Abstract
Deregulated estrogen signaling is evidently linked to breast cancer pathophysiology, although the role of signal transducer and activator of transcription (Stat)5a, integral to normal mammary gland development, is less clear. A mouse model of mammary epithelial cell-targeted deregulated estrogen receptor α (ERα) expression [conditional ERα in mammary epithelium (CERM)] was crossed with mice carrying a germ line deletion of Stat5a [Stat5a−/−] to investigate interactions between ERα and Stat5a in mammary tissue. CERM, CERM/Stat5a+/−, CERM/Stat5a−/−, Stat5a+/−, Stat5a−/− and wild-type (WT) mice were generated to test the roles of ERα and Stat5a on pubertal differentiation and cancer progression with and without exposure to the chemical carcinogen 7,12-dimethylbenz[a]anthracene (DMBA). Only CERM/Stat5a−/− mice demonstrated delayed pubertal terminal end bud differentiation. Without DMBA exposure, Stat5a loss abrogated ERα-initiated hyperplastic alveolar nodule (HAN) development and, similarly, Stat5a−/− mice did not develop HANs. However, although Stat5a loss still reduced ERα-initiated HAN prevalence following DMBA exposure, Stat5a loss without deregulated ERα was associated with an increased HAN prevalence compared with WT. Progression to ERα(+) and ERα(−) adenocarcinoma was found in all CERM-containing genotypes (CERM, CERM/Stat5a+/−, CERM/Stat5a−/−) and ERα(+) adenocarcinoma in the Stat5a−/− genotype. The mammary epithelial cell proliferative index was increased only in CERM mice independent of Stat5a loss. No differences in apoptotic indices were found. In summary, Stat5a cooperated with deregulated ERα in retarding pubertal mammary differentiation and contributed to ERα-initiated preneoplasia, but its loss did not prevent development of invasive cancer. Moreover, in the absence of deregulated ERα, Stat5a loss was associated with development of both HANs and invasive cancer following DMBA exposure.
Introduction
Deregulation of estrogen signaling is linked to breast cancer development (1). Although most human breast cancers are estrogen receptor α (ERα)-positive, ERα-negative breast cancers also appear and carry a worse prognosis. Breast cancers are believed to arise from preneoplastic lesions including atypical ductal and lobular hyperplasia and ductal carcinoma in situ, which can be markers for later invasive cancer development (2). Open questions remain on the role of deregulated estrogen signaling in ERα-negative cancers and the relationship between preneoplasia and invasive breast cancer (3).
In the mouse mammary gland, ductal hyperplasia and hyperplastic alveolar nodules (HAN) are preneoplastic lesions that mimic human preneoplasia (4). The conditional ERα in mammary epithelium (CERM) mouse model with deregulated ERα targeted to mammary epithelium is one of the few breast cancer mouse models that illustrate ERα-positivity in preneoplasia (5,6). Experiments utilizing this model have shown that cyclin D1 plays a critical survival role for mammary cells with aberrant ERα expression (7), deregulated ERα in combination with Simian Virus 40 T antigen oncogene produces ERα-positive mammary adenocarcinoma (8) and both ERα-positive and ERα-negative mammary adenocarcinomas develop when deregulated ERα is combined with Brca1 loss and p53 haploinsufficiency (9).
The polycyclic hydrocarbon 7,12-dimethylbenz[a]anthracene (DMBA) is the most commonly employed chemical carcinogen to study chemically induced mammary gland carcinogenesis in mice (10). DMBA induces the development of mammary ductal hyperplasia and HANs, as well as adenocarcinomas and adenosquamous carcinomas (4,11).
Puberty follows maturation of the hypothalamo–pituitary–gonadal axis with estrogen signaling stimulating mammary ductal elongation driven by highly proliferative mammary ductal structures called terminal end buds (TEBs) (12). TEBs are composed of proliferating cells that differentiate into myoepithelial and luminal cell lineages as the TEBs progress through the mammary fat pad. By the end of puberty, TEBs reach the end of the fat pad and differentiate into terminal ductal ends (13).
Signal transducer and activator of transcription (Stat)5a and Stat5b are mediators of the prolactin/Jak2 pathway contributing to differentiation and survival of normal mammary lobuloalveolar cells. Stat5a and Stat5b are homologs with Stat5b arising from a recent gene duplication and showing 96% protein homology with Stat5a (14). Stat5a is the predominant homolog expressed in mammary epithelial cells (15,16). During first pregnancy, Stat5a−/− mice demonstrate defective lobuloalveolar development and impaired lactation (16). Rescue of this defect with subsequent pregnancies correlates with increased Stat5b protein expression and activity (17). When both Stat5a and Stat5b are absent from mammary epithelial cells prior to pregnancy, there is no lobuloalveolar development and when Stat5a/b are conditionally deleted from alveolar cells after pregnancy, the cells undergo apoptosis (18).
Nuclear-localized Stat5a is found in 40% of human ductal carcinoma in situ lesions (19) and 76% of invasive breast cancers in association with higher levels of differentiation (20). Stat5a/b is highly activated in human breast cancers (21) and associated with a better prognosis (22). Studies in human breast cancer cell models showed that Stat5a/b reverses epithelial–mesenchymal transition and inhibits invasion (23,24) and that Stat5a expression increases with transition from preinvasive to invasive (25) and that expression of a Stat5 dominant-negative construct leads to apoptosis and a decrease in tumor size (26). Potential cross-talk between ERα and Stat5a has been shown in both normal and breast cancer cells (27–29).
In mouse models, Stat5a is a survival factor whose overexpression leads to cancer. Loss of Stat5a in the whey acidic protein-transforming growth factor alpha mouse model promotes apoptosis and decreases mammary hyperplasia development (30). Loss of one Stat5a allele in the whey acidic protein-simian virus 40 T antigen mouse model increases apoptotic rates in mammary adenocarcinomas and postpones mammary cancer progression (31). Constitutively activated Stat5 slows down involution and predisposed the multiparous mammary gland to tumor formation (32). Overexpression of Stat5a as well as a dominant-negative Stat5 construct in mammary epithelial cells gland in transgenic mice leads to mammary cancer (33) compatible with both cancer-promoting and cancer-protective activities.
This study was initiated to test the hypothesis that loss of Stat5a alters ERα-initiated preneoplasia progression and cancer development. Results revealed that Stat5a contributed to the development of ERα-initiated preneoplasia but that its loss, even in the absence of deregulated ERα expression, was associated with chemically induced carcinogenesis implying that the impact of Stat5a loss on carcinogenesis was context dependent.
Materials and methods
Mouse models, characterization and DMBA administration
Double-transgenic CERM mice carrying a mouse mammary tumor virus-long terminal repeat-reverse tetracycline responsive transactivator (rtTA) transgene (34) and a tetracycline responsive promoter-FLAG-ERα transgene (6) were crossed with Stat5a deficient (Stat5a−/−) mice with a disrupted Stat5a locus lacking the first two protein coding exons (16) to generate female CERM (n = 50), CERM/Stat5a+/− (n = 67), CERM/Stat5a−/− (n = 59), wild-type (WT) (n = 60), Stat5a+/− (n = 43) and Stat5a−/− (n = 28) mice on a C57Bl/6 background. All genotypes demonstrated similar sizes, activities, puberty onset and fertility. Puberty onset was measured by observing for vaginal opening starting at age 3 weeks until complete vaginal canalization (35). Mice were maintained on a 12 h light–dark cycle with a doxycycline-containing diet (Bio-Serv, Frenchtown, NJ) and water ad libitum. The Georgetown University Animal Care and Use Committee and Institutional Biosafety Committee approved all procedures. For genotype analysis, ear DNA was extracted (36) and 2 μl DNA was amplified in 20 μl polymerase chain reaction (PCR) reaction solution [1× PCR buffer (Sigma, St Louis, MO), 0.2 mM dNTPs (Invitrogen, Carlsbad, CA), 1 U/μl RedTaq polymerase (Sigma), forward (F) and reverse (R) primers for FLAG-ERα 0.38 μM F: 5′-CGAGCTCGGTACCCGGGTCG-3′ and R: 5′-GAACACAGTGGGCTTGCTGTTG-3′, for rtTA 0.38 μM F: 5′-ACTAAGTCATCGCGATGGAGCA-3′ and R: 5′-CTCGATGGTAGACCCGTAATTG-3′, for Stat5a−/− 0.612 μM F: 5′-GAACAAACAGATCATCCGCAA-3′ and R: 5′-TTCGTCCAGATCATCCTGATC-3′ and for Stat5aWT 0.55 μM F: 5′-CTGGATTGACGTTTCTTACCTG-3′ and R: 5′-TGGAGTCAACTAGTCTGTCTCT-3′] in a thermocycler: FLAG-ERα (35 cycles 94°C 60 s, 57°C 90 s, 72°C 120 s), rtTA (35 cycles 94°C 60 s, 55°C 90 s, 72°C 120 s) and Stat5a (94°C 30 s, 40 cycles 98°C 35 s, 58°C 60 s, 72°C 105 s). Alternatively, genotyping was performed from tails by TransnetYX (Memphis, TN) using the probe 5′-GCACTCAGCGCTGTGG-3′ with forward primer 5′-TGCCAACAAGGTTTTTCACTAGAGA-3′ and reverse primer 5′-CTCTTGATCTTCCAATACGCAACCTA-3′ to detect rtTA transgene, the probe 5′-CTTAATTCCGGAGTGT-3′ with forward primer 5′-GGAGAAGAGTTTGTGTGCCTCAA-3′ and reverse primer 5′-AAGGTGCTGGACAGAAACGT-3′ to detect FLAG-ERα transgene, two probes 5′-ACCTGTCCGGTGCCC-3′ and 5′-CCCCTCTGATAGCTCC-3′ with two forward primers 5′-GGGCGCCCGGTTCTT-3′ and 5′-GGTGGTGTCTCTGGGAAAGG 3′ and two reverse primers 5′-CCTCGTCCTGCAGTTCATTCA-3′ and 5′-ACCACAGCTGTATAAGCATCAAAGT-3′ to detect WT, Stat5a+/− and Stat5a−/− mice. Real-time reverse transcription–polymerase chain reaction (RT–PCR) of RNA extracted from mammary tissue confirmed comparable levels of FLAG-ERα messenger RNA expression in CERM, CERM/Stat5a+/− and CERM/Stat5a−/− mice and up to 2-fold increased expression levels of total ERα messenger RNA with an increased percentage of mammary epithelial cells demonstrating nuclear-localized ERα in CERM (6), CERM/Stat5a+/− and CERM/Stat5a−/− mice compared with WT. Numbers of WT, CERM, CERM/Stat5a+/−, CERM/Stat5a−/−, Stat5a+/− and Stat5a−/− mice: TEB studies at 2, 4 and 12 months (n = 5, 5, 11, 8, 5, 8; n = 19, 7, 13, 18, 17, 3; n = 10, 18, 25, 21, 8, 7, respectively); HAN prevalence in untreated mice: n = 18, 19, 25, 22, 8, 6; HAN/cancer prevalence following DMBA exposure: n = 8, 10, 9, 9, 9, 8. Numbers of WT, CERM, CERM/Stat5a+/−, CERM/Stat5a−/− for western blots (WB), immunohistochemistry and immunoprecipitation (IP)/WB, respectively: n = 0, 6, 6, 5; n = 0, 3, 0, 3; n = 1, 3, 2, 3. DMBA (D3254, Sigma) was dissolved in corn oil to a 10 μg/μl final concentration. Four-month-old mice received a single 1 mg DMBA/animal intragastric dose (10). Mice were necropsied between 10.5 and 12 months of age subsequent to weekly clinical examination until age 12 months or when tumor reached 1 cm3 or other health issues required euthanasia.
Morphology and histology
For each mouse, one abdominal mammary gland whole mount was prepared, with fixation of the second abdominal mammary gland and any tumors in 10% buffered formalin for hematoxylin and eosin-stained section preparation (6). Ductal length [measured longest two to three primary ducts from the nipple to their end close to the end of the fat pad using Metamorph Image Analysis version 6.3r2 (Downingtown, PA)], TEBs (number and size: defined as bulbous shaped terminal ends >10 000 μm2 measured by Metamorph Image Analysis version 6.3r2) and HANs (4) were evaluated on whole mounts from 2-, 4- and 12-month-old mice. Histology of hyperplasia, ductal carcinoma in situ and invasive cancers were evaluated on hematoxylin and eosin-stained sections. Images were captured using a Nikon Eclipse E800M microscope, DMX1200 camera with the ACT-1 Version 2.7 program (Nikon Corporation, Melville, NY) and assembled in Adobe Photoshop version 6.0 (San Jose, CA).
RNA expression studies
For real-time RT–PCR, total RNA was prepared from snap-frozen thoracic mammary gland tissue using Trizol extraction (Invitrogen). Complementary DNA (cDNA) was synthesized as per manufacturer's instructions (SuperScript™ Reverse Transcriptase; Invitrogen). TaqMan® Gene Expression Assays-On-Demand [Applied Biosystems (ABI), Foster City, CA] detected 18S (Hs99999901_s1), total ERα (transgene and endogenous) (Mm00433149_m1), Stat5b (Mm00839889_m1), β-casein (Mm00839664_m1), Lemd3 (Mm01156598_m1), Sstr1 (Mm00436679_s1), Stat1 (Mm00439518_m1), Stat3 (Mm00456961_m1) and transgene FLAG-tagged ERα (forward primer: 5′-CCCCGGGAGATCTGTGAAC-3′; reverse primer: 5′-TGTGAAGGGTCATGGTCATATGTTT-3′; Reporter: 5′-CCATGGACTACAAAGACG-3′). Gene amplification was performed as per manufacturer's instructions (TaqMan Universal PCR Master Mix; Roche, Basel, Switzerland) on a 7900 HT Fast Real-Time PCR System and analyzed with the ABI Prism SDS Version 2.1 program. For cDNA microarray analyses, RNA was prepared as above from thoracic mammary glands of 12-month-old WT (n = 4), CERM (n = 4) and CERM/Stat5a−/− (n = 4) mice. MOgene (St Louis, MO) prepared cDNA and performed microarrays using whole mouse genome 4x44K Agilent gene expression microarrays.
Immunohistochemistry
Immunohistochemical detection was performed on paraffin-embedded mammary gland sections following manufacturer's instructions (rabbit VectaStain or Mouse-On-Mouse kit; Vector Labs, Burlingame, CA). Antigen retrieval was performed either in a decloaker using BORG retrieval solution (Biocare Medical, Concord, CA) or by immersion at 98°C for 20 min in 10 mM citrate buffer (pH 6.0) with 0.05% Tween. Primary antibodies: Stat5a (L20 clone; sc-1081, Santa Cruz, Santa Cruz, CA), Ki67 (NCL-Ki67-MM1; Novocastra, Logan, UT), cyclin D1 (RM-9104; Neomarkers, Fremont, CA), Stat5b (sc-1656; Santa Cruz), ERα (sc-542; Santa Cruz) and progesterone receptor (PR; sc-538; Santa Cruz). Apoptotic cells were detected by terminal deoxytransferase-mediated deoxyuridine nick end-labeling assay following the ApopTag® peroxidase in situ apoptosis detection kit's instructions (Chemicon International, Billerica, MA). The numbers of mammary epithelial cells showing immunoreactivity for the ApopTag assay, nuclear-localized Ki67 for proliferation and nuclear-localized cyclin D1 were counted out of a total of 1000 cells and expressed as a percentage of positive cells.
Protein expression studies
For WB and IP/WB studies, frozen mammary gland tissue was homogenized in 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EGTA, 1% IGEPAL, 40 mM β-glycerophosphate Na, 50 mM NaF, 20 mM Na pyrophosphate, 2 mM sodium orthovanadate, 1 complete Protease Inhibitor Cocktail tablet (Roche) and centrifuged. Protein concentration was measured using BCA™ Protein Assay Kit (Pierce, Rockford, IL). Protein lysate was boiled for 3 min. Proteins were separated on 8–10% Tris–Glycine gels (Novex, Carlsbad, CA) adjacent to full Range Rainbow™ protein molecular weight markers (Amersham Bioscience, Fairfield, CT), transferred onto Immobilon-P polyvinylidene difluoride transfer membrane (Millipore, Billerica, MA), blocked in 3–5% bovine serum albumin (Sigma) followed by incubation with Stat5b (sc-1656; Santa Cruz) primary antibodies and then with anti-mouse monoclonal secondary antibody (Jackson Laboratories, Bar Harbor, MI). Proteins were detected using SuperSignal® West Dura Extended Duration Substrate (Pierce). Blots were stripped 30 min at 60°C [0.2% sodium dodecyl sulfate, 0.0625 M Tris (pH 6.7), 0.7% β-mercaptoethanol], reprobed with anti-mouse monoclonal secondary antibody to verify stripping of proteins and reprobed with actin (sc1616; Santa Cruz). For IP studies, protein lysate was prepared as above, mixed with 0.25% Na deoxycholate and 0.1% sodium dodecyl sulfate 50 min at 4°C and precleared by incubation with control isotype antibody mouse IgG1 (X0931, Dako, Carpinteria, CA) and protein A/G PLUS-Agarose beads (sc-2003; Santa Cruz) for 30 min at 4°C. Five hundred microgram protein from the precleared supernatant was incubated with 2 μg of Stat5b antibody (sc-1656, Santa Cruz) for 1 h at 4°C followed by incubation with 20 μl protein A/G PLUS-Agarose beads for 1 h at 4°C. Supernatants were loaded on gel to verify incomplete Stat5b depletion after IP. Proteins were separated on 8–10% gels (Novex). Membranes were blocked in 3–5% bovine serum albumin (Sigma) followed by incubation with 1 μg/ml AX1 anti-phosphoStat5a/b primary antibody (Advantex BioReagents, El Paso, TX) and then incubation with anti-mouse secondary antibody (Jackson Laboratories) with protein detection as above. Blots were stripped 30 min at 60°C, reprobed with anti-mouse secondary antibody to confirm protein stripping and reprobed with Stat5b antibody (sc-1656; Santa Cruz), followed by incubation with anti-mouse secondary antibody.
Statistical analysis
Statistical analyses were performed with GraphPad Prism version 4 for Windows (La Jolla, CA). One-tailed Student's t-test with Welch's correction was used to compare TEB number and nuclear-localized cyclin D1 indices between genotypes. Two-tailed Student's t-test was used to compare proliferative and apoptotic indices between genotypes. TEB, HAN and invasive cancer prevalence were compared using Fisher's Exact Test. Analysis of variance was used to compare the day of vaginal opening between genotypes. TEB number, nuclear-localized cyclin D1, proliferative and apoptotic indices, and day of vaginal opening are presented as the mean ± SEM. Statistical significance was reached when P < 0.05.
Results
Loss of Stat5a in combination with deregulated ERα led to delayed TEB differentiation
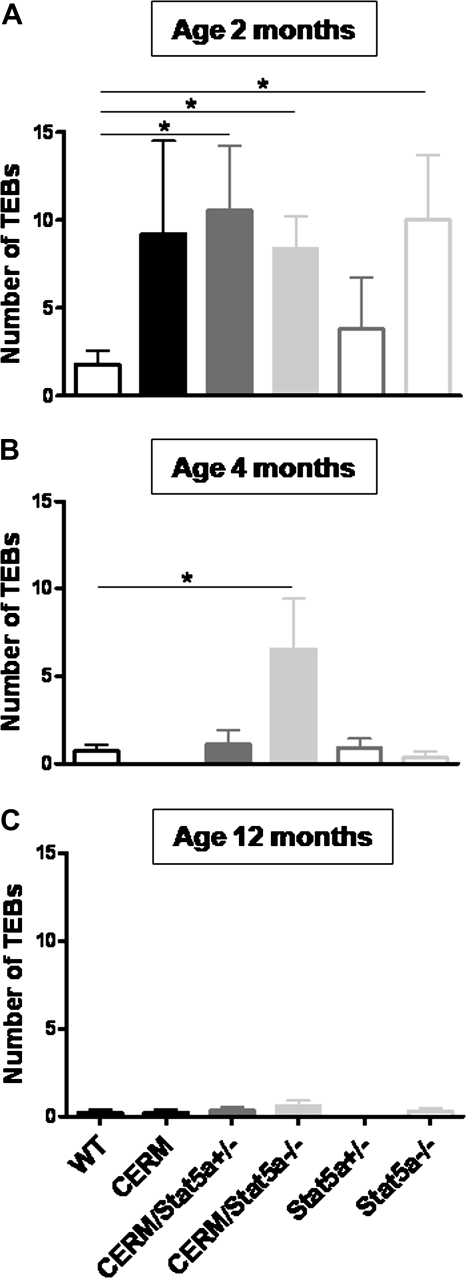
To test whether loss of Stat5a and/or deregulated ERα altered TEB differentiation, the presence, number and size of TEBs at 2, 4 and 12 months of age in CERM/Stat5a−/−, CERM/Stat5a+/−, CERM, Stat5a+/−, Stat5a−/− and WT mice were measured. Although CERM, CERM/Stat5a−/−, CERM/Stat5a+/− and Stat5a−/− mice demonstrated a higher number of TEBs compared with WT mice at 2 months of age (9.2 ± 5.3, 8.4 ± 1.8, 10.55 ± 3.7 and 10.0 ± 3.7 compared with 1.8 ± 0.8, P = 0.1, 0.005, 0.02 and 0.03, respectively), only CERM/Stat5a−/− mice demonstrated a significantly higher number of TEBs compared with WT mice at 4 months of age (CERM/Stat5a−/−: 6.6 ± 2.9 compared with WT: 0.7 ± 0.3, P = 0.03; Figure 1). By 12 months of age, the number of TEBs in CERM/Stat5a−/− mice was no longer increased. The size distribution of TEBs at any age did not significantly differ between CERM/Stat5a−/− and WT mice nor were there any differences in the length of ductal extension through the fat pad. Stat5a immunohistochemistry determined that all cells in 2-month-old CERM TEBs were nuclear Stat5a positive. The onset of vaginal opening as a marker of puberty initiation was not significantly different between CERM/Stat5a−/− (30.0 ± 1.9 days) and WT (29.7 ± 0.6 days) mice. In conclusion, the combination of Stat5a loss with deregulated ERα led to an increase in TEB number persisting through 4 months of age that was not attributable to differences in puberty onset or altered kinetics of ductal elongation.
Fig. 1.
Loss of Stat5a alleles in combination with deregulated ERα led to a delayed terminal end bud differentiation. Graphs representing the number of TEBs in WT, CERM, CERM/Stat5a+/−, CERM/Stat5a−/−, Stat5a+/− and Stat5a−/− mice at 2 (A), 4 (B) and 12 months of age (C). Horizontal lines between genotypes demonstrating statistically significant differences in TEB number (*P < 0.05); mean ± SEM indicated.
Loss of Stat5a reduced the prevalence of HANs induced by deregulated expression of ERα without altering rates of either proliferation or apoptosis
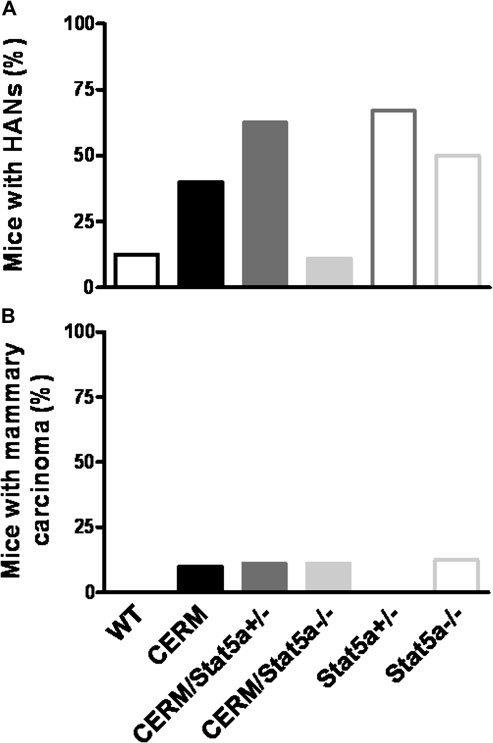
To test whether or not loss of Stat5a or deregulated ERα altered development of mammary preneoplasia, the presence or absence of HANs was compared at 12 months of age in CERM/Stat5a−/−, CERM/Stat5a+/−, CERM, Stat5a+/−, Stat5a−/− and WT mice. By itself, expression of deregulated ERα was found to increase HAN prevalence. A significantly higher percentage of CERM (37%) as compared with WT (5.5%) mice demonstrated HANs at 12 months of age (P < 0.05; Figure 2A and B) and the proliferative hyperplasia exhibited nuclear-localized Stat5a (Figure 2C). Significantly, loss of Stat5a in CERM mice reduced HAN prevalence to zero (P < 0.01) compared with CERM. Proliferative and apoptotic indices were measured to determine if changes in these measures could be correlated with the reduced preneoplasia prevalence. As shown previously (6), the proliferative index of mammary epithelial cells was statistically significantly increased in CERM mice compared with WT mice but loss of one or two Stat5a alleles did not alter this ERα-associated increase (Table I). The apoptotic index was not changed by either the presence or absence of Stat5a (WT: 1.6 ± 0.5%, CERM: 1.3 ± 0.4%, CERM/Stat5a+/−: 1.9 ± 1.2% and CERM/Stat5a−/−: 1.0 ± 0.15%, P > 0.05). A cDNA microarray analysis on whole mammary gland tissue from CERM/Stat5a−/−, CERM and WT mice was performed to determine if an altered gene signature could be defined in the CERM/Stat5a−/− mice but none was found. A reduction in β-casein gene expression was detected and confirmed by real-time RT–PCR in agreement with previous reports on Stat5a−/− mice (16). In contrast, real-time RT–PCR did not demonstrate significant changes in expression levels for other candidate genes from the array analysis including Lemd3, Sstr1, Stat1 and Stat3. In summary, loss of Stat5a reduced ERα-initiated HAN prevalence without impacting rates of cellular proliferation or apoptosis.
Fig. 2.
Deregulated ERα led to an increase in HAN prevalence, which was abrogated with loss of Stat5a alleles. (A) Graph representing the percentage of mice with HANs at 12 months of age in nulliparous WT, CERM, CERM/Stat5a+/−, CERM/Stat5a−/−, Stat5a+/− and Stat5a−/− mice. Horizontal lines between genotypes demonstrating statistically significant differences in TEB number (*P < 0.05); mean ± SEM indicated. (B) Representative mammary gland whole mounts of WT, CERM, CERM/Stat5a+/− and CERM/Stat5a−/− mice at 12 months of age (bars represent 2 mm). Inserts represent areas of mammary gland with HANs in CERM and CERM/Stat5a+/− mice or without HAN in WT and CERM/Stat5a−/− mice. Arrows indicate HANs. (C) Representative hematoxylin and eosin of HAN from CERM mouse (left panel, bar represents 100 μm). HAN is positive for nuclear-localized Stat5a and Ki67 by immunohistochemistry (right panels, bars represent 20 μm).
Table I.
Mammary epithelial cell proliferative index in WT, CERM, CERM/Stat5a+/− and CERM/Stat5a−/− mice at 12 months of age
| Genotypes | Proliferative index in mammary epithelial cells (%) | |
| CERM versus WT | 13.3 ± 2.1 versus 4.1 ± 0.9 | P < 0.05 |
| CERM versus CERM/Stat5a+/− | 13.3 ± 2.1 versus 16.1 ± 2.7 | P > 0.05 |
| CERM versus CERM/Stat5a−/− | 13.3 ± 2.1 versus 10.2 ± 2.2 | P > 0.05 |
Although loss of Stat5a reduced HAN prevalence in mice with deregulated expression of ERα exposed to DMBA, cancer development was not prevented
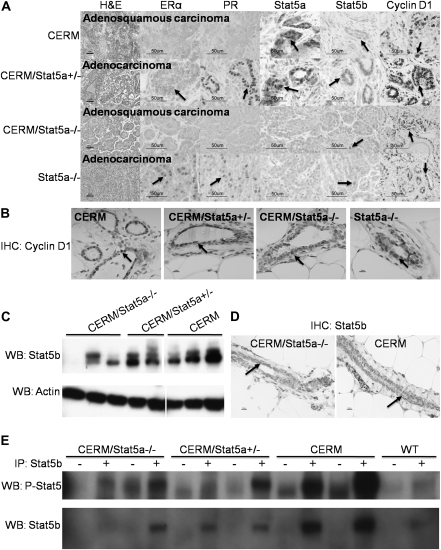
To more directly compare the carcinogenic susceptibility of mammary epithelial cells from the different genotypes, we analyzed HAN and cancer development at 12 months of age following DMBA exposure at 4 months of age in CERM/Stat5a−/−, CERM/Stat5a+/−, CERM, Stat5a+/−, Stat5a−/− and WT mice (Figure 3). Four months of age was selected for the exposure time point due to the persistent presence of TEBs in 4-month-old CERM/Stat5a−/− mice. As with the non-DMBA-exposed mice, a higher percentage of CERM (40%) compared with WT (12.5%) mice demonstrated HANs at 12 months of age and loss of Stat5a reduced HAN prevalence (11%) (Figure 3A). However, cancer development was not prevented (Figures 3B and 4). Unexpectedly, HAN prevalence also was higher in Stat5a+/− (67%) and Stat5a−/− (50%) as compared with WT following DMBA exposure and cancer developed in Stat5a−/− mice (Figures 3B and 4). The histology and expression patterns of ERα, PR, Stat5a, Stat5b and cyclin D1 were compared in the different cancers (Figure 4). The adenocarcinomas from the CERM/Stat5a+/− and Stat5a−/− mice were ERα/PR positive, whereas the adenosquamous carcinomas identified in the CERM and CERM/Stat5a−/− mice were ERα/PR-negative (Figure 4A). As expected, Stat5a was undetectable in the cancers developing in the CERM/Stat5a−/− and Stat5a−/− mice but nuclear-localized Stat5b was found in all cancers, as was cyclin D1. The percentage of cancers cells demonstrating nuclear-localized cyclin D1 (76.3 ± 9.8%; Figure 4A) was significantly higher than that found in normal appearing mammary epithelial cells (33.3 ± 4.5%, P < 0.001; Figure 4B). Since Stat5b was expressed in all cancers, levels of Stat5b expression in normal appearing mammary epithelial cells and mammary tissue from CERM, CERM/Stat5a+/− and CERM/Stat5a−/− mice were measured to determine if loss of Stat5a was directly associated with increased Stat5b expression and activity but none were found (Figure 4C–E). In conclusion, the reduction of HAN prevalence found in mice with deregulated ERα with loss of Stat5a did not prevent cancer development, and loss of Stat5a by itself without deregulated ERα was associated with an increase in HAN prevalence and cancer development after DMBA exposure.
Fig. 3.
Prevalence of HANs and mammary adenocarcinomas following DMBA exposure at 4 months of age. Graphs representing the percentage of mice with HANs (A) and mammary adenocarcinoma (B) in the mammary gland of DMBA-treated WT, CERM, CERM/Stat5a+/−, CERM/Stat5a+/−, Stat5a+/− and Stat5a−/− mice; mean ± SEM indicated.
Fig. 4.
Characterization of ERα, PR, Stat5a, Stat5b and cyclin D1 expression. (A) Representative histological sections of DMBA-induced mammary carcinomas stained with hematoxylin and eosin and immunohistochemistry for ERα, PR, Stat5a, Stat5b and cyclin D1 of DMBA-induced mammary carcinomas. Adenocarcinomas and adenosquamous carcinomas are indicated. Bars represent 50 μm. Arrows indicate epithelial cells with representative staining. (B) Representative sections of cyclin D1 labeled normal appearing ducts following DMBA exposure. Bars represent 10 μm. Arrows indicate epithelial cells with representative staining. (C) Western blot of Stat5b expression (92 kDa) using protein lysates from the mammary glands of CERM/Stat5a−/−, CERM/Stat5a+/− and CERM mice. Actin was used as loading control. Each lane represents one separate mouse. (D). Representative Stat5b immunohistochemistry in CERM and CERM/Stat5a−/− mammary glands. Arrows indicate epithelial cells with representative staining. (E). Immunoprecipitation of Stat5b (92 kDa) from mammary glands of CERM/Stat5a−/−, CERM/Stat5a+/−, CERM and WT mice was performed with (+) and without (−) Stat5b antibody. Blot was probed with phospho-Stat5, followed by western blot with Stat5b after complete stripping of proteins. Each lane represents one separate mouse.
Discussion
Only the combination of deregulated ERα and loss of Stat5a resulted in a significant delay in TEB differentiation. Stat5a is linked to terminal differentiation of mammary epithelial cells during pregnancy (15–17). Mammary epithelial transplant studies reveal an ‘ample’ TEB number in the Stat5a−/−-transplanted epithelium of 11-week-old recipient mice and showed the effects of Stat5a deficiency on differentiation are restricted to the epithelium (17). Here, we demonstrate for the first time that deregulated ERα can collaborate with Stat5a deficiency to impair differentiation of TEB mammary epithelial cells.
HANs have been likened to human hyperplastic epithelial lobular units (4). Deregulated ERα expression significantly increased HAN prevalence, nuclear-localized Stat5a was expressed in the lesions, and Stat5a loss abrogated HAN development in nulliparous mice, suggesting that Stat5a-mediated signaling pathways contribute to ERα-initiated preneoplasia. Previous studies in which Stat5a loss compromised cancer progression initiated by non-hormonal mechanisms implicated impaired mammary epithelial cell survival as a mechanism (30,31). We found apoptotic rates low in the mammary epithelial cells of mice with deregulated ERα that were not further reduced by Stat5a loss. This does not exclude the possibility that Stat5a loss may selectively impair survival of HAN progenitor cells. Stat5a also is known to mediate cell proliferation in hematopoietic cells (37,38). Deregulated ERα significantly increased mammary epithelial cell proliferation (6) but this was not reduced by loss of Stat5a in the normal appearing mammary epithelium. Therefore, Stat5a was not required for the abnormal increase in proliferation initiated by deregulated ERα, at least in the normal appearing mammary epithelial cells. It does not exclude that Stat5a loss could lead to significant changes in cell proliferation only in hyperplasia or may act to extend the cell cycle, which could explain the delay in TEB differentiation. Even though Stat5a is a transcription factor, the gene signature was not changed except for downregulation of β-casein. This does not rule out the involvement of altered gene transcription but does indicate that differences were not profound and may involve only a subpopulation of mammary epithelial cells. Loss of Stat5a has a defined role on lobuloalveolar but not ductal development (39), possibly explaining the paucity of gene expression changes in the non-pregnant mammary glands composed primarily of ductal epithelium. Moreover, the mice were not exposed to increased levels of prolactin or other growth factors through pregnancy or exogenous stimulation. It is possible that fewer gene expression changes are found when Stat5a is only basally activated in ductal cells than when Stat5a is highly activated by prolactin or other ligands in lobuloalveolar cells.
The experiments excluded changes in apoptosis, proliferation or measurable gene transcription as reasons for the absence of HANs in the CERM/Stat5a−/− mice leaving open the hypothesis that HAN precursor cells could selectively require Stat5a for their survival or proliferation. Despite being highly conserved, Stat5a and Stat5b exert non-redundant functions (40) and exhibit different potencies on similar effects (41). However, in the mammary gland, Stat5b activation through serial pregnancy has been shown to rescue the lactational defect of Stat5a−/− mice indicating that Stat5b can compensate for at least some functions of Stat5a in mammary epithelium (17). If the same were true for HAN precursor cells, then HANs would be predicted to develop in serially pregnant CERM/Stat5a−/− mice, which occurred (data not shown), supporting the concept that either of the two Stat5 homologs could support HAN development if sufficiently activated. Stat5 may contribute to the development of specific mammary cancer precursor cell lineages in a manner paralleling the established role of Stat5 in determining normal hematopoietic and mammary epithelial cell lineages (39,42–44).
The experimental results led to a conundrum. Loss of Stat5a in the context of deregulated ERα delayed TEB differentiation, a condition hypothesized to increase cancer risk (11), but Stat5a loss decreased the risk of preneoplasia development in nulliparous mice. To more rigorously test if Stat5a loss was cancer protective, 4-month-old mice were exposed to DMBA, the time point when impaired TEB differentiation was documented. HAN prevalence was reduced with loss of Stat5a in the mice with deregulated ERα but not absent. This is consistent with the notion that Stat5a might play a supportive role in survival or proliferation of HAN precursor cells but also indicated that Stat5a is not mandatory. This suggests that other factors could compensate for its absence. One compensating factor could be Stat5b (17) acting to increase cyclin D1, a Stat5a and Stat5b target gene (45–48). Nuclear-localized Stat5b was documented in all cancers and the percentage of cells with nuclear-localized cyclin D1 was significantly increased in cancers as compared with normal epithelium. The increased percentage of cells with cyclin D1 expression in the cancers is consistent with the fact that the percentage of cyclin D1-positive cells is higher in hyperplasia than normal epithelium in CERM mice (6). Cyclin D1 loss in CERM mice interrupts mammary epithelial cell survival (7). Cyclin D1 may play a role with Stat5 in development of the hyperplasias and cancers studied here.
DMBA exposure experiments revealed that Stat5a loss did not absolutely protect from cancer promotion and that the impact of Stat5a loss was context dependent. When deregulated ERα was present, Stat5a loss reduced HAN development but in the framework of normal mammary epithelial cells, Stat5a loss increased HAN development and cancer appeared. This could relate to a differentiation defect in the Stat5a−/− mice, independent from TEB differentiation, that increased susceptibility to DMBA-induced carcinogenesis (49). It also illustrated that cancer formed even though enzyme activity required for DMBA metabolism into the active form might be decreased by Stat5a loss (50). The reason Stat5a loss resulted in a different HAN prevalence following DMBA treatment dependent on the presence or absence of deregulated ERα is not clear. However, mammary cells expressing deregulated ERα demonstrate a differential dependence upon cyclin D1 for survival that is not found in normal mammary cells (7) and this may represent another example of how mammary epithelial cells with deregulated ERα react differently to genetic changes than normal mammary epithelial cells.
Both ERα/PR-positive and ERα/PR-negative mammary cancers developed in the mice with deregulated ERα, similar to when deregulated ERα is combined with Brca1 loss and p53 haploinsufficiency (9). ERα/PR-positive adenocarcinoma developed with Stat5a−/− loss and no deregulated ERα. The origins of ERα/PR-negative mammary cancers and their relationship to estrogen signaling are an active area of investigation with evidence for generation from either ERα-positive precursor (51) or ERα-negative precursor (52) cells. These mouse models are tools to further investigate the origins of ERα-positive and ERα-negative cancers.
In summary, the impact of Stat5a loss on mammary carcinogenesis was context dependent (Figure 5). Although absence of Stat5a in the background of deregulated ERα reduced the prevalence of preneoplasia, this did not extend to protection from DMBA-induced cancer. In the absence of deregulated ERα, Stat5a loss appeared to increase susceptibility to carcinogen-induced preneoplasia and was associated with cancer development. It is possible that the same interplay may be found in the human breast. This will need to be explored if and when Stat5a targeted therapies for humans become available.
Fig. 5.
Stage-dependent effects of Stat5a with respect to differentiation, hyperplasia and cancer formation in mouse models with and without ERα deregulation. Deregulated ERα (A) and loss of Stat5a demonstrated persistent TEBs by 4 months of age, abrogation of HAN development at 12 months of age and upon DMBA insult, mammary carcinoma development at 12 months of age. In contrast, loss of Stat5a without deregulated ERα (B) did not demonstrate persistent TEBs at 4 months of age; however, upon DMBA insult, it did lead to mammary carcinoma development. All groups demonstrated normal terminal ductal ends. Groups that demonstrated abnormal findings including persistent TEBs, HANs and cancer are indicated. For TEBs: normal: (≤1%), +: (1.1-2.5%), ++: (2.6-5%), +++: (5.1-7.5%), ++++: (7.6-10%). For HANs: no HAN: (0%), +: (0.1-15%), ++: (15.1-25%), +++: (25.1-35%) and ++++: (≥35.1%).
Funding
National Cancer Institute, National Institutes of Health (1RO1CA112176) to P.A.F.; Department of Defense Breast Cancer Research Program (W81XWH-05-1-0271) to A.M.M.
Acknowledgments
We thank the Histopathology, the Animal Research and the Biostatistics and Bioinformatics Shared Resources at the Lombardi Comprehensive Cancer Center for their assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- cDNA
complementary DNA
- CERM
conditional ERα in mammary epithelium
- DMBA
7,12-dimethylbenz[a]anthracene
- ERα
estrogen receptor α
- F
forward
- HAN
hyperplastic alveolar nodule
- IP
immunoprecipitation
- PCR
polymerase chain reaction
- PR
progesterone receptor
- R
reverse
- rtTA
reverse tetracycline responsive transactivator
- RT–PCR
reverse transcription–polymerase chain reaction
- Stat
signal transducer and activator of transcription
- TEB
terminal end bud
- WB
western blots
- WT
wild-type
References
- 1.Speirs V, et al. New perspectives into the biological and clinical relevance of oestrogen receptors in the human breast. J. Pathol. 2007;211:499–506. doi: 10.1002/path.2130. [DOI] [PubMed] [Google Scholar]
- 2.Allred DC, et al. Biological features of premalignant disease in the human breast. J. Mammary Gland Biol. Neoplasia. 2000;5:351–364. doi: 10.1023/a:1009573710675. [DOI] [PubMed] [Google Scholar]
- 3.Kuerer HM, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. J. Clin. Oncol. 2009;27:279–288. doi: 10.1200/JCO.2008.18.3103. [DOI] [PubMed] [Google Scholar]
- 4.Medina D. Premalignant and malignant mammary lesions induced by MMTV and chemical carcinogens. J. Mammary Gland Biol. Neoplasia. 2008;13:271–277. doi: 10.1007/s10911-008-9086-4. [DOI] [PubMed] [Google Scholar]
- 5.Allred DC, et al. The relevance of mouse models to understanding the development and progression of human breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;13:279–288. doi: 10.1007/s10911-008-9093-5. [DOI] [PubMed] [Google Scholar]
- 6.Frech MS, et al. Deregulated estrogen receptor alpha expression in mammary epithelial cells of transgenic mice results in the development of ductal carcinoma in situ. Cancer Res. 2005;65:681–685. [PMC free article] [PubMed] [Google Scholar]
- 7.Frech MS, et al. Loss of cyclin D1 in concert with deregulated estrogen receptor alpha expression induces DNA damage response activation and interrupts mammary gland morphogenesis. Oncogene. 2008;27:3186–3193. doi: 10.1038/sj.onc.1210974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilli MT, et al. Introduction of estrogen receptor-alpha into the tTA/TAg conditional mouse model precipitates the development of estrogen-responsive mammary adenocarcinoma. Am. J. Pathol. 2003;163:1713–1719. doi: 10.1016/s0002-9440(10)63529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones LP, et al. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERalpha-negative and ERalpha-positive mammary preneoplasia and cancer. Oncogene. 2008;27:794–802. doi: 10.1038/sj.onc.1210674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshava N, et al. Acceleration of mammary neoplasia in aromatase transgenic mice by 7,12-dimethylbenz[a]anthracene. Cancer Lett. 2001;167:125–133. doi: 10.1016/s0304-3835(01)00478-5. [DOI] [PubMed] [Google Scholar]
- 11.Russo IH, et al. Mammary gland neoplasia in long-term rodent studies. Environ. Health Perspect. 1996;104:938–967. doi: 10.1289/ehp.96104938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallepell S, et al. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc. Natl Acad. Sci. USA. 2006;103:2196–2201. doi: 10.1073/pnas.0510974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlin J, et al. Pubertal mammary gland development: insights from mouse models. J. Mammary Gland Biol. Neoplasia. 2006;11:283–297. doi: 10.1007/s10911-006-9024-2. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, et al. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl Acad. Sci. USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, et al. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol. Endocrinol. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, et al. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, et al. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ. 1998;9:795–803. [PubMed] [Google Scholar]
- 18.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan L, et al. Possible role of Stat5a in rat mammary gland carcinogenesis. Breast Cancer Res. Treat. 2004;88:263–272. doi: 10.1007/s10549-004-0805-2. [DOI] [PubMed] [Google Scholar]
- 20.Cotarla I, et al. Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int. J. Cancer. 2004;108:665–671. doi: 10.1002/ijc.11619. [DOI] [PubMed] [Google Scholar]
- 21.Nevalainen MT, et al. Basal activation of transcription factor signal transducer and activator of transcription (Stat5) in nonpregnant mouse and human breast epithelium. Mol. Endocrinol. 2002;16:1108–1124. doi: 10.1210/mend.16.5.0839. [DOI] [PubMed] [Google Scholar]
- 22.Nevalainen MT, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J. Clin. Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Sultan AS, et al. Co-overexpression of Janus kinase 2 and signal transducer and activator of transcription 5a promotes differentiation of mammary cancer cells through reversal of epithelial-mesenchymal transition. Cancer Sci. 2008;99:272–279. doi: 10.1111/j.1349-7006.2007.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sultan AS, et al. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 25.Rizki A, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68:1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita H, et al. Dominant-negative Stat5 inhibits growth and induces apoptosis in T47D-derived tumors in nude mice. Cancer Sci. 2004;95:662–665. doi: 10.1111/j.1349-7006.2004.tb03326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faulds MH, et al. Cross-talk between ERs and signal transducer and activator of transcription 5 is E2 dependent and involves two functionally separate mechanisms. Mol. Endocrinol. 2001;15:1929–1940. doi: 10.1210/mend.15.11.0726. [DOI] [PubMed] [Google Scholar]
- 28.Stoecklin E, et al. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. J. Steroid Biochem. Mol. Biol. 1999;69:195–204. doi: 10.1016/s0960-0760(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. ERalpha and STAT5a cross-talk: interaction through C-terminal portions of the proteins decreases STAT5a phosphorylation, nuclear translocation and DNA-binding. FEBS Lett. 2004;572:238–244. doi: 10.1016/j.febslet.2004.06.098. [DOI] [PubMed] [Google Scholar]
- 30.Humphreys RC, et al. Signal transducer and activator of transcription 5a influences mammary epithelial cell survival and tumorigenesis. Cell Growth Differ. 1999;10:685–694. [PubMed] [Google Scholar]
- 31.Ren S, et al. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- 32.Iavnilovitch E, et al. Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol. Cancer Res. 2002;1:32–47. [PubMed] [Google Scholar]
- 33.Iavnilovitch E, et al. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int. J. Cancer. 2004;112:607–619. doi: 10.1002/ijc.20484. [DOI] [PubMed] [Google Scholar]
- 34.Gunther EJ, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 35.Chehab FF, et al. Early onset of reproductive function in normal female mice treated with leptin. Science. 1997;275:88–90. doi: 10.1126/science.275.5296.88. [DOI] [PubMed] [Google Scholar]
- 36.Ren S, et al. A simplified method to prepare PCR template DNA for screening of transgenic and knockout mice. Contemp. Top. Lab. Anim. Sci. 2001;40:27–30. [PubMed] [Google Scholar]
- 37.Chretien S, et al. Erythropoietin-induced erythroid differentiation of the human erythroleukemia cell line TF-1 correlates with impaired STAT5 activation. EMBO J. 1996;15:4174–4181. [PMC free article] [PubMed] [Google Scholar]
- 38.Ilaria RL, Jr., et al. Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation. Blood. 1999;93:4154–4166. [PubMed] [Google Scholar]
- 39.Yamaji D, et al. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basham B, et al. In vivo identification of novel STAT5 target genes. Nucleic Acids Res. 2008;36:3802–3818. doi: 10.1093/nar/gkn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang JZ, et al. Signal transducer and activator of transcription (STAT)-5A and STAT5B differentially regulate human mammary carcinoma cell behavior. Endocrinology. 2010;151:43–55. doi: 10.1210/en.2009-0651. [DOI] [PubMed] [Google Scholar]
- 42.Huang X, et al. Thrombopoietin controls proliferation of embryonic multipotent hematopoietic progenitors. Genes Cells. 2009;14:851–860. doi: 10.1111/j.1365-2443.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 43.Miyoshi K, et al. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snow JW, et al. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.v99.1.95. [DOI] [PubMed] [Google Scholar]
- 45.Brockman JL, et al. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Mol. Cell. Endocrinol. 2005;239:45–53. doi: 10.1016/j.mce.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clevenger CV, et al. The role of prolactin in mammary carcinoma. Endocr. Rev. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joung YH, et al. Hypoxia activates the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer cells. Exp. Mol. Med. 2005;37:353–364. doi: 10.1038/emm.2005.45. [DOI] [PubMed] [Google Scholar]
- 48.Matsumura I, et al. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterton RT, Jr., et al. Role of the progesterone receptor (PR) in susceptibility of mouse mammary gland to 7,12-dimethylbenz[a]anthracene-induced hormone-independent preneoplastic lesions in vitro. Cancer Lett. 2002;188:47–52. doi: 10.1016/s0304-3835(02)00461-5. [DOI] [PubMed] [Google Scholar]
- 50.Park SH, et al. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J. Biol. Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- 51.Allred DC, et al. The origins of estrogen receptor alpha-positive and estrogen receptor alpha-negative human breast cancer. Breast Cancer Res. 2004;6:240–245. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dontu G, et al. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol. Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]