Abstract
Inflammatory mediators are of considerable interest as potential therapeutic targets in various cancers. Here we investigate whether interleukin (IL)-4 receptor alpha (IL4Rα), a component of the receptor complex for the T helper 2 cytokines IL4 and IL13, plays a role in colonic tumorigenesis. IL4Rα protein expression was seen in tumor cells of 28/48 human colon adenocarcinomas on a tissue microarray. In human and murine colon tumor cell lines analyzed in vitro, all of which expressed IL4Rα, treatment with exogenous ligand resulted in dose-dependent increases in proliferation. IL4 decreased apoptosis only in HCT116 cells. An orthotopic allograft model was used to determine in vivo effects of tumor cell-specific IL4Ra ablation. MC38 murine tumor cells with the IL4Ra gene knocked down showed reduced proliferation but no difference in apoptosis compared with controls after implantation in ceca of syngeneic mice. Mice null for IL4Ra and wild-type controls were treated with azoxymethane and dextran sulfate sodium to induce tumor formation. Mice with global deletion of IL4Ra had significantly fewer and smaller tumors. Reduced tumorigenicity correlated with decreased proliferation and increased apoptosis. Systemic blockade of IL4Rα–IL4 interactions with a chimeric soluble receptor protein gave similar results in the cecal implant model. Thus, IL4Rα, a component of the IL4R and IL13R, contributes to tumor formation in a mouse model of colitis-associated cancer. Proliferation appears to be directly mediated via IL4Rα on the epithelial tumor cells. Survival may be an indirect response mediated via other host cells. Our results support therapeutic targeting of IL4Rα in colon cancer.
Introduction
The function of inflammation-associated cells and molecules in cancer development is currently a topic of considerable interest (1,2). An understanding of the interplay between microenvironmental factors and tumor cells will ultimately allow development of new targeted therapies and preventative strategies for a variety of cancers. While inflammatory processes participate in many tumors, they are of particular relevance in cancers whose etiology relates to chronic inflammation. Individuals with long-standing ulcerative colitis, for example are known to have a significantly increased risk of developing colon cancer (3,4). While it is likely that the genetic mutations that initiate tumorigenesis are due to the high levels of reactive oxygen and nitrogen species present in the setting of chronic inflammation (4), it is also clear that the inflammatory setting provides tumor-promoting molecules. Both interleukin (IL)-6 and tumor necrosis factor have recently been demonstrated as inflammatory mediators that play critical roles in colitis-associated tumor development (5,6). Here, we investigate whether the IL4 receptor (IL4R) can promote colon tumor development.
IL4 is produced by several cell types including mast cells, basophils and activated T lymphocytes (7,8). It is a T helper 2 cytokine primarily known for controlling functions of B and T lymphocytes. IL4 can stimulate proliferation of these cell types as well as induce T cell differentiation and expression of a range of gene targets. Clinically, the best known roles of IL4 are in allergy, atopy and asthma where IL4 contributes to pathology (9). Another significant function for IL4 is polarization of macrophages to the ‘M2’ or alternatively activated phenotype (10). M2 macrophages are important for resolution of inflammation and wound healing, but they are also classed with the tumor-associated macrophages associated with tumor progression, especially in breast cancer (2,11). Indeed, a recent study has identified a metastasis-promoting role for IL4 in a mouse model of breast cancer, which was mediated indirectly through tumor-associated macrophage production of epidermal growth factor (12). IL4 also has diverse functions in non-hematopoietic cells, not all of which are understood. Proliferative (13), growth inhibitory (14), anti-invasive (15), pro-migratory (16) and survival (17) functions have all been ascribed to IL4 in diverse cell types. IL4 signaling is induced by binding of IL4 to the high-affinity IL4R alpha (IL4Rα) chain and its subsequent dimerization with either the common gamma receptor chain (type I complex) or the IL13Rα1 chain (type II complex) (7,8). The type II complex, found in epithelial cells, is identical to that formed by IL13 binding to IL13Rα1 with subsequent heterodimerization to IL4Rα, thus both IL4 and IL13 are ligands for the IL4R.
The availability of IL4Rα-null mice (18) allows testing of the roles of endogenous IL4R signaling in tumor development. Recently, a study of the carcinogen azoxymethane (AOM) in IL4Rα-null mice reported paradoxical roles for IL4Rα (19). In the 9-week time frame examined, AOM caused the development of putative pre-cancerous lesions, known as aberrant crypt foci. In IL4Rα-null mice, the number of aberrant crypt foci was increased compared with controls; however, the proliferative index within these lesions as well as the size was reduced in the IL4Rα-null mice (19). Since not all aberrant crypt foci become tumors, it was not clear from this study if actual tumor number would be increased or decreased in the IL4Rα-null mice. Thus, IL4Rα may represent either a therapeutic target or a beneficial molecule in colon cancer. To address this ambiguity, we utilized a model of colitis-associated colon cancer. Here we report that tumor number is significantly attenuated in IL4Rα−/− mice administered AOM and dextran sulfate sodium (DSS). Furthermore, the proliferative aspect of the enhanced tumorigenesis is apparently driven through epithelial cell IL4Rα rather than immune cell IL4Rα signaling. Our data suggest that IL4Rα is a tumor-promoting molecule that should be considered for therapeutic targeting.
Materials and methods
Reagents and antibodies
Dulbecco's modified Eagle's medium, trypsin-ethylenediaminetetraacetic acid and Trizol were purchased from Invitrogen (Carlsbad, CA). Fetal calf serum was from Atlanta Biologicals (Lawrenceville, GA). DSS (MW 36–50 000) was from MP Biomedicals (Solon, OH). Unless otherwise indicated, other chemicals were from Sigma (St Louis, MO). Antibodies were from the following companies: anti-phosphohistone H3 and anti-cleaved caspase-3 from Cell Signaling Technology (Danvers, MA); anti-beta-catenin from BD Biosciences (San Jose, CA); anti-mouse IL4Rα and β-actin from Santa Cruz Biotechnology (Santa Cruz, CA); anti-human IL4Rα and IL4R-Fc chimeric protein from R&D Systems (Minneapolis, MN) and anti-F4/80 from Invitrogen. Alexa-fluor 594 anti-rabbit, Alexa-fluor 350 anti-rat and Sytox green nuclear stain were from Invitrogen.
Cell lines and culture
The human colon tumor cell lines HCT116, HT29, DLD-1, SW480, SW620 and Caco2 were from the American Type Culture Collection. HCA7 cells were originally from Dr Susan Kirkland, University College London. The murine colon tumor lines CT26 and MC38 were from Dr D. Lee Gorden at Vanderbilt. All lines were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum at 37°C with 5% CO2. Their identity was verified both by appearance and growth curve analysis. To generate IL4Ra knockdown clones of MC38 cells, Mission lentiviral small hairpin RNA (shRNA) particles directed toward murine IL4Ra (NM_010557) were purchased from Sigma together with control non-target particles. These were used to infect MC38 cells following the manufacturer's recommended protocol. For IL4Ra knockdown in HCT116 cells, ‘Sure-Silencing’ plasmids encoding shRNA sequences directed toward four different regions of human IL4Ra (NM_000418) were purchased from SA Biosciences (Frederick, MD) and transfected into HCT116 cell using lipofectamine (Invitrogen), following manufacturer's recommendations. Two days after infection or transfection, murine or human cells were placed under selection with puromycin (10 μg/ml) or hygromycin (200 μg/ml), respectively. Once stable populations were obtained, individual clones were isolated by limiting dilution. The clones were screened for levels of IL4Rα messenger RNA by reverse transcription–polymerase chain reaction (PCR) and protein by western blotting.
Reverse transcription–PCR
RNA was isolated from confluent 60 mm plates of each cell line using Trizol (Invitrogen). The RNA from each line was reverse transcribed using Moloney Murine Leukemia Virus-Reverse Transcriptase and standard conditions. The complementary DNA was used in hot start PCR reactions with the following primers for each target: human IL4Rα forward 5′-GACCTGGAGCAACCCGTATC-3′, reverse 5′-CGTCTGCCTGTTGTGCTATG-3′; murine IL4Rα forward 5′-CAGACCCGAAGCCAGGAGTCAACC-3′, reverse 5′-GTTTGTGCAGGCAGTGAAGCAAGGG-3′ and glyceraldehyde3- phosphate dehydrogenase (multi-species) forward 5′-ACCACAGTCCATGCCATCAC-3′, reverse 5′-TCCACCACCCTGTTGCTGTA-3′. The expected sizes for the products were 335, 883 and 452 bp, respectively. The cycling conditions were 95°C for 15 min; 40 cycles of 95°C for 40 s, 50°C for 45 s and 72°C for 1 min; 1 cycle of 72°C for 7 min.
Western blotting
Protein lysates were collected from confluent 100 mm dishes of cells using radioimmunoprecipitation assay lysis buffer containing protease inhibitor (Roche Applied Science, Indianapolis, IN). Equal amounts of protein were loaded on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and run standardly except for those being probed for human IL4Rα, which required non-reducing conditions. After electrophoresis, proteins were transferred to nitrocellulose membranes, blocked with 5% milk and incubated with primary antibody at 1:1000 (β-actin) or 1:200 (human IL4Rα or mouse IL4Rα). After washing, blots were incubated with horseradish peroxidase-labeled secondary antibodies and developed using Western Lightening Reagent (Pierce/Thermo Fisher, Rockford, IL). Films were scanned and sized using Adobe Photoshop software. No alterations in background, band intensity or contrast were made.
3-(4,5-Dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide assays
Cells were plated at a density of 2–5000 cells per well on 96-well plates. Recombinant human (for HCT116 and HT29) or murine IL4 or IL13 (for CT26 and MC38) was added to replicate wells at a range of concentrations and the cells incubated for 24 h. At this time, one-fifth volumes of 5 mg/ml 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide reagent were added to each well and incubated for 4 h. Then, medium was aspirated and the precipitated formazan dye dissolved in isopropanol. Absorbance readings at 570 nm were obtained using a Dynex MRX plate reader.
Apoptosis assays
Cells were plated at 60% confluence in 6-well plates. Triplicate wells were treated with 5-fluorouracil (10 μg/ml) or equivalent volume of vehicle (dimethylsulfoxide) plus or minus 20 ng/ml species-specific recombinant IL4. After 24 h, cells were harvested by scraping and centrifugation and incubated with fluorescein isothiocyanate-annexin V (Santa Cruz) according to manufacturer's recommendations. The labeled cells were detected by flow cytometry (BD FACStar), and the percentage of positive cells determined using FlowJo software (Treestar).
Animal studies
All animal experiments were conducted following approval by the institutional animal care and use committee. IL4Rα-null mice (Balb/c) as well as wild-type Balb/c controls were purchased from (Jackson Laboratories, Bar Harbor, ME) and bred to generate sets of age- and sex-matched mice for experiments.
For the AOM/DSS protocol, 6-week-old mice (n = 12 IL4Rα−/−; n = 11 wt) were administered a single intraperitoneal injection of AOM at a dose of 12.5 mg/kg body wt. One week later, the mice were provided 3% DSS ad libitum in the drinking water. DSS exposure lasted for 4 days and mice were then returned to regular water. Consumption of DSS-containing water was measured daily and found to be similar for all cages. A second 4-day DSS exposure was started 16 days after completion of the first. Nine days after cessation of DSS, mice were euthanized and the colons harvested. The colon from anus to cecum was removed, flushed with phosphate-buffered saline and measured before being cut open longitudinally. After fixation in 10% buffered formalin, colons were stained briefly in 0.05% methylene blue, rinsed and examined under a dissecting microscope. The diameter of each visible tumor was measured using digital calipers, and this together with the total number of tumors was recorded for all specimens by an observer blinded to genotype. Individual tumors were excised and paraffin embedded.
For the colitis study, cohorts of male 8-week-old Balb/c or IL4Rα−/− mice (n = 9, both groups) were placed on a 5-day regimen of 3% DSS provided in the drinking water. The mice were weighed daily. At the end of the treatment period, the mice were euthanized, colons harvested and ‘jelly rolled’ before fixation in 10% buffered formalin and paraffin embedding to allow visualization along the length of the colon in sections.
For the allograft study, 6-week-old C57BL/6 mice were anesthetized and injected intracecally with 1 × 106 MC38-control (n = 8) or MC38-knockdown (MC38-KD) cells (n = 10) following externalization of the cecum through a small abdominal incision. After 3 weeks, the mice were euthanized and the ceca harvested, rinsed and examined for the presence of gross tumor nodules. Any tumors found were measured, excised and fixed in 10% buffered formalin before processing for paraffin embedding.
Immunostaining
Five micron formalin-fixed, paraffin-embedded tissue sections from the mouse studies, as well as a human colon and rectal cancer tissue microarray (Imgenex, San Diego, CA), were analyzed by standard immunostaining techniques. Briefly, antigen retrieval was by microwaving for 10 min in 10 mM sodium citrate, pH 6.0. Primary antibodies were incubated with the sections at the following dilutions: anti-phosphohistone H3 1:200; anti-cleaved caspase-3 1:200 and anti-human IL4Rα 1:50. After washing, sections were incubated with biotinylated secondary antibodies and then an avidin–biotin–peroxidase complex (Vectastain ABC; Vector Labs, Burlingame, CA). Signal was visualized with 3,3′-diaminobenzidine tetrahydrochloride and counterstained with Mayer's hematoxylin. Images of stained slides were captured using a Zeiss Axiophot microscope and Metamorph software. Image manipulation was limited to white balancing and size reduction. Quantification of staining was achieved by counting the number of positive cells per unit area in multiple high-powered (×40) views of each slide using ImageJ software.
For co-staining of macrophages and IL4Rα, sections were dewaxed, rehydrated and heated in 10 mM citrate as before. After blocking in 10% goat serum, sections were incubated overnight at 4°C with rat anti-mouse F4/80 (1:100) and rabbit anti-mouse IL4Rα (1:50). Following extensive washing, the sections were incubated with Alexa-fluor 350 anti-rat and Alexa-fluor 594 anti-rabbit secondaries for 30 min at room temperature, then washed, counterstained with Sytox green and coverslipped.
Statistical analyses
All analyses were performed and graphs generated using Prism 5 software (GraphPad Software, San Diego, CA). Significance was at the 95% level of confidence.
Results
Expression of the IL4Rα subunit in human and murine colon tumors
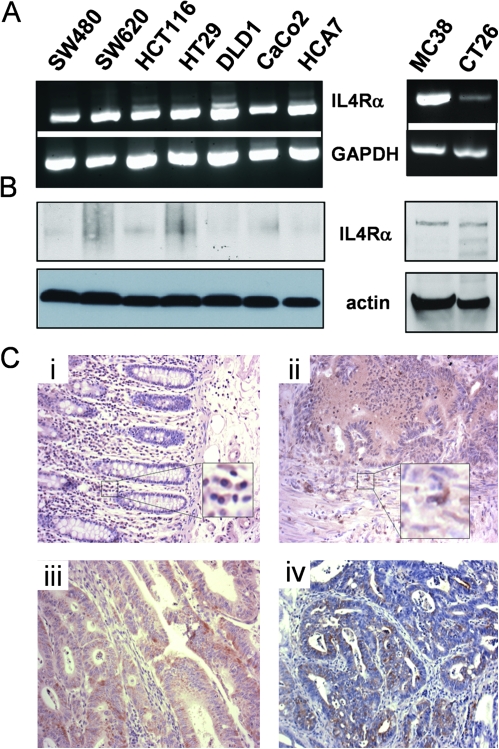
In non-hematopoietic cells, the IL4R consists of two subunits, the IL4Rα chain, which is also part of the receptor complex on hematopoietic cells, and the IL13Rα2 chain. Based on reports in the literature that several epithelial cancers including breast, brain and colon had up-regulated IL4R expression (20–22), we examined a panel of human and murine colon tumor cell lines for IL4Rα expression at both the RNA and the protein levels. All the human cell lines expressed IL4Rα (Figure 1A). The two murine lines represent carcinogen-induced tumors from a Balb/c (CT26) and a C57BL/6 (MC38) mouse. Both murine cell lines expressed detectable IL4Rα (Figure 1B). We then also examined IL4Rα expression in human colon cancer tumor sections on a commercially available tissue microarray. Clear expression was noted in tumor cells in 60% (29/48) of the colon carcinoma cores stained (Figure 1Cii and iii). Murine colon tumors showed similar positivity in tumor epithelium (Figure 1Civ). Epithelium in normal human colon was negative; however, positive staining was visible in infiltrating cells consistent with myeloid cells (Figure 1Ci). Similar IL4Rα-positive staining infiltrating cells were also visible in tumor sections (Figure 1Cii). Double immunofluoresence of the murine macrophage marker F4/80 and IL4Rα confirmed that tumor-associated macrophages visible in tumors from murine models of colon cancer express IL4Rα (supplementary Figure 1 is available at Carcinogenesis Online). Overall, these analyses demonstrate that expression of IL4Rα is widespread in human colon cancer and in both human and murine colon cancer cell lines.
Fig. 1.
IL4Rα is expressed in human and murine colon cancer cells. (A) IL4Rα (upper panels) and glyceraldehyde3- phosphate dehydrogenase (GAPDH) (lower panels) transcripts in a panel of human (left) and murine (right) colon tumor cell lines as detected by reverse transcription–PCR. (B) IL4Rα (upper panels) and β-actin (lower panels) protein detected by western blotting in lysates from a panel of human (left) and murine (right) cell lines. Note: The antibody for human IL4Rα recognizes only the non-reduced protein; hence, the bands are not as sharp as for the murine protein. (C) Examples of the staining pattern for IL4Rα (brown) seen in tissue sections of (i) normal human colon; (ii, iii) human colon adenocarcinoma on a tissue microarray and (iv) murine AOM-induced colon tumor. Insets show infiltrating cells with positive staining for IL4Rα. Nuclei are counterstained blue with hematoxylin.
IL4 stimulates proliferation of murine and human colon tumor cells in vitro
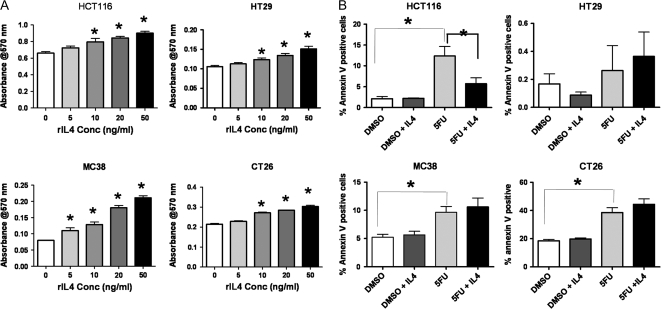
Since all colon tumor cell lines tested expressed IL4Rα, we tested whether this was a constituent of a functional receptor that could respond to exogenously supplied IL4. We used four cell lines, two human and two murine to perform these analyses. MC38 and CT26 murine colon tumor cells and HCT116 and HT29 human colon cells all showed dose-responsive increases in proliferation after treatment with recombinant mouse or human IL4, respectively (Figure 2A). We also examined the ability of exogenously added recombinant IL4 to inhibit cell death induced by addition of the chemotherapeutic agent 5-fluorouracil (Figure 2B). Surprisingly, only HCT116 cells showed a significant survival response after addition of 20 ng/ml recombinant IL4 (Figure 2B). An additional human colon cancer cell line, SW620, behaved similarly to HT29 cells with a proliferative response to IL4, but no survival response (data not shown). Together these results suggest that proliferative effects are the dominant autonomous response of colon epithelial tumor cell lines to exogenous IL4.
Fig. 2.
Colon tumor cell lines respond directly to exogenous IL4 in vitro. (A) 3-(4,5-Dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide assays showing the response of four different cell lines, both human (HCT116 and HT29) and mouse (CT26 and MC38), to increasing concentrations of species-specific recombinant IL4. Values shown are for 24 h treatment. Asterisks indicate results significantly higher than the corresponding control (0 ng/ml), P ≤ 0.05 by analysis of variance (ANOVA) with Dunnett's post hoc test. (B) Annexin V assays for cells undergoing apoptosis in response to 24 h exposure to the chemotherapeutic agent, 5-fluorouracil (5FU), in the presence or absence of 20 ng/ml species-specific IL4. Asterisks indicate P ≤ 0.05 by ANOVA with Dunnett's post hoc test.
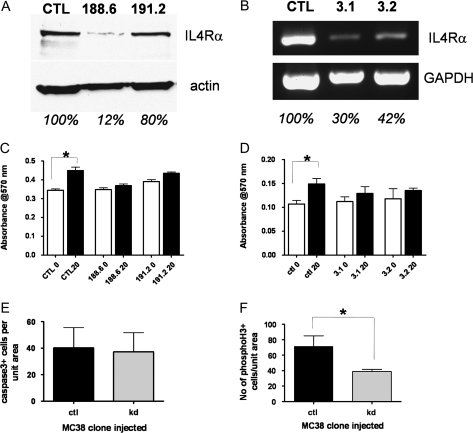
To ensure that effects we were seeing were mediated through IL4Rα, we generated a series of MC38 and HCT116 clones with IL4Ra knocked down using sets of specifically targeted shRNAs to murine or human IL4Ra, respectively. At least two different shRNA sequences specific to either human or murine IL4Ra resulted in some knockdown as determined by western blotting or reverse transcription–PCR (Figure 3A and B). When these clones were compared with control shRNA clones in the proliferation assays, exogenous IL4 was no longer effective (Figure 3C and D). The control shRNA clones behaved similarly to the parental lines.
Fig. 3.
IL4Rα knockdown in MC38 and HCT116 cells abrogates proliferative response to IL4. (A) Western blotting of IL4Rα protein (upper panel) and β-actin (lower panel) in MC38 cells infected with control or IL4Ra-targeted shRNA lentivirus. Results are shown for two different targeting sequences. The percentage of expression of IL4Rα, based on actin normalization, is shown below each lane. (B) Reverse transcription–PCR of IL4Rα transcript (upper panel) and GAPDH (lower panel) in HCT116 cells transfected with control or IL4Ra-targeted shRNA plasmids. Results are shown for two different clones. The percentage of expression of IL4Rα, based on GAPDH normalization, is shown below each lane. (C) 3-(4,5-Dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide (MTT) assay of MC38-control and knockdown clones with (black bars) or without (white bars) 24 h treatment with 20 ng/ml murine IL4. Only the control clone showed a significant response. (D) MTT assay of HCT116 control and knockdown clones with or without 20 ng/ml human IL4. (E) Cleaved caspase-3 immunostaining in cecal tumors from mice injected with MC38-control (n = 4) or MC38-knockdown cells (n = 5). Shown are the mean number of positive cells per unit tumor area per mouse, P = 0.95 by Mann–Whitney test. (F) Phosphohistone H3 immunostaining in cecal tumors from mice injected with MC38-control (n = 4) or MC38-knockdown cells (n = 5). Shown are the mean number of positive cells per unit tumor area per mouse, *P = 0.03 by Mann–Whitney test.
Tumor cell IL4Rα signaling is responsible for tumor proliferation but not survival in vivo
Multiple cell types, including tumor cells (20), fibroblasts (13), macrophages (12) and lymphocytes (7) have been reported to express IL4Rα. Indeed, by immunohistochemical analysis in tumor sections, cells whose appearance is consistent with myeloid cells stain positive for IL4Rα (Figure 1Ci and ii). Moreover, in a breast cancer model, the tumor-promoting effects of IL4 were shown to be mediated through macrophages and not tumor cells (12). Our in vitro data suggest that proliferation, but not necessarily survival, can be a direct response of tumor cells to IL4. To address whether tumor cell-expressed IL4Rα played a role in tumor development in vivo, we used an orthotopic allograft model. Clones of MC38 cells expressing a non-targeting shRNA (control) or an IL4Rα-specific shRNA (MC38-KD) were injected in the cecum of immunocompetent syngeneic C57BL/6 mice. The knockdown clone used had an 88% reduction in level of IL4Rα protein expression by western blotting compared with the control (Figure 3A). After 3 weeks, the mice were euthanized and the ceca harvested. Gross tumor measurements showed no significant difference in tumor size, although the range was large (1.91–11.35 mm diameter for control tumors; 1.78–9.03 mm diameter for MC38-KD tumors). Histological examination of tumor sections showed that some tumors had significant inflammatory infiltrates so that tumor diameters were not accurate reflections of tumor size. We therefore analyzed levels of apoptosis and proliferation in tumor cells, as determined by histological appearance, to assess whether the reduced level of IL4Rα in the knockdown cells had any effect on these parameters. As shown in Figure 3E, apoptosis levels as measured by immunostaining for cleaved caspase-3 were not different between the two sets of tumor types. Proliferation, as measured by immunostaining for phosphohistone H3, was, however, significantly reduced in the MC38-KD tumors compared with controls (Figure 3F). These results are similar to the in vitro data and indicate that signaling through colon tumor cell IL4Rα is important for proliferation but not survival.
Expression of IL4Rα contributes to tumor development in a mouse model of colon cancer
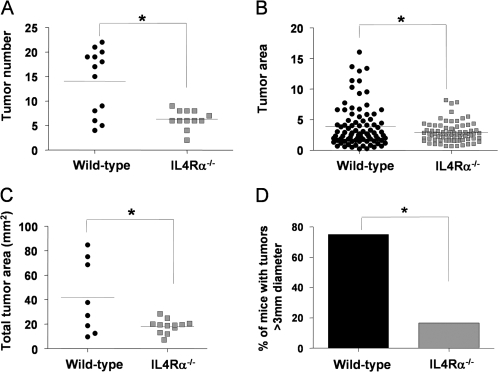
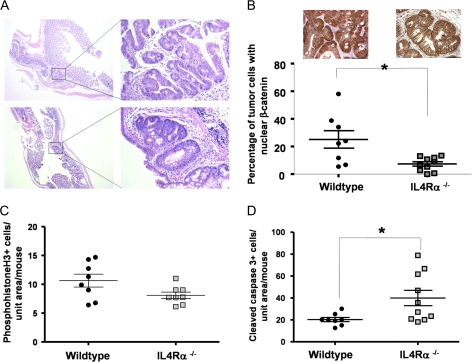
Having established that IL4Rα is indeed expressed and functional in human and murine colon tumors, we next investigated whether its expression played a role in endogenous tumor development initiated by a carcinogen. We used mice on the Balb/c background in which IL4Ra had been genetically ablated. This is a global knockout; thus, neither epithelial nor hematopoietic/bone marrow-derived cells expressed IL4Rα. The mice are phenotypically normal unless challenged with worm infestation in which case they show an inability to clear the parasite and succumb to overwhelming disease burden (23). We used the carcinogen AOM combined with the colitis-inducing agent DSS, which has been shown to promote AOM-induced cancers in order to produce definite tumors. Cohorts of IL4Rα-null and wild-type age- and sex-matched mice all on the Balb/c background received a single injection of AOM at 5–6 weeks of age. Two weeks later, they were administered 3% DSS in the drinking water for a 4-day period. After 16 days back on normal water, they were returned to the 3% DSS for another 4 days. Following this second round of DSS, we observed weight loss in several of the wild-type but none of the IL4Rα-null mice. We therefore decided to terminate the experiment at this stage to prevent excessive morbidity. The mice were killed and the colons removed, rinsed and fixed in formalin. There was no significant difference in colon length, an indicator of inflammation-induced tissue damage, between the wild-type and IL4Rα-null mice. Following brief staining with methylene blue, the colons were evaluated for tumor number and size by an observer blinded to genotype. As shown in Figure 4, the absence of IL4Rα resulted in significantly fewer tumors as well as significantly smaller tumors. There was a 75% decrease in the number of tumors >3 mm in diameter. Histologically, the tumors were generally similar and could be classified as adenomas (Figure 5A). Immunostaining for beta-catenin showed that nuclear localization was significantly more pronounced in the wild-type tumors (Figure 5B), indicating that the AOM-initiated tumors had acquired mutations in the Wnt/adenomatous polyposis coli pathway.
Fig. 4.
Tumor number and size are significantly reduced in IL4Rα−/− mice following AOM/DSS treatment. (A) Total tumor number per wild-type (n = 13) or IL4Rα−/− (n = 12) colons, *P = 0.0008, Mann–Whitney test. (B) Individual tumor area for wild-type (n = 86 tumors from eight mice) versus IL4Rα−/− (n = 75 tumors from 12 mice), *P = 0.018, Mann–Whitney test. (C) Total tumor area per mouse for wild-type (n = 8) versus IL4Rα−/− (n = 12), *P = 0.015, Mann–Whitney test. (D) Incidence of large tumors (>3 mm diameter) in wild-type (six of eight mice) versus IL4Rα−/− (2 of 12 mice), *P = 0.019, Fisher's exact test.
Fig. 5.
Nuclear β-catenin is less prevalent, proliferation is decreased and apoptosis increased in colon tumors from IL4Rα-null mice. (A) Hematoxylin- and eosin-stained tumors from wild-type (top) and ILRα−/− (bottom) mice. Images on the left are taken with a ×2.5 objective, bar = 1 mm. Boxes indicate the region magnified on the right, bar = 100 μm. (B) Quantification of the percentage of tumor cells per mouse examined in which the β-catenin was localized to the nucleus. At least 500 cells were evaluated per mouse, *P = 0.02, Mann–Whitney test. Inset panels show sample immunostaining for β-catenin in tumor cells present in colonic lesions from wild-type (left) and IL4Rα−/− (right) mice. Bar = 65 μm. (C) Phosphohistone H3 immunostaining in colon tumors from wild-type (n = 8) and IL4Rα−/− (n = 8) mice. Shown are the number of positive cells per unit tumor area per mouse, P = 0.06, Mann–Whitney test. (D) Cleaved caspase-3 immunostaining in colon tumors from wild-type (n = 8) and IL4Rα−/− (n = 10) mice. Shown are the number of positive cells per unit tumor area per mouse, *P = 0.02, Mann–Whitney test.
Since it was possible that the reason tumors formed less readily in the IL4Rα-null mice might be because the inflammation-promoting agent DSS is less effective on this genetic background, we performed an acute study of response to DSS in wild-type versus IL4Rα-null mice. There was no significant difference in weight loss between the two genotypes during the course of treatment. At termination, colon lengths were similar. Histological evaluation indicated similar levels of inflammation and ulceration between the two sets of mice (supplementary Figure 2 is available at Carcinogenesis Online).
Tumors from IL4Rα-null mice show decreased proliferation and increased apoptosis
The decreased size of tumors that formed in IL4Rα−/− mice suggested that proliferation and/or tumor cell apoptosis may be altered. We used immunostaining for phosphohistone H3, a marker of late metaphase, and for cleaved caspase-3, the predominant executioner caspase, to assess levels of proliferation and apoptosis, respectively. There was reduced proliferation in the tumors from IL4Rα−/− mice (Figure 5C), which approached statistical significance (P = 0.06). On the other hand, levels of apoptosis were significantly higher in the IL4Rα−/− tumors (Figure 5D), suggesting that reduced tumor cell survival is a major contributing factor to the decrease in tumor size.
Systemic inhibition of IL4Rα–IL4 interactions recapitulates genetic ablation
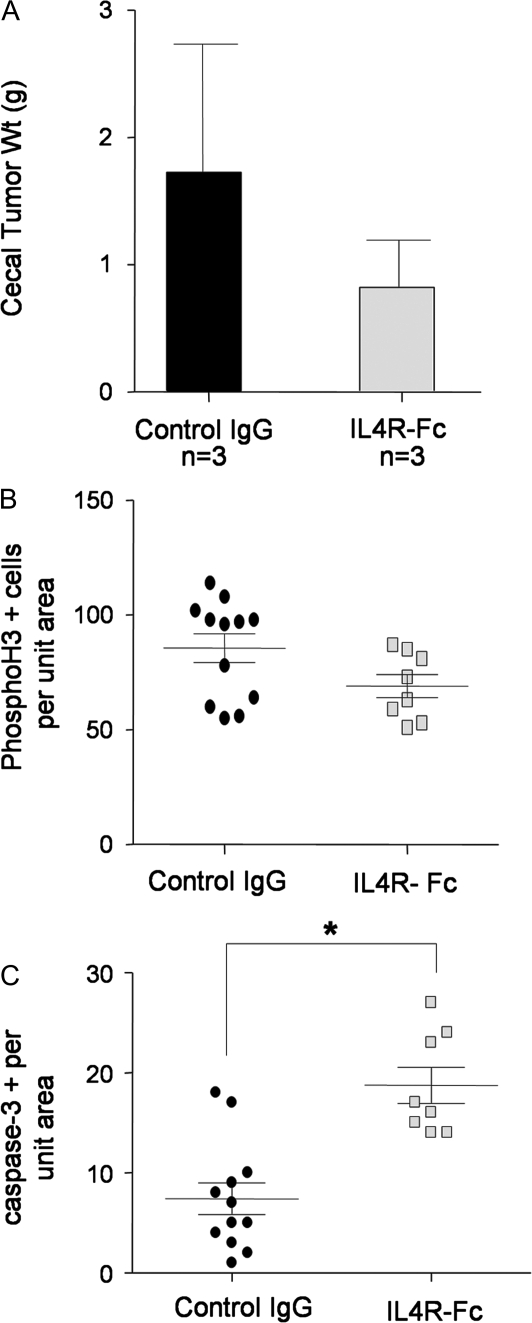
Since the proliferation and apoptosis results obtained with the carcinogen model (Figure 5C and D) were opposite to that obtained with the implanted tumor model (Figure 3E and F), we wondered whether the difference lay in the fact that IL4Rα was globally ablated in the AOM/DSS model versus knocked down only in tumor cells in the cecal implant model. To test this, we performed a pilot study using the cecal implant model and an IL4 blocking chimeric protein. IL4R-Fc is a fusion protein consisting of the extracellular domain of murine IL4Rα fused to the human Fc domain. It behaves as a decoy receptor binding IL4 and thus preventing it from interacting with transmembrane IL4Rα localized on cells. MC38 colon tumor cells were implanted into the ceca of syngenic C57BL/6 mice. The following day, mice were randomized to receive either IL4R-Fc or immunoglobulin G control given systemically. Treatment was repeated every 3 days for 2 weeks. At the end of the treatment period, the mice were euthanized and the cecal tumors harvested. While the tumor size, as determined by wet weight, trended toward reduced size in the IL4R-Fc-treated mice, that was not significantly different because of high variability (Figure 6A). When sections of the tumors were analyzed for proliferation and apoptosis by phosphohistone H3 and cleaved caspase-3 immunostaining, respectively, proliferation was reduced but not significantly (Figure 6B), whereas apoptosis was significantly increased in tumors from mice treated with IL4R-Fc (Figure 6C). These results echoed those obtained in IL4Rα−/− mice following tumor induction by AOM/DSS. Thus, global attenuation of IL4Rα results in reduced tumor proliferation and significantly increased tumor cell apoptosis. In contrast, tumor cell-specific IL4Rα attenuation results in decreased proliferation but no effect on tumor cell apoptosis.
Fig. 6.
IL4Rα blockade leads to reduced proliferation and increased apoptosis of tumor cells in an implanted tumor model. (A) Wet weight of MC38 cecal tumors from control immunoglobulin G (IgG) and IL4R-Fc-treated mice. (B) Phosphohistone H3 immunostaining in MC38 cecal tumors from control IgG and IL4R-Fc-treated mice. Shown are the number of positive cells per unit tumor area per section, P = 0.07, Mann–Whitney test. (C) Cleaved caspase-3 immunostaining in MC38 cecal tumors from control IgG and IL4R-Fc-treated mice. Shown are the number of positive cells per unit tumor area per section, *P = 0.003, Mann–Whitney test.
Discussion
Cytokines and their receptors have recently been demonstrated as important contributors to tumor development with both IL6 and tumor necrosis factor α being particularly associated with colon cancer (5,6). Here we have focused on a possible role for IL4Rα, a component of the receptor for both IL4 and IL13. In a mouse model of colitis-associated colon cancer induced by AOM and DSS exposure, mice with global deletion of IL4Rα had significantly reduced tumor number. The tumors that did form in the absence of IL4Rα were significantly smaller in size than their wild-type counterparts. The smaller size correlated with decreased proliferation and increased apoptosis within the tumors. When we examined the effects of exogenous IL4 on a panel of human and murine colon tumor cell lines, we could measure a significant proliferative response in all cell lines. Notably, apoptosis induced by a chemotherapeutic drug was only affected by IL4 treatment in one cell line. Tumor cells with IL4Rα expression silenced by shRNA, which were injected orthotopically into the ceca of wild-type mice, showed significantly reduced proliferation but no apoptosis differences compared with control cells. Together, our data point to proliferation as an autonomous function of IL4Rα in colonic epithelial tumor cells, whereas tumor cell death is mediated by host cell IL4Rα. These data support IL4Rα as a potential therapeutic target in colon cancer.
IL4Rα expression has been reported in multiple cell types including lymphocytes (7), macrophages (12), fibroblasts (13) and epithelial cells (14,20). Up-regulation of receptor expression is also a frequent occurrence in several types of cancer (20–22). In our expression analysis, we found that all colon tumor cell lines examined expressed IL4Rα, although at different levels. The variation was more evident for the protein than for the messenger RNA transcript (Figure 1) and may reflect generation of a soluble form of the receptor by some of the cell lines. In human colonic adenocarcinoma tissue sections, the majority of the staining for IL4Rα was seen on tumor cells. Occasional stromal cell expression was also detected, mostly in what appeared to be infiltrating cells. A recent study has demonstrated a role for myeloid IL4R in tumor metastasis in a mouse model of breast cancer (12). Strikingly, that study showed no effect of IL4 on primary tumor growth and development. One potential reason for the disparate results between that study and the data reported here is the type of model used. The breast cancer model was the mouse mammary tumor virus- polyoma virus middle T transgenic mouse where the strong polyoma virus middle T oncogene is targeted to the mammary epithelium. It is known that the PyVmT oncogene activates Src and phosphoinositide 3-kinase pathways (24). Since IL4 has also been reported to act, at least in part, through phosphoinositide 3-kinase activity (7,8), it is possible that the strong oncogene signaling rendered IL4R signaling redundant. In the carcinogen-induced model used here, the genetic lesions caused by AOM exposure typically activate Ras and Wnt pathway signaling (25), which are distinct from IL4R-activated pathways (8).
A key finding from this study was that proliferation, but not always survival, was an epithelial cell autonomous response to IL4. Several recent studies have concluded that tumor cell survival is a major function for IL4 (20,26), but since tumor formation in vivo was the end point, tumor autonomous versus host-mediated effects of IL4 were impossible to distinguish. In our AOM/DSS study, differences in tumor cell survival correlated significantly with tumor development, but since both tumor and stromal cells were deficient in IL4Rα, it is unclear which cell types were responsible for this result. Similarly, systemic blockade of IL4Rα–IL4 interactions with a chimeric protein in an implanted tumor model reduced tumor cell survival. One example from the literature where in vitro analysis did identify a cell autonomous activation of survival signaling was in colon cancer stem cells (27). It is possible that the survival effect of IL4 in vitro was only evident in our hands in the HCT116 cell line because of a difference in abundance of cells with cancer stem cell characteristics in this line compared with the other three examined. Alternatively, other aspects of the survival signaling pathway may be functional in the HCT116 cells and not the other cell lines. One other consideration is that tumor cell lines have already acquired the characteristics of cancer, which include resistance to apoptosis (28). Therefore, the survival response that can be activated by IL4 may already be active in some cell lines. Nevertheless, three of the four cell lines examined showed a significant apoptotic response to treatment with 5-flurouracil, indicating that apoptotic resistance was not optimal.
While our in vitro studies were conducted predominantly with IL4, it should be remembered that the receptor complex of which IL4Rα is a part also binds IL13. This may be particularly relevant in the setting of colitis-associated colon cancer, as IL13 has been reported to be significantly involved in the pathogenesis of ulcerative colitis (29). The colon tumor cell lines all showed some endogenous production of IL13, although at low levels (data not shown). While there are differing affinities of the IL4R complex for IL4 and IL13 (30,31) and a recent study has suggested that IL4 is the preferred ligand for the IL4R on monocytes/macrophages (32), in patients with colitis, local concentrations of IL13 may be sufficiently high to cause significant receptor activation.
In conclusion, our studies have identified a tumor-promoting role for IL4Rα in a mouse model of colitis-associated colon cancer. While both proliferation and tumor cell survival can be affected by the presence of IL4R, these processes may be mediated by different cell types within the tumor microenvironment. Since IL4R is already a clinically relevant target in other pathologies such as allergy and atopy, drugs targeting it are already in clinical development. Our data suggest that colon cancer is a rational setting for additional use of such drugs.
Funding
Vanderbilt-Ingram Cancer Center Transition to B.F. from National Institutes of Health Cancer Center Support Grant (5P30CA068485 to J.Pietenpol); Vanderbilt University Medical Center Discovery Program to B.F.; T32 CA106183 to F.L.K.
Supplementary material
Supplementary Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AOM
azoxymethane
- DSS
dextran sulfate sodium
- IL
interleukin
- IL4R
IL4 receptor
- IL4Rα
IL4R alpha
- MC38-KD
MC38-knockdown
- PCR
polymerase chain reaction
- shRNA
small hairpin RNA
References
- 1.Coussens LM, et al. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Pohl C, et al. Chronic inflammatory bowel disease and cancer. Hepatogastroenterology. 2000;47:57–70. [PubMed] [Google Scholar]
- 4.Itzkowitz SH, et al. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G7–G17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 5.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popivanova BK, et al. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J. Clin. Invest. 2008;118:560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelms K, et al. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 8.Kelly-Welch AE, et al. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300:1527–1528. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 9.Li-Weber M, et al. Regulation of IL4 gene expression by T cells and therapeutic perspectives. Nat. Rev. Immunol. 2003;3:534–543. doi: 10.1038/nri1128. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FO, et al. Macrophage activation and polarization. Front. Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 11.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 12.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumour properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.OuYang Z, et al. Interleukin-4 stimulates proliferation of endometriotic stromal cells. Am. J. Pathol. 2008;173:463–469. doi: 10.2353/ajpath.2008.071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toi M, et al. Inhibition of colon and breast carcinoma cell growth by interleukin-4. Cancer Res. 1992;52:275–279. [PubMed] [Google Scholar]
- 15.Uchiyama A, et al. Interleukin 4 inhibits hepatocyte growth factor-induced invasion and migration of colon carcinomas. J. Cell. Biochem. 1996;62:443–453. doi: 10.1002/(SICI)1097-4644(19960915)62:4%3C443::AID-JCB2%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Kanai T, et al. Regulatory effect of interleukin-4 and interleukin-13 on colon cancer cell adhesion. Br. J. Cancer. 2000;82:1717–1723. doi: 10.1054/bjoc.2000.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francipane MG, et al. Crucial role of interleukin-4 in the survival of colon cancer stem cells. Cancer Res. 2008;68:4022–4025. doi: 10.1158/0008-5472.CAN-07-6874. [DOI] [PubMed] [Google Scholar]
- 18.Barner M, et al. Differences between IL-4R alpha-deficient and IL-4-deficient mice reveal a role for IL-13 in the regulation of Th2 responses. Curr. Biol. 1998;8:669–672. doi: 10.1016/s0960-9822(98)70256-8. [DOI] [PubMed] [Google Scholar]
- 19.Ko CW, et al. Lack of interleukin-4 receptor alpha chain-dependent signalling promotes azoxymethane-induced colorectal aberrant crypt focus formation in Balb/c mice. J. Pathol. 2008;214:603–609. doi: 10.1002/path.2316. [DOI] [PubMed] [Google Scholar]
- 20.Todaro M, et al. Apoptosis resistance in epithelial tumours is mediated by tumour-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 21.Obiri NI, et al. Expression of high-affinity IL-4 receptors on human melanoma, ovarian and breast carcinoma cells. Clin. Exp. Immunol. 1994;95:148–155. doi: 10.1111/j.1365-2249.1994.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi BH, et al. In situ expression of interleukin-4 (IL-4) receptors in human brain tumours and cytotoxicity of a recombinant IL-4 cytotoxin in primary glioblastoma cell cultures. Cancer Res. 2001;61:8058–8061. [PubMed] [Google Scholar]
- 23.Herbert DR, et al. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity. 2004;20:623–635. doi: 10.1016/s1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- 24.Guy CT, et al. Induction of mammary tumours by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi M, et al. Gene mutations and altered gene expression in azoxymethane-induced colon carcinogenesis in rodents. Cancer Sci. 2004;95:475–480. doi: 10.1111/j.1349-7006.2004.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. Endogenous interleukin-4 promotes tumour development by increasing tumour cell resistance to apoptosis. Cancer Res. 2008;68:8687–8694. doi: 10.1158/0008-5472.CAN-08-0449. [DOI] [PubMed] [Google Scholar]
- 27.Todaro M, et al. Colon cancer stem cells dictate tumour growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, et al. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 29.Bouma G, et al. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 30.LaPorte SL, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terabe M, et al. Role of IL-13 in regulation of anti-tumour immunity and tumour growth. Cancer Immunol. Immunother. 2004;53:79–85. doi: 10.1007/s00262-003-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junttila IS, et al. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J. Exp. Med. 2008;205:2595–2608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.