Abstract
Industrial melanism in the peppered moth (Biston betularia) is an iconic case study of ecological genetics but the molecular identity of the gene determining the difference between the typical and melanic (carbonaria) morphs is entirely unknown. We applied the candidate gene approach to look for associations between genetic polymorphisms within sixteen a priori melanisation gene candidates and the carbonaria morph. The genes were isolated and sequence characterised in B. betularia using degenerate PCR and from whole-transcriptome sequence. The list of candidates contains all the genes previously implicated in melanisation pattern differences in other insects, including aaNAT, DOPA-decarboxylase, ebony, tan, tyrosine hydroxylase, yellow and yellow2 (yellow-fa). Co-segregation of candidate gene alleles and carbonaria morph was tested in 73 offspring of a carbonaria male-typical female backcross. Surprisingly, none of the sixteen candidate genes was in close linkage with the locus controlling the carbonaria-typical polymorphism. Our study demonstrates that the ‘carbonaria gene’ is not a structural variant of a canonical melanisation pathway gene, neither is it a cis-regulatory element of these enzyme-coding genes. The implication is either that we have failed to characterize an unknown enzyme-coding gene in the melanisation pathway, or more likely, that the ‘carbonaria gene’ is a higher level trans-acting factor which regulates the spatial expression of one or more of the melanisation candidates in this study to alter the pattern of melanin production.
Introduction
Industrial melanism in the peppered moth Biston betularia remains the textbook example of a rapid evolutionary response to a dramatically altered environment. Essentially unknown before the first official recording from 1848 Manchester (England), by the turn of the 19th century the black (carbonaria) morph had largely if not entirely replaced the light-speckled (typical) form in the most polluted parts of UK [1]. Following 1960s legislation to control smoke pollution, reverse selection has reduced carbonaria to a rarity [2]. Yet, despite its celebrity status within evolutionary biology, the molecular genetic and developmental control of the carbonaria-typical polymorphism is unknown. All that is known on this topic is that the trait is controlled by a dominant allele at a single Mendelian locus [3].
One approach to identifying genetic switches is to look for associations between the trait of interest and molecular genetic variation within known genes in the biochemical pathway. This candidate gene approach has been applied successfully to discover the genetic basis of melanism in mammals and other vertebrates, implicating several different mutations within the melanocortin-1-receptor gene (Mc1r) [4], [5]. In arthropods, the process of melanisation is variously used for purposes of crypsis, sexual signalling [6], and thermoregulation [7] but also plays a major role in immune response [8], wound healing and cuticular hardening [9]. Melanisation as a visual signal in adults is mainly a genetically determined developmental trait, whereas melanisation as defence is an induced (phenotypic) response.
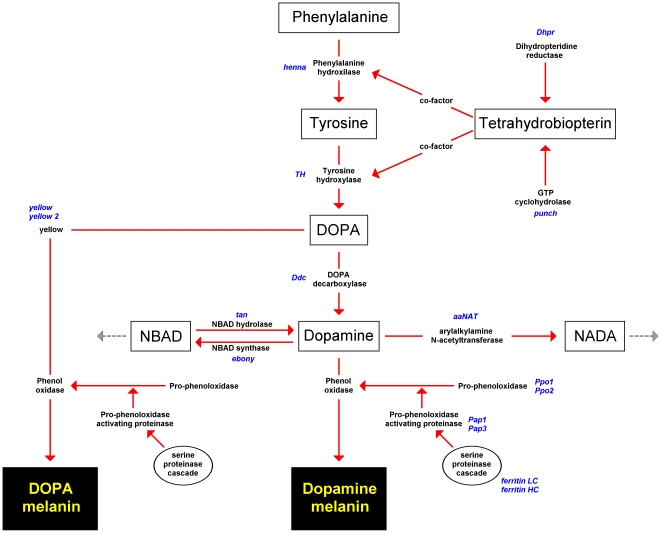
The biosynthesis of melanin as a pigment has been well characterized, predominantly in Drosophila [10], [11] and Lepidoptera [12], [13], [14], [15], [16], [17]. The universal mechanism which forms the core of the melanisation pathway involves a conserved pattern of substrate substitutions catalysed by a number of distinct genes. It begins with the two-step transformation of phenylalanine to tyrosine, then DOPA (Figure 1). These two transformations are mediated by phenylalanine hydroxylase (PAH or henna) and tyrosine hydroxylase (TH or pale) respectively. Tetrahydrobiopterin (BH4) is an essential cofactor (electron donor) for henna and TH, and is in turn dependent on guanosine triphosphate-cyclohydrolase I (GTPCHI or punch) and dihydropteridine reductase (Dhpr). DOPA is subsequently transformed into either DOPA-melanin or dopamine-melanin. The formation of DOPA-melanin is catalyzed by one or more members of the yellow gene family and by phenoloxidase (PO). Transition from DOPA to dopamine-melanin involves the formation of dopamine by DOPA decarboxilase (Ddc) and subsequently dopamine-melanin by PO. Dopamine is also utilized by NBAD synthase (ebony or BAS) and arylalkylamine N-acetyl transferase (aaNAT) to produce pigments other than melanin. In the swallowtail butterfly, Papilio glaucus, this alternative use of substrate lowers the dopamine titer to levels that prevent wing melanisation [18]. NBAD hydrolase (tan) catalyses the reaction in the opposite direction to ebony, increasing Dopamine concentration [19].
Figure 1. Insect melanisation biochemical pathway.
The initial substrate, intermediate metabolites and two types of melanin are shown in rectangles. The enzymes that catalyze the different reactions are in black and the genes that code for these enzymes are italicized in blue. The genes in this figure correspond with the candidate genes that have been isolated and examined. The serine proteinase cascade is represented as a single cluster of events. A more comprehensive representation of this cascade is produced in Cerenius and Söderhäll [8]. The interrupted arrows pointing away from NBAD and NADA indicate that these are not the final metabolites. (Figure adapted from De Gregorio et al., True, and Futahashi & Fujiwara [16], [17], [20]).
Immune-defence melanisation in arthropods is triggered by an intrusion of microbes, fungi, or other foreign objects, or by physical damage (wounds) [8], [20], [21]. Melanin encapsulates and immobilizes these harmful intrusions, or forms a barrier that replaces damaged cuticle, and at the same time, quinone intermediates that are highly toxic to pathogens are released as a by-product of the enzymatic processes involved [8]. The immune response proceeds through a series of biochemical transformations of proteolytic enzymes in a process named the serine proteinase cascade (reviewed in [8]). This cascade has branches with specific genes corresponding with the different types of intrusions and its activity can be locally reduced or blocked by serine proteinase inhibitors (serpins) [20], [22]. The cascade ultimately feeds into the core of the melanisation pathway (Figure 1) by transforming pro-phenol oxidase (Ppo) to PO by pro-phenoloxidase activating proteinases (Pap or Ppae) [23], [24], [25]. This complexity of gene interactions makes it difficult to explore all representatives of the serine proteinase cascade as melanisation candidate genes. Moreover, it is often not possible to identify orthologs of genes in different species by means of protein sequence comparison alone, because serine proteinases and serpins belong to very large gene families in which highly similar members (i.e. paralogs) often have completely unrelated functions [26], [27], [28], [29]. Ferritin, which plays a role in the serine proteinase cascade in Manduca sexta [30], is an exception because it is coded by two distinct subunits that are not easily confused with paralogs.
The premise of the candidate gene approach as a means of identifying causal mechanisms, rather than downstream consequences, is that the functional polymorphism resides either within the candidate gene itself, or is closely linked to it (e.g. cis-regulatory element). The current study examines 16 genes as potential candidates controlling the carbonaria-typical polymorphism in B. betularia. The genes that are involved with pigment-related melanisation, i.e., those in the core of the pathway, are expected to be more relevant with regard to industrial melanism than those that are triggered by an immune response. A number of these pigment candidates stand out because they have been shown to play a major role in melanisation patterns in insects including Lepidoptera. Melanisation in Papilio glaucus is caused by an unidentified sex-linked gene, and coincides with Ddc expression patterns but can also be down-regulated by ebony [15], [18]. Ddc and PO have been identified as key enzymes for cuticular melanisation in Manduca sexta [31], and in Papilio xuthus, Ddc, punch and TH are up-regulated in melanising patches of the larval cuticle [12], [16]. In Drosophila melanogaster, yellow has been implicated in melanin pattern formation on larval mouth parts and adult wings, mediated either through insertion of a transposable element closely downstream of yellow [32] or by a wing-specific cis-regulatory element [10]. The difference between the morphs in B. betularia could also be considered as variation in pattern rather than melanisation per se because the typical form has black patches, which define an underlying pattern, whereas carbonaria is completely black, except for two small white spots where the forewing attaches to the thorax. Thus the potential of yellow to regulate the distribution of melanin across wings and cuticle makes it a very relevant candidate gene. Tan is another promising melanisation candidate because it can counteract the reduction of Dopamine, a melanin substrate, by ebony [19].
To determine whether a candidate gene is likely to be directly involved in B. betularia melanisation, we tested whether polymorphisms in the candidate genes co-segregate with the melanic phenotype in a carbonaria-typical backcross. Polymorphisms within a candidate gene must be in full linkage disequilibrium within this cross if the gene is directly responsible, that is, the marker allele distribution should be identical to the phenotype distribution in the offspring. If a cis-regulatory element of a melanisation pathway gene causes the phenotypic switch, we would expect the allele distribution of the candidate gene to be closely similar to the phenotype distribution.
Results and Discussion
Sixteen melanisation candidate genes, as indicated in Figure 1, were isolated either by targeted PCR (11 genes) or by a transcriptome approach (5 genes). They cover all of the core melanisation pathway genes with the possible exception of some members of the yellow family, and additionally include some of the immune response genes. All genes were identified based on a blastx search [33] against the GenBank database (www.ncbi.nlm.nih.gov). The best hits in GenBank are presented in Table 1. The B. betularia sequences are available in GenBank under accession numbers GU980199-GU980212, GU953216-GU953231, and GS923573. In principle, the ‘best hit’ was considered the ortholog but, additionally, the difference between the best hit and the second-best hit within a species and the proportion of positive amino acid (aa) matches were taken into account to confirm sequence identity. The existence of B. betularia specific paralogs of the melanisation genes cannot be entirely excluded, but is highly unlikely because recently derived paralogs are usually revealed through sequence polymorphisms that do not follow Mendelian segregation, i.e. variation that does not represent allelic variation within a gene. We did not encounter such patterns in any of the genes presented here, though it is the case that Pap3 and the yellows belong to families known to have high duplication rates.
Table 1. NCBI blastx ‘best hits’.
| Gene | B. betularia Accession # | Blast hit accession number | Blast hit species | Blast hit gene name | e-value | positives; gaps |
| aaNAT | GU953216 | NP_001073122.1 | Bombyx mori | arylalkylamine N-acetyltransferase | 7e-112 | 236/261 (90%); 0 |
| Ddc | GU953217 | BAB68545.1 | Mamestra brassicae | dopa decarboxylase | 0.0 | 440/457 (96%); 0 |
| Dhpr | GU953218 | XP_001652256.1 | Aedes aegypti | dihydropteridine reductase | 4e-46 | 121/146 (82%); 0 |
| ebony | GU953219 | BAE43845.2 | Papilio xuthus | ebony | 1e-80 | 343/394 (87%); 1 |
| ferritin LC | GU953220 | AAF44717.1 | Manduca sexta | ferritin | 3e-41 | 168/182 (92%); 0 |
| ferritin HC | GU953221 | AAG41120.1 | Galleria mellonella | 26kDa ferritin subunit | 4e-09 | 65/67 (97%); 0 |
| punch | GU953222 | BAH11149.1 | Bombyx mori | GTP cyclohydrolase I isoform A | 6e-95 | 184/194 (94%); 2 |
| henna | GU953223 | BAE66652.1 | Papilio xuthus | phenylalanine hydroxylase | 8e-92 | 167/172 (97%); 0 |
| Pap1 | GU953224 | AAX18636.1 | Manduca sexta | prophenoloxidase-activating proteinase-1 | 2e-151 | 305/379 (80%); 1 |
| Pap3 | GU953225 | AAX18637.1 | Manduca sexta | prophenoloxidase-activating proteinase-3 | 8e-32 | 85/115 (73%); 2 |
| Ppo1 | GU953226 | NP_001037335.1 | Bombyx mori | phenoloxidase subunit 1 precursor | 0.0 | 424/469 (90%); 0 |
| Ppo2 | GU953227 | ABM65701.1 | Heliothis virescens | prophenoloxidase-2 | 0.0 | 617/690 (89%); 1 |
| tan | GU953228 | XP_001599569.1 | Nasonia vitripennis | N/A (conserved hypothetical protein) | 7e-79 | 233/361 (64%); 11 |
| TH | GU953229 | BAE43824.1 | Papilio xuthus | tyrosine hydroxylase | 0.0 | 538/561 (95%); 1 |
| yellow | GU953231 | NP_001037434.1 | Bombyx mori | yellow-y | 0.0 | 442/532 (83%); 26 |
| yellow2 | GU953230 | NP_001037424.1 | Bombyx mori | yellow2/yellow-fa | 1e-145 | 289/349 (83%); 5 |
Blastx results for the 16 candidate genes in the GenBank database. The hits with the highest e-values are listed. The positives and gaps are listed to provide a context for the e-values because a low e-value does not necessarily reflect low similarity. The ‘Low complexity regions filter’ was disabled for the henna search. The tan ortholog of Nasonia vitripennis is not named, instead the Apis mellifera hit with slightly lower score confirms the identity of this gene (acc. # XP_623115.1; Apis mellifera tan; Expect = 2e-74, Positives = 235/353 (66%), Gaps = 11/353 (3%)). Yellow2 blastx reveals two identical B. mori sequences that have been named differently (NP_001037424.1 = yellow2, ABC96695.1 = yellow-fa).
The proportion of positive matches for tan relative to the ‘best hit’ were lower than for the rest of the genes (64%), but it is evident that the B. betularia sequence is the proper ortholog because tan has no paralogs in any of the examined insect genomes (species specific blast: Bombyx mori, Tribolium castaneum, Apis melifera, D. melanogaster, Anopheles gambiae, Nasonia vitripennis). The relatively low proportion of positive aa matches in tan is due to the moderately conserved nature of this gene in insects and not a result of mis-identification. The aa sequence of tan in different insect species is aligned in fasta-alignment Text S1. When these blast searches were performed the contiguous sequence of tan in B. mori was not deposited in any of the sequence databases and therefore had to be assembled from different sequences (fasta-alignment Text S2). More recently, however, the full sequence has become pubclicly available on GenBank.
The Pap3 search revealed an unambiguous best hit in Manduca sexta, with Pap2 and a number of hemolymph proteinases as alternatives within the same species, but with considerably less similarity. As Pap3 belongs to the extensive serine proteinase family there is greater potential of hitting paralogs if the true ortholog is not in the database. This seems unlikely however, as the serine proteinases of M. sexta have been thoroughly surveyed [23], [34], [35], [36], [37], suggesting that they should be well covered in GenBank. The yellow genes also belong to a gene family, but this family is smaller and better characterized (at least in B. mori) than the serine proteinases [38]. The ortholog status of the two B. betularia genes based on aa sequence is unambiguous, but members of this gene family have not always been named consistently. The B. betularia yellow gene is known as yellow and yellow-y in B. mori; the B. betularia yellow2 gene is named both yellow2 and yellow-fa in B. mori.
Ppo1 and Ppo2 are much alike and have reciprocal saturated e-values (0.0), but the proportion of positive matches and presence of aa regions that are diagnostic allow them to be identified without question. The relatively low e-value for ferritinHC reflects the short length of the available B. betularia sequence (97 aa), but the similarity within this short stretch is very high and unambiguous. The remaining genes did not raise any issues because they have high e-values, a high percentage of positive matches and no obvious paralogs. This is also the case for Pap1 because, unlike Pap3, it is a very distinct gene [22].
Polymorphisms were found for all the 16 genes in the father of the carbonaria/typical backcross. Most of these polymorphisms were inside introns, some in coding regions and one in a closely linked BAC-end sequence (Table S1). The genes tan, TH and henna displayed a hemizygous genotype distribution, with heterozygotes restricted to the males and one of either alleles in the individual females. This is consistent with the WZ/ZZ sex determination system that is generally found in Lepidoptera and it reveals that these three genes are on the Z-chromosome in B. betularia, which corresponds with their location in B. mori.
The segregation patterns of the polymorphisms in all of the 16 candidate genes were significantly different from the carbonaria/typical phenotype distribution, within a progeny sample of 38 carbonaria and 35 typicals (Table 2). This demonstrates, somewhat surprisingly, that none of the genes examined, or linked regulatory units are primarily responsible for the melanisation switch in B. betularia. As our study includes essentially all the structural gene candidates (with the possible exception of undetected members of the yellow family), the result suggests that the genetic switch controlling the carbonaria polymorphism is more likely to be a higher level trans-regulatory gene [39]. Pigmentation studies in drosophilids are revealing a diversity of mechanisms regulating variation in epidermal melanin deposition, both within and between species. Thus, variation in a transcription factor binding site in the cis-regulatory element of yellow, together with downregulation of ebony, mediate species-specific wing pattern melanisation [10]; and sequence variation either within or closely linked to tan and ebony underlie intraspecific polymorphism in body colour within Drosophila americana [40]. In other cases, pigment patterning appears to be effected through the altered expression of regulatory genes themselves, possibly through variation in enhancer sequences [39].
Table 2. Test of association between genetic variation in 16 melanisation gene candidates and offspring phenotype.
| locus | allele A | allele B | X 2 value | ||
| carb | typ | carb | typ | ||
| aaNAT | 15 | 20 | 23 | 15 | 0.516545 |
| Ddc | 14 | 21 | 21 | 17 | 0.533008 |
| Dhpr | 16 | 15 | 22 | 20 | 0.999927 |
| ebony | 21 | 19 | 17 | 16 | 0.999844 |
| ferritinHC | 18 | 17 | 20 | 18 | 0.999712 |
| ferritinLC | 18 | 17 | 20 | 18 | 0.999712 |
| henna | 16 | 20 | 22 | 15 | 0.648496 |
| punch | 16 | 14 | 22 | 21 | 0.998396 |
| Pap1 | 20 | 15 | 18 | 20 | 0.873806 |
| Pap3 | 18 | 17 | 13 | 25 | 0.182833 |
| Ppo1 | 27 | 20 | 11 | 15 | 0.673701 |
| Ppo2 | 28 | 19 | 10 | 16 | 0.393203 |
| tan | 20 | 20 | 18 | 15 | 0.98527 |
| TH | 20 | 20 | 18 | 15 | 0.98527 |
| yellow | 21 | 14 | 17 | 21 | 0.636792 |
| yellow2 | 22 | 12 | 16 | 23 | 0.252829 |
Distribution of 38 carbonaria and 35 typical offspring among the paternal alleles (A and B) at each candidate locus.
In contrast to the success of the candidate gene approach applied to melanism in vertebrates, particularly through Mc1r [4], [5] but also tyrosinase-related protein 1 [41], Agouti [42], and K locus [43], the same strategy has been far less useful as a means to identifying polymorphisms controlling melanism in insects [44], [45]. A rare exception is for abdominal and thoracic trident pigmentation in Drosophila melanogaster which co-segregates with nucleotide variation in and around ebony [46], [47]. This is in spite of the fact that melanic patterns are frequently associated with differential expression of one or more genes from the same set of candidates: ebony, tan, TH, Ddc or yellow [10], [12], [15], [40], [47], [48]. The current impression that vertebrate melanism is frequently the result of variation within structural, rather than regulatory, genes, goes some way to explaining this apparent difference, as structural genes forming part of a biosynthetic pathway are easier to target. Why the regulatory machinery of pigment melanin production should be more complex or less conserved in insects is not obvious from a comparison of vertebrate and invertebrate melanogenesis [9], and it may be that a restricted set of conserved regulatory melanisation or patterning genes for insects will eventually emerge.
Other than ruling out specific candidate genes, the present study does not bring us any closer to finding the ‘carbonaria gene’. Having effectively exhausted the a priori list of promising melanisation candidates, we are currently in the process of constructing a linkage map of B. betularia to identify the region that controls this famous polymorphism. The B. betularia-specific coding sequences that were isolated for this candidate gene analysis will be useful in future gene expression studies, and the expanded set of degenerate primers (Table S2) serves as a resource for applying similar approaches to other insects.
Materials and Methods
Experimental design
The offspring of a cross between a homozygous typical female (daughter of a wild pairing collected in Morkery Wood, Linconshire, UK) and a heterozygous carbonaria/typical male (wild caught in Greasby, Cheshire, UK) were used to examine whether candidate genes coincide with the carbonaria phenotype. Caterpillars were reared on oak leaves and adult phenotypes were scored after eclosion of the pupae. All procedures were performed following our institutional animal husbandry guidelines. DNA of half a thorax was extracted with a standard phenol-chloroform procedure [49]. Sixteen genes were screened for co-segregation of polymorphisms with the carbonaria phenotype. Eleven of these were isolated using degenerate primer PCR with cDNA template, and sequences of the remaining five were generated as part of a B. betularia transcriptome sequencing project. Polymorphisms were usually obtained from introns since these are more likely to include genetic variation than exons. However, in the case of henna, for which no internal polymorphism could be found, the end-sequence of a BAC clone containing this gene was used as a closely linked marker for genotyping.
Primer design
A number of the degenerate primers used were designed by Mitchell et al. [50] and some by Hartzer et al. [51], sometimes slightly modified; the remainder were newly designed (Table S2). Nucleotide sequences of orthologous candidate genes in Hexapoda were obtained from NCBI (www.ncbi.nlm.nih.gov), ButterflyBase (http://butterflybase.ice.mpg.de) and SilkDB (http://silkworm.genomics.org.cn) and aligned based on their aa sequences to identify conserved regions. Blast searches [33] were performed to identify paralogs. If present, highly similar domains of paralogs were aligned to the target gene alignments to make sure that only gene-specific, rather than gene-family specific, consensus regions were used for primer design. The primers were designed to cover synonyms of codon triplets over the full length of the recognition site, except for the ferritinHC primers which have a long 5′ region based on the Bombyx mori sequence and only a relatively short degenerate 3′ match within a wide range of insect species. All primers based on B. betularia specific sequence were designed with Oligo 6 [52]. Their names start with ‘Bb’ in Table S2. Introns were identified either by a trial and error approach with randomly placed primer pairs or with primers that were positioned based on the assumption of conserved intron position between B. betularia and B. mori. The intron positions in B. mori were identified by comparing coding sequence and genomic sequence from the SilkDB database.
RNA extraction and cDNA synthesis for gene-targeting
Total RNA was extracted from half a thorax of an adult in 1 ml of Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturers protocol. The cDNA was synthesized from 1 µg total RNA with 200 units Superscript III reverse transcriptase (Invitrogen), 40 units RNAseOUT (Invitrogen), 1X first-Strand Buffer, 5 µM DTT and 50 pmol oligoT18, or 50 pmol M13-polyTv, or 2 pmol gene-specific primer (GSP) in a 20 µl reaction. Primer sequences are available in Table S2. The oligoT18-primed cDNA was generated to serve as template to obtain internal coding regions of target genes, the M13-polyTv-primed cDNA was used for 3′ RACE and the cDNA strands generated with GSPs were used for 5′ RACE. First strand transcription involved one hour at 50°C for oligoT18-primed and M13-polyTv-primed cDNA and one hour at 55°C for cDNA generated with GSPs. The reverse transcriptase was subsequently heat inactivated at 70°C for 15 min.
Gene-specific sequence targeting
Internal regions were amplified with degenerate consensus primers based on gene regions conserved in a wide range of insects (cds 1st fragment primers in Table S2). The initial internal sequence was sometimes extended with an additional degenerate primer outside the initial sequence and a B. betularia-specific primer from within (cds extension primers in Table S2), or alternatively, two additional degenerate primers targeting a non-overlapping fragment were used to cover more internal sequence (cds additional primers in Table S2). The gap between two non-overlapping fragments was bridged by PCR (cds bridging primers in Table S2). The sequences were extended by means of rapid amplification of cDNA ends (RACE). Some genes were completed in both directions using 5′ RACE and 3′ RACE, and others only in the 3′ direction with 3′ RACE. FerritinHC contained an informative polymorphism within the initial part of the sequence and was not extended any further in either direction. The initial, extended and additional internal regions were amplified in 15 µl reaction volumes containing 0.6 units AmpliTaq Gold (Applied Biosystems), 1X AmpliTaq buffer I, 0.35 µM of each primer, 0.2 mM of each dNTP, 0.5 µl oligoT18-primed cDNA. Cycling regime was 9 min 94°C, 11 cycles of 30s 94°C, 30s Tatd, 50s 72°C, 30 cycles of 30s 94°C, 30s Ta, 50s 72°C, with Tatd either [55°C →50°C (−0.5°C per cycle)], [52°C →47°C (−0.5°C per cycle)] or [50°C →45°C (−0.5°C per cycle)] and Ta = 50°C, 47°C, 45°C respectively. PCR cycle details are provided per primer combination in Table S2.
PCRs were inspected on 2% agarose gels and if needed, the product of interest (based on expected size) was isolated with the Qiagen gel extraction kit (Qiagen GmbH, Hilden, Germany). The original concentration of the gel-extracted amplicons were restored by 10 PCR cycles with the same Ta and reaction mix as used for the previous PCR, except for the template, which was 2 µl gel-extract in this case. Gaps between segments of coding sequence were filled in with 15 µl PCRs using primers on either side of the gaps with 0.5 µl oligoT18-primed cDNA as template, 0.6 units AmpliTaq Gold, 1X AmpliTaq buffer I, 0.35 µM of each primer, 0.2 mM of each dNTP and the 35X57HS (hot-start) cycling conditions described in Table 3. The partial B. betularia Ddc sequence deposited in GenBank (accession number EU032788) was not available at the time these experiments were performed.
Table 3. PCR conditions for different experiments.
| PCR experiment | polymerase | Initial denaturation | cycle step 1 denaturation | cycle step 2 annealing | cycle step 3 extension | number of cycles |
| 35X57 | Amplitaq | 3 min 94°C | 30 s 94°C | 30 s 57°C | 50 s 72°C | 35 |
| 35X57HS | Amplitaq Gold | 9 min 94°C | 30 s 94°C | 30 s 57°C | 50 s 72°C | 35 |
| 38X60HS | Amplitaq Gold | 9 min 94°C | 30 s 94°C | 30 s 60°C | 60 s 72°C | 38 |
| 45X60HS | Amplitaq Gold | 9 min 94°C | 30 s 94°C | 30 s 60°C | 60 s 72°C | 45 |
| 35X68LR | Advantage 2 | 1 min 95°C | 15 s 95°C | 6 min 68°C | 35 | |
| 21X68LR | Advantage 2 | 1 min 95°C | 15 s 95°C | 6 min 68°C | 21 |
3′ RACE
The template used for 3′ RACE was M13-polyTv-primed cDNA, which has a synthetic extension of the poly-A tail that acts as a target for the M13 primer. The PCR primer combination consists of a forward GSP positioned in the internal sequence and the M13 primer. PCRs were performed in 15 µl reaction volumes containing 0.6 units AmpliTaq Gold, 1X Amplitaq buffer I, 0.35 µM of each primer, 0.2 mM of each dNTP, 0.5 µl M13-polyTv-primed cDNA and 38X60HS cycling profile (Table 3). A nested primer was used to sequence the PCR products and primer walking (PW) was used to extend the sequence towards the 3′ end for PCR products that were larger than a single reliable sequence read. The three consecutive GSPs used for 3′ RACE are specified as 3R_PCR, 3R_SEQ, and 3R_PW in Table S2 (although 3R_PW was only needed for a few genes).
5′ RACE
5′ RACE used cDNA that was primed with an antisense GSP. The gene-specific cDNA was cleaned with QIAquick PCR purification spin columns (Qiagen) to remove all dNTPs used in first-strand synthesis (to allow the construction of an uninterrupted G-tail subsequently). Then 5 µl of cleaned gene-specific cDNA was combined with 6.5 µl H2O, heated at 65°C for 5 min and chilled on ice immediately after. The 3′end of the cDNA (corresponding with the 5′ end of mRNA) was extended with a poly-G stretch by including the chilled cDNA in a 20 µl reaction mix containing 15 units Terminal deoxynucleotidyl transferase (TdT) (Invitrogen), 1X TdT buffer and 1 mM dGTP. The tailing reaction mix was incubated at 37°C for 30 min and heat inactivated for 3 min at 80°C. The G-tailed cDNA was PCR amplified with a mix of three primers. The first primer is M13_polyC, which has a 3′ poly-C region to match the synthesised poly-G tail and a 5′ M13 region to act as primer recognition site. The second primer is an M13 primer that acts as a forward primer once the M13_polyC is incorporated during the initial PCR cycles. The third primer is a gene-specific antisense primer that is nested relative to the first-strand synthesis primer. These nested GSPs are named 5R_PCR primers in Table S2. PCRs were performed in 15 µl containing 0.6 units AmpliTaq Gold (Applied Biosystems), 1X AmpliTaq buffer I, 0.15 µM M13_polyC primer, 0.35 µM of M13 primer, 0.35 µM of 5R_PCR primer, 0.2 mM of each dNTP and 2 µl G-tailed gene specific cDNA and 45X60HS cycling profile (Table 3). The PCR products were sequenced with nested sequencing primers, which are named 5R_SEQ primers in Table S2. Pap1, Ppo2 and yellow required primer walking to obtain the full sequence, the primers used are named 5R_PW in Table S2.
Transcriptome sequence
The transcriptome was constructed from a 5th instar larva using the Clontech SMART™ PCR cDNA Synthesis Kit (Mountain View, CA, USA), following the ‘first-strand cDNA synthesis’ and ‘cDNA Amplification by LD PCR’ instructions. Total RNA was extracted in Trizol (as described above). The cDNA synthesis was performed for 1 hour at 42°C in 10 µl with 1X First-Strand Buffer, 200 units Superscript II, 1.2 µM 3′ SMART CDS Primer II A, 1.2 µM SMART II A Oligonucleotide, 2 mM DTT, 1 mM of each dNTP, and 0.5 µg total RNA. Amplification took place in 200 µl containing 1X Advantage 2 PCR Buffer, 0.2 mM of each dNTP, 0.24 µM 5′ PCR Primer II A and 1X Advantage 2 Polymerase Mix. PCR was performed with the 21X68LR program (Table 3). The transcriptome was sequenced on a 454 FLX Titanium (Roche, Branford, CT, USA) and assembled with Newbler software (Roche). A blastx search [33] against the B. mori annotated genes in SilkDB was used to identify the melanisation candidates.
Localization of introns
Primers were positioned in presumed neighboring exons (based on the assumption that intron positions are conserved between B. betularia and B. mori) to amplify the intron between them. The intron in punch was obtained with a forward B. betularia-specific primer within partial cds from the transcriptome and a degenerate reverse primer within the next exon. FerritinHC required a trial-and-error primer-pair approach because the intron position in B. betularia differed from that in B. mori. With the exception of the Pap1 intron amplification, PCRs were performed in 15 µl containing 0.6 units AmpliTaq (Applied Biosystems), 1X AmpliTaq buffer I, 0.35 µM of each primer, 0.2 mM of each dNTP and 25 ng of gDNA as template with 35X57 cycle conditions (Table 3). PAP1 required a long-range PCR polymerase due to the large size of the target intron. The reaction was performed in 30 µl containing 1X Advantage 2 PCR Buffer, 0.2 mM of each dNTP, 0.21 µM of each primer, 1X Advantage 2 Polymerase Mix and 50 ng gDNA template with the 35X68LR two-step PCR conditions described in Table 3. Dhpr does not contain introns in B. mori and two proximal primers were included in this experiment to test whether introns are also absent in B. betularia.
BAC identification
A 5X coverage BAC library, supplied with superpools and matrixpools for PCR-based BAC identification, was constructed by Amplicon Express (Pullman, WA, USA). DNA from the clone that was positive for henna was isolated with the BACMAX™ DNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA). End sequences were obtained by Sanger sequencing with T7 Promoter Primer and BAC-R primer (primer sequences in Table S2).
Genotyping
The parental sequences were screened for single nucleotide polymorphisms (snps) and for insertion-deletions (indels) that were heterozygous in the carbonaria father of the cross and homozygous in the typical mother. These polymorphisms give male-informative (MI) backcross segregation in the offspring. PCR-RFLP was used to assay snps within restriction endonuclease recognition sites for ferritinHC (TaqI), TH (MseI), ebony (HpyCH4IV) and PpoI (HaeIII), whereas ferritinLC contains an indel that is large enough to distinguish the two alleles unambiguously on a 2% agarose gel. Offspring genotypes at the remaining polymorphic loci were established by Sanger sequencing.
Genotype-phenotype co-segregation
Co-segregation between the carbonaria phenotype and each of the candidate gene′s genotypes was assessed by simple inspection and more formally using a chi-square test. To account for stochastic deviations from an exact 1∶1 ratio of paternal alleles in the offspring, for each candidate gene locus, the expected values for carbonaria versus typical offspring were calculated with respect to the actual frequencies of each paternal allele (A or B) in the offspring sample. This test is conservative because the confidence value refers to whether the association between genotype and phenotype is larger than would be expected by chance. In reality, given the single locus dominant nature of the carbonaria polymorphism, we expect a 100% match between offspring phenotype and genotype for a marker locus tightly linked to the functional polymorphism, and a marginally weaker association if linkage is weaker (e.g. cis-regulatory element).
Supporting Information
Insect amino acid alignment of tan.
(0.00 MB TXT)
Assembly of tan coding sequence in Bombyx mori based on different resources. The tan sequence for Bombyx mori is wrongly predicted in SilkDB and Kaikobase and the sequences in ButterflyBase and SilkBase are incomplete. This alignment assembles tan for B. mori using the combined information in these databases and a 3'RACE sequence produced especially for this assembly.
(0.01 MB TXT)
Table of polymorphisms used to genotype the melanisation candidates.
(0.05 MB DOC)
Table of primers used to isolate and genotype the candidate genes.
(0.21 MB DOC)
Acknowledgments
John True was involved in early discussions about melanisation gene candidates. Nicola Edmonds provided technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the Natural Environment Research Council (NE/C003101/1), http://www.nerc.ac.uk/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cook LM. The rise and fall of the Carbonaria form of the peppered moth. Q Rev Biol. 2003;78:399–417. doi: 10.1086/378925. [DOI] [PubMed] [Google Scholar]
- 2.Saccheri IJ, Rousset F, Watts PC, Brakefield PM, Cook LM. Selection and gene flow on a diminishing cline of melanic peppered moths. Proc Natl Acad Sci USA. 2008;105:16212–16217. doi: 10.1073/pnas.0803785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook LM, Muggleton J. The peppered moth, Biston betularia (Linnaeus, 1758)(Lepidoptera:Geometridae): a matter of names. Ent Gazette. 2003;54:211–221. [Google Scholar]
- 4.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- 5.Majerus ME, Mundy NI. Mammalian melanism: natural selection in black and white. Trends Genet. 2003;19:585–588. doi: 10.1016/j.tig.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Wiernasz DC. Female choice and sexual selection of male wing melanin pattern in Pieris occidentalis (Lepidoptera). Evolution. 1989;43:1672–1682. doi: 10.1111/j.1558-5646.1989.tb02617.x. [DOI] [PubMed] [Google Scholar]
- 7.Talloen W, Dyck HV, Lens L. The cost of melanization: Butterfly wing coloration under environmental stress. Evolution. 2004;58:360–366. doi: 10.1111/j.0014-3820.2004.tb01651.x. [DOI] [PubMed] [Google Scholar]
- 8.Cerenius L, Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 9.Sugumaran M. Complexities of cuticular pigmentation in insects. Pigment Cell Melanoma Res. 2009;22:523–525. doi: 10.1111/j.1755-148X.2009.00608.x. [DOI] [PubMed] [Google Scholar]
- 10.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Change caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 11.Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- 12.Futahashi R, Fujiwara H. Melanin-synthesis enzymes coregulate stage-specific larval cuticular markings in the swallowtail butterfly, Papilio xuthus. Dev Genes Evol. 2005;215:519–529. doi: 10.1007/s00427-005-0014-y. [DOI] [PubMed] [Google Scholar]
- 13.Hiruma K, Riddiford LM. Granular phenoloxidase involved in cuticular melanization in the tobacco hornworm: regulation of its synthesis in the epidermis by juvenile hormone. Dev Biol. 1988;130:87–97. doi: 10.1016/0012-1606(88)90416-2. [DOI] [PubMed] [Google Scholar]
- 14.Hori M, Hiruma K, Riddiford LM. Cuticular melanization in the tobacco hornworm larva. Insect Biochem. 1984;14:267–274. [Google Scholar]
- 15.Koch PB, Keys DN, Rocheleau T, Aronstein K, Blackburn M, et al. Regulation of dopa decarboxylase expression during color pattern formation in wild-type and melanic tiger swallowtail butterflies. Development. 1998;125:2303–2313. doi: 10.1242/dev.125.12.2303. [DOI] [PubMed] [Google Scholar]
- 16.Futahashi R, Fujiwara H. Expression of one isoform of GTP cyclohydrolase I coincides with thelarval black markings of the swallowtail butterfly, Papilio xuthus. Insect Biochem Mol Biol. 2006;36:63–70. doi: 10.1016/j.ibmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 17.True JR. Insect melanism: the molecules matter. TREE. 2003;18:640–647. [Google Scholar]
- 18.Koch PB, Behnecke B, ffrench-Constant RH. The molecular basis of melanism and mimicry in a swallowtail butterfly. Current Biology. 2000;10:591–594. doi: 10.1016/s0960-9822(00)00494-2. [DOI] [PubMed] [Google Scholar]
- 19.True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, et al. True et al 2005 Drosophila tan Encodes a Novel Hydrolase Required in Pigmentation and Vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, et al. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 21.Hultmark D. Immune reactions in Drosophila and other insects: a model for innate immunity. Trends in Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 22.Zou Z, Wang Y, Jiang H. Manduca sexta prophenoloxidase activating proteinase-1 (PAP-1) gene: organization, expression, and regulation by immune and hormonal signals. Insect Biochem Mol Biol. 2005;35:627–636. doi: 10.1016/j.ibmb.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta S, Wang Y, Jiang H. Manduca sexta prophenoloxidase (proPO) activation requires proPO-activating proteinase (PAP) and serine proteinase homologs (SPHs) simultaneously. Insect Biochem Mol Biol. 2005;35:241–248. doi: 10.1016/j.ibmb.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SY, Kwon TH, Hyun JH, Choi JS, Kawabata SI, et al. In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur J Biochem. 1998;254:50–57. doi: 10.1046/j.1432-1327.1998.2540050.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Lee SY, Cerenius L, Söderhäll K. Properties of the prophenoloxidase activating enzyme of the freshwater crayfish, Pacifastacus leniusculus. Eur J Biochem. 2001;268:895–902. doi: 10.1046/j.1432-1327.2001.01945.x. [DOI] [PubMed] [Google Scholar]
- 26.Barrett AJ, Rawlings ND. Families and Clans of Serine Peptidases. Archives of Biochemistry and Biophysics. 1995;318:247–250. doi: 10.1006/abbi.1995.1227. [DOI] [PubMed] [Google Scholar]
- 27.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors, structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 28.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan A, Giri AP, Gupta VS. Structural and functional diversities in lepidopteran serine proteases. Cellular and Molecular Biology Letters. 2006;11:132–154. doi: 10.2478/s11658-006-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Beck MH, Wang Y, Jiang H, Strand MR. The Viral Protein Egf1.0 Is a Dual Activity Inhibitor of Prophenoloxidase-activating Proteinases 1 and 3 from Manduca sexta. J Biol Chem. 2008;283:21325–21333. doi: 10.1074/jbc.M801593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiruma K, Riddiford LM. The molecular mechanisms of cuticular melanization: the ecdysone cascade leading to dopa decarboxylase expression in Manduca sexta. Insect Biochem Mol Biol. 2009;39:245–253. doi: 10.1016/j.ibmb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Biessmann H. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1985;82:7369–7373. doi: 10.1073/pnas.82.21.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta Hemolymph Proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorman MJ, Wang Y, Jiang H, Kanost MR. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase-activating proteinase 3 in an insect innate immune response proteinase cascade. J Biol Chem. 2007;282:11742–11749. doi: 10.1074/jbc.M611243200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 37.Tong Y, Jiang H, Kanost MR. Identification of Plasma Proteases Inhibited by Manduca sexta Serpin-4 and Serpin-5 and Their Association with Components of the Prophenol Oxidase Activation Pathway. J Biol Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia A-H, Zhou Q-X, Yu L-L, Li W-G, Yi Y-Z, et al. Identification and analysis of yellow protein family genes in the silkworm, Bombyx mori. BMC Genomics. 2006;7:195. doi: 10.1186/1471-2164-7-195. doi: 10.1186/1471-2164-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends in Genetics. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 40.Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, et al. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science. 2009;326:540–544. doi: 10.1126/science.1176980. [DOI] [PubMed] [Google Scholar]
- 41.Gratten J, Beraldi D, Lowder BV, McRae AF, Visscher PM, et al. Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc Royal Soc B. 2007;274:619–626. doi: 10.1098/rspb.2006.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingsley EP, Manceau M, Wiley CD, Hoekstra HE. Melanism in Peromyscus is caused by independent mutations in Agouti. PLoS ONE. 2009;4:e6435. doi: 10.1371/journal.pone.0006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llopart A, Elwyn S, Lachaise D, Coyne JA. Genetics of a difference in pigmentation between Drosophila yakuba and Drosophila santomea. Evolution. 2002;56:2262–2277. doi: 10.1111/j.0014-3820.2002.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 45.Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. PNAS. 2003;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pool JE, Aquadro CF. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol. 2007;Ecol16:2844–2851. doi: 10.1111/j.1365-294X.2007.03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi A, Takahashi K, Ueda R, Takano-Shimizu T. Natural variation of ebony gene controlling thoracic pigmentation in Drosophila melanogaster. Genetics. 2007;177:1233–1237. doi: 10.1534/genetics.107.075283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Futahashi R, Sato J, Meng Y, Okamoto S, Daimon T, et al. yellow and ebony Are the Responsible Genes for the Larval Color Mutants of the Silkworm Bombyx mori. Genetics. 2008;180:1995–2005. doi: 10.1534/genetics.108.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saccheri IJ, Bruford MW. DNA fingerprinting in a butterfly, Bicyclus anynana (Satyridae). Heredity. 1993;84:195–200. [Google Scholar]
- 50.Mitchell A, Mitter C, Regier JC. More Taxa or More Characters Revisited: Combining Data from Nuclear Protein-Encoding Genes for Phylogenetic Analyses of Noctuoidea (Insecta: Lepidoptera). Syst Biol. 2000;49:202–224. [PubMed] [Google Scholar]
- 51.Hartzer KL, Zhu KY, Baker JE. Phenoloxidase in larvae of Plodia interpunctella (Lepidoptera: Pyralidae): molecular cloning of the proenzyme cDNA and enzyme activity in larvae paralyzed and parasitized by Habrobracon hebetor (Hymenoptera: Braconidae). Arch Insect Biochem Physiol. 2005;59:67–79. doi: 10.1002/arch.20056. [DOI] [PubMed] [Google Scholar]
- 52.Rychlik W. West Cascade, CO USA.: Molecular Biology Insights; 2000. OLIGO: primer analysis software. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Insect amino acid alignment of tan.
(0.00 MB TXT)
Assembly of tan coding sequence in Bombyx mori based on different resources. The tan sequence for Bombyx mori is wrongly predicted in SilkDB and Kaikobase and the sequences in ButterflyBase and SilkBase are incomplete. This alignment assembles tan for B. mori using the combined information in these databases and a 3'RACE sequence produced especially for this assembly.
(0.01 MB TXT)
Table of polymorphisms used to genotype the melanisation candidates.
(0.05 MB DOC)
Table of primers used to isolate and genotype the candidate genes.
(0.21 MB DOC)