Abstract
Runx1 transcription factor is highly expressed at a CD4/CD8-double-negative (DN) stage of thymocyte development but is down-regulated when cells proceed to the double-positive (DP) stage. In the present study, we examined whether the down-regulation of Runx1 is necessary for thymocyte differentiation from the DN to DP stage. When Runx1 was artificially over-expressed in thymocytes by Lck-driven Cre, the DN3 population was unaffected, as exemplified by proper pre-T-cell receptor expression, whereas the DN4 population was perturbed as shown by the decrease in the CD27hi sub-fraction. In parallel, the growth rate of DN4 cells was reduced by half, as measured by bromodeoxyuridine incorporation. These events impaired the transition of DN4 cells to the DP stage, resulting in the drastic reduction of the number of DP thymocytes. The Runx1 gene has two promoters, a proximal and a distal promoter; and, in thymocytes, endogenous Runx1 was mainly transcribed from the distal promoter. Interestingly, only distal, but not proximal, Runx1 over-expression exhibited an inhibitory effect on thymocyte differentiation, suggesting that the distal Runx1 protein may fulfil a unique function. Our collective results indicate that production of the distal Runx1 protein must be adequately down-regulated for thymocytes to transit from the DN to the DP stage, a critical step in the massive expansion of the T-cell lineage.
Keywords: double negative, double positive, Runx1, thymocyte development, transgenic mice
Introduction
Runx1 and Runx3 transcription factors play critical roles during thymocyte development (see ref. 1 for review). The role of each Runx protein is dependent on its expression pattern during the different stages of thymocyte differentiation. For example, Runx1 protein is produced during the CD4/CD8 double-negative (DN), CD4/CD8 double-positive (DP), CD4 single-positive (SP) and CD8 SP lineages of thymocyte differentiation.2 Hence, Lck-mediated targeting of Runx1 was reported to cause defects in the differentiation of DN thymocytes, and CD4-mediated targeting of Runx1 disrupted the positive selection of CD4 SP cells.3,4 On the other hand, Runx3 protein is produced mainly in CD8 SP thymocytes,2,5 where it suppresses the transcription of CD4, a costimulatory molecule of T-cell receptors (TCRs), and that of ThPok, a transcription factor specifying the CD4 lineage.6,7 Runx3 also activates the transcription of CD8.2 Targeting Runx3 resulted in the abolition of CD8 SP cells in the thymuses of targeted mice.7
It is worth noting that a substantial amount of Runx1 protein is expressed at the DN stage, in contrast to the Runx3 protein, which can also be detected but to a lesser extent than Runx1.2,7,8 In accordance with its expression profile in DN cells, the targeting of Runx3 had only a marginal effect such as a partial de-repression of CD4 expression,7,8 whereas a Runx1 deficiency resulted in severe defects in the differentiation of DN thymocytes. However, the observed defects varied from one report to another, namely the developmental block at either the DN1 to DN2, DN2 to DN3, or DN3 to DN4 stages of differentiation.3,4,9 This variation may be the result of the different targeting methods used, for example, the use of different promoter-driven Cre-transgenic mice. In any case, the targeting studies established clearly that Runx1 is essential for the correct differentiation of DN thymocytes.
Expression of the Runx1 gene is initiated from two distinct promoters, distal and proximal.10,11 In thymocytes, the majority of Runx1 transcripts represent transcription from the distal promoter.12 Distal and proximal Runx1 transcripts encode Runx1 proteins that are identical except for the last 19 amino acid residues (in the distal isoform) and the last five amino acids (of the proximal isoform), at the N-terminal end of the protein. Runx1 protein (mainly the distal isoform) is detected throughout the stages of thymocyte differentiation. Its level of expression is highest at the DN stage, and it is then substantially down-regulated at the DP stage.2 This down-regulation of the Runx1 protein occurs in parallel with changes in Runx1 transcripts.7 The implications of this phenomenon are unknown. The studies cited above used a gene knockout approach to evaluate the significance of Runx1 protein expression in the DN subset, but provided no explanations for the possible consequences of Runx1 down-regulation.
In the present study, we focused on the issue of whether the down-regulation of Runx1 protein is a necessary step in the correct progression of thymocytes from the DN to the DP stage. For this purpose, transgenic Runx1 was artificially over-expressed in thymocytes, with the expectation that it would disturb the physiological down-regulation of Runx1. We observed that the over-expressed Runx1 protein is an inhibitory factor during the DN to DP transition. Interestingly, this inhibitory effect was observed only with over-expression of the distal isoform, but not the proximal isoform, suggesting that this effect was unique to the distal isoform of the Runx1 transcription factor.
Materials and methods
Cell culture
The three T-cell lines EL-4, TK-1 and 1200M, and MEL cells (an erythroleukaemic cell line) were grown in RPMI-1640 medium (Gibco/Invitrogen, Carlsbad, CA), whereas NIH3T3 fibroblasts were cultured in Dulbecco’s modified Eagle’s medium. Both media were supplemented with 10% [volume/volume (v/v)] fetal bovine serum (FBS).
Plasmids
The plasmids dRunx1-HA and pRunx1-HA representing the murine Runx1 coding region of the distal (d) and proximal (p) isoforms, respectively, each contained a haemagglutinin (HA) tag fused to the C-terminal of Runx1. A BglII–EcoRI fragment harbouring Runx1-HA was cleaved from each plasmid and cloned into the EcoRV site of the plasmid pCAG-CAT-oligo. This CAT (chloramphenicol acetyltransferase) plasmid harbours a CAG (chicken β-actin) promoter-driven CAT-SV40pA element flanked by loxP sites.13 The resulting plasmids were designated as pCAG-loxP-CAT-loxP-dRunx1-HA and pCAG-loxP-CAT-loxP-pRunx1-HA, respectively.
Mice
A KpnI–Not I fragment containing pCAG-loxP-CAT-loxP-Runx1-HA was purified and microinjected into fertilized eggs from C57BL/6 mice. Transgenic founders were identified and crossed to C57BL/6 mice. The presence, or absence, of the transgene was examined by polymerase chain reaction (PCR) using genomic DNA as a template. The sense and antisense primers were as follows: for dRunx1, 5′-ATGGCTTCAGACAGCATTTTTGAG-3′ and 5′-ATGCGTATCCCCGTAGATGCC-3′; for pRunx1, 5′-TCCCCCGGGCTTGGTCTGATCATC-3′ and 5′-ATGCGTATCCCCGTAGATGCC-3′, respectively. Proximal Lck-Cre-transgenic mice were provided by J. Takeda.14
Flow cytometry
Cells were liberated from the mouse thymus and suspended in phosphate-buffered saline (PBS) containing 1% (v/v) FBS. Cell surface proteins were labelled by incubating single-cell suspensions of 2 × 106 cells with appropriately diluted monoclonal antibodies (mAbs) on ice for 30 min. Intracytoplasmic proteins were labelled by fixing and permeabilizing the surface-labelled cells using a FIX & PERM kit (Caltag Laboratories, Burlingame, CA), followed by incubation with a second mAb on ice for 30 min. The following fluorescein-conjugated mAbs were used: fluorescein isothiocyanate- (FITC-) Thy-1.2, FITC-TCR-β, FITC-CD3ε, FITC-CD25, FITC-HSA (heat stable antigen), FITC-Annexin V, phycoerythrin- (PE-) CD8a, PE-CD44, PE-CD25, PE-CD27, PE-TCR-γδ, Cychrome-CD4, Cychrome-CD8a, Cychrome-CD44 (these 14 mAbs were from BD Pharmingen, San Jose, CA), RED613-CD4 and RED613-CD8a (these two mAbs were from GibcoBRL, Gaithersburg, MD). The labelled cells were separated with an analytical EPICS-XL flow cytometer (Beckman Coulter, Miami, FL), and the data were analysed with expo32 software (Beckman Coulter).
Fractionation of thymocytes
Various thymocyte subsets were purified from a total thymocyte suspension using Auto-MACS (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany) or FACStar (Becton Dickinson, Mountain View, CA). The subsets prepared in this way corresponded to DN1/2, DN3, DN4 and DP fractions. The DN3 was further sub-divided into DN3S and DN3L based on cell size. The purity of each isolated fraction was > 95% as judged by flow cytometry.
Immunoblot analysis
Protein was extracted from 1 × 106 cells and dissolved in 80 μl of 9 m urea, 2% (v/v) Triton-X-100, 1% [weight/volume (w/v)] dithiothreitol and 20 μl of 10% (w/v) lithium dodecylsulphate. Protein extract, equivalent to 2 × 105 cells, was loaded in one lane, separated through a sodium dodecyl sulphate 8% (w/v) polyacrylamide gel, and electroblotted on to a polyvinylidene difluoride (PVDF) membrane (BioRad, Hercules, CA). The filter was blocked by immersing it in PBS containing 5% (w/v) skimmed milk at 4° overnight. The primary antibodies used were anti-HA mAb 3F10 (Roche Diagnostics, Indianapolis, IN), anti-β-actin mAb (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-panRunx-peptide rabbit serum.15 Anti-rat immunoglobulin G or anti-rabbit immunoglobulin G were used as the secondary antibodies. Immunocomplexes were detected using an ABC kit (Vector Laboratories, Burlingame, CA) and exposed to X-ray film (Fujifilm, Tokyo, Japan).
Southern blot analysis
Genomic DNA was prepared from the tails of mice by proteinase K digestion, phenol–chloroform extraction and ethanol precipitation. DNA was digested with EcoRI, electrophoresed through a 0·8% (w/v) agarose gel and processed for Southern blot analysis as described previously.16 The hybridization probe contained the Runt domain sequence of murine Runx1 complementary DNA (cDNA).
Reverse transcription–polymerase chain reaction analysis
RNA was extracted from cells using Isogen (Nippon Gene, Tokyo, Japan) and cDNA was synthesized from RNA using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The PCR amplification was performed for 25 cycles using a cDNA template and LA-Taq polymerase (Takara, Ohtsu, Japan). The following sense and anti-sense primers were used to detect transcripts: for distal Runx1, 5′–ATGGCTTCAGACAGCATTTTTGAGTCATTT–3′ and 5′–ACTGTCATTTTGATGGCTCTATGGTAGGT–3′; for proximal Runx1, 5′–ATGCGTATCCCC-GTAGATGCCAGCAC–3′, and 5′–ACTGTCATTTTGATG-GCTCTATGGTAGGT–3′; for β-actin, 5′–GATGACGATA-TCGCTGCGCTG–3′ and 5′–GTACGACCAGAGGCATACAGG–3′; and for pTα, 5′-TCACACTGCTGGTAGATGGAAGG-3′ and 5′-CATCGAGCAGAAGCAGTTTGA-3′.
Northern blot analysis
Poly(A)+ RNA was selected from the RNA fraction using Oligo-dT-Latex (Takara), and 2 μg was separated on a 1% (w/v) agarose gel containing 2·2 m formaldehyde. Electronic transfer, hybridization and washing procedures were as described previously.17 Specific distal or proximal Runx1 probes corresponding to 5′ untranslated regions of each Runx1 transcript were radioactively labelled and used for detection of each Runx1. Sequences of each probe were as follows: for proximal Runx1, 5′-CGCATCACAACAAGCCGATTGAGTAAGGACCCTGAAAACAGCTCCT-ACTAGACGGCGACAGGGGCTCGGATCTTCTGCAAG-CTGCTCCCGGGAGACCAACATACAAGTTCAGAAGC-CTTTATTACTACCGGAGGGTTGTGGGGGTAGGAGACTAAATTACCATCAGTCCCGGACTGAGATCTAGTTAC-ACGGA&CGCATCACAACAAGCCGATTGAGTAAGGA-CCCTGAAAACAGCTCCTACTAGACGGCGACAGGGG-CTCGGATCTTCTGCAAGCTGCTCCCGGGA CGCA-3′, and for distal Runx1, 5′-AAACAACCACAGAACCACAAGTTGGTAGCCTGGCAGTGTCAGAAGTGTAAGCCCAGCACAGTGGTCAGCAGGCAGGACGAATCACACTG-AATGCAAACCACAGGCTTTCGCAGAGCGGTGAAAGA-AATTATAGAATCCCCCGCCTTCAGGTAGTAGGTGCG-TTTTCGAAAGGAAACGATGGCTTCAGACAGCAÛAAACAACCACAGAACCACAAGTTGGTAGCCTGGCAGTGT-3′.
Detection of TCR-β recombination
A DN3 thymocyte subset was purified as described above, and genomic DNA was prepared from cells using Isogen. The frequency of DJ and/or V(D)J recombination was evaluated by PCR amplification using specific primers and 6·25 ng, 25 ng and 100 ng of genomic DNA as templates. The PCR cycle numbers were 28 for Rag2, 30 for DJ recombination and 33 for V(D)J recombination. The PCR products were separated on agarose gels and electroblotted onto PVDF membranes. The filters were processed for Southern blot hybridization using specific oligonucleotides as labelled probes. The sequences of the PCR primers and those of the oligonucleotide probes were as described previously.18,19 Specifically, table 1 in ref. 19 is convenient to find the sequences. Recombination detected represented Dβ2-Jβ2, Vβ2-Jβ2, Vβ4-Jβ2, Vβ10-Jβ2 and Vβ14-Jβ2, respectively.
Bromodeoxyuridine labelling of cells
One milligram of 5-bromo-2-deoxyuridine (BrdU) in PBS was injected intraperitoneally into a mouse, and the thymus was excised after 4 hr. Cell surface proteins were fluorescently labelled as described above, and the cells with incorporated BrdU were then detected using a BrdU Flow Kit (BD Biosciences, San Diego, CA). Briefly, 2 × 106 cells were fixed, permeabilized, digested with 300 μg/ml DNAse I in PBS for 1 hr at 37°, and incubated with FITC-anti-BrdU for 20 min in the dark. Labelled cells were analysed by flow cytometry as described above.
Results
Expression of Runx1 transcript and protein decreases accompanying the DN to DP differentiation of thymocytes
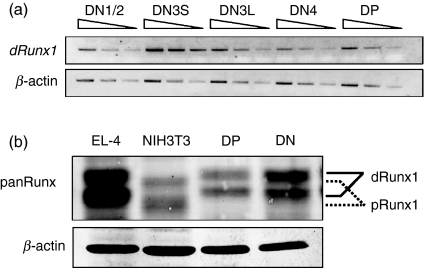
We first examined the expression profiles of Runx1 transcript by semi-quantitative reverse transcription (RT-) PCR analysis (Fig. 1a). The DN as well as DP fractions were prepared from thymuses of C57BL/6 mice. The DN fraction was further sub-divided into DN1/2, DN3 and DN4, using CD25 and CD44 as fractionation markers. DN3S (CD44− CD25hi, small) and DN3L (CD44− CD25low, large) represent sub-fractions of DN3 before and after pre-TCR mediated selection. The amount of distal Runx1 transcript was maximal at the DN3S stage and decreased markedly at the subsequent stages such as DN3L, DN4 and DP. We note that, when primers specific to proximal Runx1 transcript were used, PCR products were detected only after extensive cycles of amplification (data not shown). This is in accordance with the literature describing scarce expression of proximal Runx1 in thymocytes.12
Figure 1.
Expression profiles of Runx1 transcript and protein in the double-negative (DN) and double-positive (DP) fractions prepared from thymocytes of wild-type C57BL/6 mice. (a) Semi-quantitative reverse transcription–polymerase chain reaction (RT-PCR) analysis of distal Runx1 transcript levels during thymocyte differentiation. RNA was extracted from DN1/2, DN3S, DN3L, DN4 and DP fractions each, and converted to complementary DNAs (cDNA). An increasing amount of cDNA was used for PCR as indicated. Transcript of β-actin served as a control. (b) Immunoblot analysis of Runx1 protein in thymocytes. Protein extracts were prepared from the DN and DP fractions as well as from EL-4 and NIH3T3 cell lines, and processed for immunoblot detection using anti-panRunx antibody. A slight but significant difference in the migration corresponded to distal and proximal Runx1 polypeptides as indicated. β-actin served as a loading control.
Figure 1(b) shows immunoblot analysis of DN and DP cells, using anti-panRunx antibody. The detected doublet bands of Runx1 corresponded to unphosphorylated and phosphorylated forms, as reported.20 In agreement with the result of the RT-PCR, the amount of Runx1 protein was higher at the DN stage and decreased at the DP stage. To confirm which of the distal and proximal Runx1 proteins was expressed in thymocytes, lysates were prepared from EL-4 and NIH3T3 cells and probed in parallel, because these cell lines expressed solely distal or proximal Runx1 transcripts, respectively (Fig. S1). As expected, doublets detected for thymocytes co-migrated with doublets in EL-4 that corresponded to distal Runx1. Taking the above findings together, levels of distal Runx1 transcript and protein were high at the DN stage (DN3S) and decreased at the DP stage of thymocytes.
Establishment of Runx1-transgenic mouse lines
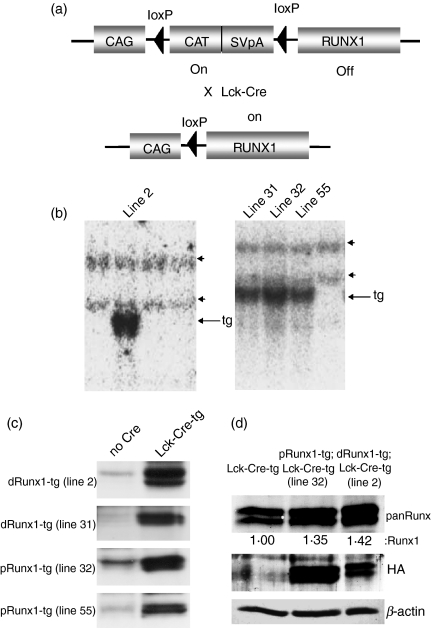
The purpose of this study was to elucidate the functional significance of the Runx1 down-regulation that accompanies the differentiation of thymocytes from the DN to DP stages. We tried to counter the Runx1 down-regulation by artificially over-expressing distal or proximal Runx1 and examining its effect on thymocyte differentiation. We generated transgenic (tg) mice that harboured pCAG-loxP-CAT-loxP-Runx1-HA in the genome (Fig. 2a, upper). To screen for a positive mouse line, genomic DNA extracted from the tails of mice was prepared for Southern blot analysis (Fig. 2b). A probe recognizing the Runt domain of Runx1 was used for detection of the transgene (indicated by arrows) as well as the endogenous gene (arrowheads). We managed to establish four lines of transgenic mice, including lines 2 and 31 harbouring distal Runx1-tg and lines 32 and 55 harbouring proximal Runx1-tg.
Figure 2.
Establishment of Runx1 transgenic mouse lines. (a) Schematic illustration of the Runx1-transgene in the mouse genome. In a Runx1 single-transgenic mouse (upper), the CAG promoter-driven transcript of the CAT gene is polyadenylated by an SV40pA element, hence impeding Runx1 expression. On the other hand, in a Runx1 and Cre double-transgenic mouse (lower), the CAT-SV40pA element is deleted, so allowing CAG promoter-driven expression of Runx1. (b) Detection of the Runx1-transgene by Southern blot analysis. Genomic DNAs were prepared from each mouse line, digested by EcoRI and processed for Southern blot analysis using the Runt domain of Runx1 as a hybridization probe. Arrows and arrowheads indicate the Runx1 transgene and endogenous Runx1 gene, respectively. Lines 2 and 31 harbour distal Runx1, whereas lines 32 and 55 harbour the proximal Runx1 isoform. (c) Detection of transgene-derived Runx1-haemagglutinin (HA) protein by immunoblot analysis. Protein extracts were prepared from thymocytes and processed for immunoblot detection using an anti-HA antibody. The left-hand column indicated by ‘no Cre’ represents Runx1 single tg mice, whereas the right-hand column indicated by ‘Lck-Cre-tg’ represents Runx1;Lck-Cre double tg mice. dRunx1 and pRunx1 denote distal and proximal Runx1, respectively. (d) Immunoblot detection of Runx1 protein expressed in the double-negative (DN) thymocytes. The DN fractions were purified from Lck-Cre-tg, distal Runx1-tg;Lck-Cre-tg (line 2) and proximal Runx1-tg;Lck-Cre-tg (line 32) thymuses, respectively, and cell lysates were processed for immunoblot using anti-panRunx, anti-HA and anti-β-actin antibodies each. Levels of Runx1 protein were quantified by a densitometer, and presented as the ratios relative to that in the Lck-Cre-tg cells as 1.00.
Runx1-HA was not expressed in Runx1-tg mice because the transcription of Runx1 from the CAG promoter was hindered by the presence of the CAT gene and the SV40-derived poly A (pA) addition element (Fig. 2a, upper). We then crossed Runx1-tg mice with Lck-Cre-tg mice. In the T lymphocytes of Runx1-tg;Lck-Cre-tg double-transgenic mice, the CAT-SV40pA element was cleaved off by Lck-driven Cre recombinase, permitting the CAG (chicken β-actin, not Lck) promoter to drive the expression of Runx1-HA (Fig. 2a, lower part). Protein extract was prepared from the thymuses of the transgenic mice and processed for immunoblot analysis using an anti-HA antibody (Fig. 2c). Runx1-HA-positive bands were not detected in the single-transgenic Runx1-tg thymocytes (left-hand column), but were evident in the thymocytes from the Runx1-HA-tg;Lck-Cre-tg double-transgenic animals (right-hand column).
Essentially similar thymocyte phenotypes were obtained from the two distal mouse lines (2 and 31) and the two proximal mouse lines (32 and 55) so the results obtained from lines 2 and 32 are shown as being representative. First, an extent of Runx1 over-expression was evaluated for these two lines (Fig. 2d). The DN fractions were purified from Lck-Cre-tg, distal Runx1-tg;Lck-Cre-tg and proximal Runx1-tg;Lck-Cre-tg thymuses, respectively, and processed for immunoblot analyses using anti-panRunx antibody. Levels of Runx1 protein were 1·4-fold increased in both types of Runx1-tg cells as compared with the non-Runx1-tg cells. This confirmed the additive expression of transgene-derived protein.
Over-expression of distal, but not proximal, Runx1 reduces the number of thymocytes
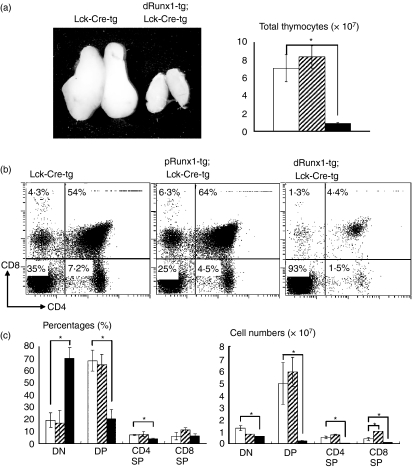
We observed that the macroscopic size of the thymus from distal Runx1-tg;Lck-Cre-tg mouse was much smaller than that from Lck-Cre-tg mouse (Fig. 3a). The total number of thymocytes in the distal Runx1-tg;Lck-Cre-tg mice (closed box) was reduced to one-ninth of that in the Lck-Cre-tg mice (open box), whereas the total number of thymocytes in the proximal Runx1-tg;Lck-Cre-tg (shaded box) was not affected (n = 3 for each genotype). In Fig. 3(a), statistical significance of difference was tested using Student’s t-test, and is indicated, if any, by an asterisk (P < 0·05).
Figure 3.
Effects of Lck-driven Runx1 over-expression on thymocyte differentiation. (a) Gross appearance of thymuses and total numbers of thymocytes. (b) Thymocytes were isolated from the transgenic mice indicated and processed for the flow cytometric analysis of CD4 and CD8 expression. Numbers given in each quadrant indicate the percentages of cells in each subset. Representative profiles are shown here. (c) Comparison of the percentages and cell numbers of double-negative (DN), double-positive (DP), CD4 single-positive (SP) and CD8 SP subset. In (a) and (c), the genotypes of mice were Lck-Cre-tg (open bars), proximal Runx1-tg;Lck-Cre-tg (shaded bars) and distal Runx1-tg;Lck-Cre-tg (closed bars), respectively. Mean ± SD (n = 3) are shown for each thymocyte subset and each transgenic mouse. Significances of difference were statistically tested by Student’s t-test, and if detected between the compared genotypes, they are indicated by brackets with *(P < 0·05). (b) CD4 repression by Runx1 over-expression was not observed in the present study, unlike the case of Runx1 introduction into a thymocyte culture by retrovirus.37 (c) The number of CD8 SP cells was significantly increased in proximal Runx1-tg;Lck-Cre-tg compared with Lck-Cre-tg thymuses (P < 0·05). This observation supports our previous report of CD2-driven, proximal Runx1-tg thymuses.5
To examine the effect of Runx1 over-expression, we performed flow cytometric analysis of CD4 and CD8 expression in thymocytes (Fig. 3b). As the thymocytes of Lck-Cre-tg mice contained an unusually high proportion of DN cells (20% on average) compared with a wild-type thymus (around 5%, data not shown), we used the Lck-Cre-tg mice as controls throughout this study when analysing Runx1-tg;Lck-Cre-tg thymuses. A remarkable difference in the DP fractions was observed between the distal Runx1-tg;Lck-Cre-tg and control mice. The proportion of DP cells was dramatically reduced in the distal Runx1-tg;Lck-Cre-tg thymus compared with the single Lck-Cre-tg thymus (4·4% compared to 54%). Additionally, the percentages of CD4 SP and CD8 SP cells were also significantly lower in the distal Runx1-tg;Lck-Cre-tg thymus (1·5% and 1·3%, respectively) compared with the Lck-Cre-tg thymus (7·2% and 4·3%, respectively).
From the analysis of the absolute cell counts (Fig. 3c), we found that in the distal Runx1-tg;Lck-Cre-tg thymus, the reduction in the total number of thymocytes (Fig. 3a) was mainly the result of the massive reduction in the number of DP cells (Fig. 3c). Although the percentage of DN cells increased substantially in the distal Runx1-tg;Lck-Cre-tg thymus, the actual number of DN cells decreased to half that of the control. It is therefore likely that over-expression of distal Runx1 impairs the DN to DP transition. In contrast to the results from distal Runx1-tg;Lck-Cre-tg, the percentages of each DN and DP subset were fairly similar between proximal Runx1-tg;Lck-Cre-tg and the control Lck-Cre-tg thymuses.
DN differentiation is perturbed in the distal Runx1-tg;Lck-Cre-tg thymocytes
To characterize the DN cells in more detail, we performed three-colour flow cytometric analysis of CD4, CD8 and Thy-l.2, and found that the ratios of Thy-1.2-positive versus Thy-1.2-negative DN cells were similar in the Runx1-tg and control thymuses. Furthermore, the Thyl.2+-gated fraction demonstrated proportions of DN, DP and SP subtypes that were essentially identical to those of non-gated thymocytes (Fig. S2). We also examined TCR-γδ expression and found that the percentages of TCR-γδ+ cells in the DN fractions were similarly low in both Lck-Cre-tg and distal Runx1-tg;Lck-Cre-tg thymuses (Fig. S3a). These results confirm that the DN cells, illustrated in Fig. 3, mostly reflect authentic T lymphocytes of αβ lineage.
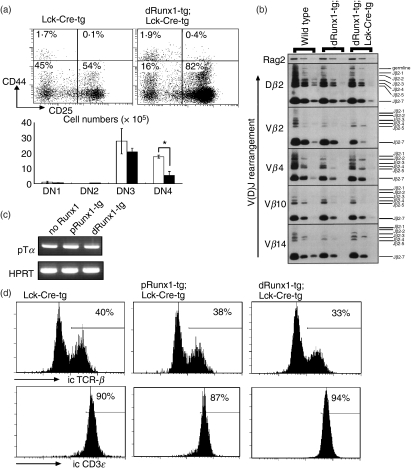
The DN population can be subdivided into four sub-stages designated as DN1 (CD44+ CD25−), DN2 (CD44+ CD25+), DN3 (CD44− CD25+) and DN4 (CD44− CD25−). To examine the differentiation of DN cells, we carried out four-colour flow cytometric analysis using CD4/CD8, Thy-1.2, CD44 and CD25. The CD44 and CD25 expression profiles of the Thy-1.2+-gated DN fractions are shown in Fig. 4(a). In the distal Runx1-tg;Lck-Cre-tg thymus, the percentages of DN1 and DN2 cells were normal. However, the percentages of DN3 and DN4 cells were increased and decreased, respectively, compared with the Lck-Cre-tg thymus. When the absolute cell number of each DN subset was counted, the DN4, but not DN3, cell numbers were substantially decreased in the distal Runx1-tg;Lck-Cre-tg thymus. This suggests that over-expression of distal Runx1 probably causes a defect at the DN4 rather than the DN3 stage.
Figure 4.
Effects of Lck-driven Runx1 over-expression on double-negative (DN) thymocyte differentiation. (a) Sub-staging of DN thymocyte differentiation. Thymocytes from distal Runx1-tg;Lck-Cre-tg (closed bars) and Lck-Cre-tg mice (open bars) were processed for flow cytometric analysis of CD4/CD8, Thy-1.2, CD44 and CD25. The data shown here were gated for the Thy-1.2+ CD4− CD8− fraction that represents authentic, DN T lymphocytes. Sub-staging of DN cells was based on the levels of CD44 and CD25 expression. The representative expression profiles are shown as well as the mean ± SD values (n = 3) of the absolute number of cells at each DN sub-stage. Significances of difference were statistically tested by Student’s t-test, and if detected between the compared genotypes, they are indicated by a bracket with *(P < 0·05). (b) Detection of TCRβ gene recombination in DN thymocytes. The DN3 subset of cells was purified by cell-sorting from C57BL/6 (wild-type), distal Runx1-tg and distal Runx1-tg;Lck-Cre-tg mice, and genomic DNA was prepared. Polymerase chain reaction (PCR) was performed using different amounts of template DNA and specific oligonucleotides as primers. PCR products were processed for Southern blot analysis. The patterns of V(D)J recombination detected are as indicated. Rag2 served as a control. (c) Expression of pTα in thymocytes. RNA was prepared from total thymocytes, and processed for reverse transcription-PCR. Transcripts representing pTα and HPRT were amplified. (d) DN3 subset was purified from thymocytes, stained for intracytoplasmic T-cell receptor-β (TCR-β) and CD3ε and processed for flow cytometric analysis. Numbers indicate the proportions of TCR-β+ and CD3ε+ cells in each DN3 subset.
Pre-TCR expression proceeds appropriately in the distal Runx1-tg;Lck-Cre-tg thymocytes
The DN3 stage is a critical step in early thymocyte differentiation as DJ and V(D)J rearrangements take place at this stage to form the pre-TCR complex.21 Cells that fail to form a functional pre-TCR die by apoptosis. We investigated whether pre-TCR expression proceeded correctly in the distal Runx1-tg;Lck-Cre-tg cells. Genomic DNA was prepared from the purified DN3 fraction and used as a template for PCR amplification. Specific PCR primers and hybridization probes were designed to detect various patterns of DJ and V(D)J segment recombination.18,19 As far as the gene combinations examined were concerned, the DJ and V(D)J segments of TCRβ were properly rearranged in the DN3 cells from distal Runx1-tg;Lck-Cre-tg, distal Runx1-tg and wild-type thymuses (Fig. 4b).
The expression of the pre-TCR complex itself was also examined. The major components of the pre-TCR complex include pTα, TCR-β and CD3ε. Transcripts of pTα were detected in thymocytes from distal Runx1-tg;Lck-Cre-tg mice (Fig. 4c) and intracytoplasmic TCR-β and CD3ε expression was detected by flow cytometric analysis (Fig. 4d). Both TCR-β and CD3ε were expressed to a similar extent in DN3 thymocytes from the distal Runx1-tg;Lck-Cre-tg, proximal Runx1-tg;Lck-Cre-tg and Lck-Cre-tg mice. Based on the observations illustrated in Fig. 4, it is not likely that over-expression of distal Runx1 affects the process of pre-TCR expression at the DN3 stage.
DN4 differentiation as probed by CD27-expression is impaired in the distal Runx1-tg;Lck-Cre-tg thymocytes
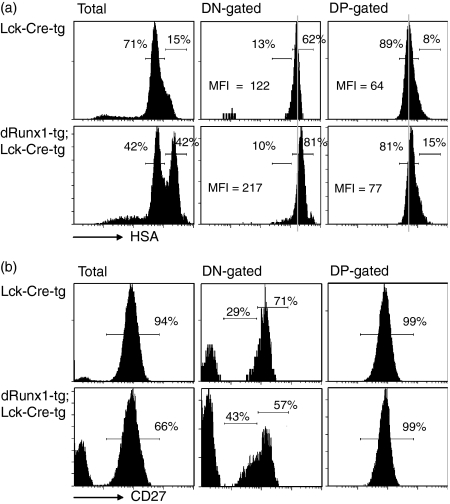
We then examined the differentiation status of DN thymocytes using other markers such as HSA (Fig. 5a) and CD27 (Fig. 5b). In non-gated, distal Runx1-tg;Lck-Cre-tg thymocytes, a substantial peak of HSAhi population was observed in addition to an HSAlo population. Furthermore, mean fluorescence intensity of HSA in the DN fraction was higher in the distal Runx1-tg;Lck-Cre-tg compared with that in the Lck-Cre-tg (217 versus 122). This suggests that the majority of the distal Runx1-tg;Lck-Cre-tg DN cells were highly immature.
Figure 5.
Flow cytometric analyses of HSA/CD27 expression in the double-negative (DN) and double-positive (DP) fractions. Thymocytes were prepared from Lck-Cre-tg and distal Runx1-tg;Lck-Cre-tg mice, and processed for flow cytometric analyses of CD4, CD8 and HSA/CD27. The DN-gated and DP-gated fractions were analysed for their fluorescence intensities of HSA (a) and CD27 (b). In (a), the percentages of HSAhi and HSAlo sub-fractions and the mean fluorescence intensities of HSA are indicated, whereas the numbers seen in (b) indicate the percentages of CD27med/hi sub-fractions. Note that the percentage of CD27med/hi is assumed to be 100 for the DN fractions in (b).
CD27 is a newly reported DN marker whose expression is low at the DN3S stage (CD27lo) and increases sharply at the DN3L (CD27med) and DN4 stages (CD27hi).22 Although the DN1/2 cells also express high CD27, they constitute < 3% of the entire DN population, and contribute little to the CD27med/hi population. These CD27med and CD27hi populations were substantially increased and decreased, respectively, in the DN fraction from distal Runx1-tg;Lck-Cre-tg thymus (43% and 57% each) compared with those from Lck-Cre-tg thymus (29% and 71% each). Alteration of CD27 expression pattern was analogous to the CD25 and CD44 profile in the sense that the percentage of DN3 increased and that of DN4 decreased in the DN cells from distal Runx1-tg;Lck-Cre-tg thymus (see flow cytometry analyses in Fig. 4a). Collectively, the alterations of HSA and CD27 expressions observed in the distal Runx1-tg;Lck-Cre-tg thymocytes suggest an impairment of thymocyte differentiation at the DN3L/DN4 stage.
Over-expression of distal Runx1 reduces the growth activity of thymocytes at the DN4 stage and impairs the DN to DP transition
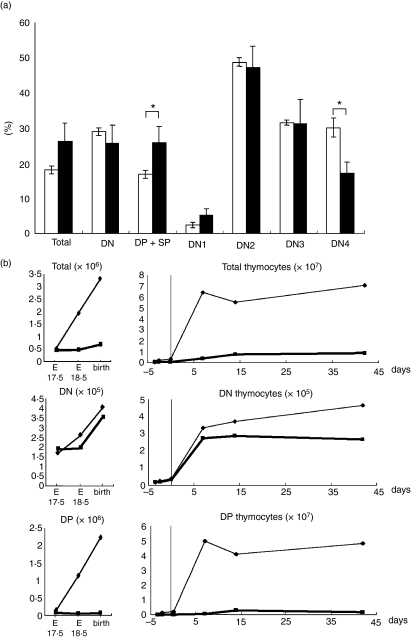
As the DN4 stage was likely to be perturbed by Runx1 over-expression, we examined the degree of cell proliferation in Runx1-tg thymuses. Eight-week-old mice were injected intraperitoneally with BrdU, and their thymuses were harvested after 4 hr. Thymocytes were processed for four-colour flow cytometric analysis of BrdU, CD4/8, CD25 and CD44 (Fig. 6a). For the DN2 and DN3 fractions, the percentages of BrdU+ cells were comparable between the distal Runx1-tg;Lck-Cre-tg (closed bars) and Lck-Cre-tg thymocytes (open bars). However, in the DN4 cells of the distal Runx1-tg;Lck-Cre-tg thymus, the percentage of BrdU+ cells was reduced to nearly half that of the control. An increase of the BrdU+ percentage in the DP + SP fraction might reflect a compensatory growth enhancement in the distal Runx1-tg;Lck-Cre-tg thymuses. An essentially similar result was obtained when 1-week-old mice were used for the same experiments (data not shown). We conclude therefore that over-expression of distal Runx1 impairs the ability of DN4 thymocytes to proliferate. It must be noted that the extent of apoptosis, as measured by Annexin V staining, was not enhanced in the distal Runx1-tg;Lck-Cre-tg thymocytes compared with the Lck-Cre-tg thymocytes (Fig. S3b).
Figure 6.
Effects of Runx1-tg on the growth and expansion of thymocytes. (a) Incorporation of bromodeoxyuridine (BrdU) into various subsets of thymocytes. Lck-Cre-tg mice (open boxes) and distal Runx1-tg;Lck-Cre-tg mice (closed boxes) were each injected with BrdU intraperitoneally. Thymocytes were prepared after 4 hr and processed for flow cytometric analysis of BrdU, CD4/CD8, CD25 and CD44. Percentages of BrdU+ cells in various subsets are shown as the mean ± SD values (n = 3). Significances of difference were statistically tested by Student’s t-test, and if detected between the compared genotypes, they are indicated by brackets with *(P < 0·05). (b) Ontogeny of DN and DP thymocytes during mouse development. Thymuses were taken from mice at embryonic day 17·5 (E17.5), E18.5, at birth, 1, 2 and 6 weeks after birth, and processed for flow cytometric analysis of CD4 and CD8 expression. The numbers of total, DN and DP cells are shown in distal Runx1-tg;Lck-Cre-tg (thick lines) and Lck-Cre-tg mice (thin lines).
Finally, we analysed thymocyte differentiation during mouse development. Thymuses were harvested at embryonic day (E) 17·5, E18·5, at birth and 1, 2 and 7 weeks after birth, from distal Runx1-tg;Lck-Cre-tg and Lck-Cre-tg mice, and processed for flow cytometric analysis with CD4, CD8 and Thy-1.2. The numbers of Thy-1.2+ DN and DP cells were plotted together with the development and growth of the mice (Fig. 6b). There were slightly fewer DN cells in the distal Runx1-tg;Lck-Cre-tg thymuses (thick line) than in the Lck-Cre-tg thymuses (thin line) throughout the developmental stages examined. In contrast, the number of DP cells from the distal Runx1-tg;Lck-Cre-tg thymuses was remarkably diminished when compared with the Lck-Cre-tg cells. This difference was particularly marked 1 week after birth. This was because, during this period, the total cell number (consisting mainly of DP cells) increased dramatically in the control thymus but not in the Runx1-tg;Lck-Cre-tg thymus. Taken collectively, the results illustrated in Fig. 6 indicated that over-expression of distal Runx1 appears to severely disrupt the transition of thymocytes from the DN4 to DP stages.
Discussion
Previous studies have demonstrated that the abolition of Runx1 expression causes severe impairments in the differentiation of DN thymocytes, highlighting an indispensable role for Runx1 at this particular stage of T-cell development.3,4,9 These targeting studies evaluated the significance of Runx1 expression in DN cells; however, the Runx1 protein is then down-regulated during the transition from the DN to the DP stage. In this study, we investigated whether the down-regulation of Runx1, that accompanies the DN to DP differentiation, has biological significance. This was assessed using conditional Runx1-transgenic mice to disrupt the down-regulation of Runx1. Lck-Cre-mediated deletion has been shown to be initiated at DN2 and completed by DN3.23 Therefore, the onset of Runx1 transgene expression can be manipulated to start at the DN2 stage by mating Runx1-tg mice with Lck promoter-driven Cre-transgenic mice. When Runx1 over-expression was initiated at such an early DN stage, it resulted in a marked impairment in the transition of cells from the DN to DP stages.
The DN-cell population is not a homogeneous entity, but rather comprises a population of developing cells with different characteristics divided into four subgroups (DN1 to DN4). For example, DN1 cells retain the potential to differentiate into B-cell and myeloid lineages, but they lose/reduce this capability once they have entered the DN2 stage.24–29 The cells at a DN3 stage undergo a critical β-selection checkpoint. In the distal Runx1-tg;Lck-Cre-tg DN3 thymocytes, rearrangement of the TCRβ locus and pre-TCR expression did not appear to be affected.
On the other hand, in the same distal Runx1-tg;Lck-Cre-tg thymuses, the number of DN4 cells was substantially decreased, and the proportion of CD27hi (DN4) population was remarkably reduced. In parallel, an extent of cell growth, as measured by BrdU incorporation, was decreased by nearly half in these double-transgenic DN4 cells. Hence, the differentiation of distal Runx1-tg;Lck-Cre-tg thymocytes was probably perturbed during the DN4 stage and/or the DN4 to DP transition. The DN to DP transition is the step when cells rapidly undergo massive expansion as seen in the thymocyte ontogeny. Disruption in the normal processing of this transition is a likely explanation for the paucity of DP cells in distal Runx1-tg;Lck-Cre-tg thymuses (for example, if the normal growth rate decreased by half during each of four cell divisions, the resulting cell number would be one-sixteenth that of the Lck-Cre-tg control thymus).
In this study we have shown that the over-expression of distal Runx1 is deleterious for the DN4 thymocytes. In contrast, vav-promoter driven, over-expression of distal Runx1 is reported to be oncogenic in the T-cell lineage and to cause lymphoma in mice.30,31 A clue for the apparently discrepant effects of Runx1 over-expression on thymocytes might be found in a study of CD2-Runx2-tg thymuses by Vaillant et al.32 There, as in the present study, Runx2 over-expression causes anti-proliferative effects and a differentiation block at the DN to DP intermediate stage, but, unlike the present case, simultaneously and eventually brings about predisposition to lymphoma development. On the other hand, we previously reported that cell division in TCR and Runt (a dominant interfering form of Runx1) double-transgenic thymocytes was also moderately impaired during the DN4 to DP transition.33 Considering together the previous reports and the present results, for the cells to pass safely through the critical DN4 point, a dosage of Runx1 must be adequately regulated within a rather narrow window.
The transcription of the Runx1 gene is initiated either from the distal or proximal promoters.10,11 This feature is conserved in all vertebrate Runx genes examined, and it is shared by all three members of the Runx gene family (Runx1, Runx2 and Runx3). The distal and proximal Runx1 proteins are identical to each other, except for the extreme N-terminal 19 (distal) and five (proximal) amino acid residues. Hence, the abolition of DP thymocytes by the over-expression of distal Runx1, but not proximal Runx1, can be ascribed solely to the differences in the N-termini of the two Runx1 isoforms. In fact, the distal Runx1 protein binds to a Runx consensus binding site with a two-fold to three-fold higher affinity than does the proximal Runx1 protein.12 In addition, the distal and proximal forms of the Runx3 protein exhibit different activities in assays using Runx site-dependent reporter plasmids, in which only the N-terminal region of the distal, but not the proximal, Runx3 protein possesses transactivation capability.34 Furthermore, the selective loss or transgenic over-expression of the proximal or distal Runx2 isoforms have been shown to cause differential effects on bone development in mice.35,36 It is conceivable that the 19 amino acids of distal Runx1 (highly conserved with Runx2/3) also possess a unique transcriptional modulating activity, which influences DN-cell differentiation. It must be noted that the major species of Runx1 expressed and/or down-regulated in the thymus is a distal Runx1. This observation is in concordance with our observation that over-expression of distal, but not proximal, Runx1 catastrophically impaired DN-cell development.
Acknowledgments
We would like to express our thanks to E.V. Rothenberg for helpful discussions. We also thank J. Takeda for providing us with Lck-Cre-transgenic mice. This work was supported in part by a research grant from the Japan Science and Technology. M.S. is a participant in the Global COE Program ‘Network Medicine’ at Tohoku University.
Disclosures
None.
Supporting Information
Additional Supporting information may be found in the online version of this article:
Figure S1. Northern blot analysis of Runxl transcripts in various mouse cell lines.
Figure S2. (a) Total numbers of Thy-1.2+ gated thymocytes. (b) Comparison of the percentages and cell numbers of Thy-1.2+ gated DN, DP, CD4 SP and CD8 SP subsets.
Figure S3. Expression profiles of TCR-γδ and Annexin-V in the DN and DP cells from distal Runxl-tg;Lckl-Cre-tg and Lck-Cre-tg thymi.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Kohu K, Kubo M, Ichikawa H, Ohno S, Habu S, Sato T, Satake M. Pleitropic roles of Runx transcription factors in the differentiation and function of T lymphocytes. Curr Immunol Rev. 2008;4:101–15. [Google Scholar]
- 2.Sato T, Ohno S, Hayashi T, Sato C, Kohu K, Satake M, Habu S. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–28. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–57. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K, Abe N, Watanabe T, Obinata M, Ito M, Sato T, Habu S, Satake M. Overexpression of AML1 transcription factor drives thymocytes into the CD8 single-positive lineage. J Immunol. 2001;167:4957–65. doi: 10.4049/jimmunol.167.9.4957. [DOI] [PubMed] [Google Scholar]
- 6.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–5. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 7.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 8.Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA. 2003;100:7731–6. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talebian L, Li Z, Guo Y, et al. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–40. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–9. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 12.Telfer JC, Rothenberg EV. Expression and function of a stem cell promoter for the murine CBFalpha2 gene: distinct roles and regulation in natural killer and T cell development. Dev Biol. 2001;229:363–82. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 13.Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci USA. 1995;92:160–4. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahama Y, Ohishi K, Tokoro Y, Sugawara T, Yoshimura Y, Okabe M, Kinoshita T, Takeda J. Functional competence of T cells in the absence of glycosylphosphatidylinositol-anchored proteins caused by T cell-specific disruption of the Pig-a gene. Eur J Immunol. 1998;28:2159–66. doi: 10.1002/(SICI)1521-4141(199807)28:07<2159::AID-IMMU2159>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Kanto S, Chiba N, Tanaka Y, et al. The PEBP2β/CBFβ–SMMHC chimeric protein is localized both in the cell membrane and nuclear subfractions of leukemic cells carrying chromosomal inversion 16. Leukemia. 2000;14:1253–9. doi: 10.1038/sj.leu.2401821. [DOI] [PubMed] [Google Scholar]
- 16.Okada H, Watanabe T, Niki M, et al. AML1−/− embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 1998;17:2287–93. doi: 10.1038/sj.onc.1202151. [DOI] [PubMed] [Google Scholar]
- 17.Chiba N, Watanabe T, Nomura S, Tanaka Y, Minowa M, Niki M, Kanamaru R, Satake M. Differentiation dependent expression and distinct subcellular localization of the protooncogene product, PEBP2β/CBFβ, in muscle development. Oncogene. 1997;14:2543–52. doi: 10.1038/sj.onc.1201109. [DOI] [PubMed] [Google Scholar]
- 18.Senoo M, Wang L, Suzuki D, Takeda N, Shinkai Y, Habu S. Increase of TCR Vβ accessibility within Eβ regulatory region influences its recombination frequency but not allelic exclusion. J Immunol. 2003;171:829–35. doi: 10.4049/jimmunol.171.2.829. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki D, Wang L, Senoo M, Habu S. The positional effect of Eβ on Vβ genes of TCRβ chain in the ordered rearrangement and allelic exclusion. Int Immunol. 2005;17:1553–60. doi: 10.1093/intimm/dxh333. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Kurokawa M, Ueki K, et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–79. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol. 1999;11:135–42. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- 22.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 2002;16:869–79. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 24.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–7. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 25.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–45. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg EV. Cell lineage regulators in B and T cell development. Nat Immunol. 2007;8:441–4. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- 27.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor–ligand interactions. J Exp Med. 2004;200:469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–72. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 30.Blyth K, Slater N, Hanlon L, Bell M, Mackay N, Stewart M, Neil JC, Cameron ER. Runx1 promotes B-cell survival and lymphoma development. Blood Cells Mol Dis. 2009;43:12–9. doi: 10.1016/j.bcmd.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Wotton S, Stewart M, Blyth K, Valliant F, Kilbey A, Neil JC, Cameron ER. Proviral insertion indicates a dominant oncogenic role for Runx1/AML-1 in T-cell lymphoma. Cancer Res. 2002;62:7181–5. [PubMed] [Google Scholar]
- 32.Vaillant F, Blyth K, Andrew L, Neil JC, Cameron ER. Enforced expression of Runx2 perturbs T cell development at a stage coincident with β-selection. J Immunol. 2002;169:2866–74. doi: 10.4049/jimmunol.169.6.2866. [DOI] [PubMed] [Google Scholar]
- 33.Sato T, Ito R, Nunomura S, Ohno S, Hayashi K, Satake M, Habu S. Requirement of transcription factor AML1 in proliferation of developing thymocytes. Immunol Lett. 2003;89:39–46. doi: 10.1016/s0165-2478(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 34.Chung DD, Honda K, Cafuir L, McDuffie M, Wotton D. The Runx3 distal transcript encodes an additional transcriptional activation domain. FEBS J. 2007;274:3429–39. doi: 10.1111/j.1742-4658.2007.05875.x. [DOI] [PubMed] [Google Scholar]
- 35.Kanatani N, Fujita T, Fukuyama R, et al. Cbf β regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296:48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Z, Awad HA, Liu S, Mahlios J, Zhang S, Guilak F, Mayo MS, Quarles LD. Selective Runx2-II deficiency leads to low-turnover osteopenia in adult mice. Dev Biol. 2005;283:345–56. doi: 10.1016/j.ydbio.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–70. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.