Abstract
SSBP proteins bind and stabilize transcriptional cofactor Lim Domain Binding protein1 (LDB1) from proteosomal degradation to promote tissue specific transcription through an evolutionarily conserved pathway. The human SSBP2 gene was isolated as a candidate tumor suppressor from a critical region of loss in chromosome 5q14.1. By gene targeting, we demonstrate increased predisposition to B cell lymphomas and carcinomas in Ssbp2−/− mice. Remarkably, loss of Ssbp2 causes increased LDB1 turnover in the thymus, a pathway exploited in Trp53−/− Ssbp2−/− mice to develop highly aggressive, immature thymic lymphomas. Using T cell differentiation as a model, we report a stage specific up regulation of Ssbp2 expression which in turn regulates LDB1 turnover under physiological conditions. Furthermore, transcript levels of pTα, a target of LDB1 containing complex, and a critical regulator T cell differentiation is reduced in Ssbp2−/− immature thymocytes. Our findings suggest disruption of the SSBP2 regulated pathways may be an infrequent but critical step in malignant transformation of multiple tissues.
Introduction
The single-stranded DNA binding proteins (SSBPs [SSBP2, 3 and 4]), interact with the transcriptional cofactor LIM domain binding protein (LDB1) through a highly conserved amino terminal motif (Chen et al., 2002; van Meyel et al., 2003). LDB1 in turn binds to the LIM domain of LIM Homeodomain (LHX) and LIM only (LMO) proteins with high affinity to assemble multi-protein complexes(Agulnick et al., 1996; Goardon et al., 2006). The activity of LDB1-containing complexes is also regulated by the E3 ubiquitin ligase ring finger protein 12 (RNF12/RLIM), which promotes rapid turnover of LDB1(Bach et al., 1999). As LIM domains do not bind DNA but mediate protein-protein interactions, target genes are recognized by the homeodomain in LHX or by LMO interaction with E box binding basic helix loop helix (bHLH) transcription factors(Goardon et al., 2006). Despite the genetic evidence for stringent regulation of these LIM domain-containing complexes, the contribution of individual SSBP members in regulating LDB1 levels in vivo is unknown.
Human SSBP2 was isolated as a candidate myeloid leukemia suppressor from a critical region of loss at 5q13-14(Castro et al., 2002). Independent of the gene discovery, global expression profiling revealed loss of SSBP2 expression in a subset of therapy induced acute myeloid leukemia (AML) with 5q deletion (Qian et al., 2002). Inducible expression of SSBP2 in the AML cell line U937 resulted in growth arrest and partial differentiation(Liang et al., 2005). Furthermore, adenoviral oncoprotein E1B55K directly binds and sequesters SSBP2 to juxtanuclear aggresomes, suggesting a broader role for disruption of normal SSBP2 function during transformation (Fleisig et al., 2007). Accumulating evidence suggests a tumor suppressor function for SSBP2: (i) Retroviral integration-mediated disruption leading to reduced expression of SSBP2 appeared to cooperate with homeobox B4 (HOXB4)-induced AML in a primate (Zhang et al., 2008). (ii) A reciprocal translocation resulting in an SSBP2-JAK2 fusion capable of functioning as a dominant negative SSBP2 allele was detected as the sole anomaly in an adult with B precursor lymphoblastic leukemia (Poitras et al., 2008). (iii) Finally, SSBP2 is frequently silenced through epigenetic mechanisms in prostate cancer in contrast to benign prostatic hyperplasia (Liu et al., 2008).
Mice hypomorphic for Ssbp3 resemble Lhx1 nulls with aberrant head structures and embryonic lethality demonstrating SSBP3-LDB1- LHX1 interactions to be essential for normal development. Whether Ssbp2, with a >80% overall identity to Ssbp3 and 100% identity in the LDB1 interaction domain, is necessary for embryonic development is unknown. Thus, the roles of Ssbp2 in embryonic development and tumor suppression are important issues to resolve. Here we report Ssbp2 to be dispensable for development. However, genetic ablation of Ssbp2 causes a broad range of oncogenic events, supporting its in vivo tumor suppressor activity.
Materials and methods
Targeted disruption of murine Ssbp2 gene
A mouse Ssbp2 cDNA clone was used to screen a lambda phage mouse (129/SvEv) genomic library to obtain part of the gene. A targeting vector was designed to replace exon 1 with PGK-Neo flanked by 3.1 kbp 5’ and 3.7 kbp 3’ genomic sequences from the Ssbp2 locus and thymidine kinase for double selection. Targeted recombinants were verified by Southern blot analysis with unique 5′ and 3′ probes outside the targeted locus (Fig. 1A). Mouse embryonic stem (ES) transfected with the targeting vector underwent double selection prior to microinjection into C57/BL6 blastocysts. Chimeric offsprings were crossed with C57/BL6 mice and germ line transmission verified. Ssbp2−/− mice were crossed to Trp53−/− mice in a BALB6xC57/BL6 mixed background, a gift from Koch and Lozano (Koch et al., 2007), to generate Ssbp2+/− Trp53+/− mice, which were then bred to generate double null mice. Genotyping for Trp53 was performed by polymerase chain reaction (PCR) as described previously (Jacks et al., 1994). All mice were maintained in a conventional specific-pathogen-free (CSPF) facility according to NIH guidelines using an approved Animal Care and Use Committee protocol. The mice were monitored on a daily basis and euthanized when moribund.
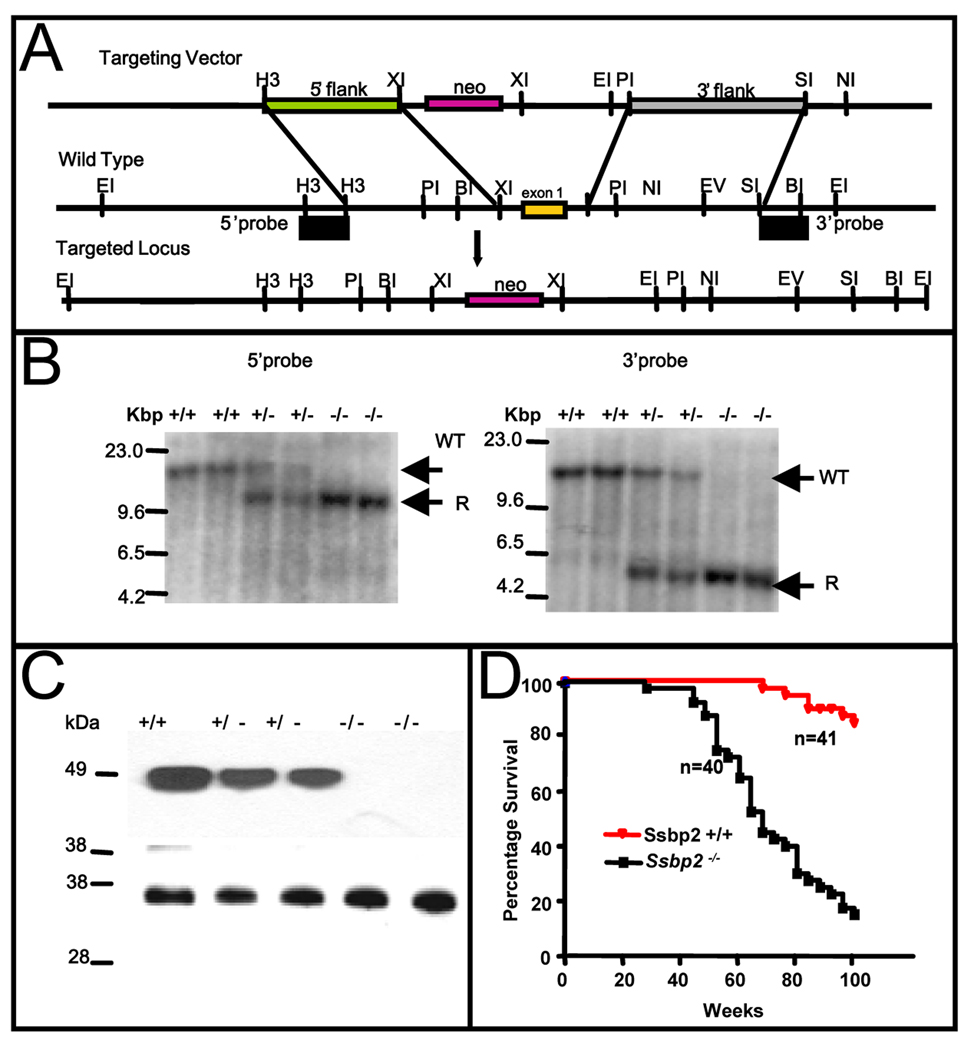
Figure 1. Targeted disruption of the murine Ssbp2 locus.
(A) Partial restriction maps of the targeting vector, mouse Ssbp2 genomic locus and the expected targeted allele are depicted. Ssbp2 exon 1 containing the initiation ATG was replaced with a neomycin resistance gene in the plasmid vector pPNT1. Unique single copy fragments flanking the integration are shown. The restriction sites indicated are: BI (BamH1), EI (Eco RI), EV (Eco RV), H3 (HindIII) NI (NotI), PI (PstI), SI (SpeI) and XI (XhoI). The probes used were derived from the 5’ HindIII-HindIII and 3’ SpeI-BamHI fragments. These probes detected different sized wild type and rearranged fragments containing the inserted neo casette in DNAs digested with Eco RI (B) Southern blot analysis with 5’ probe or 3’ probe of Eco RI-digested tail DNA from littermates obtained from intercrossing Ssbp2+/− mice. Germline DNA yielded a 13.9 kbp fragment when cleaved with EcoRI. The knockout allele gave 10.1 kbp and 4.5 kbp fragments, respectively, with the 5’ and 3’ probes. Unique wild type (WT) and recombinant (R) fragments are denoted. (C) Immunoblotting of nuclear lysate from Ssbp2+/+, Ssbp2+/− and Ssbp2−/− mice. 30 µg of protein aliquots from nuclear lysates of 4 week-old mice thymi were separated on a 4–12% NuPage gel, transferred to a Hybond-P PVDF membrane, and probed with SSBP2 antibody. Lower panel shows Ponceau staining carried out to visualize overall protein loading. (D) Kaplan-Meier survival curve showing the percent survival of Ssbp2 −/− (n=41) and Ssbp2 +/+ (n=40) cohort mice as a function of age in weeks. Both groups of mice were monitored for greater than two years and sacrificed when moribund. Survival of SSBP2+/+ and SSBP2−/− differed significantly (P < 0.0001).
Immunohistochemistry
Separate sections were immunostained after antigen retrieval over a steaming citrate (0.01M, pH6) bath, after which anti-CD3ζ monocolonal antibody(1:100, sc-1239, Santa Cruz) or anti-B220 monocolonal antibody(1:100, sc-19597, Santa Cruz) and biotinylated link universal antibody (DAKO) and DakoCytomation LSAB+System-HRP staining reagents (k0690, DAKO) were applied.
Flow cytometry
Single-cell suspensions were prepared from thymi or tumors by dispersing the organs between two glass slides, passing them through mesh filters. Cells were briefly suspended in hypotonic buffer (0.15mM NH4Cl, 1mM KHCO3, and 300µM EDTA) to lyse red blood cells prior to staining with the following conjugated antibodies: anti-CD4-Tri-Color, anti-CD8a-FITC anti-CD3-PE from In Invitrogen. To characterize the double negative population, cells were stained with antibodies for the lineage markers Ter119, Gr1, CD11b, CD3, CD4, and CD8 (Becton Dickinson) and the lineage-negative cells were stained with anti-CD25-PE and anti-CD44-FITC (Invitrogen). Flow cytometry was performed on either a Becton Dickson’s (BD) Fluorescence activated cell sorter (FACS)-Caliber or FACS-Aria instrument. FACS data were analyzed with WinMDI 2.8 software.
Western Blotting
Immunoblots were developed with rabbit polyclonal antibodies to SSBP2 (1:10,000) as detailed previously (Liang et al., 2005). Goat antibody to LDB1 (1:250), goat antibody to LMO2 (1:250) and goat antibody to E2A (1:250) were used under the same conditions. The mouse monoclonal antibody against human histone H1 (Santa Cruz Biotechnology, Santa Cruz, CA) was used as loading control.
Short term culture of T Lymphocytes
Thymi were collected from 6-week old mice and placed in 3 ml of tissue culture medium. Short term culture conditions were identical to those described by Baseta and Stutman (Baseta & Stutman, 2000). Single cell suspensions were prepared by passage through an 18G needle and cell strainer (BD Falcon #352350). Cells were cultured for an hour in a 100mm dish at a density of 2.2 × 106/ml prior to treatment with cycloheximide, harvested, washed with PBS. 10 µg of nuclear extracts were resolved in 10% NuPAGE gels (Invitrogen) and transferred to PVDF membranes for immunoblotting.
Reverse Transcriptase coupled Polymerase Chain Reaction (RT-PCR) Total RNA was prepared by QIAGEN’s RNeasy Micro Kit (No.74004) or Mini Kit according to manufacturer’s instructions. For first-strand cDNA synthesis, 1 µg of total RNA, 20pmol Oligo(dT)12–18 and 200 units SuperScript II Reverse transcriptase (Invitrogen) were mixed in a final volume of 20µl. 1µl cDNA thus generated was added to the 20µl PCR mixture containing 1xTaqMan Gene Expression Assay primers (Applied Biosystems) and 1xTaqMan Universal PCR Master Mix (4324018, Applied Biosystems). Each sample was amplified in triplicate. The PCR consisted of 40 cycles of denaturation at 95 °C for 15 sec, annealing and amplification at 60 °C for 60 sec in an ABI7900HT Sequence Detection System Machine (Applied Biosystems). The specific number for TaqMan Gene Expression Assay primers are: Ssbp2: Mm00452502_m1. Ldb1: Mm00440147_m1. pTα: Mm00478363_m1. Lmo2: Mm00493153_m1. 18S rRNA primers ( 4319413E) were used as internal control. The specific position and length of all these PCR products can be found on Applied Biosystems Website.
Results
Ssbp2 null mice are viable and fertile but have a shortened lifespan
The murine Ssbp2 gene, spanning 17 exons, is highly homologous to the human gene. To completely disrupt expression, a targeting vector was generated by replacing the first exon of Ssbp2 (encoding the initiation Met), with a neomycin resistance gene as detailed in Materials and Methods. The wild type and targeted alleles were distinguished by single copy probes flanking the 5’ and 3’ ends of the targeted locus (Fig.1A). As shown in Fig.1B, Southern blotting experiments verified accurate targeting. Absence of Ssbp2 expression was further confirmed by immunoblotting. In heterozygous mice, SSBP2 levels were reduced to about 50%, whereas no protein was detected in null animals (Fig.1C).
Mice homozygous for the Ssbp2 mutation (Ssbp2−/−) were born at a lower frequency (~20%) than the predicted Mendelian ratio of 25% in the mixed S129/SvEv xC57/BL6 mixed background (Supplementary Table I). This slight reduction in live births indicated a low penetrant lethality phenotype. Nonetheless, the general appearance of null mice was indistinguishable from that of wild type and heterozygous littermates in this mixed background. Total necropsies performed at three and six months of age were unremarkable. Additionally, there was no compensatory increase in Ssbp3 expression in the Ssbp2−/− mice (Y. Wang and L. Nagarajan unpublished results). Thus, despite extensive overlap in expression and >80% overall identity, targeted disruption of Ssbp2 yields an apparently different phenotype.
To investigate whether loss of Ssbp2 contributed to malignant transformation or other adult onset abnormalities, we monitored a cohort of Ssbp2+/+ (n = 41), and Ssbp2−/− (n = 40) mice for a period of two years. Kaplan-Meier analysis showed a significant decrease in overall survival for null mice compared to wild-type littermates, with 50% of null mice dead by 68 weeks of age (Fig.1D).
Ssbp2 null mice exhibit enhanced predisposition to lymphomas and carcinomas
To identify the cause of this premature lethality, total necropsies were performed on 13 moribund mice (Supplementary Table IIA). Histopathological analysis revealed disseminated lymphomas involving multiple organs in 6 of 13 mice (435, 486, 536, 572, 576 and 625). There were carcinomas in two out of 13 mice, involving the Hardian gland (559) and lung (637), and one mouse (373) had an adenoma of the lung. Other significant findings were polyarteritis, glomerular nephropathy, anemia, and degenerative joint disease. Additionally, total necropsies of 12 Ssbp2−/− “non-morbid” mice between 60–80 weeks of age (median age 68.1 weeks) revealed malignancies in six animals (Supplementary Table IIB). Four mice in this subset had microscopic lymphomas in multiple organs (414, 498, 538, and 617), one had squamous cell carcinoma at hock in tibia (616) and the sixth mouse had an adenocarcinoma of jejunum and adenoma of the lung (575). Mouse 498 also had an unclassified sarcoma in the abdomen. Analogous to the moribund mice, chronic glomerular nephropathy and systemic polyarteritis were other significant findings in these mice. Thus, 62% of moribund and 50% of non-moribund Ssbp2−/− mice developed full-blown malignancies between 60–80 weeks of age. Frequently, the lymphomas were accompanied by lymphopenia and pancytopenia (Supplementary Table III).
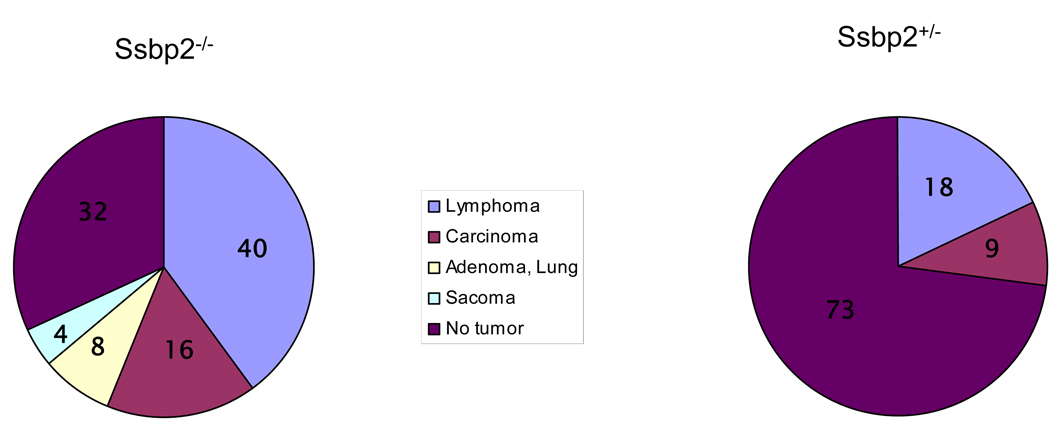
To determine whether Ssbp2+/− mice also show enhanced predisposition to malignancy we evaluated 11 age matched mice by total necropsy. Of these one mouse had a lymphoma and the second mouse had both a lymphoma and sarcoma. In contrast, no tumors were detected in age matched, wild type mice. The pie chart (Fig.2) summarizes the tumor spectrum in these mice.
Figure 2. Tumor spectrum in Ssbp2 null mice.
A) Pie chart depicts relative incidence of malignancies seen in Ssbp2−/− mice. A total of 14 tumors from 25 mice ( Supplementary Table II) was used to determine the frequencies. B) Eleven age matched Ssbp2+/− mice were sacrificed for total necropsy. Three malignancies were found in two mice. Numbers inside each slice denote the percentage of specific tumor type.
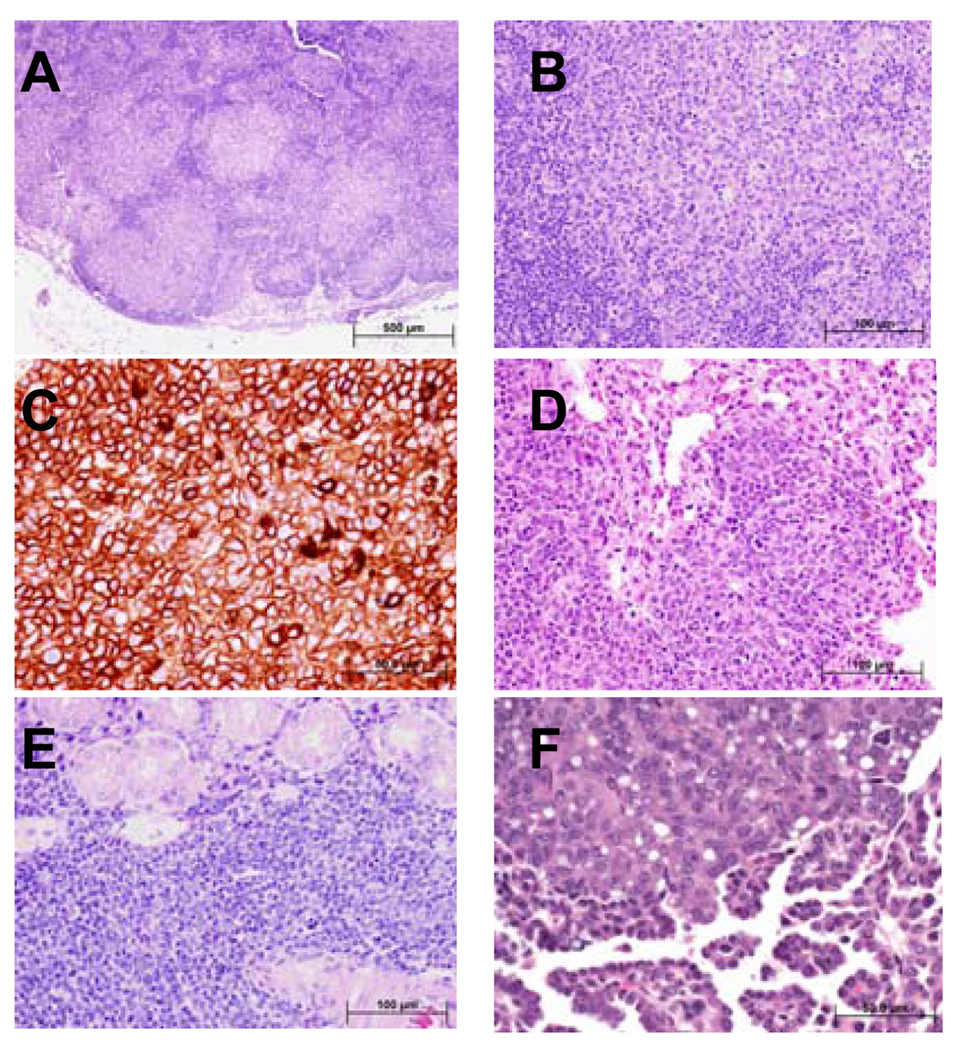
Figure 3A shows a representative lymph node from mouse 538, in which malignant lymphoid follicles effaced the normal nodal architecture. At higher magnification, the lymphoma cells appeared intermediate to large in size and contained abundant eosinophilic cytoplasm (Figure 3B). To better characterize the phenotype of the malignant lymphocytes, the tumors were examined for expression of the B and T cell markers B220 and CD3, respectively. Immunohistochemical staining detected strong B220 expression, supporting a B cell phenotype (Figure 3C). In contrast, this tumor was negative for CD3 (data not shown). The lymphomas had spread widely at the time of detection and had infiltrated parenchymal organs as shown in representative sections from the lung (mouse 576, Figure 3D) and salivary gland (mouse 617, Figure 3E). These findings are consistent with a high-grade large B cell lymphoma. Carcinomas constituted the second most common malignancy. Fig. 3F is an example of an aggressive bronchioalveolar carcinoma from mouse 637.
Figure 3. Ssbp2−/− mice develop disseminated high-grade large B-cell lymphoma and other malignancies.
A total of 25 moribund, age matched non-morbid mice were necropsied (Table II). (A) Histological sections from mandibular lymph node demonstrate total effacement of the normal architecture by neoplastic lymphoid follicles that lack the normal structure of the reactive lymphoid follicles including distinctive germinal centers. The neoplastic lymphoid follicles are arranged in a back-to-back pattern without significant interfollicular lymphoid tissue. (B) At higher magnification the cells within the neoplastic follicles are intermediate to large in size with round vesicular nuclei, occasional prominent 1–3 nucleoli, and abundant eosinophilic cytoplasm. Patchy increased mitotic figures are present. C. Immunohistochemical staining where the neoplastic lymphocytes are strongly positive for B220 supporting a B-cell immunophenotype. The histological and cytological features are most consistent with high-grade large B-cell lymphoma of follicle center cell origin. D Sections from the lung showing that most of the alveolar spaces are lacking because of extensive infiltration of the alveolar walls by the high-grade large B-cell lymphoma cells. The cytological features of the lymphoma cells are similar to the ones described in the lymph node shown in B. E. Extensive interstitial and diffuse infiltration of salivary gland by malignant lymphoma cells. The neoplastic lymphoma cells focally invade the glandular epithelial structures. The cytological features of the lymphoma cells are similar to the ones described in the lymph node shown in (B). F. Ssbp2−/− mice also developed carcinomas such as this low-grade adenocarcinoma of lung. The tumor is composed of bland glandular structures lined with cuboidal to low columnar cells that invade the pulmonary stroma. Bars denote the magnification.
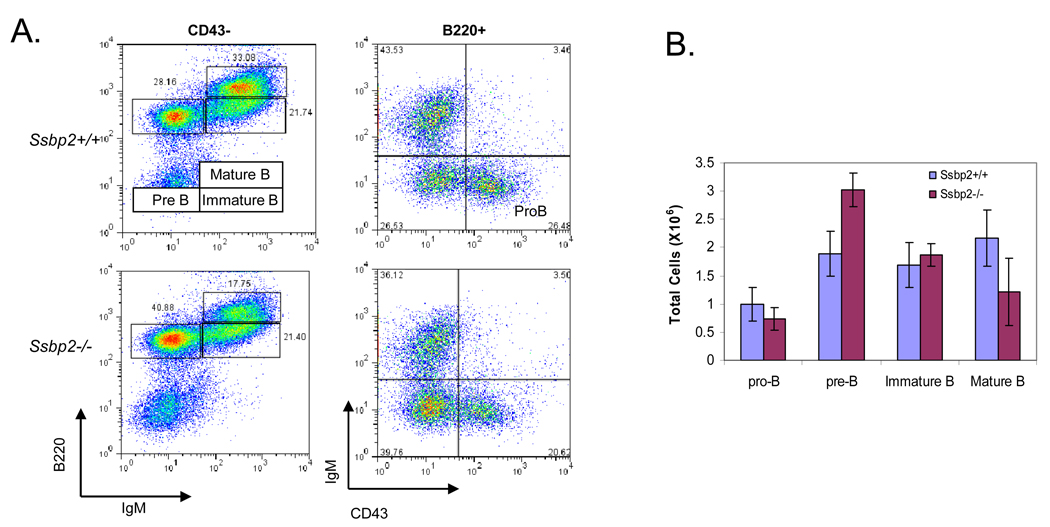
As B cell lymphomas were the most common malignancy, we investigated the status B cell differentiation in Ssbp2−/− null bone marrow. Of the four stages of B cell differentiation evaluated (proB, preB, immature and mature) a significant expansion of the pre B cell compartment was detected in the marrows from six week old Ssbp2−/− mice (Fig.4). The findings raise the possibility of a critical role for Ssbp2 in preB- immature B transition.
Figure 4. Expansion of the preB cell compartment in Ssbp2−/− bone marrow.
A. FACS Analysis of Pro-B cells (B220+CD43+IgM−), preB cells (B220+CD43−IgM−), immature B cells (B220low CD43− IgM+), and mature B cells (B220 hi CD43− IgM+ ) in bone marrow harvests cells from wild type and Ssbp2−/− mice. Data shown are representative flow cytometric analyses of five independent experiments. B. Summary of results shown in Fig. 4A.
Ssbp2 deficiency accelerates aggressive thymic lymphomas in Trp53 null mice
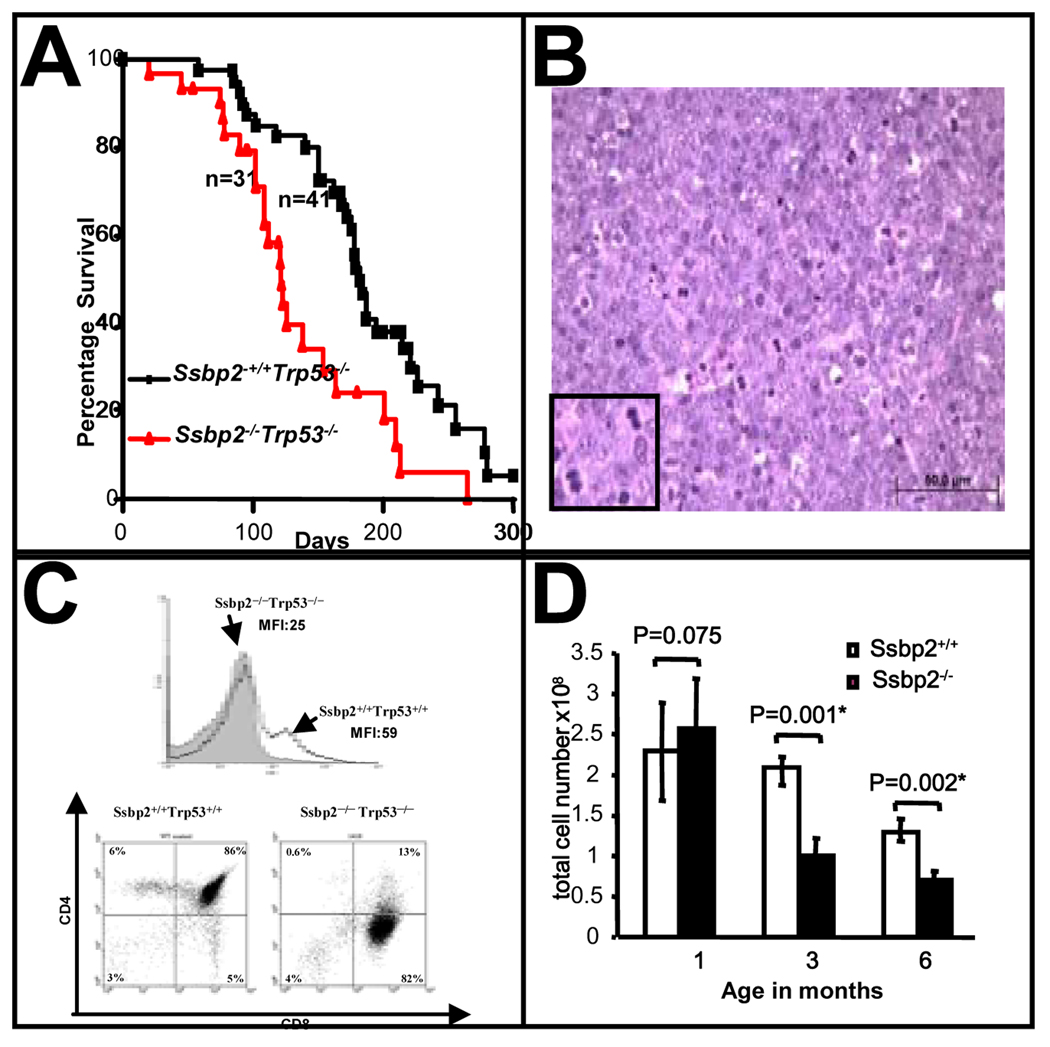
While the Ssbp2−/− mice were on tumor watch, we were uncertain of the tumorigenic phenotype. Furthermore, the tumors from Ssbp2−/− mice were frequently microscopic. To determine if loss Ssbp2−/− would cooperate with a known tumor suppressor Trp53−/− we derived double null mice. For these studies, we used Trp53 null mice in BALB6xC57/BL6 background mice which are predisposed to mammary carcinomas rather than lymphomas (Koch et al, 2007). We did not readily detect an exacerbation of carcinomas or B cell lymphomas in this mixed background. Unexpectedly, there was a 2.4 fold increase in thymic lymphomas. Twenty of 31 (65%) double null mice developed thymic lymphomas in contrast to eight of 30 (27%) Trp53−/− mice. As shown in Fig. 5A, the median survival of Ssbp2−/− Trp53−/− mice was shortened to 122 days from 182 days for the Trp53−/− mice (p=0.0019). Upon necropsy, large thymic masses infiltrating into the thoracic cavity were seen, and histologic examination revealed high grade tumors with abundant mitoses (Fig.5B).
Figure 5. Loss of Ssbp2 exacerbates aggressive thymic lymphomas in Trp53−/− mice.
(A) Kaplan- Meir survival plot of Ssbp2 −/− Trp53−/−, Ssbp2 +/+ Trp53−/− mice. Cohorts of Ssbp2 −/− Trp53−/− (n=31), Ssbp2 +/+ Trp53−/− (n=41) were monitored daily for 300 days for symptoms of disease and moribund mice were euthanized. The median survival was 122 days for the Ssbp2 −/− Trp53−/− mice compared to 182 days for the Ssbp2+/+ Trp53−/− mice. Statistical analysis (by Prism3 software) confirmed the difference in survival between the two groups to be significant (p=0.0019).
(B) Histological characteristics of thymic lymphomas in Ssbp2 −/− Trp53−/− mice Hematoxylin-Eosin stain of thymic tumor from a double knockout mouse. The lymphoma is composed of large lymphoid cells with high mitotic index. Inset shows representative mitotic nuclei. The neoplastic lymphocytes are round with vesicular nuclei, single centrally located nucleoli, and scant eosinophilic cytoplasm. Bar denotes the scale.
(C) Ssbp2 −/− Trp53 −/− thymic lymphomas are CD3lo ,CD4lowCD8+ (A) The gray shading shows expression of CD3 in the Ssbp2 −/−, Trp53−/− thymic lymphoma cells, compared with that of the Ssbp2 +/+, Trp53+/+ mouse. (B) The right panel is a representative for Ssbp2 −/−, Trp53−/− thymic lymphoma, in which most of the cells are CD8+, CD4 low. The left panel is a wild type control (Ssbp2 +/+, Trp53+/+) thymus in which most of the cells are CD4+,CD8+.
(D) Thymic hypoplasia is accelerated in Ssbp2−/− mice
Thymi from Ssbp2+/+ and Ssbp2−/− mice were harvested at the indicated time points, minced, single cells isolated and counted. Each group includes a minimum of four mice.
The striking exacerbation of large thymic lymphomas in the double null mice, together with the relative abundance of SSBP2 transcripts in the normal thymus (Castro et al., 2002) provided an excellent model for further characterization of tumors lacking SSBP2. T cell maturation is orchestrated in a stepwise regulatory process, with two major checkpoints at the double negative 3 –double negative 4 (DN3-DN4) and double positive – single positive (DP-SP) transitions (Rothenberg, 2007a). More importantly, LDB1-, LMO2- and basic helix-loop-helix factor (bHLH)-containing transcriptional complexes are central to this process(Rothenberg, 2007b). In the transition from the DN3 to DN4 stage, cells that do not have a functional T cell receptor β (TCR β) protein are eliminated, while auto-reactive T cells are selected against in the DP to SP transition. First, to determine how a potentially impaired differentiation program impacted transformation, we examined the tumors for surface markers. Most tumors that developed in the Ssbp2−/− Trp53−/− mice were low for surface CD3 (CD3lo) and either CD4loCD8+ or CD4−CD8+ single positive (Figure 5C, Supplementary Table IV), in contrast to the Trp53−/− thymic lymphomas that were CD8hiCD4hi double positive, CD4+ high, CD8+ high and varied CD3 expression (Supplementary Table IV). Unlike normal CD8+ thymocytes or DP lymphoma cells, the CD8+ population from these tumors expressed low levels of CD3, abundant LMO2 (Supplementary Fig.1), suggesting the notion of an immature cell of origin. The thymic lymphomas used for the LMO2 analyses were from mice 1521 (21.4% CD8+, 42% CD4+CD8+) and 1386(59.4% CD8+, 6.4% CD4+CD8+ ) for the double null mice and 1320 (0.5% CD8+, 95.6% CD4+CD8+ ) and 1332(72.5% CD8+, 23.6% CD4+CD8+ ) for the Trp53 null mice. Of these, the Trp53−/− tumor 1320 is double positive, a stage at which LMO2 is not expressed. But in mouse 1332 despite the high CD8 expression the LMO2 levels were not high in contrast to tumors from mice 1521 and 1386. These findings also raise the possibility of an overall increase in LMO2 expression in the double null thymi or an expansion of the LMO2 expressing pre T population in the premalignant thymus. However, LMO2 levels in four weeks old thymi from Ssbp2−/− Trp53−/− were not higher than those of single nulls or wild type ( Wang and Nagarajan unpublished results). Thus the target cell for transformation in the Ssbp2−/− Trp53−/− appears to be an LMO2 expressing immature thymocyte.
It is also possible that expression of CD4, a known transcriptional target of E2A- and LDB1-containing complexes, as well as other T cell specific differentiation markers, was specifically inhibited or delayed in Ssbp2 null mice. A subtle alteration in the T cell differentiation program in either the DP stage or DN4→DP transition, resulting in a clone that would normally be eliminated, appears to be the target of transformation in the absence of Trp53. To determine whether Ssbp2 plays a stage-specific role in T cell maturation, we examined its expression in cells selected by cell surface phenotype. Although Ssbp2 expression is detectable throughout T cell differentiation, there is a dramatic increase at the DP stage and a subsequent decrease at the SP stage (Supplementary Fig.2), suggesting a role for Ssbp2 in DP cell homeostasis or DN4-DP transition.
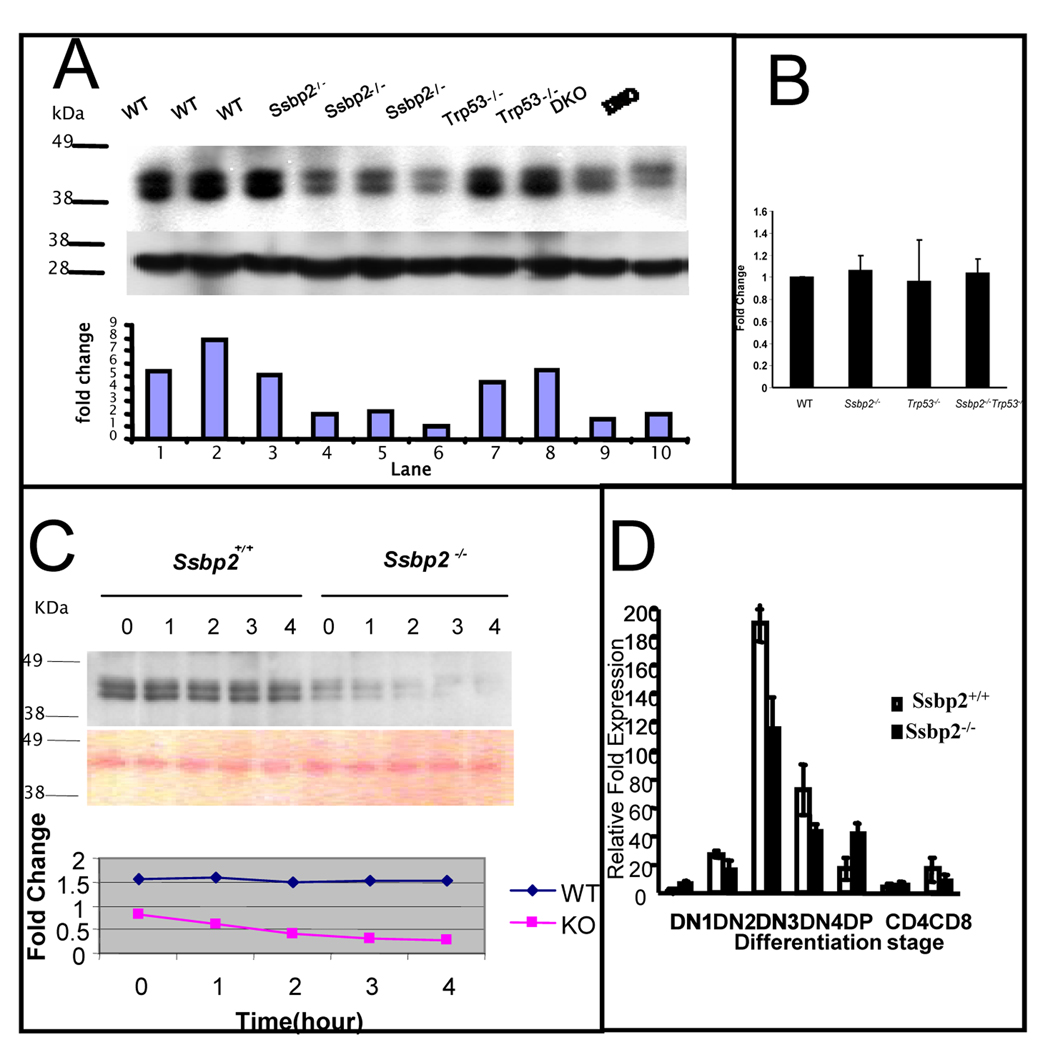
When examined for histological alterations, the Ssbp2−/− thymi were normal with distinct cortical and medullary zones. While their sizes were similar at 1 month, Ssbp2−/− thymi were smaller at 3 and 6 months of age. This reduction in thymic size was reflected in a substantial decrease in cellularity at 3 and 6 months of age (Fig.5D). However, the percentage of DP or CD4, SP cells estimated by FACS analysis was not significantly different between the Ssbp2−/− mice and wild type littermates (data not shown). Thus, genetic ablation of Ssbp2 appears to accelerate thymic atrophy analogous to oncogenic models of stem cell leukemia hematopoietic transcription factor (SCL)/LMO1 double transgenics, SCL transgenic in an E2A and HEB heterozygous background, and E2A-HLF transgenic mice (O'Neil et al., 2004; Smith et al., 1999). If the cooperation between Ssbp2 and Trp53 is mediated by a dominant Ssbp2 dependent mechanism, the thymocytes from Ssbp2−/− and Ssbp2−/−Trp53−/− mice should express this phenotype. If homozygous loss of Ssbp2 was necessary and sufficient to enhance LDB1 turnover under physiological conditions, it may be causally linked to the accelerated lymphomagenesis. As shown in Fig.6A, LDB1 protein levels were diminished by > 50% in the Ssbp2−/− thymocytes. Importantly, LDB1 expression was considerably reduced in double null thymocytes. In contrast, LDB1 levels in Trp53−/− null thymocytes were > 80% of that in wild type cells. Fig. 6B demonstrates that the differences in LDB1 protein levels were not due to transcriptional down-regulation of Ldb1, as its mRNA did not vary by >10% between the strains. To confirm that the decrease in LDB1 levels was due to enhanced turnover, when the thymocytes from wild type and null mice were placed in short term culture and protein synthesis inhibited with cycloheximide, LDB1 levels decreased rapidly in null thymocytes. The estimated half life of LDB1 in null thymocytes was 2 hours compared to > 4 hours in wild type thymocytes (Fig. 6C). In contrast to LDB1, E47 protein abundance and half life were unaltered in the Ssbp2−/− thymocytes (Supplementary Fig.3). These findings thus establish that SSBP2 is a direct regulator of LDB1 protein levels in vivo.
Figure 6. Decreased LDB1 half life in Ssbp2−/− thymocytes underlies impaired T cell differentiation.
(A) LDB1 levels are decreased in the thymi of Ssbp2−/− mice. Nuclear proteins from 4 weeks old wild type, Ssbp2 −/−, Trp53−/− and Ssbp2 −/− Trp53−/− mice thymi were separated on a 4–12% NuPage gel, transferred to nitrocellulose and probed with Ldb1 antibody. Lower panel shows Ponceau staining to denote equal loading. The signals were analyzed by IMAGE analysis software and the ratio for each sample is denoted. Representative samples from triplicate experiments are shown.
(B) Ldb1 transcript levels are not altered in Ssbp2−/− thymocytes
Ssbp2 transcripts in four weeks old thymi were estimated by Real-time PCR. Results show that Ldb1 transcript levels were similar between all the four genotypes tested. Data represent average of two separate experiments each with triplicate reactions.
(C) LDB1 half life is shortened in the absence of Ssbp2
Short term cultured thymocytes from wild type and Ssbp2−/− mice were treated with cycloheximide for the indicated lengths of time and whole cell lysates isolated were examined for LDB1 levels by immunoblotting. Protein loading was quantified by Ponceau staining. The signals were analyzed by IMAGE analysis software and the ratios at various time points plotted to determine LDB1 half life.
(D) preTα expression is decreased in Ssbp2−/− thymus. Thymocytes from four weeks old mice were divided into seven subsets based on maturity. Real-time PCRs were performed on purified cell populations using specific primers for pTα. For each cDNA pool, transcript levels were normalized against 18sRNA within that sample. Two separate experiments, each with triplicate reactions were done.
To determine whether the altered LDB1 stability affected transcriptional targets in vivo, we examined the surrogate T cell receptor (TCR) preTα, a direct transcriptional target of SCL-, LMO2-, and LDB1-containing complexes (Tremblay et al., 2003). In fact, preTα expression was significantly reduced in the DN3 and DN4 stages of Ssbp2−/− thymocytes (Fig.6D), analogous to E2A and HEB null mice, suggesting SSBP2 may be a limiting component of LMO2-, E2A-, and HEB-containing complexes. In contrast, preTα regulation in DP thymocytes appeared to be independent of SSBP2, reminiscent of E2A-null DP thymocytes which are characterized by decreased expression in DN stages and not DP cells (Tremblay et al., 2003). Impaired pre TCR signaling in Trp53−/− mice causes transformation, (Haks et al., 1999). In aggregate these results suggest homozygous loss of Ssbp2 provides a window for rapid transformation in the absence of Trp53.
Discussion
We establish the tumor suppressor activity of SSBP2 in this report. The development of multiple malignancies, including both lymphomas and carcinomas, in the Ssbp2−/− mice (Fig. 2 and 3, supplementary Table II), demonstrates the in vivo suppressor activity. While B cell lymphoma was the predominant malignancy observed, the development of carcinomas and sarcomas underscore how LMO/LHX-LDB1-SSBP interaction might play a fundamental role in inhibiting neoplastic transformation in multiple tissues.
Although the precise levels of LHX, LMO, and LIM-binding proteins have been shown to be important for a number of developmental programs, accumulating evidence implicates the context dependent function of altered stochiometry of these complexes during oncogenesis. Together with the established oncogenic activity of LMO1 and LMO2 in human T cell lymphomas, LMO3 in neuroblastoma, and LMO4 in breast cancer and carcinoma of the oral cavity, (Aoyama et al., 2005; Nam & Rabbitts, 2006; Sum et al., 2005), over expression of LHX2 in chronic myeloid leukemia (CML) blast crisis (Wu et al., 1996) our findings reveal another component of a pathway affected in multiple malignancies. Preferential retroviral integrations into the Ssbp3 locus were identified when Trp53−/− mice were challenged with retroviruses(Uren et al., 2008). LDB1 and RNF12, are also cofactors for estrogen receptor α (ERα) regulated target genes. In contrast to LHX mediated transcription, LDB1 inhibits ERα mediated activation of target genes. More importantly, elevated LDB1 expression correlated with ER, PR (progesterone receptor) positivity and impaired differentiation in primary breast tumors (Johnsen et al., 2009). Inactivation of SSBP2 and inactivation/ activation of related members SSBP3 and 4 may be a common mechanism to perturb normal differentiation regulated by LMO, HLH containing complexes during oncogenesis.
Ssbp2, a suppressor of B cell lymphogenesis
The data presented here reveal SSBP2 to be a critical tumor suppressor in B lymphoid neoplasms. Consistent with this notion, deletions of 5q13-14 have been reported in a distinct subset of human diffuse large B cell lymphomas (DLBCL) lacking BCL6 rearrangements (Cerretini et al., 2006). Moreover, global expression profiling to distinguish Burkitt lymphoma from DLBCL identified high SSBP2 expression in Burkitt lymphoma compared to DLBCL(Dave et al., 2004). Thus, the predisposition of Ssbp2−/− mice to B cell lymphoma provides the first clues on the significance of this pathway in DLBCL. A regulatory function for the LDB1-interacting LIM domain protein LMO2 has not been established in pro B-lymphocytes, although aberrant expression of LMO2 is a marker for human DLBCL (Natkunam et al., 2008). Of note, the E box binding proteins E2A, LYL1, and HEB, all, potentially, members of LMO2- and LDB1-containing transcriptional complexes, are also critical regulators of B cell differentiation. Mice with targeted disruption of LYL1, HEB and E2A showed impaired B cell differentiation (reviewed in (Murre, 2005, Capron et al., 2006). Our preliminary analyses of bone marrow derived B cells show a modest increase in pre B cells from 6 weeks old Ssbp2−/− mice (Fig.4) recapitulating some of the alterations seen in the b-HLH null mice. Further studies inpurified strains will determine how the subtle alteration in B cell program contributes to lymphomagenesis.
Loss of Ssbp2 cooperates with Trp53 deletion through decreased LDB1 stability
Thymic lymphomas are significantly exacerbated in the absence of Trp53 (Fig 5 and Fig 6). Abundant LMO2 expression in these tumors suggests these cells to be immature thymocytes (DN). Interestingly LMO2, LHX2 levels are stabilized in an LDB1 dependent manner in erythroid and pituitary cells (Cai et al., 2008; Gungor et al., 2007; Xu et al., 2007). Abundant LMO2 in the context of rapidly turning over LDB1 raises a possible SCL/HEB mediated stabilization during specific stages of differentiation(Lecuyer et al., 2007). Our findings implicate compromised checkpoint responses in the absence of Ssbp2 and Trp53 to underlie the accelerated transformation seen in these cells. Indeed, mice with aberrant TCRβ rearrangement due to inactivation of the cis element Eβ (Eβ−/−, EβR/R) show impaired differentiation. Loss of Trp53 in these mice induces CD8+ T cell lymphomas (Haines et al., 2006).
In conclusion, Ssbp2 is a potent in vivo tumor suppressor and modulator of LDB1 activity. Although this gene was isolated as a candidate AML suppressor, the Ssbp2-null mice did not develop overt myeloid leukemia. Likewise, we did not find prostate malignancies in the null mice although the human ortholog is silenced by methylation. Ssbp2 expression is very high, in hematopoietic stem and progenitor cells, a common target for myeloid transformation (J. Li, Y. Wang and L. Nagarajan unpublished results). Indeed, SSBP2 global expression profiling reveals decreased SSBP2 transcripts in primary acute myelogenous leukemia initiating cells (LICs) in contrast to normal hematopoietic stem cells (Majeti et al., 2009). Since thymic lymphomas became evident only in the context of Trp53 loss, lack of myeloid or prostatic malignancies might reflect the absence of cooperating mutations. Future studies will establish the myeloid leukemogenic potential Ssbp2 null bone marrow and the relevant leukemogenic mutations.
Supplementary Material
Acknowledgements
We thank Xiaoping Ma and Mayur Patel for expert technical assistance, Samuel Pfaff and Dong Er Zhang for housing the Ssbp2−/− mice in the early stages of these investigations, Richard Behringer, present and former members of the Nagarajan laboratory for helpful discussions, Kim-Anh Vu for assistance with the histology photographs. These studies were supported by HL744409 to L.N. The genotyping facility and veterinary services were supported by NCI core grant CA16672.
Footnotes
Conflict of Interest
The authors do not have any competing financial interest in the results presented.
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. Nat Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- Baseta JG, Stutman O. J Immunol. 2000;165:5621–5630. doi: 10.4049/jimmunol.165.10.5621. [DOI] [PubMed] [Google Scholar]
- Cai Y, Xu Z, Nagarajan L, Brandt SJ. Biochem Biophys Res Commun. 2008;373:303–308. doi: 10.1016/j.bbrc.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron C, Lecluse Y, Kaushik AL, Foudi A, Lacout C, Sekkai D, Godin I, Albagli O, Poullion I, Svinartchouk F, Schanze E, Vainchenker W, Sablitzky F, Bennaceur-Griscelli A, Dumenil D. Blood. 2006;107:4678–4686. doi: 10.1182/blood-2005-08-3145. [DOI] [PubMed] [Google Scholar]
- Castro P, Liang H, Liang JC, Nagarajan L. Genomics. 2002;80:78–85. doi: 10.1006/geno.2002.6805. [DOI] [PubMed] [Google Scholar]
- Cerretini R, Noriega MF, Narbaitz M, Slavutsky I. Eur J Haematol. 2006;76:284–293. doi: 10.1111/j.1600-0609.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, Ogryzko VV, Dawid IB, Westphal H. Proc Natl Acad Sci U S A. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, Miller TP, LeBlanc M, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Connors JM, Lansdorp PM, Ouyang Q, Lister TA, Davies AJ, Norton AJ, Muller-Hermelink HK, Ott G, Campo E, Montserrat E, Wilson WH, Jaffe ES, Simon R, Yang L, Powell J, Zhao H, Goldschmidt N, Chiorazzi M, Staudt LM. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- Fleisig HB, Orazio NI, Liang H, Tyler AF, Adams HP, Weitzman MD, Nagarajan L. Oncogene. 2007;26:4797–4805. doi: 10.1038/sj.onc.1210281. [DOI] [PubMed] [Google Scholar]
- Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. Embo J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proc Natl Acad Sci U S A. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines BB, Ryu CJ, Chang S, Protopopov A, Luch A, Kang YH, Draganov DD, Fragoso MF, Paik SG, Hong HJ, DePinho RA, Chen J. Cancer Cell. 2006;9:109–120. doi: 10.1016/j.ccr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Haks MC, Krimpenfort P, van den Brakel JH, Kruisbeek AM. Immunity. 1999;11:91–101. doi: 10.1016/s1074-7613(00)80084-9. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Johnsen SA, Gungor C, Prenzel T, Riethdorf S, Riethdorf L, Taniguchi-Ishigaki N, Rau T, Tursun B, Furlow JD, Sauter G, Scheffner M, Pantel K, Gannon F, Bach I. Cancer Res. 2009;69:128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch JG, Gu X, Han Y, El-Naggar AK, Olson MV, Medina D, Jerry DJ, Blackburn AC, Peltz G, Amos CI, Lozano G. Mamm Genome. 2007;18:300–309. doi: 10.1007/s00335-007-9028-2. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Lariviere S, Sincennes MC, Haman A, Lahlil R, Todorova M, Tremblay M, Wilkes BC, Hoang T. J Biol Chem. 2007;282:33649–33658. doi: 10.1074/jbc.M703939200. [DOI] [PubMed] [Google Scholar]
- Liang H, Samanta S, Nagarajan L. Oncogene. 2005;24:2625–2634. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- Liu JW, Nagpal JK, Sun W, Lee J, Kim MS, Ostrow KL, Zhou S, Jeronimo C, Henrique R, Van Criekinge W, Moon CS, Califano JA, Trink B, Sidransky D. Clin Cancer Res. 2008;14:3754–3760. doi: 10.1158/1078-0432.CCR-07-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Proc Natl Acad Sci U S A. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Nam CH, Rabbitts TH. Mol Ther. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, Connors JM, Gratzinger D, Rosado M, Zhao S, Pohlman B, Wongchaowart N, Bast M, Avigdor A, Schiby G, Nagler A, Byrne GE, Levy R, Gascoyne RD, Lossos IS. J Clin Oncol. 2008;26:447–454. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Shank J, Cusson N, Murre C, Kelliher M. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, Morton CC. Genes Chromosomes Cancer. 2008;47:884–889. doi: 10.1002/gcc.20585. [DOI] [PubMed] [Google Scholar]
- Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM. Proc Natl Acad Sci U S A. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. Nat Immunol. 2007a;8:441–444. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Curr Opin Hematol. 2007b;14:322–329. doi: 10.1097/MOH.0b013e3281de72a8. [DOI] [PubMed] [Google Scholar]
- Smith KS, Rhee JW, Naumovski L, Cleary ML. Mol Cell Biol. 1999;19:4443–4451. doi: 10.1128/mcb.19.6.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum EY, Segara D, Duscio B, Bath ML, Field AS, Sutherland RL, Lindeman GJ, Visvader JE. Proc Natl Acad Sci U S A. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M, Herblot S, Lecuyer E, Hoang T. J Biol Chem. 2003;278:12680–12687. doi: 10.1074/jbc.M209870200. [DOI] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, Wessels L, Adams DJ, van Lohuizen M, Berns A. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel DJ, Thomas JB, Agulnick AD. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- Wu HK, Heng HH, Siderovski DP, Dong WF, Okuno Y, Shi XM, Tsui LC, Minden MD. Oncogene. 1996;12:1205–1212. [PubMed] [Google Scholar]
- Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XB, Beard BC, Trobridge GD, Wood BL, Sale GE, Sud R, Humphries RK, Kiem HP. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.