolysis reaction.11 Extraction of the reaction mixture followed by LC-MS analysis revealed the formation of a single product (Figure 1B) and the accompanying decrease of 2. The product eluted with the same retention time and has the same mass fragmentation pattern as the standard 1 (Figure S4A). In contrast, reactions either without LovD or with a S76A active site mutant of LovD did not afford any 1, demonstrating the essential catalytic role of LovD in facilitating the acyltransfer. Importantly, synthesis of 1 confirmed that the entire range of LovF functions can be reconstituted in vitro, including activities of all six catalytic domains, as well as the proposed interactions with LovD.
To quantify the rate of synthesis of 1 using α-S-methylbutyryl-LovF (MB-LovF) as an acyl donor, different concentrations of LovF were first treated with malonyl-CoA and all the co-factors for one hour to preload the ACP domain with α-S-methylbutyrate. LovD and 2 were then added and the initial velocities of the reactions were measured by HPLC (Figure S5). The reaction displayed Michaelis-Menten kinetics and the kinetic parameters were determined as shown in Table 1. The catalytic efficiency (kcat/Km) of LovD towards MB-LovF was 1.29 min−1µM−1. To determine the effects of protein-protein interaction on the acyltransfer reaction, we also measured the reaction kinetics of LovD towards α-S-methylbutyryl-CoA (MB-CoA, Figure S6) and α-S-methylbutyryl-S-N-acetylcysteamine (MB-SNAC). Compare to that of MB-LovF (Table 1), LovD displayed a ~20,000-fold and ~800,000-fold attenuation in kinetic efficiency towards MB-CoA and MB-SNAC, respectively. Both Km and kcat were significantly improved when the methylbutyrate side chain is attached to LovF. These results demonstrate that protein-protein interactions between LovF and LovD play a key role in facilitating rapid offloading of the diketide substrate from LovF to LovD and ensure efficient biosynthesis of 1. This is analogous to the inter-module transfer of polyketide intermediates in bacterial type I PKSs, in which protein-protein interactions significantly lowers the Km of the acyltransfer reaction.12 The lower Km is critical for the ping-pong bi-bi reaction, as LovD can be inhibited by 2 when the acyl donor substrate binds with a high Km, such as in the case of MB-CoA and MB-SNAC.11
Table 1.
Kinetic parameters of LovD towards MB-SNAC, MB-CoA, MB- ACP, and MB-LovF
| kcat (min−1) | Km (mM) | kcat/Km (min−1 µM−1) | |
|---|---|---|---|
| MB-LovF | 48.4 ± 3.6 | 0.039 ± 0.008 | 1.29 |
| MB-ACP | ND1 | ND | 0.32 |
| MB-CoA | 0.17 ± 0.01 | 2.5 ± 0.31 | 6.9 × 10−5 |
| MB-SNAC | 0.024 ± 0.001 | 9.2 ± 0.9 | 1.6 × 10−6 |
ND: Not determined separately
To verify that LovD interacts with LovF through the ACP domain, we cloned and expressed the ACP domain of LovF from Escherichia coli expression strain BL21(DE3)/pXK64 (Figure S1C). The apo-form of the standalone ACP was confirmed by ESI to be 100% apo (Figure S7). The broad spectrum Ppant transferase Sfp was used to prepare α-S-methylbutyryl-LovF-ACP (MB-ACP) using apo-LovF ACP and MB-CoA.13 The conversion from apo to holo was monitored by mass spectrometry (Figure S8). MB-ACP was prone to spontaneous hydrolysis of the thioester after preparation. As a result, the individual kcat and Km values were not determined and only the catalytic efficiency was measure at low MB-ACP concentrations. As shown in Table 1, LovD displayed comparable efficiency towards MB-ACP when compared to that of MB-LovF, indicating the protein-protein interaction between the LovF ACP domain and LovD is sufficient to afford high catalytic efficiency. There remains the possibility that LovD makes additional contacts with the LovF megasynthase, which may account for the four-fold difference in kcat/Km between MB-LovF and MB-ACP. We further determined LovD is highly specific towards LovF ACP. When the methylbutyryl modified-versions of heterologous ACPs were presented to LovD and 2, including OxyC (type II PKS), DEBS ACP3 (bacterial type I) and ACPp (E. coli FAS), we were unable to detect any trace of 1 in the reaction mixture.
We then tested the product profiles of LovF-LovD system when different cofactors were excluded from the reaction (Figure 1C). The identities of new products were verified by comparing to authentic standards synthesized from acyl-CoA and LovD (Figure S4B-D). When SAM was removed as a means to disable the MT domain, no 1 was detected; however, a new product with retention time and mass fragmentation consistent with C8-butyryl-MJ acid 3 was synthesized. Considering methylation is the first tailoring step after diketide synthesis by LovF, formation of 3 suggests that LovF KR, DH and ER are not specific towards α-methylated intermediates. When NADPH was excluded from the reaction, we observed synthesis of the known shunt metabolite monacolin X 4, which was previously isolated from Monascus ruber.14 Lastly, when both SAM and NADPH were excluded, C8-acetoacetyl-MJ acid 5 was synthesized, albeit at much lower conversions. As both 4 and 5 are synthesized from LovD-mediate transfer of LovF-diketide intermediates, it is interesting that none of these acyl intermediates were transferred by LovD when all cofactors are present and only 1 was synthesized (Figure 1B). This indicates that in the presence of all cofactors, only the completely tailored, α-S-methylbutyryl-ACP is accessible by LovD. Two possible mechanisms may account for this phenomenon. First, the methyltransfer, ketoreduction, dehydration and enoyl reduction steps may take place very rapidly following exit of the acetoacetyl-ACP from the KS active site. The rate of these modification steps may be further enhanced by the in cis interactions between acyl-ACP and these built-in domains. Second, based on the X-ray crystal structure of mammalian FAS15, which shares the same domain architecture of HR-PKSs, the acyl-ACP may be inaccessible by LovD during the tailoring steps. When no additional tailoring reactions are possible, the flexible ACP domain may swing outward, exposing the acyl group for product release mediated by LovD.
In summary, we showed protein-protein interactions between LovF and LovD facilitate highly efficient release and transfer of the diketide product to an accepting acyl substrate. Recently, a PLP-dependent chain releasing mechanism using an oxoamine synthase was reported during the biosynthesis of fumonisins.4 Together these examples illustrate the different strategies nature employs to release highly reduced polyketides from megasynthase assembly lines.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (1R21HL091197 and 1R01GM85128) to YT. We thank Prof. Da Silva for the BJ5464-NpgA strain.
Footnotes
Supporting Information Available: Experimental details are available free of charge via the internet at http://pubs.acs.org.
References
- 1.Cox RJ. Org Biomol Chem. 2007;5:2010. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Science. 1999;284:1368. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 3.Cox RJ, Glod F, Hurley D, Lazarus CM, Nicholson TP, Rudd BA, Simpson TJ, Wilkinson B, Zhang Y. Chem Commun (Camb) 2004:2260. doi: 10.1039/b411973h. [DOI] [PubMed] [Google Scholar]
- 4.Gerber R, Lou L, Du L. J Am Chem Soc. 2009 doi: 10.1021/ja8091054. [DOI] [PubMed] [Google Scholar]
- 5.a) Akey DL, Kittendorf JD, Giraldes JW, Fecik RA, Sherman DH, Smith JL. Nat Chem Biol. 2006;2:537. doi: 10.1038/nchembio824. [DOI] [PubMed] [Google Scholar]; b) Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM. Proc Natl Acad Sci U S A. 2001;98:14808. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Boddy CN, Schneider TL, Hotta K, Walsh CT, Khosla C. J Am Chem Soc. 2003;125:3428. doi: 10.1021/ja0298646. [DOI] [PubMed] [Google Scholar]
- 6.a) Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Chem Biol. 2001;8:189. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]; b) Ma SM, Zhan J, Xie X, Watanabe K, Tang Y, Zhang W. J Am Chem Soc. 2008;130:38. doi: 10.1021/ja078091o. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Science. 2008;320:243. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Smith L, Hong H, Spencer JB, Leadlay PF. Chembiochem. 2008;9:2967. doi: 10.1002/cbic.200800585. [DOI] [PubMed] [Google Scholar]; b) Moldenhauer J, Chen XH, Borriss R, Piel J. Angew Chem Int Ed Engl. 2007;46:8195. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]; c) Strieter ER, Koglin A, Aron ZD, Walsh CT. J Am Chem Soc. 2009;131:2113. doi: 10.1021/ja8077945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KM, DaSilva NA. Yeast. 2005;22:431. doi: 10.1002/yea.1221. [DOI] [PubMed] [Google Scholar]
- 9.Lee KM, DaSilva NA, Kealey J. 2009 Submitted. [Google Scholar]
- 10.a) Dorrestein PC, Kelleher NL. Nat. Prod. Rep. 2006;23:893. doi: 10.1039/b511400b. [DOI] [PubMed] [Google Scholar]; b) Meluzzi D, Zheng WH, Hensler M, Nizet V, Dorrestein PC. Bioorg Med Chem Lett. 2008;18:3107. doi: 10.1016/j.bmcl.2007.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie X, Watanabe K, Wojcicki WA, Wang CC, Tang Y. Chem Biol. 2006;13:1161. doi: 10.1016/j.chembiol.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Wu N, Tsuji SY, Cane DE, Khosla C. J Am Chem Soc. 2001;123:6465. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 13.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 14.Endo A, Hasumi K, Nakamura T, Kunishima M, Masuda M. J Antibiot (Tokyo) 1985;38:321. doi: 10.7164/antibiotics.38.321. [DOI] [PubMed] [Google Scholar]
- 15.Maier T, Leibundgut M, Ban N. Science. 2008;321:1315. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.