Abstract
Purpose
Gain-of-function mutations in BRAF, NRAS, or KIT are associated with distinct melanoma subtypes with KIT mutations and/or copy number changes frequently observed among melanomas arising from sun-protected sites, such as acral skin (palms, soles, and nail bed) and mucous membranes. GAB2 has recently been implicated in melanoma pathogenesis, and increased copy numbers are found in a subset of melanomas. We sought to determine the association of increased copy numbers of GAB2 among melanoma subtypes in the context of genetic alterations in BRAF, NRAS, and KIT.
Experimental Design
A total of 85 melanomas arising from sun-protected (n = 23) and sun-exposed sites (n = 62) were analyzed for copy number changes using array-based comparative genomic hybridization and for gain-of-function mutations in BRAF, NRAS, and KIT.
Results
GAB2 amplifications were found in 9% of the cases and were associated with melanomas arising from acral and mucosal sites (P = 0.005). Increased copy numbers of the KIT locus were observed in 6% of the cases. The overall mutation frequencies for BRAF and NRAS were 43.5% and 14%, respectively, and were mutually exclusive. Among the acral and mucosal melanomas studied, the genetic alteration frequency was 26% for GAB2, 13% for KIT, 30% for BRAF, and 4% for NRAS. Importantly, the majority of GAB2 amplifications occurred independent from genetic events in BRAF, NRAS, and KIT.
Conclusions
GAB2 amplification is critical for melanomas arising from sun-protected sites. Genetic alterations in GAB2 will help refine the molecular classification of melanomas.
Several melanoma subtypes are recognized based on anatomic site, sun exposure characteristics, and histopathologic features. In recent years, the identification of distinct genetic aberrations among melanoma subtypes has resulted in improved classification (1), with the ultimate goal of developing treatment strategies based on molecular characteristics. However, the identification of additional genetic events is necessary to refine the current melanoma classification and develop novel therapeutic agents.
The mitogen-activated protein kinase pathway is a key regulator of melanoma cell proliferation with extracellular signal-regulated kinase activation seen in majority of melanomas (2). BRAF and NRAS mutations, leading to constitutive activation of this pathway, occur in ~50% and ~15% of melanomas, respectively (3), and have been associated with melanomas arising from nonchronically sun-exposed sites, most of which are located on trunk and extremities (1, 4). Mutations in these genes are mutually exclusive in melanoma. In contrast, BRAF and NRAS mutations are rare in melanomas arising from sun-protected areas, such as acral, mucosal, and uveal melanomas, suggesting genetic events or mechanisms other than oncogenic mutations in BRAF and NRAS leading to extracellular signal-regulated kinase activation in these melanoma subtypes (4, 5). Amplification of the KIT locus on 4q12 and activating mutations in the KIT gene have recently been identified in a subset of melanomas (7% amplification and 3% mutation frequency) of which the majority consisted of melanomas on acral and mucosal sites (6).
GAB2 is a scaffolding protein that mediates interactions with various signaling pathways, such as RAS-ERK and PI3K-AKT signaling (7, 8). We recently identified GAB2 as a critical molecule for melanoma tumor progression and metastasis by activating AKT signaling (9). Importantly, we found increased copy numbers and gene amplification of GAB2 in a subset of melanomas. In this study, we examined the clinical correlates of increased DNA copy numbers of GAB2 and its relation to genetic aberrations in BRAF, NRAS, and KIT.
Materials and Methods
Study population
We studied 85 frozen melanoma tumor samples, consisting of 23 cases arising from sun-protected sites (acral and mucous membranes) and 62 arising from head, neck, trunk, and extremities. Sixty-four cases were part of a previously published data set (9). Clinical data, such as gender, age at diagnosis, primary tumor depth, and primary tumor site, were available for the majority of the cases. All patients, except two, had died of metastatic disease. The tumors were obtained from the archives of Fachklinik Hornheide and Columbia University. The study was approved by the institutional review board of Columbia University.
Translational Relevance.
GAB2 amplifications and overexpression are recently described in a subset of melanomas as a novel genetic event. GAB2 overexpression accelerates tumor growth potential and confers a metastatic phenotype. In an attempt to identify a group of tumors that may potentially benefit from therapeutic targeting of GAB2, we examined whether GAB2 amplifications are unique to a distinct subtype of melanoma. Our findings clearly indicate that GAB2 amplifications are observed among the acral and mucosal melanomas and are independent from genetic alterations in BRAF, NRAS, and KIT, suggesting its critical role in a distinct subset of melanomas and its use in molecular classification of melanomas.
Experimental methods
Array-based comparative genomic hybridization was done using arrays with ~19,000 RPCI-11 BAC clones providing an average resolution of 150 kb (10). Hybridization was carried out on 1 μg of genomic DNA, labeled by random priming, and analyzed as described previously (11). A log2 ratio exceeding its flanking regions by 0.5 and/or an absolute log2 ratio exceeding 0.9 was used to define amplification. Hierarchical clustering was done using Cluster 3.0 software, whereas TreeView software was used for the generation of the graph.
Mutation analysis was done for the entire coding region of GAB2 and for the hotspot regions of BRAF (exon 15), NRAS (exons 2 and 3), and KIT (exons 11, 13, 17, and 18) using PCR amplification. Purified PCR products were sequenced using a Big Dye Terminator cycle sequencing kit and an ABI Prism 310 automated sequencer system (Applied Biosystems).
Fluorescent in situ hybridization was done as described previously (12). Briefly, RP11-653J20 and RP11-444N24 BAC clones were obtained from Invitrogen. DNA was prepared from BAC clones using standard methods and labeled by nick translation using spectrum red or spectrum green dUTP fluorochromes. Spectrum red or spectrum green – labeled centromeric probes were used to enumerate chromosome numbers (Vysis). Hybridization signals were scored on at least 200 interphase nuclei from tissue sections on 4′,6-diamidino-2-phenylindole – counterstained slides.
Immunohistochemistry was done on paraffin-embedded tissue arrays and tissue sections using standard protocols with an antibody against GAB2 (26B6; Cell Signaling). Staining intensity levels were scored from 0 to 3, in which a staining intensity of 2 or greater was considered positive. A sample with known positive staining was used as an external positive control. Lack of protein expression in the epidermis served as a negative control.
Statistical methods
Fisher's exact test and Pearson's χ2 test were used to assess the statistical significance of two categorical variables, such as the association of genetic alterations with primary tumor site. P values of <0.05 were considered significant.
Results
Patient characteristics and the frequencies of genetic alterations in BRAF, NRAS, KIT, and GAB2 are summarized in Table 1. Based on previous literature (13), primary tumor site was categorized into two groups: sun-protected sites including palms, soles, and mucous membranes and sun-exposed sites including head, neck, trunk, and extremities. There were 73% of tumors arising from sun-exposed and 27% from sun-protected sites. All of the primary tumors were intermediate thick (1–4 mm) or thick (>4.0 mm).
Table 1.
Patient characteristics and frequencies of genetic alterations
| Variable | Case (n = 85) |
|---|---|
| Median age at diagnosis (y) | 54 |
| Gender (n) | |
| Male | 40 |
| Female | 28 |
| DU | 17 |
| Primary tumor site (n) | |
| Head and neck | 5 |
| Trunk and extremities | 57 |
| Acral and mucosal sites | 23 |
| Primary tumor thickness (n) | |
| <1.0 mm | 0 |
| 1–4 mm | 35 |
| >4.0 mm | 35 |
| DU | 15 |
| Genetic alteration frequency, n (%) | |
| BRAF mutation | 37 (43.5) |
| NRAS mutation | 12 (14) |
| KIT amplification | 5 (6) |
| GAB2 amplification | 8 (9) |
Abbreviation: DU, data unavailable.
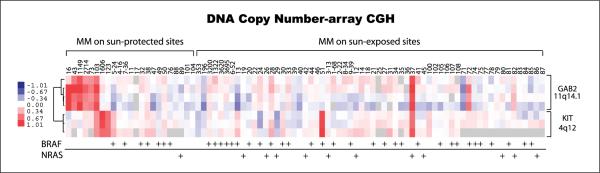
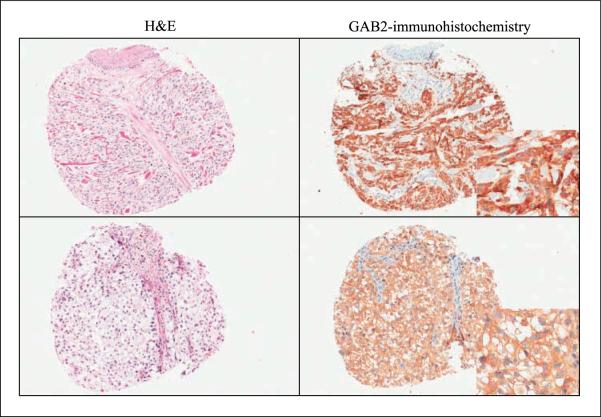
In an attempt to examine GAB2 amplifications in the context of other genetic events, a total of 85 melanoma tumor samples were evaluated for activating mutations in BRAF, NRAS, and KIT and for copy number changes using array-based comparative genomic hybridization. Mutations in BRAF and NRAS were mutually exclusive, and the mutation frequencies were 43.5% (37 of 85) for BRAF and 14% (12 of 85) for NRAS. Increased copy numbers of the KIT locus on 4q12 were found in 5 of the 85 (6%) cases. GAB2 located on 11q14.1 was amplified in 8 of the 85 (9%) cases (Table 1; Fig. 1). Of the eight cases with GAB2 amplifications, two cases coexisted with KIT amplifications and two cases occurred together with a BRAF or an NRAS mutation. GAB2 amplifications occurred independent from KIT, BRAF, and NRAS in five of the eight cases, all of which were melanomas arising from sun-protected sites (Table 2; Fig. 1). Mutation analysis of the hotspot regions of the KIT gene and the entire coding region of the GAB2 gene failed to detect any sequence variations. Increased copy numbers of the GAB2 locus were confirmed by fluorescent in situ hybridization analysis. All amplified cases, except one case that could not be examined due to limited tissue, showed increased GAB2 protein expression by immunohistochemistry (Table 2; Fig. 2).
Fig.1.
GAB2 amplifications in melanoma by array-based comparative genomic hybridization. Supervised hierarchical clustering of copy number data shows clustering among melanomas arising from sun-protected sites (acral and mucosal melanoma subtypes).
Table 2.
GAB2-amplified cases by array-based comparative genomic hybridization
| Case no. | Anatomic site | FISH |
IHC |
Mutations |
||
|---|---|---|---|---|---|---|
| GAB2 | GAB2 | BRAF | NRAS | KIT | ||
| 16 | Acral | Amplified | Positive | WT | WT | WT |
| 43 | Acral | Amplified | Positive | WT | WT | WT |
| 1149 | Genitalia | Amplified | Positive | WT | WT | WT |
| 2714 | Acral | Amplified | Positive | WT | WT | WT |
| 73 | Acral | Amplified | Positive | WT | WT | WT |
| 103 | Acral | ND | ND | WT | WT | WT |
| 37 | Face | Amplified | Positive | WT | Q61K | WT |
| 72 | Leg | Amplified | Positive | V600E | WT | WT |
Abbreviations: FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; WT, wild type; ND, not done due to limited tissue.
Fig.2.
Representative examples of cases with GAB2 amplifications that are wild-type for BRAF, NRAS, and KIT. These cases show high levels of GAB2 protein expression by immunohistochemistry. Note the absence of staining of the overlying epidermis as a negative control (top).
We then examined genetic alterations in GAB2, BRAF, NRAS, and KIT in relation to clinical variables such as primary tumor site. GAB2 amplifications were associated with melanomas arising from sun-protected sites (acral and mucous membranes; P = 0.005; Fig. 3). Although there were a limited number of cases with KIT amplification, they were more frequent, but not statistically significant, in melanomas arising from sun-protected sites compared with sun-exposed sites (P = 0.135). Supervised hierarchical clustering of the melanomas by anatomic site showed clustering of GAB2 and KIT amplifications among the acral and mucosal melanomas (P =0.001;Figs. 1 and 3).
Fig.3.
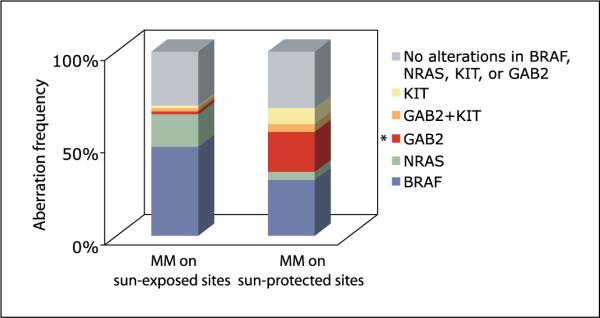
Frequency distributionof genetic alterations in GAB2,KIT, BRAF, and NRAS among two groups of melanoma. Sun-protected sitesinclude cases occurring onacral sites (palms and soles) and mucous membranes. Sun-exposed sites include melanomas arising from the head, neck, trunk, and extremities. BRAF and NRAS mutations were mutually exclusive. Two cases, one occurring on a sun-protected site and the other on a sun-exposed site, showed increased copy numbers in both GAB2 and KIT. **, P = 0.005, statistically significant association of GAB2 amplification with melanomas arising from sun-protected sites (acral and mucosal melanomas).
Among the 23 acral and mucosal melanomas studied, copy number changes in GAB2 were found in 26% (6 of 23) and KIT in 13% (3 of 23) of the cases (Fig. 3). Mutations in BRAF were identified in 30% (7 of 23) of the cases and NRAS, in 4%(1 of 23) of the cases. None of these genetic alterations were found in the remaining 31%. Among the 62 melanomas arising from head and neck, trunk, and extremities, mutations in BRAF or NRAS accounted for 72% of the cases (30 of 62 in BRAF and 11 of 62 in NRAS). GAB2 and KIT alterations were rare in this group, each accounting for 3% (Fig. 3).
Discussion
Melanomas arising from sun-protected sites represent a unique subset; BRAF and NRAS mutations are found at a lower frequency, whereas genetic aberrations in KIT, CDK4, and CYCLIN D1 are observed at a higher frequency compared with melanomas occurring on other anatomic sites (1, 13). In this study, we first highlight a novel subset of melanomas characterized by GAB2 amplifications and show its association with melanomas arising from sun-protected sites. The finding that GAB2 amplifications occurred in acral and mucosal melanoma cases in the absence of mutations in BRAF and NRAS suggests that GAB2 may be critical in oncogenic transformation of a subset of BRAF/NRAS wild-type melanomas. Second, our study implicates a role for GAB2 in refining molecular classification of melanomas and its potential use in therapeutic decision-making in the pharmacogenomics era. Finally, we validate previous observations that genetic aberrations in acral and mucosal melanomas differ from other sites suggesting different mechanistic routes among melanoma subtypes.
Mutations and/or copy number increases in KIT are common among acral (36%) and mucosal (39%) melanomas (6). Imatinib, which inhibits the tyrosine kinase activity of KIT, is currently being tested in clinical trials for melanomas that harbor gain-of-function mutations in KIT. Similarly, amplifications of CDK4 occur in a small subset of melanomas and are found frequently among acral and mucosal subtypes (1, 14). Melanomas harboring CDK4 amplifications in the absence of mutations in BRAF or NRAS, but coexisting with KIT alterations, have been described (15). In our series of 85 melanomas, we found only one case (1%, 1 of 85) with a CDK4 locus amplification in which the primary tumor was located on the trunk. Our study is consistent with previous reports that CDK4 locus amplifications are observed in a small subset of melanomas (14). Amplification and overexpression of CYCLIN D1, located on 11q13.2, is well characterized in human tumors including melanoma (16). Increased copy numbers of CYCLIN D1 are found in 11% of primary melanomas and are found at a higher frequency among acral melanomas (44.4%) compared with other melanoma subtypes (16). We have previously reported that GAB2 amplification is observed independent of CYCLIN D1 locus in melanoma (9). Additional studies with larger cohorts of acral and mucosal melanomas will be critical to validate genomic alterations in this subtype.
Although efforts in correlating genetic alterations with clinical correlates such as melanoma subtypes characterized by sun exposure characteristics provide insights into melanoma pathogenesis, it is likely that, due to genetic heterogeneity, such a correlation may not be perfect and, therefore, the ultimate classification scheme will be based on molecular rather than clinical characteristics.
Acknowledgments
Grant support: NIH grants AR050273 and CA133925 (J.T. Celebi).
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 2.Cohen C, Zavala-Pompa A, Sequeira JH, et al. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clin Cancer Res. 2002;8:3728–33. [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–90. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 5.Takata M, Goto Y, Ichii N, et al. Constitutive activation of the mitogen-activated protein kinase signaling pathway in acral melanomas. J Invest Dermatol. 2005;125:318–22. doi: 10.1111/j.0022-202X.2005.23812.x. [DOI] [PubMed] [Google Scholar]
- 6.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 7.Gu H, Neel BG. The “Gab” in signal transduction. Trends Cell Biol. 2003;13:122–30. doi: 10.1016/s0962-8924(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 8.Bentires-Alj M, Gil SG, Chan R, et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–21. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- 9.Horst B, Gruvberger-Saal S, Hopkins B, et al. Gab2-mediated signaling promotes melanoma metastasis. Am J Pathol. 2009;174:1524–33. doi: 10.2353/ajpath.2009.080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osoegawa K, Mammoser AG, Wu C, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–96. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak NJ, Miecznikowski J, Moore SR, et al. Challenges in array comparative genomic hybridization for the analysis of cancer samples. Genet Med. 2007;9:585–95. doi: 10.1097/gim.0b013e3181461c4a. [DOI] [PubMed] [Google Scholar]
- 12.Scotto L, Narayan G, Nandula SV, et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer. 2008;7:58. doi: 10.1186/1476-4598-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25:1606–20. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 14.Muthusamy V, Hobbs C, Nogueira C, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2006;45:447–54. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- 15.Smalley KS, Contractor R, Nguyen TK, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008;68:5743–52. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauter ER, Yeo UC, von Stemm A, et al. Cyclin D1is a candidate oncogene in cutaneous melanoma. Cancer Res. 2002;62:3200–6. [PubMed] [Google Scholar]