Abstract
Previous studies from our laboratory have indicated that overexpression of the epidermal growth factor receptor pathway substrate 8 (EPS8) enhances cell proliferation, migration and tumorigenicity in vivo, although the mechanisms involved remain unexplored. A microarray screen to search for potential mediators of EPS8 identified upregulation of multiple cell cycle-related targets such as the transcription factor FOXM1 and several of its reported downstream mediators, including cdc20, cyclin B1, cyclin A, aurora-B kinase and cdc25C in cells with elevated EPS8, as well as matrix metalloproteinase-9, which we reported previously to be upregulated by EPS8-dependent mechanisms. Cells engineered to overexpress FOXM1 showed increased proliferation, similar to EPS8-overexpressing cells. Conversely, targeted knockdown of FOXM1 in EPS8-overexpressing cells reduced proliferation. Cotransfection of EPS8 with a FOXM1-luciferase reporter plasmid into 293-T- or SVpgC2a-immortalized buccal keratinocytes demonstrated that EPS8 enhances FOXM1 promoter activity, whereas chromatin immunoprecipitation assays revealed elevated levels of acetylated histone H3 associated with the FOXM1 promoter in cells expressing high levels of EPS8. Treatment of EPS8-overexpressing cells with inhibitors of phosphoinositide 3-OH kinase or AKT reduced expression of FOXM1 and aurora-B kinase, a transcriptional target of FOXM1. Overexpression of EPS8 induced expression of the chemokine ligands CXCL5 and CXCL12 in a FOXM1-dependent manner, which was blocked by LY294002 or a dominant-negative form of AKT. Additionally, overexpression of FOXM1 enhanced cell migration, whereas targeted knockdown of CXCL5 or inhibition of AKT reduced migration of EPS8-expressing cells. These data suggest that EPS8 enhances cell proliferation and migration in part by deregulating FOXM1 activity and inducing CXC-chemokine expression, mediated by PI3K- and AKT-dependent mechanisms.
Introduction
Rapid progression through the cell cycle is one key feature of neoplastic disease, which contributes to enhanced tumor growth. In many epithelial cancers, signal transduction through the epidermal growth factor receptor (EGFR) is elevated beyond that found in normal cells as a result of receptor overexpression, constitutive activation or increased availability of ligand such as transforming growth factor-α (1–4). Activation involves receptor dimerization and transphosphorylation of a number of tyrosine residues in the cytoplasmic region of the EGFR, thereby facilitating recruitment of Src homology 2- or phosphotyrosine-binding-domain-containing proteins, such as Grb2, Shc and Crk. Signaling downstream of the receptor may involve multiple pathways, including those regulated by phospholipase C-γ, phosphoinositide 3-OH kinase (PI3K) and Src, in addition to the well-characterized Ras-Raf-MEK-ERK pathway. EGFR activation has also been reported to result in activation of c-jun N-terminal kinases in a Rac-dependent manner (5,6). Resultant stimulation of these biochemical cascades may contribute to enhanced growth and motility, characteristics associated with tumor progression, and all have been reported to be deregulated in a wide range of human cancers.
Additional mechanisms have been documented to mediate some actions of the EGFR. Epidermal growth factor receptor pathway substrate 8 (EPS8) (7) is a 97 kDa protein that exists as a dimer (8) and binds to the juxtamembrane region of the EGFR (9). Overexpression of EPS8 has been shown to enhance mitogenic signaling by epidermal growth factor (EGF) (10), as well as EGF-dependent transformation of NIH3T3 fibroblasts overexpressing the EGFR (11), and is constitutively tyrosine phosphorylated in some human tumor cell lines (11). EPS8 contains a Src homology 3 domain, and it interacts with a number of cellular binding partners, including RN-tre (12), Shc, E3B1 (Abi-1) (13), Dvl-1 (14) and Irsp53 (15). Interaction of RN-tre with EPS8 impacts on Rab5 activity and, thus, inhibits EGFR internalization (16), whereas complex formation between EPS8, E3B1 and the guanine nucleotide exchange factor Sos-1 has been shown to activate the small GTPase Rac1, resulting in actin remodeling and cell motility (17–19).
Enhanced mitogenesis is a feature of cells overexpressing EPS8 (10), and EPS8 is reportedly required to mediate growth-promoting effects of the Src oncoprotein (20); however, it is unclear what biochemical mechanisms are involved, although more rapid transit through the cell cycle mediated by effects on cell cycle regulators might be predicted. Our recent studies used a model system of squamous carcinoma cells expressing differing levels of EPS8 (21). We found that overexpression of EPS8 enhanced cell growth in vitro and was sufficient to confer a tumorigenic phenotype on non-tumorigenic cells in orthotopic transplantation assays. Furthermore, EPS8 enhanced the expression and activity of matrix metalloproteinase-9 in a PI3K- and AKT-dependent manner, although we found no evidence to suggest elevated activity of c-jun N-terminal kinase in EPS8-overexpressing cells, even though Rac activation of c-jun N-terminal kinase has been reported in other model systems (5). Similarly, activity of ERK1/2, key mediators of the EGFR proliferative response, was unaltered between cells expressing low and high levels of EPS8 [(22), H.Wang and W.A.Yeudall, unpublished data]. In the present study, we sought to determine potential downstream mediators of EPS8-dependent proliferation.
Methods
Cell lines and culture conditions
HN4 cells, derived from a primary squamous cell carcinoma of the head and neck, and HN12 cells, derived from a synchronous lymph node metastasis, and derivative cell lines, were cultured as described previously in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 0.4 μg/ml hydrocortisone at 37°C in 95% air/5% CO2 (22). Saos-2 and 293-T cells were obtained from ATCC (Manassas, VA). SVpgC2a immortalized keratinocytes have been described previously (23).
Growth factors and inhibitors
Recombinant human EGF was purchased from Austral Biologicals (San Ramon, CA), diluted in Dulbecco’s modified Eagle’s medium containing 0.1% bovine serum albumin and used to treat cells at a final concentration of 2.5 nM (22,24). LY294002 was purchased from Sigma–Aldrich (St Louis, MO) and used at a concentration of 10 μM, as determined previously (22). The AKT inhibitor 1L6-hydroxymethyl-chiro-inositol-2-(R)-2-O-methyl-3-O-octadecyl-sn-glycerocarbonate (Merck 124005) was purchased from EMD Biosciences (San Diego, CA) and used at a concentration of 20 μM, at which these cells show no noticeable signs of toxicity.
Antibodies
Antibodies that recognize ERK2 (sc-54), FOXM1 (sc-500), FOXM1 (sc-502) and actin (sc-1616) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). EPS8 (E-18220) antibody was purchased from BD Transduction Laboratories (San Diego, CA). Anti-p-AKT (4058), which recognizes phospho-S473, and anti-GSK-3β (9322), which recognizes phospho-S9, were obtained from Cell Signaling Technology (Danvers, MA). Anti-AKT1 (559028) was purchased from BD Biosciences Pharmingen (Mississauga, Ontario, Canada). Horseradish peroxidase-conjugated anti-goat, anti-rabbit and anti-mouse secondary antibodies were obtained from MP Biomedical (Aurora, OH).
Plasmid constructions and transfections
A plasmid encoding human FOXM1 (MGC-9577) was obtained from ATCC. short hairpin RNA (shRNA) sequences targeting FOXM1 were designed as previously reported and cloned into the pSirenRetroQ plasmid (BD Clontech, San Diego, CA). Controls of ‘scrambled’ nucleotide sequences with the same base composition were similarly treated. Nucleotide sequences are given in supplementary Table 2 (available at Carcinogenesis Online). FOXM1 promoter-luciferase and expression plasmids were as described previously (25). EPS8, wild-type AKT and dominant-negative form of AKT (dnAKT) expression plasmids were as described previously (21,26). All plasmids were sequence-verified prior to use. HN4, HN12 and derivative cell lines were nucleofected (Lonza, Rockville, MD) with 2 μg of plasmid DNA. Forty-eight hours later, puromycin was added to a final concentration of 1 μg/ml and cells selected for stable expression. Transient transfection of SVpgC2a, 293-T and Saos-2 cells was accomplished using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. To generate recombinant GSK-3β for use as a substrate, a complementary DNA encoding the first 50 amino acids of human GSK-3β was obtained by polymerase chain reaction (PCR), cloned into the pGEX4T plasmid and recombinants used to express GSK-3β as a glutathione S-transferase fusion protein. The shRNA plasmid targeting CXCL5, pSirenRetroQ-shCXCL5 (24), and the CXCL5 promoter-luciferase plasmid [a generous gift from Dr A.C.Keates, Harvard Medical School (27)] have been described previously.
Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (qRT–PCR) was performed using an ABI 7500 Fast system (Applied Biosystems, Rockville, MD) and a SYBR green-based procedure, as described previously (24). Oligonucleotide pairs for use as PCR primers were designed using the Primerbank database (http://pga.mgh.harvard.edu/primerbank/index.html) (28). Primer sequences are listed in supplementary Table 3 (available at Carcinogenesis Online). Complementary DNA for use as template was reverse transcribed from 1 μg total cellular RNA as described previously (29). Serial dilutions were made using previously generated PCR products, assigned arbitrary values corresponding to the dilutions and used to construct relative standard curves for each gene target. Data were normalized to actin as an internal standard.
Cell proliferation assays
To determine proliferation, 1 × 104 HN12 or 2 × 104 HN4 cells, or derivative cell lines, were plated in triplicate in 12-well culture plates. On each of six consecutive days, cells were trypsinized and counted using a hemocytometer. Alternatively, cells were grown on glass coverslips, pulsed with 10 μM 2-bromodeoxyuridine (BrdU) for 30 min and then harvested and stained with anti-BrdU antibody and detected with a horseradish peroxidase-conjugated secondary antibody using 3, 3′-diaminobenzidine as a substrate. Stained cells were counted in 20 random high-power fields.
Cell cycle analysis
Distribution of cells in each phase of the cell cycle was determined following propidium iodide staining, essentially as described (30), using a Guava Easycyte-Mini flow cytometer (Guava Technologies, Hayward, CA).
Western blot analysis
Analysis of protein expression was carried out as described previously (31). Blots were blocked in 5% skimmed milk in TTBS [10 mM Tris–HCl (pH 7.6), 0.5% Tween 20, 150 mM NaCl] for 1 h at ambient temperature, then incubated for 1 h with primary antibodies diluted 1:1000 in blocking buffer (1:5000 for anti-EPS8), washed 3× in TTBS, incubated with horseradish peroxidase-conjugated secondary antibodies and detected by ECL (Amersham Biosciences, Piscataway, NJ). For immunoblotting of FOXM1, a modified lysis buffer was used, comprising 10 mM Tris (pH 7.4), 1% sodium dodecyl sulfate (SDS), 1 mM sodium orthovanadate and protease inhibitors (32).
AKT activity assays
As a complement to the analysis of AKT activity by measuring phosphorylation at serine-473, we developed and used an in vitro kinase assay. Briefly, total AKT was immunoprecipitated from 500 μg of cell lysates overnight at 4°C and immune complexes captured on Protein A-Sepharose beads. After washing in lysis buffer (3×) and two washes in kinase assay buffer [25 mM Tris–HCl (pH 7.5), 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2], beads were resuspended in 1× kinase assay buffer containing 200 μM adenosine triphosphate and 2 μg of glutathione-S-transferase–GSK-3β as substrate and incubated at 30°C for 30 min. Reactions were terminated by heating in 1× sodium dodecyl sulfate–polyacrylamide gel electrophoresis sample buffer, then resolved in 12% polyacrylamide gels and western blotted with anti-phospho-GSK-3β antibody. Data were normalized using total AKT protein after western blotting with AKT antibody.
Luciferase reporter assays
To determine promoter activity, cells were cultured to 60% confluence, then transfected with plasmids in which the promoter (in this case, FOXM1 or CXCL5) drives expression of Firefly luciferase, together with a Renilla luciferase plasmid to facilitate normalization. Forty-eight hours later, cells were harvested, lysates prepared and luciferase activity determined by standard procedures using a commercially available kit (Dual Luciferase Assay System; Promega, Madison, WI). FOXM1 expression constructs were used as positive control to activate the reporter plasmid.
Cell migration assays
Cell migration was carried out using wound-healing (scratch) assays. Briefly, cells were trypsinized, plated in triplicate in 12-well cell culture plates and incubated at 37°C until cells were completely confluent. At this time, a sterile pipette tip was used to scratch across the surface of the plate, removing the complete layer of cells within the scratch area. Following cell removal, each well was washed once with phosphate-buffered saline and then replaced with growth medium. Immediately following, the width of the scratch was measured at three specific points under a ×5 objective using a light microscope and AxioVision software (Carl Zeiss Microimaging, Thornwood, NY). Cells were incubated at 37°C for a total of 6 h, at which time the scratch width was measured at the same position as at time 0.
Chromatin immunoprecipitation assays
To cross-link protein and DNA, cell cultures were incubated in 2% formaldehyde for 10 min at ambient temperature and then 200 mM glycine was added for a further 10 min. Cells were washed in cold phosphate-buffered saline, scraped and centrifuged. Pellets were resuspended in lysis buffer containing protease inhibitors and then sheared by multiple passages through a 27.5 gauge needle followed by 25 min of sonication on ice. Following centrifugation, the protein content of the supernatants was determined and equal amounts used for immunoprecipitation with anti-acetylated histone H3, anti-FOXM1 or IgG as a control, overnight at 4°C. Immune complexes were captured using Protein A-Sepharose, then washed sequentially in RIPA buffer [150 mM NaCl, 50 mM Tris (pH 8), 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40], high salt buffer [500 mM NaCl, 50 mM Tris (pH 8), 0.1% SDS, 1% NP-40], twice in LiCl buffer [250 mM LiCl, 50 mM Tris (pH 8), 0.5% sodium deoxycholate, 1% NP-40] then twice in TE buffer. Protein was eluted from beads in fresh elution buffer (20% SDS, 10 mM dithiothreitol, 100 mM NaHCO3), cross-linking reversed overnight at 65°C in the presence of NaCl and then samples were ethanol precipitated. Following centrifugation, pellets were resuspended in TE buffer and incubated sequentially with 50 μg/ml RNase A (30 min) and 100 μg/ml proteinase K (1 h). Samples were phenol extracted, ethanol precipitated and the pellets washed in 70% ethanol, dried and resuspended in sterile water. Quantitative PCR was carried out, as described previously, using three independent oligonucleotide primer pairs that target regions of the FOXM1 and CXCL5 promoters. Nucleotide sequences are given in supplementary Table 4 (available at Carcinogenesis Online).
Statistical analysis
Data obtained from migration, reporter and qRT–PCR assays were analyzed by t-test using the SPSS v.13 software package (SPSS, Chicago, IL). A value of P < 0.05 was considered to be statistically significant.
Results
FOXM1 is elevated in cells expressing EPS8
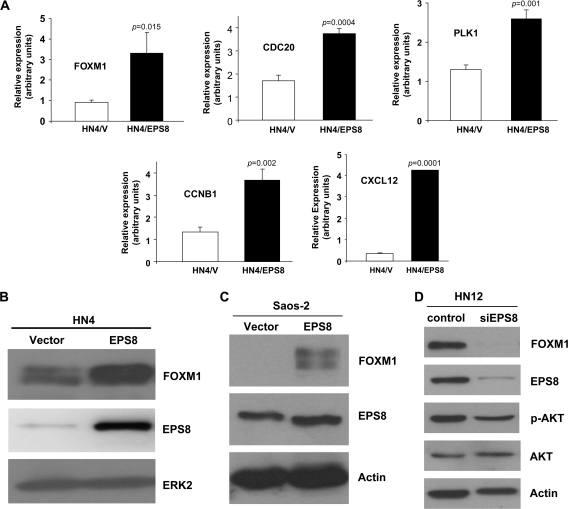
To begin to determine mechanisms that mediate the effects of EPS8, we performed microarray experiments (see supplementary Data, available at Carcinogenesis Online) using RNA derived from EPS8- and vector-transfected HN4 squamous carcinoma cells. Among the gene products that showed elevated expression in the presence of EPS8 was the forkhead transcription factor FOXM1 (Figure 1A and B), together with a cohort of known FOXM1 targets (supplementary Table 1 is available at Carcinogenesis Online). These included cell cycle regulators CDC20, CDC2, CDC25B and CDC25C phosphatases, cyclins A and B and aurora-B kinase. Additional molecules associated with mitotic progression, such as aurora-A kinase, polo-like kinase-1, centromere protein-A, -E and -F, and stimulators of angiogenesis and motility including vascular endothelial cell growth factor and CXCL12 were also identified and independently validated (Figure 1A; supplementary Table 1 is available at Carcinogenesis Online). Using the Database for Annotation, Visualization and Integrated Discovery software (http://david.abcc.ncifcrf.gov/) (33,34), which enables the discovery of enriched biological themes within gene/protein lists and the generation of gene annotation tables, EPS8-overexpressing cells were found to be enriched for genes whose products play roles in cell cycle progression and mitotic spindle function (supplementary Figures 1 and 2 are available at Carcinogenesis Online). Further analysis using Ingenuity Pathway Analysis software revealed a network of gene products including FOXM1 and other cell cycle-related molecules (supplementary Figure 3 is available at Carcinogenesis Online). Taken together, these data suggest that overexpression of EPS8 leads to elevated expression of a cohort of cell cycle regulators, which may begin to explain the enhanced proliferative response of EPS8-expressing cells.
Fig. 1.
FOXM1 is upregulated by EPS8. (A) HN4/V and HN4/EPS8 cells were cultured in the absence of serum, RNA prepared, reverse transcribed and used as template in qRT–PCR experiments with primers for the indicated genes. Relative expression is indicated, normalized to actin as an internal standard. Bar = standard deviation. (B) HN4 cells were stably transfected with an EPS8 expression plasmid or empty vector as control, cell lysates prepared and analyzed by western blotting with the indicated antibodies. (C) Saos-2 cells were transiently transfected with an EPS8 expression plasmid or empty vector and analyzed by western blotting with the indicated antibodies. (D) HN12 cells were transiently transfected with small interfering RNA targeting EPS8, or a non-targeting control, and analyzed by western blotting with the indicated antibodies.
Although our primary interest was in examining the effects of EPS8 overexpression, as seen in many different tumor types, we also found that elevated levels of EPS8 resulted in gene expression changes similar or different to those elicited by EGF. For example, EPS8 overexpression enhanced the basal levels of CEP55 and RANBP1, as well as potentiating their expression in the presence of EGF (supplementary Figure 4 is available at Carcinogenesis Online). In contrast, the expression of WNT4 is enhanced by EPS8 whereas, at least in this cell line, the overall effect of EGF is to suppress WNT4 (supplementary Figure 4 is available at Carcinogenesis Online) and, thus, the effects of EPS and EGF appear to be antagonistic, perhaps due to differential activation of signaling pathways downstream of the EGFR. In addition, EPS8 elevates expression of TCOF1 under basal conditions (supplementary Figure 4 is available at Carcinogenesis Online), yet expression in the presence of EGF is similar with or without higher levels of EPS8. Taken together, the data suggest that overexpression of EPS8 can result in gene expression changes that are both similar and distinct from those arising from EGFR stimulation.
FOXM1 enhances cell proliferation
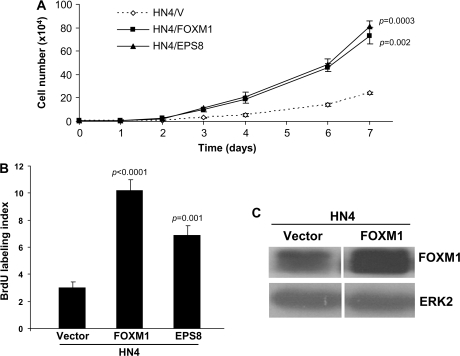
Our previous studies reported that EPS8 overexpression leads to enhanced cell proliferation (21) and our experiments, described above, identified elevated expression of the transcription factor FOXM1, a key regulator of cell cycle progression, and several of its targets in HN4 cells expressing EPS8. Similar induction of FOXM1 was observed in Saos-2 cells transfected with an EPS8 expression plasmid (Figure 1C). Conversely, RNA interference (RNAi)-mediated knockdown of EPS8 in HN12 cells, which express high endogenous levels of EPS8, led to a reduction in FOXM1 protein levels (Figure 1D). Thus, we chose to focus our attention on the possible role of FOXM1 as a downstream mediator of EPS8. To address this, we generated stable transfectants of FOXM1 in HN4 cells and compared their proliferative potential with cells overexpressing EPS8 over a 6-day time course. As shown in Figure 2A, HN4 cells expressing FOXM1 proliferated at a rate similar to cells transfected with EPS8 and higher than vector-transfected controls. BrdU-labeling studies provided additional support for these observations (Figure 2B). Western blotting of cell lysates demonstrated elevated FOXM1 expression in EPS8- and FOXM1-overexpressing cells (Figures 1B–D and 2C). This close relationship suggests that FOXM1 is a likely candidate to mediate the proliferative functions of EPS8 on cell growth.
Fig. 2.
FOXM1 enhances proliferation of HNSCC cells. (A) HN4 cells expressing FOXM1, EPS8 or an empty vector control were seeded in 12-well tissue culture plates and the cell number determined by daily trypsinization and counting. (B) HN4 cells expressing FOXM1, EPS8 or an empty vector control were pulsed with BrdU, then immunostained with anti-BrdU antibodies and counted. Bar = standard error of the mean. (C) HN4 cells transfected with a FOXM1 expression plasmid or empty vector as control were lysed and western blotted with the indicated antibodies.
EPS8 stimulates FOXM1 promoter activity
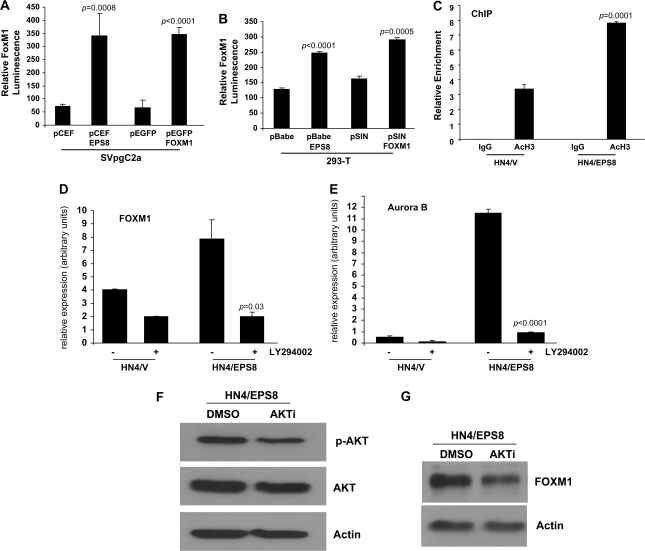
To characterize the effects of EPS8 on FOXM1 further, cells were transfected with EPS8 expression plasmids together with a FOXM1 promoter-luciferase reporter plasmid and luciferase activity determined. As shown in Figure 3A, in the presence of EPS8, the FOXM1 promoter was activated up to 5-fold compared with empty vector controls in SVpgC2a immortalized buccal keratinocytes. Similarly, EPS8 produced an elevation of FOXM1 promoter activity in 293-T cells (Figure 3B). Chromatin immunoprecipitation experiments were undertaken to determine the relative amounts of acetylated histone H3 associated with the FOXM1 promoter. As indicated in Figure 3C, levels are increased 2.5-fold in HN4/EPS8 cells compared with control cells transfected with empty vector alone. The data support a role for EPS8 in mediating transactivation of the FOXM1 promoter.
Fig. 3.
EPS8 induces FOXM1 expression via PI3K and AKT. SVpgC2a cells (A) or 293-T cells (B) were transiently transfected with the plasmids indicated, together with FOXM1B-luciferase promoter and β-galactosidase plasmids. Forty-eight hours later, luciferase assays were performed as described in Methods. FOXM1B luminescence is shown, normalized to β-galactosidase. (C) Chromatin immunoprecipitation (ChIP) was performed using antibody that recognizes acetylated histone H3, or IgG as control, as described in Methods. Quantitative PCR was carried out using three sets of primers specific for the FOXM1 promoter. Data are shown for one primer set and are representative of data obtained with the other primers. (D and E) HN4/V and HN4/EPS8 cells were cultured in the presence of LY294002 (10 μM) or an equivalent volume of solvent (dimethyl sulfoxide). Twenty-four hours later, total RNA was prepared, reverse transcribed and used as template for qRT–PCR experiments using primers for FOXM1 (D) and Aurora-B kinase (E). (F and G) HN4/EPS8 cells were incubated in the presence of AKT inhibitor (AKTi) or solvent. Twenty-four hours later, cell lysates were prepared and western blotted with (F) p-AKT or (G) FOXM1 antibodies.
EPS8 activates FOXM1 through a PI3K-dependent mechanism
As we had found previously that EPS8 could activate AKT through stimulation of PI3K (21), we sought to determine if a similar mechanism might be responsible for the elevated expression of FOXM1. Thus, cells were treated with the PI3K inhibitor, LY294002, RNA prepared and FOXM1 expression determined by qRT–PCR. Figure 3D shows that FOXM1 transcript levels were reduced in inhibitor-treated HN4/EPS8 cells by 5-fold compared with controls, suggesting regulation by PI3K signaling. Expression of the FOXM1 target aurora-B kinase was also repressed in the presence of LY294002 (Figure 3E), suggesting that EPS8 regulates expression of aurora-B kinase in these cells. Similarly, treatment of cells with a specific AKT inhibitor resulted in inhibition of AKT, as judged by reduced phosphorylation at serine-473 (Figure 3F), and concomitant reduction in FOXM1 protein levels (Figure 3G). Taken together, these data support a PI3K- and AKT-dependent mechanism for regulation of FOXM1 by EPS8.
FOXM1 RNAi reduces the effects of EPS8 overexpression
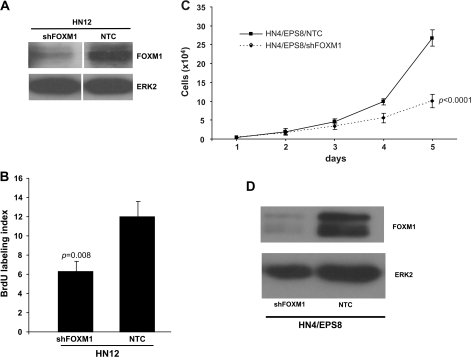
HN12 cells, which express elevated levels of EPS8 (22), were derived from a synchronous lymph node metastasis, which occurred in the patient from which HN4 cells were derived (35). The rapid growth of these cells is inhibited by reduction of EPS8 (21). Therefore, in light of our observations linking EPS8 and FOXM1, we were keen to determine if inhibiting FOXM1 would have a similar effect. HN12 cells were transfected with plasmids encoding FOXM1 shRNA or a non-targeting control, selected in puromycin and FOXM1 expression assessed by western blot. Figure 4A shows substantial knockdown of FOXM1 achieved in HN12 cells compared with control. In parallel, cells were pulsed with BrdU and the number of stained cells determined by microscopy. As shown in Figure 4B, a 50% reduction in BrdU incorporation was found in HN12 cells in which FOXM1 is inhibited by RNAi. We performed similar knockdown experiments in HN4/EPS8 cells. As shown in Figure 4C and D, the rate of proliferation of HN4/EPS8 cells is reduced when FOXM1 expression is inhibited, consistent with the notion of FOXM1 as a mediator of the growth-promoting effects of EPS8. RNAi-mediated knockdown of FOXM1 in HN4/EPS8 cells also led to a reduction in expression of multiple cell cycle regulators (supplementary Figure 5 is available at Carcinogenesis Online).
Fig. 4.
FOXM1 RNAi inhibits cell growth in EPS8-overexpressing cells. (A) HN12 cells transfected with a non-targeting control (NTC) or FOXM1 shRNA plasmid were lysed and western blotted with FOXM1 antibody or ERK-2 as a loading control. (B) The indicated cells were pulsed with BrdU, then immunostained with anti-BrdU antibodies and counted as described in Methods. Bar = standard error of the mean (SEM). (C). HN4/EPS8 cells were stably transfected with the FOXM1 shRNA plasmid or a non-targeting control. Cell proliferation was determined by daily counting. Bar = SEM. (D) Protein lysates were prepared from the cells described in (C) and western blotted with the indicated antibodies.
EPS8 upregulates expression of CXCL5 via FOXM1-dependent mechanisms
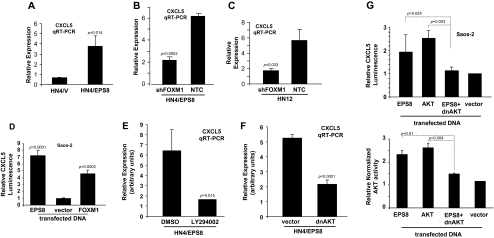
Previously, we have shown that some chemokines are key factors in squamous carcinogenesis (24,36). For example, CXCL5 is highly expressed in HN12 cells but not in HN4 cells, which have low levels of EPS8. Furthermore, our microarray screen had indicated upregulation of CXCL12 in HN4/EPS8 cells (supplementary Table 1 is available at Carcinogenesis Online). Therefore, to test whether CXCL5 expression might be dependent on EPS8, we determined the expression of this chemokine in cells in which EPS8 or FOXM1 had been overexpressed and/or inhibited by RNAi. Figure 5A demonstrates that expression of EPS8 in HN4 cells induces expression of CXCL5. Furthermore, shRNA-mediated knockdown of FOXM1 in HN4/EPS8 cells (Figure 5B) or in HN12 cells (Figure 5C) led to a reduction in CXCL5 expression, as well as CXCL8 (supplementary Figure 6 is available at Carcinogenesis Online). Moreover, cotransfection of a FOXM1 expression plasmid with a CXCL5 promoter plasmid resulted in a 4.5-fold enhancement of activity compared with control-transfected cells (Figure 5D). Taken together, these data suggest that CXCL5 expression is enhanced by EPS8 in a FOXM1-dependent manner.
Fig. 5.
EPS8 induces expression of CXCL5 in a PI3K- and AKT-dependent manner. (A) HN4/EPS8 and vector control cells were used to prepare RNA, reverse transcribed and subjected to qRT–PCR for CXCL5 and actin. (B) RNA was prepared from HN4/EPS8 cells transfected with FOXM1 shRNA or control plasmids, and CXCL5 expression determined by qRT–PCR, relative to actin. (C) RNA was prepared from HN12 cells transfected with FOXM1 shRNA or control plasmids and subjected to qRT–PCR for CXCL5. Data are shown normalized to actin. (D) HN4 cells were cotransfected with a CXCL5 promoter-luciferase plasmid, together with FOXM1 expression plasmid, or empty vector control, and a Renilla luciferase plasmid as internal control. Forty-eight hours later, luciferase activity was determined as described in Methods. (E) HN4/EPS8 cells were treated with LY294002 or vehicle as control, and CXCL5 expression determined by qRT–PCR relative to actin. (F) HN4/EPS8 cells were transfected with a plasmid expressing dominant-negative form of AKT (dnAKT) (K179M) or control, and CXCL5 expression determined as in (E). (G) Saos-2 cells were cotransfected with the CXCL5 promoter-luciferase plasmid, together with the indicated expression plasmids. Forty-eight hours later, luciferase activity was determined and is shown relative to Renilla luciferase as an internal control (upper panel). Parallel cultures were lysed, AKT immunoprecipitated and subjected to in vitro kinase assay as described in Methods. Relative AKT activity is shown, which represents GSK-3β phosphorylation normalized to AKT expression.
CXCL5 is regulated by a PI3K- and AKT-dependent mechanism
As we and others have demonstrated that EPS8 can transduce signals through the PI3K and AKT pathway (21,37), we investigated the effects of blocking these kinases on chemokine expression. First, HN4/EPS8 cells were treated with LY294002 and CXCL5 expression determined by qRT–PCR. As shown in Figure 5E, compared with vehicle control, CXCL5 expression was reduced 4-fold in the presence of inhibitor. In parallel, cells were transiently transfected with a plasmid encoding a dnAKT or empty vector as control. Results of qRT–PCR experiments (Figure 5F) indicate that dnAKT results in a decrease in CXCL5 expression. In addition, Saos-2 cells were transfected with the CXCL5 promoter-luciferase plasmid, together with plasmids encoding EPS8, AKT, dnAKT or empty vector, and luciferase activity measured. Figure 5G shows that EPS8 or AKT can stimulate CXCL5 promoter activity, whereas the dnAKT plasmid negates the effect of EPS8, reducing CXCL5 promoter activity to that seen in vector controls. This is paralleled by changes in AKT activity, as measured by in vitro kinase assay using recombinant GSK-3β as a substrate (Figure 5G). Together, these data support our observations that EPS8 stimulates CXCL5 expression in a PI3K- and AKT-dependent manner.
EPS8 and FOXM1 enhance HNSCC motility
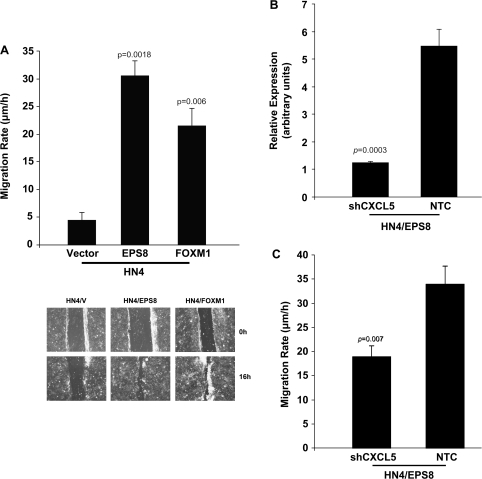
Previously, in independent studies, we showed that EPS8 and CXCL5 enhance cell migration (21,24). Given our present data and the recent observations of others (38), we determined whether FOXM1 could enhance migration of HN4 cells. As indicated in Figure 6A, HN4/EPS8 cells were more migratory than controls, consistent with our previous results (21). Additionally, HN4/FOXM1 cells exhibited a more motile phenotype, consistent with a role for FOXM1 in upregulating chemokine expression (38).
Fig. 6.
EPS8 and FOXM1 enhance cell motility. (A) The indicated cells were cultured to confluence, a scratch made in the monolayer and migration determined, as described in Methods. Upper panel: migration speed is indicated. Bar = standard deviation. Lower panel: pictorial representation of scratch assay. (B) HN4/EPS8 cells stably transfected with a shRNA plasmid targeting CXCL5 or a non-targeting control (NTC) were analyzed for CXCL5 expression by qRT–PCR. (C) Cell motility was measured as described in (A).
To investigate the possibility that chemokines mediate the promigratory effects of EPS8 and FOXM1, we inhibited CXCL5 expression using shRNA (Figure 6B). A statistically significant, although incomplete, reduction in migration was observed in HN4/EPS8 cells with CXCL5 knockdown compared with non-targeting controls (Figure 6C). Taken together, the data suggest that CXCL5 plays a role in mediating motility in cells that express elevated levels of EPS8.
Discussion
Elevated expression of EPS8 enhances growth in a number of cell culture model systems (10,11,21) and also endows tumorigenicity in vivo to non-tumorigenic human cells (21), suggesting a central role in cancer biology. In the present study, we investigated potential mechanisms of EPS8-induced cell growth, beginning with a genome-wide screen. While we found upregulation of matrix metalloproteinase-9, consistent with our previous observations (21), we also identified a cohort of genes that were upregulated in EPS8-overexpressing cells, and which are likely to contribute to the mitogenic effects of EPS8. Among these was the transcription factor FOXM1, a key regulator of cell cycle progression (39), together with several of its target genes encoding B-type cyclins, CDC20, polo-like kinase-1, centromere protein-A, -E and -F and aurora-B kinase. Several genes that we identified as being upregulated in EPS8-overexpressing cells were also found by Laoukili et al. (40), who profiled genes that were overexpressed in osteosarcoma cells transfected with FOXM1. These authors reported elevated expression of cyclin B2, centromere protein-F, NEK-2 and polo-like kinase-1, established regulators of G2/M, when FOXM1 was overexpressed. In addition, the centrosomal protein CEP55 and the DNA helicase HELLS were recently identified as novel (direct and indirect, respectively) targets of FOXM1 and shown to be upregulated in a FOXM1-dependent manner, as well as being found to correlate with HNSCC progression (41). Both these molecules were also present in our list of EPS8-upregulated genes. This provides further support for our current observations that EPS8 lies upstream of FOXM1. Furthermore, we found similar proliferation rates between EPS8- and FOXM1-overexpressing cells, suggesting that many of the growth-promoting effects of EPS8 may be mediated to some extent by FOXM1. Chromatin immunoprecipitation assays also revealed an enrichment of acetylated histone H3 associated with the FOXM1 promoter in cells overexpressing EPS8, which is indicative of regions of chromatin that are permissive for gene expression. The mechanism of FOXM1 activation by EPS8 is currently under investigation in our laboratory.
A recent study has highlighted the importance of FOXM1 in oral neoplasia. Gemenetzidis et al. (41) found that FOXM1 expression is high in oral carcinomas and is also elevated in dysplastic lesions, suggesting that this may be an early event in malignant progression. Moreover, FOXM1 was reported to induce genomic instability in a non-random manner, as expression of FOXM1 in primary oral keratinocytes gave rise to significant copy number abnormalities. As we have now identified a link between EPS8 and FOXM1, this raises the possibility that EPS8-dependent signaling may enhance malignant progression by similar mechanisms. As nicotine is documented to stimulate FOXM1 expression (41), it may be that cells expressing elevated EPS8 may be more susceptible to undergo malignant progression when nicotine is present in the environment. Indeed, our own studies indicate that EPS8 expression is also upregulated in the presence of nicotine (supplementary Figure 7 is available at Carcinogenesis Online).
We found that expression of chemokine ligands CXCL5 and CXCL12 was increased in HN4/EPS8 cells compared with vector controls. These data are consistent with our previous observations, which documented high levels of CXCL5 in HN12 cells derived from a synchronous lymph node metastasis that occurred in the same patient from whom HN4 cells were obtained (42). Furthermore, we found that CXCL5 was critical for enhancing cell proliferation and migration, as well as tumorigenicity (24). In a recent study, FOXM1-transgenic mice developed chemically induced lung tumors that were associated with a persistent inflammatory reaction and concomitant expression of CXC- and CC-chemokines (of which CXCL5 was the most highly expressed at over 21-fold higher than normal tissues), as well as cell cycle regulators and matrix remodeling enzymes (38). In the present study, our observations might be explained by EPS8 driving expression of FOXM1, which activates transcription from the CXCL5 promoter, although it remains to be established if chemokine expression is stimulated by direct binding of FOXM1. Using chromatin immunoprecipitation, we have thus far been unable to demonstrate a direct interaction of FOXM1 with the CXCL5 promoter (data not shown), although a consensus FOXM1 binding site is present in the CXCL5 promoter (supplementary Figure 8 is available at Carcinogenesis Online). We are currently pursuing this line of investigation.
At least some of the characteristics of cells expressing high levels of EPS8 are probably to be due to EPS8-FOXM1-CXCL5 signal transduction and would be consistent with EPS8 inducing a tumorigenic phenotype in non-tumorigenic HN4 cells (21,24). Using shRNA, we were able to reduce CXCL5 expression by greater than 5-fold in HN4/EPS8 cells; however, a reduction in migration of around only 2-fold was observed. Treatment with a specific inhibitor of AKT produced a further significant reduction in cell migration (supplementary Figure 9 is available at Carcinogenesis Online). This suggests that mediators additional to CXCL5 play a role in migration. Possible candidates include chemokines such as CXCL8 and CXCL12, which we also found to be elevated in EPS8- and FOXM1-overexpressing cells. Our previous studies have reported CXCL8 as a regulator of HNSCC migration and proliferation (36) and reports from other laboratories indicate this to be a general mechanism in tumor progression (43–46). Similarly, CXCL12 is well recognized as an important mediator of cancer cell migration (47,48). As FOXM1 regulates expression of these chemokines, it seems likely that they may synergize to mediate some of the effects of EPS8.
Our results also indicate that the PI3K–AKT pathway mediates EPS8-dependent upregulation of FOXM1 to a considerable extent. AKT has been reported to be activated in many different tumor types, including head and neck cancers (49–51) and is thought to be a key regulator of oncogenic progression. Of relevance to the data presented herein, at least in some cells chemokines may activate AKT downstream of their cell surface receptors (52–57). Thus, the potential exists for an EPS8-induced autostimulatory loop, in which EPS8-AKT-FOXM1 signaling induces secretion of chemokines, which are then able to trigger receptor-mediated activation of AKT.
Our previous work showed that overexpression of EPS8 was sufficient to convert non-tumorigenic HN4 cells to a tumorigenic phenotype (21). There may be several reasons for this, including enhanced proliferation and motility. However, an additional important component of tumor growth is the ability of tumor cells to stimulate the development of a blood supply to sustain the growing cell mass. In our microarray screen, we found that expression of vascular endothelial cell growth factor, a principal mediator of angiogenesis, was elevated in cells expressing EPS8. A recent study has now linked FOXM1 to angiogenesis and tumor progression in gastric carcinoma. Li et al. (58) found that overexpression of FOXM1 stimulated growth and metastatic spread of gastric cancer cells and also increased tumor vascularity. This was found to occur by FOXM1-mediated transcriptional activation of the vascular endothelial cell growth factor promoter. Although we did not address potential angiogenic activity of EPS8 in the present study, it may be that EPS8 is also able to stimulate neovascularization via FOXM1, in addition to effects on growth and motility of tumor cells. These studies are ongoing in our laboratory.
In a previous study, Laoukili et al. (40) found upregulation of tissue plasminogen activator and urokinase plasminogen activator, as well as a kelch-domain-containing protein, KLHL1, in FOXM1-transfected cells. In our microarray screen to identify mediators of EPS8 function, we found upregulation of urokinase plasminogen activator and two kelch-like proteins, KLHL1 and KLHL32, in EPS8-overexpressing cells (supplementary Table 1 is available at Carcinogenesis Online). The role of urokinase plasminogen activator in tumor cell metastasis is well documented (59–62) and elevated expression would be consistent with a role for this molecule in EPS8-dependent tumor progression. Similarly, kelch proteins have been identified as mediators of cell motility and invasion. In a model system of rat F208 cells, Krp1 was identified as being upregulated following v-fos- and v-ras-mediated transformation (63) and was found to colocalize with F-actin and enhance pseudopod formation. Krp1 has also been shown to enhance mesenchymal invasion in vitro, independent of integrin and Rho-ROCK signaling (64). Previously, we showed that overexpression of EPS8 enhances cell migration and invasion in vitro and tumorigenicity in vivo (21). In the present study, we found that FOXM1-overexpression also enhances cell migration, in addition to effects on proliferation. These observations raise the possibility that EPS8 and FOXM1 may mediate cell migration through a series of common downstream mediators.
Funding
Supported in part by the Commonwealth Health Research Board of the State Council of Higher Education for Virginia through a grant to W.A.Y. and by the intramural programs of the National Institute of Dental and Craniofacial Research.
Supplementary material
Supplementary Data, Tables 1–4 and Figures 1–9 can be found at http://carcin.oxfordjournals.org/
Supplementary Material
Acknowledgments
We thank Sumitra Deb and Catherine Vaughan for their advice and help with chromatin immunoprecipitation experiments.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BrdU
2-bromodeoxyuridine
- dnAKT
dominant-negative form of AKT
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EPS8
epidermal growth factor receptor pathway substrate 8
- PCR
polymerase chain reaction
- PI3K
phosphoinositide 3-OH kinase
- qRT–PCR
quantitative real-time polymerase chain reaction
- SDS
sodium dodecyl sulfate
- RNAi
RNA interference
- shRNA
short hairpin RNA
References
- 1.Ozanne B, et al. Over-expression of the EGF receptor is a hallmark of squamous cell carcinomas. J. Pathol. 1986;149:9–14. doi: 10.1002/path.1711490104. [DOI] [PubMed] [Google Scholar]
- 2.Salomon DS, et al. Loss of growth responsiveness to epidermal growth factor and enhanced production of alpha-transforming growth factors in ras-transformed mouse mammary epithelial cells. J. Cell Physiol. 1987;130:397–409. doi: 10.1002/jcp.1041300313. [DOI] [PubMed] [Google Scholar]
- 3.Prime SS, et al. Epidermal growth factor and transforming growth factor alpha characteristics of human oral carcinoma cell lines. Br. J. Cancer. 1994;69:8–15. doi: 10.1038/bjc.1994.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prigent SA, et al. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J. Biol. Chem. 1996;271:25639–25645. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- 5.Coso OA, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 6.Minden A, et al. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 7.Fazioli F, et al. Identification and biochemical characterization of novel putative substrates for the epidermal growth factor receptor kinase. J. Biol. Chem. 1992;267:5155–5161. [PubMed] [Google Scholar]
- 8.Kishan KV, et al. The SH3 domain of Eps8 exists as a novel intertwined dimer. Nat. Struct. Biol. 1997;4:739–743. doi: 10.1038/nsb0997-739. [DOI] [PubMed] [Google Scholar]
- 9.Castagnino P, et al. Direct binding of eps8 to the juxtamembrane domain of EGFR is phosphotyrosine- and SH2-independent. Oncogene. 1995;10:723–729. [PubMed] [Google Scholar]
- 10.Fazioli F, et al. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matoskova B, et al. Constitutive phosphorylation of eps8 in tumor cell lines: relevance to malignant transformation. Mol. Cell. Biol. 1995;15:3805–3812. doi: 10.1128/mcb.15.7.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matoskova B, et al. RN-tre specifically binds to the SH3 domain of eps8 with high affinity and confers growth advantage to NIH3T3 upon carboxy-terminal truncation. Oncogene. 1996;12:2679–2688. [PubMed] [Google Scholar]
- 13.Biesova Z, et al. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene. 1997;14:233–241. doi: 10.1038/sj.onc.1200822. [DOI] [PubMed] [Google Scholar]
- 14.Inobe M, et al. Identification of EPS8 as a Dvl1-associated molecule. Biochem. Biophys. Res. Commun. 1999;266:216–221. doi: 10.1006/bbrc.1999.1782. [DOI] [PubMed] [Google Scholar]
- 15.Funato Y, et al. IRSp53/Eps8 complex is important for positive regulation of Rac and cancer cell motility/invasiveness. Cancer Res. 2004;64:5237–5244. doi: 10.1158/0008-5472.CAN-04-0327. [DOI] [PubMed] [Google Scholar]
- 16.Lanzetti L, et al. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 17.Scita G, et al. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- 18.Scita G, et al. An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J. Cell Biol. 2001;154:1031–1044. doi: 10.1083/jcb.200103146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Disanza A, et al. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat. Cell Biol. 2004;6:1180–1188. doi: 10.1038/ncb1199. [DOI] [PubMed] [Google Scholar]
- 20.Leu TH, et al. Participation of p97Eps8 in Src-mediated transformation. J. Biol. Chem. 2004;279:9875–9881. doi: 10.1074/jbc.M309884200. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, et al. Role for EPS8 in squamous carcinogenesis. Carcinogenesis. 2009;30:165–174. doi: 10.1093/carcin/bgn252. [DOI] [PubMed] [Google Scholar]
- 22.Yeudall WA, et al. Uncoupling of epidermal growth factor-dependent proliferation and invasion in a model of squamous carcinoma progression. Oral Oncol. 2005;41:698–708. doi: 10.1016/j.oraloncology.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni PS, et al. Characterization of human buccal epithelial cells transfected with the simian virus 40 T-antigen gene. Carcinogenesis. 1995;16:2515–2521. doi: 10.1093/carcin/16.10.2515. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki H, et al. Downregulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006;66:4279–4284. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]
- 25.Teh MT, et al. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- 26.Li W, et al. Protein kinase C-alpha overexpression stimulates Akt activity and suppresses apoptosis induced by interleukin 3 withdrawal. Oncogene. 1999;18:6564–6572. doi: 10.1038/sj.onc.1203065. [DOI] [PubMed] [Google Scholar]
- 27.Keates AC, et al. ZBP-89, Sp1, and nuclear factor-kappa B regulate epithelial neutrophil-activating peptide-78 gene expression in Caco-2 human colonic epithelial eells. J. Biol. Chem. 2001;276:43713–43722. doi: 10.1074/jbc.M107838200. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, et al. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeudall WA, et al. MTS1/CDK4I is altered in cell lines derived from primary and metastatic oral squamous cell carcinoma. Carcinogenesis. 1994;15:2683–2686. doi: 10.1093/carcin/15.12.2683. [DOI] [PubMed] [Google Scholar]
- 30.Wrighton KH, et al. Aberrant p53 alters DNA damage checkpoints in response to cisplatin: downregulation of CDK expression and activity. Int. J. Cancer. 2004;112:760–770. doi: 10.1002/ijc.20446. [DOI] [PubMed] [Google Scholar]
- 31.Jakus J, et al. Growth inhibitory concentrations of EGF induce p21 (WAF1/Cip1) and alter cell cycle control in squamous carcinoma cells. Oncogene. 1996;12:2369–2376. [PubMed] [Google Scholar]
- 32.Wonsey DR, et al. Loss of the forkhead transcription factor FOXM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 33.Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 34.Huang DW, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Cardinali M, et al. Tyrosine phosphorylation as a marker for aberrantly regulated growth-promoting pathways in cell lines derived from head and neck malignancies. Int. J. Cancer. 1995;61:98–103. doi: 10.1002/ijc.2910610117. [DOI] [PubMed] [Google Scholar]
- 36.Christofakis EP, et al. Roles of CXCL8 in squamous cell carcinoma proliferation and migration. Oral Oncol. 2008;44:920–926. doi: 10.1016/j.oraloncology.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Innocenti M, et al. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang IC, et al. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27:4137–4149. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 39.Costa RH. FOXM1 dances with mitosis. Nat. Cell Biol. 2005;7:108–110. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 40.Laoukili J, et al. FOXM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 41.Gemenetzidis E, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS ONE. 2009;4:e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazaki H, et al. Growth factor-sensitive molecular targets identified in primary and metastatic head and neck squamous cell carcinoma using microarray analysis. Oral Oncol. 2006;42:240–256. doi: 10.1016/j.oraloncology.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Itoh Y, et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine. 2005;29:275–282. doi: 10.1016/j.cyto.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Luppi F, et al. Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation. Lung Cancer. 2007;56:25–33. doi: 10.1016/j.lungcan.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 45.Metzner B, et al. Overexpression of CXC-chemokines and CXC-chemokine receptor type II constitute an autocrine growth mechanism in the epidermoid carcinoma cells KB and A431. Oncol. Rep. 1999;6:1405–1410. doi: 10.3892/or.6.6.1405. [DOI] [PubMed] [Google Scholar]
- 46.Schraufstatter IU, et al. IL-8 activates endothelial cell CXCR1 and CXCR2 through Rho and Rac signaling pathways. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001;280:L1094–L1103. doi: 10.1152/ajplung.2001.280.6.L1094. [DOI] [PubMed] [Google Scholar]
- 47.Singh S, et al. CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab. Invest. 2004;84:1666–1676. doi: 10.1038/labinvest.3700181. [DOI] [PubMed] [Google Scholar]
- 48.Rehman AO, et al. SDF-1{alpha} promotes invasion of head and neck squamous cell carcinoma by activating NF-{kappa}B. J. Biol. Chem. 2008;283:19888–19894. doi: 10.1074/jbc.M710432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molinolo AA, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin. Cancer Res. 2007;13:4964–4973. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- 50.O-Charoenrat P, et al. Signaling pathways required for matrix metalloproteinase-9 induction by betacellulin in head-and-neck squamous carcinoma cells. Int. J. Cancer. 2004;111:174–183. doi: 10.1002/ijc.20228. [DOI] [PubMed] [Google Scholar]
- 51.Sodhi A, et al. Akt plays a central role in sarcomagenesis induced by Kaposi's sarcoma herpesvirus-encoded G protein-coupled receptor. Proc. Natl Acad. Sci. USA. 2004;101:4821–4826. doi: 10.1073/pnas.0400835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandrasekar B, et al. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I kappa B kinase, and nuclear factor-kappa B and induces cell-cell adhesion and aortic smooth muscle cell proliferation. J. Biol. Chem. 2004;279:3188–3196. doi: 10.1074/jbc.M311660200. [DOI] [PubMed] [Google Scholar]
- 53.Bonacchi A, et al. Signal transduction by the chemokine receptor CXCR3: activation of Ras/ERK, Src, and phosphatidylinositol 3-kinase/Akt controls cell migration and proliferation in human vascular pericytes. J. Biol. Chem. 2001;276:9945–9954. doi: 10.1074/jbc.M010303200. [DOI] [PubMed] [Google Scholar]
- 54.Limatola C, et al. Signaling pathways activated by chemokine receptor CXCR2 and AMPA-type glutamate receptors and involvement in granule cells survival. J. Neuroimmunol. 2002;123:9–17. doi: 10.1016/s0165-5728(01)00472-6. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, et al. Diverse signaling pathways through the SDF-1/CXCR4 chemokine axis in prostate cancer cell lines leads to altered patterns of cytokine secretion and angiogenesis. Cell. Signal. 2005;17:1578–1592. doi: 10.1016/j.cellsig.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, et al. Chemokine receptor 7 activates phosphoinositide-3 kinase-mediated invasive and prosurvival pathways in head and neck cancer cells independent of EGFR. Oncogene. 2005;24:5897–5904. doi: 10.1038/sj.onc.1208740. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J. Biol. Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 58.Li Q, et al. Critical role and regulation of transcription factor FOXM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr. Pharm. Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 60.Dano K, et al. Plasminogen activation and cancer. Thromb. Haemost. 2005;93:676–681. doi: 10.1160/TH05-01-0054. [DOI] [PubMed] [Google Scholar]
- 61.Lester RD, et al. uPAR induces epithelial mesenchymal transition in hypoxic breast cancer cells. J. Cell Biol. 2007;178:425–436. doi: 10.1083/jcb.200701092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dass K, et al. Evolving role of uPA/uPAR system in human cancers. Cancer Treat. Rev. 2008;34:122–136. doi: 10.1016/j.ctrv.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Spence HJ, et al. Krp1, a novel kelch related protein that is involved in pseudopod elongation in transformed cells. Oncogene. 2000;19:1266–1276. doi: 10.1038/sj.onc.1203433. [DOI] [PubMed] [Google Scholar]
- 64.Spence HJ, et al. AP-1 differentially expressed proteins Krp1 and fibronectin cooperatively enhance Rho-ROCK-independent mesenchymal invasion by altering the function, localization, and activity of nondifferentially expressed proteins. Mol. Cell. Biol. 2006;26:1480–1495. doi: 10.1128/MCB.26.4.1480-1495.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.