Abstract
Salmonella enterica serovar Typhimurium can enter non-phagocytic cells, such as intestinal epithelial cells, by virtue of a Type Three Secretion System (TTSS) encoded in the Salmonella Pathogenicity Island 1 (SPI-1), which translocates bacterial effector molecules into the host cell. Salmonella can also be taken up by dendritic cells (DCs). Although the role of SPI-1 in non-phagocytic cell invasion is well established, its contribution to invasion of phagocytic cells has not been evaluated. Here, we have tested the invasive capacity of a S. Typhimurium strain lacking a key component of its TTSS-1 (ΔInvC) leading to defective translocation of SPI-1-encoded effectors. Whereas this mutant Salmonella strain was impaired for invasion of non-phagocytic cells, it was taken up by DCs at a significantly higher rate than wild-type Salmonella. Similar to wild-type Salmonella, the ΔInvC mutant strain retained the capacity to avoid antigen presentation to T cells. However, mice infected with the ΔInvC mutant strain showed higher survival rate and reduced organ colonization. Our data suggest that, besides promoting phagocytosis by non-phagocytic cells, SPI-1 modulates the number of bacteria that enters DCs. The SPI-1 could be considered not only as an inducer of epithelial cell invasion but as a controller of DC entry.
Keywords: bacteria/bacterial immunity, dendritic cells, phagocytosis
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a facultative intracellular pathogen that produces a typhoid-like disease in mice, resembling the typhoid fever produced by S. Typhi in humans.1Salmonella enters the host via oral ingestion and colonizes the small intestine, entering preferentially via M cells located at the Peyer’s patches.2,3 The M cells are specialized phagocytic cells that sample intestinal antigens and deliver them to the antigen-presenting cells that underlie the epithelium in Peyer’s patches.1 The invading Salmonella that succeeded at translocating across the intestinal epithelial layer can reach the subepithelial compartment where they interact most efficiently with dendritic cells (DCs) and macrophages that reside under Peyer’s patches.4Salmonella can also invade non-phagocytic epithelial cells by promoting cytoskeletal rearrangements that cause membrane ‘ruffles’ that engulf bacteria.5 The capacity to induce its own phagocytosis is achieved by means of a sophisticated Type III secretion system (TTSS) encoded in the Salmonella Pathogenicity Island 1 (SPI-1) (TTSS-1), which promotes the secretion of SPI-1-encoded effector proteins into the host cell cytoplasm.6–9 The SPI-1 codes not only for the TTSS-1, but also for a number of effector proteins that contribute mainly to the initial interaction of these bacteria with the intestinal epithelium, as well as to triggering apoptosis of infected cells.10–12
The TTSS-1 is a conserved multi-protein secretion apparatus. The central piece of this system is a supramolecular structure known as the needle complex.13–15 The needle structure itself protrudes outward from the base and consists of a straight tube of 80 nm in length across which effectors proteins are driven into the host cell cytoplasm.16 A highly conserved adenosine triphosphatase (ATPase) provides the energy to secrete effector proteins through TTSS-1. This ATPase presents a significant similarity in amino acid sequence to the catalytic β subunit of the F0F1 ATPases.17Salmonella ATPase, known as InvC, plays a central role in effector secretion and hence in bacterium virulence.17,18 This molecule recognizes chaperone–effector complexes and induces their disassembly.19 Furthermore, InvC induces the unfolding of the cognate secreted protein, so allowing the ‘naked’ and unfolded effectors to translocate across the TTSS-1.19–21
Salmonella can also invade DCs, professional antigen-presenting cells that protrude prolongations between the epithelial cells of the intestine.22–24 It is thought that DC invasion enables Salmonella to shuttle across the epithelium barrier.24–27 Although Salmonella possesses at least three ways of invading host cells and entering the host,28 little is known about the relative contribution of each method of invasion to the infection process. Further, although the role of TTSS-1 in Salmonella internalization into non-phagocytic cells has been well characterized,26,29,30 whether TTSS-1 contributes to the invasion of DCs remains unknown. Dendritic cells are key elements for the generation of an efficient adaptive immune response against bacterial pathogens, such as Salmonella,25 and are considered the link between innate and adaptive immunity. These cells can capture pathogens at the site of infection and process their antigens to produce peptides that are presented to T cells on major histocompatibility complex (MHC) molecules.31 The outcome of this sequence of events is the activation of pathogen-specific T cells that ultimately contribute to preventing the proliferation of microbes in host tissues.27,32–38
Another pathogenicity island of Salmonella, SPI-2, encodes for a second TTSS (TTSS-2) and for additional specific effector proteins.39,40 In contrast to SPI-1, expression of SPI-2 is required for intracellular survival and evasion of the adaptive immune response.26,36,41–43 We and others have previously observed that virulent Salmonella can evade adaptive immunity by preventing DCs from activating T cells.34,36,37,44–47 Virulent Salmonella seems capable of impairing the presentation of bacteria-derived antigens to T cells.36,48,49
Here, we have evaluated the role of SPI-1 in invasion of DCs and non-phagocytic cells by using a mutant strain of S. Typhimurium in which the invC gene has been substituted for a kanamicin-resistance cassette. As a result of this mutation, the ΔInvC strain was rendered unable to secrete effector proteins because of the lack of a functional TTSS-1. Although the ΔInvC mutant strain failed to invade non-phagocytic cells, it showed a significantly increased capacity to invade DCs compared with the wild-type (WT) Salmonella strain. In contrast, WT and ΔInvC strains were found to be equally successful at avoiding T-cell activation. However, infection experiments showed that the ΔInvC strain was attenuated in vivo. Extended survival and reduced organ colonization rates were observed for mice infected with the ΔInvC strain compared with mice challenged with the WT Salmonella strain. The in vivo experiments underscore the importance of DCs during bacterial colonization and their role as a carrier of bacteria. These data support the notion that SPI-1 is determinant for differentially modulating the entry into phagocytic and non-phagocytic host cells.
Materials and methods
Mice
C57BL/6 WT mice were obtained originally from The Jackson Laboratory (Bar Harbor, ME) and used at 6–8 weeks of age. The OT-I and OT-II transgenic mice expressing a specific T-cell receptor for H-2Kb/OVA257–264 and I-Ab/OVA323–337 respectively, were kindly provided by Dr R. Steinman (The Rockefeller University, New York, NY). All mice were maintained and manipulated in specific pathogen-free conditions and animal work was performed according to institutional guidelines at the animal facility of the Pontificia Universidad Católica de Chile (Santiago, Chile).
Cell lines
L-cells (murine fibroblast cell line) and MLE-12 (murine alveolar epithelium cell line, kindly provided by Dr Marcela Hermoso from Universidad de Chile) were grown in RPMI-1640 medium (Gibco, Invitrogen, Carlsbad, CA) supplemented with glutamine and 5% fetal bovine serum. The macrophage J774.3 cell line used in this study was kindly provided by Dr María Inés Becker (Biosonda SA, Santiago, Chile). J774.3 cells were routinely grown in high-glucose Dulbecco’s modified Eagle’s minimum essential medium (Gibco) supplemented with 10% fetal bovine serum (HyClone, Thermo Scientific, Waltham, MA) and 1 mm HEPES (Gibco). The L, MLE-12 and J774.3 cell lines were incubated at 37° and were used in gentamicin protection assays and laser confocal microscopy analyses.
DCs preparation and drug treatments
Dendritic cells were differentiated from bone marrow precursors of C57BL/6 mice. Cells were incubated in complete RPMI-1640 medium supplemented with 5% fetal calf serum, 2 mm glutamine, 1 mm non-essential amino acids, 1 mm pyruvate, 1 mm HEPES and 10 ng/ml of recombinant murine granulocyte–macrophage colony-stimulating factor (Peprotech, Rocky Hill, NJ). All cell culture media and supplements were purchased from Gibco (Invitrogen). Culture medium was replaced every 2 days. After 6 days, the phenotype of DCs was analysed by flow cytometry for the expression of surface markers CD11c, I-Ab, H-2Kb, CD80, CD86 and CD40. Routinely, over 75% of cells were CD11c+ and showed an immature phenotype. Before infection, DCs were left untreated or were treated with the following drugs: 10 μg/ml caspase inhibitor VI Z-VAD FMK for 30 min (Calbiochem, EMD Chemicals, Gibbstown, NJ); 1 mmα-1-lipoic acid for 30 min (Sigma-Aldrich, St Louis, MO); 5 μg/ml cytochalasin D for 10 min (Sigma-Aldrich) or 0·1 μm wortmannin for 30 min (Sigma-Aldrich). Upon incubation, DCs were washed three times with 1 ml phosphate-buffered saline (PBS) and infected at 37° with bacteria in 1 ml of culture medium for gentamicin protection assays, confocal microscopy and flow cytometry analyses. To induce DC maturation, cells were treated with 2 μg/ml of lipopolysaccharide (LPS; Sigma) in PBS for 30 hr before infection.
Bacterial strains
Salmonella Typhimurium (ATCC 14028s; American Type Culture Collection, Manassas, VA) was used as the parental strain in all experiments. The S. Typhimurium strain carrying a deletion of the invC gene was generated by Eichelberg and colleagues18 by inserting a kanamycin-resistance cassette by double homologous recombination. The aphT-interrupted invC gene was transduced to the 14028s strain using phage P22 HT105/1 int-201.18,50 The disruption of the invC gene in the chromosome of S.Typhimurium was confirmed by polymerase chain reaction (PCR; Fig. S1a). The primers used in these assays that aligned up and downstream from the exchanged site had the following sequences: 5′-AGTCGGTCGCTAATGACATG-3′ and 5′-AATTCTGGTCAGGGAATGCA-3′. avrA was used as a PCR control and this sequence was amplified with the following primers: 5′-CGTAATGAAATCGTACCAGAGG-3′ and 5′-ACCCGATTGCGGTAGGAAT-GA-3′. To complement ΔInvC Salmonella, the invC gene was PCR amplified with primers: 5′-GGCCATGGGAATGAAAACACCTCGTT-3′ and 5′-GGAAGCTTTTAATTCTGGTCAGCG-3′, cloned in pCR2.1®-TOPO (Invitrogen), digested with restriction enzimes NcoI and HindIII (New England Biolabs, Ipswich, MA) and ligated in NcoI–HindIII-digested pKK233.2 cloning vector, using T4 DNA ligase (New England Biolabs). The pKK233.2-invC was incorporated to ΔInvC Salmonella by electroporation and complemented strains were selected using 100 μg/ml ampicillin. Ovalbumin (OVA)-expressing and green fluorescent protein (GFP)-expressing S. Typhimurium were generated as previously described.34 A S. Typhimurium strain lacking whole SPI-1 was provided by Dr Carlos A. Santiviago, Universidad de Chile, Santiago, Chile. Bacteria were grown overnight in Luria–Bertani broth at 37° and recombinant bacteria were selected using 100 μg/ml ampicillin (for GFP-expressing S. Typhimurium, and OVA-expressing S. Typhimurium). For infection of DCs, overnight bacterial cultures were diluted 1/100 and grown until exponential phase [optical density at 600 nm (OD600) = 0·5–0·7]. To quantify the LPS content in bacterial inoculates, the Limulus amoebocyte lysate endotoxin quantification assay (Cambrex, East Rutherford, NJ) was used according to the manufacturer’s instructions.
Western blotting
Protein secretion by the SPI-1-encoded TTSS was evaluated by Western blot detection of SopE in supernatants and pellets of bacterial cultures. Salmonella Typhimurium WT and S. Typhimurium ΔInvC were grown until optical density was equal to 0·6. Bacteria were centrifuged at 6000 g and the supernatant was concentrated after serial centrifugations at 300 g with Centriprep filter tubes (Millipore, Billerica, MA). Finally, the pellet and the concentrated supernatant were analysed by Western blot with SopE effector-specific antibodies. Consistently with a deficiency on the function of TTSS-1, the SopE protein could not be detected in culture supernatants of the ΔInvC mutant strain (Fig. S1b). Ovalbumin expression in S. Typhimurium WT-pKK-OVA and S. Typhimurium ΔInvC-pKK-OVA was also confirmed by Western blot analyses, using rabbit immunoglobulin G (IgG) anti-OVA (data not shown).
Gentamicin protection assays
To evaluate the survival of each Salmonella strain inside DCs, J774.3 cells, MLE-12 cells and L cells, gentamicin protection assays were used.51 Overnight bacteria cultures were subcultured until they reached exponential phase (OD600 = 0·5–0·7); they were then washed and resuspended on ice-cold PBS. The DCs, MLE-12 cells or L cells were infected with S. Typhimurium WT or ΔInvC at a multiplicity of infection (MOI) equal to 25 for 1 hr. Cells were washed and incubated for the indicated times with 50 μg/ml gentamicin (Sigma-Aldrich) to kill extracellular bacteria. Gentamicin treatment leads to 100% extracellular bacterial death after 2 hr of incubation (data not shown). Cell viability was determined by exclusion of trypan blue. To recover intracellular bacteria, 10 000 live cells were counted and lysed for 15 min with 0·1% Triton-X-100 in PBS. The lysed cells were seeded on Luria–Bertani agar plates and incubated for 12–16 hr at 37° to count intracellular bacteria as colony-forming units (CFUs). Data from gentamicin protection assays were normalized as the percentage of recovered CFUs relative to the maximum amount obtained in each experiment (defined as 100%). For experiments with DCs, the maximum CFU amount was observed for the ΔInvC mutant strain. In contrast, for epithelial cells the maximum CFU values were observed for the WT Salmonella strain.
Laser confocal microscopy
To evaluate Salmonella entrance into DCs or L cells, these cells were seeded on round cover slips (250 000 cells per cover slip) in complete RPMI-1640 medium and were infected with GFP-expressing WT S. Typhimurium or ΔInvC (MOI = 50). After 1 hr of infection, cells were washed twice with ice-cold PBS and incubated in complete RPMI-1640 medium supplemented with 50 μg/ml gentamicin (to kill extracellular bacteria) for a period of 3 hr. Cells were fixed and permeabilized for 10 min at − 20° with methanol (100%) and blocked for 1 hr with PBS plus 3% bovine serum albumin (BSA). After incubation, cover slips were washed three times, mounted on microscope slides and analysed on a fluoview FV100 Olympus confocal microscope. Quantitative analyses were performed by counting the number of cells containing intracellular bacteria relative to the total number of cells in the field. Intracellular bacteria were distinguished from extracellular bacteria by 0·5 μm Z-stack analyses. This methodology provided equivalent data to the detection of extracellular bacteria using antibodies (not shown). At least 10 random fields were selected and quantified by independent observers. At least 300 cells were analysed per experiment.
Antigen presentation assays
C57BL/6 bone marrow-derived DCs were prepared as previously described. At day 5 of culture, DCs were pulsed for 1 hr with OVA-expressing WT S. Typhimurium or ΔInvC strain at an MOI of 25. As a control, DCs were pulsed with 10 μg/ml OVA protein, OT-I and OT-II peptides and with either free or IgG-coated S. Typhimurium WT or ΔInvC, as described previously.46 As an additional control, S. Typhimurium WT- or ΔInvC-infected DCs were simultaneously pulsed with 10 μg/ml purified OVA. Viability of DCs was analysed by Trypan Blue exclusion and staining with propidium iodide by flow cytometry. After infection, the DCs were washed and treated with gentamicin to eliminate extracellular bacteria, as described above. After an additional 12 hr of incubation, DC viability was determined by trypan blue exclusion and different amounts of live DCs were co-cultured with either OT-I or OT-II T cells (105 T cells/well), obtained from lymph nodes (LNs) of the respective transgenic mice. After 24 hr of DC–T-cell co-culture, CD69 expression was measured by flow cytometry and interleukin-2 (IL-2) release was measured by cytokine enzyme-linked immunosorbent assay (ELISA) as described previously.34,52,53
Cytokine ELISA
Interleukin-2 release by DCs was measured 24 hr after infection with WT S. Typhimurium or ΔInvC. Uninfected controls were included for each experiment. Previously, ELISA plates (Maxisorb, Nunc, eBioscience, San Diego, CA) were coated with 50 ng/well of purified anti-IL-2 antibodies (Clone JES6-1A12, BD Pharmingen, San Diego, CA) in 50 μl PBS. Then, plates were blocked with PBS-BSA 3% and 200 μl supernatant from infected or uninfected DCs was added to each well before overnight incubation at 4°. After this time, wells were washed twice with PBS and 25 ng/well of anti-IL-2-biotin (Clone JES6-5H4, BD Pharmingen, San Diego, CA) in PBS-BSA 1% was added. Finally, plates were washed and incubated with streptavidin-horseradish peroxidase (BD Pharmingen, San Diego, CA). 3-3′-5-5′-Tetramethyl-benzidine, final concentration 100 μg/ml (Sigma-Aldrich) was used as a colorimetric substrate. The enzymatic reaction was stopped with 4 m H2SO4, and absorbance was recorded at 450 nm. Recombinant IL-2 (BD Pharmingen) was used as standard for cytokine quantification.
Infection experiments
Mice (6–8 weeks of age) were orally inoculated with 20 μl PBS containing 105 CFU of exponentially grown (OD600 = 0·5–0·7) WT or ΔInvC S. Typhimurium strains, using a micropipette. Uninfected control mice received an equivalent volume of PBS. For survival experiments, mice were inspected daily. To monitor organ colonization, mesenteric LNs, spleen and liver were recovered at days 4 and 7 post-infection and mechanically disrupted. Single-cell suspensions were lysed with 0·25% Triton-X-100 for 15 min to recover the intracellular bacteria. Bacterial loads (CFUs) were determined by seeding 10 000 tissue cells on Luria–Bertani agar plates.
For in vivo FLT3-L assays, mice were daily injected intraperitoneally with 10 μg recombinant human FLT3-L (kindly provided by Dr Christian Münz, University of Zürich, Switzerland) in saline solution for 7–9 days before infection with WT or ΔInvC Salmonella strains and at day 7 post-infection, bacterial loads (CFU) in mesenteric LNs, spleen and liver were determined as described above.
To detect Peyer’s patch DCs infected with S. Typhimurium, mice were orally infected with 107 CFU of either ΔInvC or WT S. Typhimurium strains expressing GFP. After 5 hr of infection, Peyer’s patches were recovered from small intestine, mechanically disrupted to obtain single-cell suspensions and treated as described below for flow cytometry analyses.
Flow cytometry
All data acquisition were performed on a FACScalibur II flow cytometer (BD Biosciences, Mountain View, CA). For determination of CD69 expression, DCs were pulsed with Salmonella as mentioned above and co-cultured with T cells for 48 hr. After this time, CD69 expression in T cells was determined by staining with a phycoerythrin-conjugated anti-CD69 (clone H1.2F3; BD Pharmingen, San Diego, CA) and allophycocyanin (APC) -conjugated anti-CD4 (clone RM4-5; BD Pharmingen, San Diego, CA) or APC-conjugated anti-CD8 monoclonal antibody (clone 53-6.7; BD Pharmingen, San Diego, CA). To analyse by FACS the DC population infected by either WT or ΔInvC Salmonella, cells were infected for 90 min with each Salmonella strain expressing GFP and stained with an APC-conjugated anti-mouse CD11c antibody (clone HL3; BD Pharmingen, San Diego, CA). Samples were acquired on a FACScalibur flow cytometer and data were analyzed using Winmdi software (http://facs.scripps.edu).
Results
Generation of a Salmonella ΔInvC mutant strain
To evaluate the role of TTSS-1 and the SPI-1-encoded effectors in the capacity of S. Typhimurium to interact with phagocytic and non-phagocytic cells, a mutant strain was generated by disruption of the invC gene, as described in the Materials and methods section. We tested whether the ΔInvC Salmonella mutant strain was able to induce DC death. With this aim, we measured phosphatidylserine redistribution by annexin-V staining as a parameter of apoptosis in response to bacterial challenge. In addition, we evaluated the cell membrane integrity on infected cells by using propidium iodide staining as described in the Materials and methods section. As shown in Fig. S1c, DCs infected with the ΔInvC mutant showed significantly lower annexin-V and propidium iodide staining at 2, 4, 18 and 24 hr than did DCs infected with WT Salmonella. These results are consistent with previous observations relative to the involvement of the TTSS-1 in the apoptosis induced by Salmonella on host cells.54,55
ΔInvC Salmonella fails to enter non-phagocytic cells but shows increased DCs invasion
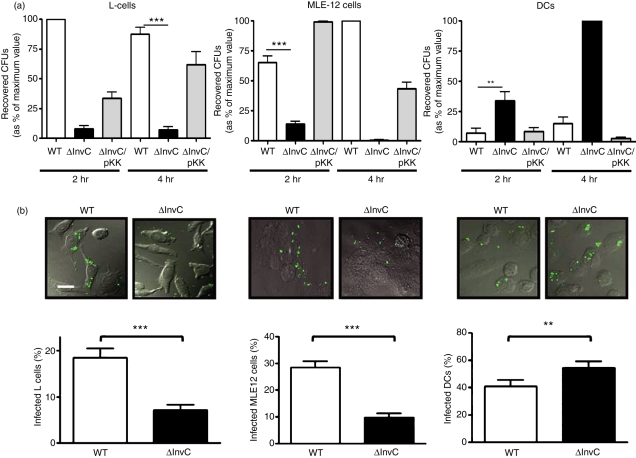
To evaluate the uptake of Salmonella strains by phagocytic and non-phagocytic cells, we used gentamicin protection assays to measure the capacity of WT and ΔInvC S. Typhimurium strains to invade either DCs (phagocytic), L or MLE-12 cells (fibroblasts and epithelium, both non-phagocytic cell types). Consistent with previous studies,18,56 WT Salmonella was significantly more efficient at invading non-phagocytic L and MLE-12 cells than was ΔInvC strain (Fig. 1a). However, the opposite pattern was observed in DCs, in which ΔInvC Salmonella entered with significantly higher efficiency than did WT Salmonella (Fig. 1a). Complementation with the wild-type invC gene under the control of a constitutive promoter reduced the entry to DCs of the ΔInvC Salmonella strain to levels equivalent to those shown by the WT Salmonella strain (Fig. 1a). Similar results were obtained with an S. Typhimurium strain lacking the entire SPI-1 (Fig. S2). Furthermore, experiments performed in J774.3 macrophages suggested that the increased invasive capacity of the ΔInvC strain was restricted to DCs (Fig. S3).
Figure 1.
ΔInvC Salmonella fails to enter non-phagocytic cells but shows increased entry into dendritic cells (DCs). (a) Intracellular survival rates for each Salmonella strain in L cells, MLE-12 cells or DCs. Cells were infected with wild-type (WT), ΔInvC or ΔInvC/pKK233.2-invC (ΔInvC/pKK) Salmonella strains (multiplicity of infection = 25) and, at the indicated times, intracellular bacteria were released from DCs and seeded on Luria–Bertani agar plates. After 12 hr of incubation at 37°, colonies were counted. Colony-forming units (CFUs) were expressed as a percentage of the maximum value for each experiment (as described in the Material and methods section). (b) Internalization of bacteria analysed by confocal microscopy. L cells, MLE-12 cells and DCs and were infected with either WT or ΔInvC Salmonella (multiplicity of infection = 50). Bacterial internalization was quantified for L cells, MLE-12 cells and DCs by analysing several random fields and counting bacteria-infected cells in at least 300 cells per bacterial strain (lower graphs). Representative microphotographs are shown above each graphic. Bar in the microphotograph represents 10 μm. Data are means of two or three independent experiments and bars represent SE (Student’s t-test: *P < 0·05; **P < 0·01; ***P < 0·001).
To confirm the invasion results obtained from the gentamicin protection assays, DCs, L cells and MLE-12 cells infected with WT and ΔInvC Salmonella were visualized by confocal laser microscopy. While WT Salmonella was found inside non-phagocytic cells (L and MLE-12 cells), ΔInvC Salmonella remained outside (Fig. 1b, upper panels). In contrast, an inverse pattern was observed for phagocytic DCs, in which significantly more ΔInvC Salmonella entered than did WT Salmonella (Fig. 1b, upper right panel). Confocal data quantification confirmed these observations (Fig. 1b, lower panels). As a control, DCs were infected with heat-killed WT and ΔInvC Salmonella. Similar amounts of both bacterial strains were found inside DCs (data not shown), suggesting that the increased entry of ΔInvC Salmonella into DCs was the result of an active process displayed by this bacterium.
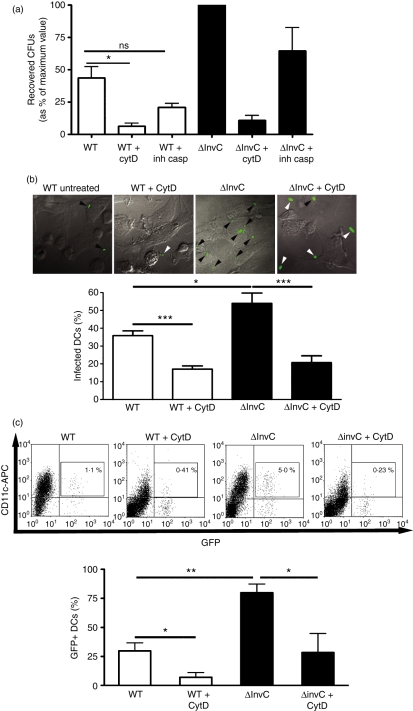
Although WT S. Typhimurium induced higher levels of DC death after infection than did the ΔInvC strain (Fig. S1), reduced entry of WT Salmonella strain to DCs was unlikely to be the result of WT Salmonella-induced DC death, because equivalent invasion patterns were obtained in the presence of a pan-caspase inhibitor (Fig. 2a).
Figure 2.
Enhanced ΔinvC Salmonella entry to dendritic cells (DCs) is not the result of reduced DC death and requires cytoskeletal rearrangement. (a) DCs were treated with the pan-caspase inhibitor VI Z-VAD FMK (casp inh) for 30 min or with cytochalasin D (CytD) for 10 min and then infected with either wild-type (WT) or ΔinvC Salmonella. After 2 hr, DCs were lysed and intracellular bacteria were seeded on Luria–Bertani agar to determine intracellular colony-forming units (CFUs). CFUs were expressed as a percentage of the maximum value for each experiment (as described in the Material and methods section). Data are means of three independent experiments and bars are SE. (b) DCs were treated with cytochalasin D for 10 min and then infected with either WT or ΔinvC Salmonella expressing green fluorescent protein (GFP). After 1 hr of infection and 3 hr of gentamicin treatment, DCs were washed, fixed and analysed by confocal microscopy to detect bacteria-infected DCs. Representative microphotographs are shown. Bar graph shows the mean of infected DCs in three independent experiments (at least 300 cells analysed). (c) DCs treated or not with cytochalasin D were infected with either WT or ΔinvC Salmonella expressing GFP. After 2 hr, DCs were stained with an allophycocyanin-conjugated anti-mouse CD11c antibody and infected cells (CD11c+/GFP+) were detected by flow cytometry. Representative dot plots are shown and the bar graph shows percentages of infected CD11c+/GFP+ cells. Data are means of three independent experiments. Bars are SE. Student’s t-test: *P = 0·05; **P = 0·01; ***P = 0·001.
To evaluate whether cytoskeletal rearrangement was required for the uptake of both Salmonella strains by DCs, cells were treated with cytochalasin D, an actin-polymerization inhibitor. As a result, a significant reduction for bacterial entry was observed for both WT and ΔInvC Salmonella (Fig. 2a). Consistent with these data, confocal microscopy assays showed reduced numbers of infected DCs after cytochalasin D treatment (Fig. 2b). Similar results were obtained when the amount of Salmonella-infected DCs was determined by flow cytometry (Fig. 2c).
Our findings reveal a new feature for Salmonella invasion of phagocytic cells, which seems to be negatively modulated by SPI-1 function. In addition, our data suggest that as SPI-1 is determinant for the capacity of Salmonella to enter non-phagocytic host cells, it might contribute to modulating the invasion of phagocytic cells, such as DCs. Consistent with this notion is the observation that infection with virulent WT Salmonella seems to reduce actin polymerization in phagocytic cells, as suggested by phalloidin staining (data not shown).
Increased ΔInvC Salmonella uptake by DCs is phosphoinositide 3-kinase-dependent
Next, we performed experiments to explore the potential mechanism responsible for the inhibition of WT Salmonella entry to DCs. First, dextran-fluorescein isothiocyanate (FITC) uptake assays were carried out to evaluate whether infection with WT Salmonella results in a global inhibition of the phagocytic activity. These assays showed no significant differences in dextran-FITC uptake between DCs infected with either WT or ΔInvC Salmonella strains, whether live or heat-killed (Fig. S4). Furthermore, uptake of dextran-FITC was equivalent for infected cells and for uninfected DCs (Fig. S4). As a positive control for phagocytosis inhibition, LPS-treated DCs and DCs incubated at 4° were included in all experiments (Fig. S4). Furthermore, the amount of LPS in the bacterial inoculum was 10 ng/ml, which was about 200 times lower than the amount of LPS used to induce DC maturation in our experiments (data not shown). These data suggest that WT Salmonella did not inhibit the bystander uptake of a soluble molecule nor did it block phagocytosis because of its LPS content.
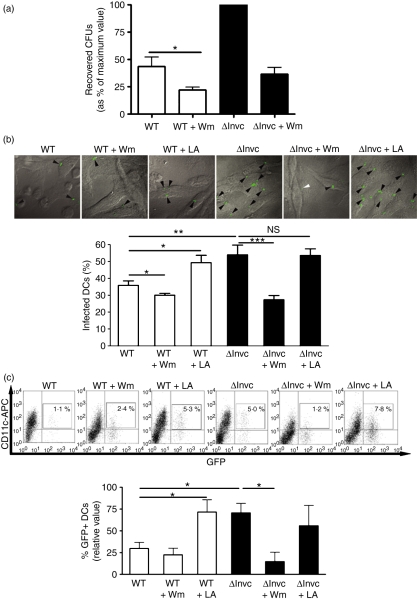
To evaluate whether the reduced entrance of WT Salmonella in DCs was the result of an active inhibition of the phosphoinositide 3-kinase (PI3-K) activity, a crucial enzyme involved in phagocytosis, we assessed bacterial uptake by DCs in gentamicin protection assays under conditions of PI3-K inhibition or induction. We observed that in the presence of the PI3-K inhibitor wortmannin, DCs exhibited a significantly reduced uptake for both WT and ΔInvC Salmonella (Fig. 3a). Equivalent data were obtained by confocal laser microscopy analyses of infected DCs (Fig. 3b).
Figure 3.
Increased entry of ΔinvC Salmonella to dendritic cells (DCs) requires phosphoinositide 3-kinase (PI3-K) activity. (a) DCs were treated for 30 min with Wortmannin (Wm) and then infected with either wild-type (WT) or ΔinvC Salmonella. After 2 hr of infection, DCs were lysed and intracellular bacteria were seeded on Luria–Bertani agar to determine amounts of intracellular colony-forming units (CFUs). Graph shows means of relative amounts of CFUs, expressed as a percentage of the maximum value for each experiment (as described in the Material and methods section). Bars represent SE. (b) DCs treated for 30 min with Wm or α-1-lipoic acid (LA) were infected for 2 hr with WT or ΔinvC Salmonella strains expressing green fluorescent protein (GFP). Then, DCs were fixed and analysed by confocal microscopy. Representative microphotographs (100 × magnification) for each treatment are shown. The graph shows the quantification of infected cells in three independent experiments. At least 300 cells for each treatment were analysed per experiment. Bars represent SE. (c) DCs were treated for 30 min with either Wm or LA and then infected with Salmonella strains expressing GFP. After 2 hr of infection, CD11c+/GFP+ cells were quantified by flow cytometry. Representative dot plots are shown and the bar graph shows mean values from three independent experiments. Student’s t-test: *P = 0·05; **P = 0·01; ***P = 0·001.
On the contrary, treatment of DCs with α-1-lipoic acid, a PI3-K activator, caused a significant increase in the uptake of WT Salmonella by DCs (Fig. 3b). These data suggest that the induction of PI3-K activity was able to overcome the inhibition of DC phagocytosis caused by WT Salmonella.
The observations made with the gentamicin protection assays were corroborated by flow cytometry analyses of DCs infected with Salmonella-GFP strains in the presence of the above PI3-K inhibitor and activator molecules. As shown in Fig. 3(c), the percentage of infected DCs was reduced after treatment with wortmannin and augmented after α-1-lipoic acid treatment. These results suggest that Salmonella uptake by DCs is PI3-K dependent. Modulation of phagocytosis by the WT Salmonella strain seems to be an active process that requires targeting of PI3-K by SPI-1-derived effector molecules.
SPI-1 activity is not required for Salmonella to survive inside DCs or to prevent antigen presentation to specific T cells
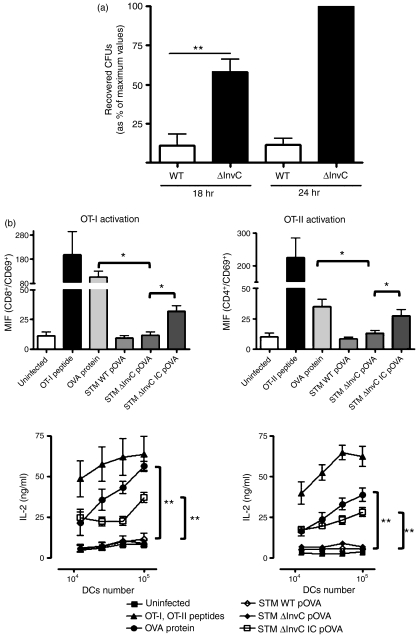
We have already demonstrated that ΔInvC Salmonella is able to invade DCs more efficiently than the WT strain. To assess if the mutant strain was able to survive inside DCs, gentamicin protection assays were performed at 18 and 24 hr post-infection. We observed that ΔInvC Salmonella was capable of surviving and proliferating inside DCs as efficiently as did the WT Salmonella strain (Fig. 4a). These results are consistent with the observation that an intact SPI-2 and not SPI-1 is necessary for intracellular survival.57
Figure 4.
ΔInvC Salmonella retains the capacity to keep dendritic cells (DCs) from activating T cells. (a) DCs were infected with wild-type (WT) or ΔInvC Salmonella for 1 hr and then treated with 50 μg/ml gentamicin. After 18 and 24 hr, DCs were lysed and intracellular Salmonella was seeded in Luria–Bertani agar to determine the colony-forming units (CFUs). Graph shows means of relative amounts of CFUs, expressed as a percentage of the maximum value for each experiment (as described in the Material and methods section) and bars are SE. (b) DCs were infected either with WT or ΔInvC Salmonella expressing ovalbumin (OVA) and co-cultured with OT-I or OT-II T cells. CD69 expression (upper panels) and interleukin-2 (IL-2) secretion (lower panels) was determined after 12 hr. As positive controls, DCs were pulsed with 10 μg/ml of OVA or the respective OT-I and OT-II peptides (2·5 ng/ml). To control that while being inside DCs, bacteria express OVA, DCs were pulsed with immunoglobulin G-opsonized ΔInvC Salmonella expressing OVA. The observed restoration of DCs capacity to activate OVA-specific T cells indicates that impairment of T-cell activation by ΔInvC Salmonella expressing OVA is not the result of a lack of OVA expression by the bacteria. Data shown are means of three independent experiments. Bars represent SE. Student’s t-test: *P < 0·05, **P < 0·01.
It has been previously suggested that WT S. Typhimurium can impair the DC capacity to activate CD4+ and CD8+ T cells in a SPI-2-dependent manner.34,36,43,46,58–61 Whether SPI-1 function is required for interfering with antigen presentation by DCs remains unknown. To approach this question, we evaluated the capacity of DCs infected with OVA-expressing ΔInvC Salmonella to activate OVA-specific CD4+ (OT-II) and CD8+ (OT-I) T cells. In agreement with previous reports, we observed that DC infection with either WT or ΔInvC Salmonella failed to prevent the processing and presentation of exogenously added OVA protein (Fig. S5). However, as shown in Fig. 4(b), DCs infected either with OVA-expressing WT or ΔInvC Salmonella failed to activate both CD4+ and CD8+ T cells, as demonstrated by low CD69 expression (Fig. 4b, upper panels) and low IL-2 secretion (Fig. 4b, lower panels). In addition, as a result of the previously observed restoration of T-cell activation when DCs internalize Salmonella via Fcγ receptors,34,46,62 DCs pulsed with IgG-opsonized WT or ΔInvC Salmonella were included as controls. As shown in Fig. 4(b), IgG opsonization partially restored the DC capacity to activate OVA-specific T cells, as shown by CD69 up-regulation and IL-2 secretion, which suggests that bacterial strains were expressing OVA while inside DCs. Similar results were obtained when heat-killed Salmonella strains expressing OVA were used as a control (data not shown36). These data are consistent with the notion that SPI-1 and TTSS-1 function would not be required for evading processing of bacterial antigens by DCs nor for preventing T-cell activation.
ΔInvC Salmonella is only partially attenuated in vivo
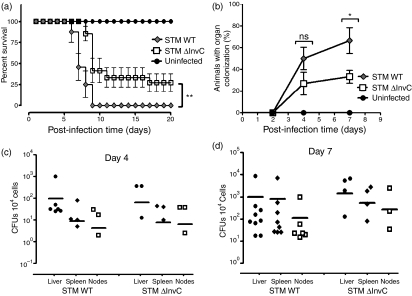
Our data suggest that the ΔInvC mutation caused an increase in the capacity of Salmonella to invade DCs (Fig. 1a,b). It is likely that these alterations could influence systemic dissemination of bacteria and survival of infected mice. To approach this question, mice were orally infected with WT or ΔInvC Salmonella and survival was determined daily. As shown in Fig. 5(a), extended survival was observed for mice infected with ΔInvC Salmonella. When organ colonization efficiency was determined, a significantly lower number of mice infected with ΔInvC Salmonella showed colonized organs at days 4 and 7 post-infection, as compared with mice infected with WT Salmonella (Fig. 5b). However, no significant differences in CFUs were observed for those mice showing organ colonization after challenge with either WT or ΔInvC Salmonella after 4 and 7 days post-infection (Fig. 5c,d). These data suggest that, although ΔInvC Salmonella is less efficient at disseminating throughout the host, once it has entered host cells this mutant strain seems able to proliferate and colonize internal tissues with an efficiency similar to that of WT Salmonella.
Figure 5.
ΔInvC Salmonella is only mildly attenuated in vivo. (a) Mice infected with ΔInvC Salmonella showed extended survival as compared to mice infected with wild-type (WT) Salmonella. C57BL/6 mice were orally infected with 105 bacteria and survival was registered daily. Mice treated with vehicle were included as controls. (b) Mice infected with ΔInvC Salmonella showed reduced organ colonization at 4 and 7 days post-infection, as compared to mice infected with WT Salmonella. To determine organ colonization capacity, mice were killed at day 4 or 7 after infection and their liver, spleen and mesenteric lymph nodes were lysed and plated for intracellular bacterial count. (c, d) Bacterial colonization for individual organs at 4 days (c) and 7 days (d) post-infection with either WT or ΔInvC Salmonella. Survival and organ colonization experiments were repeated at least three times for each strain. At least four mice per group were included in each experiment. Bars represent SE (Student’s t-test: *P < 0·05; **P < 0·01).
An increase in the frequency of DCs improves organ colonization by ΔInvC Salmonella
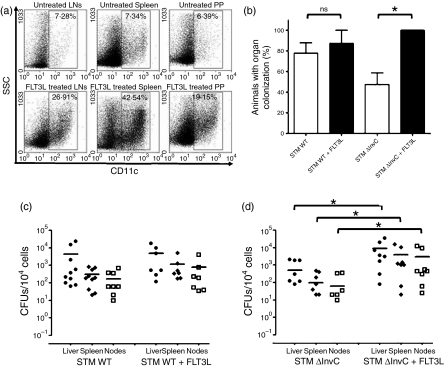
As mentioned above, Salmonella can shuttle across the epithelium barrier by invading the protruding prolongations of DCs between the epithelial cells of the intestine epithelium.22,26 To evaluate whether entry through intestinal DCs can contribute to Salmonella dissemination throughout the host, frequency of these cells was increased by treating mice with FLT3L before infection.63 As shown in Fig. 6(a), the percentage of DCs in Peyer’s patches, spleen and MLNs was significantly increased in FLT3L-treated mice.
Figure 6.
An increase in dendritic cell (DC) frequency by FLT3 ligand treatment improves organ colonization by ΔInvC Salmonella. (a) Representative dot plots showing the percentage of CD11c positive cells in spleen, mesenteric lymph nodes and Peyer’s patches obtained from control (upper pannels) and FLT3 ligand-treated mice (lower pannels). (b) FLT3 ligand-treated mice showed an increased colonization of organs at 7 days post-infection only when they were infected with ΔInvC Salmonella strain, compared with mice infected with WT Salmonella. (c, d) Bacterial colonization in liver, spleen and lymph nodes of control and FLT3 ligand-treated mice at 7 days post-infection with either WT (c) or ΔInvC (d) Salmonella. Data shown are means of two independent experiments, including at least five mice per group. Bars represent SE (Student’s t-test: *P ≤ 0·05).
Although treatment with FLT3L did not alter significantly the number of mice with organ colonization by WT Salmonella, it caused a significant increase of mice with ΔInvC Salmonella in organs after 7 days post-infection (Fig. 6b). When analysing the CFUs found in the organs of colonized mice, no significant differences were observed between FLT3L-treated and not treated mice infected with WT Salmonella (Fig. 6c). However, CFUs in the liver of FLT3L-treated mice were significantly higher than in untreated mice, after challenge with ΔInvC Salmonella (Fig. 6d).
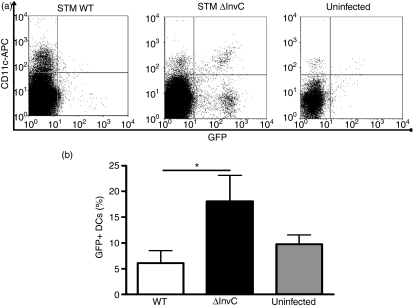
The observation that an increase in the total number of intestinal DCs enhanced systemic ΔInvC Salmonella dissemination would suggest that these cells might be the preferred entrance route for this mutant strain. To address this hypothesis, mice were orally infected for 5 hr with either WT or ΔInvC Salmonella expressing GFP. Then, Peyer’s patch DCs were obtained and analysed by flow cytometry to quantify the amount of bacteria captured by DCs. As shown in Fig. 7, significantly more CD11c+/GFP+ cells were detected in Peyer’s patches from animals infected with ΔInvC Salmonella. These results support the notion that ΔInvC Salmonella relies on intestinal DCs to reach internal organs and spread systemically.
Figure 7.
Mice infected with ΔInvC Salmonella show higher numbers of bacteria-containing Peyer’s patch dendritic cells (PP DCs). Mice were orally infected with 107 colony-forming units (CFU) of either wild-type (WT) or ΔInvC Salmonella expressing green fluorescent protein (GFP). After 6 hr of infection, PPs were recovered, single cell suspensions were obtained and DCs were stained with an allophycocyanin-conjugated anti-mouse CD11c antibody. (a) Representative dot plots are shown for PP DCs obtained from mice infected with WT Salmonella, ΔInvC Salmonella or from uninfected mice. (b) Graph shows the percentage of CD11c+/GFP+ cells in PPs after Salmonella infection. Data are means of three independent experiments, including at least three mice per group, and bars represent SE. Student’s t-test: *P = 0·05.
Discussion
In this study we have shown that SPI-1 activity can differentially modulate the entry of Salmonella to non-phagocytic versus phagocytic cells. Our data suggest that whereas SPI-1 was required for the invasion of non-phagocytic cells, such as L and MLE-12 cells, its function was needed to prevent massive entrance of Salmonella to phagocytic DCs. Apparently, translocation of Salmonella effectors via SPI-1 was not required for intracellular survival of bacteria nor for evading antigen presentation on MHC molecules to T cells. In vivo, SPI-1 deletion caused only minor attenuation and mutant strains remained capable of causing systemic infection and colonizing internal organs.
Consistent with previous studies,26,56,64–67 we observed that an InvC deficiency caused a significant impairment of bacterial entrance to non-phagocytic cells, as compared with WT Salmonella. Lack of InvC is thought to render Salmonella unable to translocate the SPI-1-derived effector molecules required to induce phagocytosis by epithelial cells.18,50 In contrast to what was observed in non-phagocytic cells, deletion of InvC led to a significant enhancement in bacterial entry to phagocytic DCs. It is likely that increased internalization of Salmonella was the result of an increased capacity of murine DCs to capture bacteria when they lacked a functional TTSS-1. Our data support the notion that the TTSS-1 or the effector proteins translocated by it can inhibit DC uptake of Salmonella via phagocytosis.
Furthermore, we observed that the reduced entry of WT Salmonella to DCs was dependent of PI3-K activity, because inhibition of PI3-K by wortmannin prevented the excessive entry of the ΔInvC Salmonella strain to DCs. Consistent with this, induction of PI3-K activity by α-1-lipoic acid treatment led to an enhanced entry of WT Salmonella into DCs. It is therefore likely that an effector molecule translocated by SPI-1 could, directly or indirectly, be responsible for suppressing PI3-K activity during infection, which would modulate bacterial phagocytosis.
These findings suggest that virulent Salmonella possesses a mechanism for modulating DC phagocytosis, equivalent to what has been shown for other pathogenic enterobacteria, such as enteropathogenic Escherichia coli (EPEC) and Yersinia enterocolitica.68,69 For EPEC, specific virulence factors were identified as responsible for inhibiting phagocytosis in host cells.70
It has been reported that Salmonella strains defective for invasion genes are impaired at entering M cells56 and RAW264.7 macrophages,10 both of them phagocytic cell types. Consistent with this notion, we observed that the entry of ΔInvC strain to J774.3 macrophages was decreased compared with the WT strain (data not shown). In contrast, we observed that the non-invasive ΔInvC Salmonella strain showed an increased entry to DCs. This observation would suggest that inhibition of phagocytosis by SPI-1 might be specific for DCs. It is possible that such a mechanism might contribute to Salmonella pathogenesis by modulating entry to DCs and the inflammatory response induced by them. Fewer replicating bacteria inside DCs could ensure intracellular survival and migration to internal tissues from the site of infection, by exploiting perhaps the capacity of DCs to move from the site of infection to LNs and the spleen.
However, despite being professional antigen-presenting cells capable of activating naive T cells, DCs infected with virulent Salmonella are rendered unable to efficiently present bacteria-expressed antigens to T cells. It is thought that by avoiding fusion with lysosomes, Salmonella residing inside phagosomes can evade processing and presentation of antigens on MHC molecules to T cells.26,36,37,43,45 Interference with antigen presentation was shown to be dependent on the activity of a TTSS and effector proteins encoded in the SPI-2.36,39,40,71 Consistent with this notion, we observed that the ΔInvC strain remained capable of avoiding T-cell activation when inside DCs, suggesting a normal function of SPI-2 for this strain.
Salmonella has several ways of invading intestinal mucosa: (i) invasion of epithelial cells,41 (ii) capture by M cells,2,72 (iii) paracellular translocation,73 and (iv) capture by DCs.22,24,72,74 It seems that this last mechanism, which is dependent on MyD88 signalling,24,75 is not the main route of entry and dissemination of virulent Salmonella. However, translocation by DCs seems to be fundamental for Salmonella strains lacking TTSS-1 to invade epithelial mucosa and disseminate through liver and spleen. Our results support this notion, because we detected a higher number of Peyer’s patch DCs infected with ΔInvC strain after an oral infection. In addition, ΔInvC strain was able to cause a delayed systemic disease when compared with the WT strain. Remarkably, mice that were successfully colonized by ΔInvC strains showed a similar extent of internal organ colonization as WT Salmonella.
However, the ΔInvC strain was attenuated in mice and showed reduced efficiency at causing systemic colonization compared with the WT virulent Salmonella. These results suggest that the ΔInvC mutant is only partially attenuated for in vivo infection because invasion of intestinal mucosa probably depends on the number of intercallating DCs available for Salmonella entrance along the intestine.
In agreement with this notion, when the frequency of DCs in the intestine was increased after FLT3 ligand treatment the dissemination rate of ΔInvC strain was enhanced. Our data are consistent with a previous study showing enhanced dissemination of Listeria after FLT3 ligand treatment.76 Entry via DCs seems to rely on the capacity of intracellular bacterial pathogens to avoid degradation by DCs. Because the ΔInvC strain remains able to avoid antigen presentation by DCs – as shown by our antigen presentation assays – it is likely that an increase in the frequency of DCs in the intestinal mucosa could improve dissemination of these non-invasive bacteria. However, our experiments cannot rule out the possibility that other intestinal cells could also be affected after FLT3 ligand treatment in vivo, which might account for a higher capacity of the ΔInvC strain to disseminate from the intestine to deeper organs. Our in vitro experiments suggest that the treatment with this drug does not increase the capacity of the ΔInvC strain to invade epithelial cells or macrophages and it does not promote intestinal damage (data not shown); however, further analyses are required to determine if other DC-independent mechanisms are also involved in the spreading of the ΔInvC strain from intestine.
Experiments performed on DC-depleted mice have suggested that invasion through intercalating DCs might not be the main invasion method for virulent Salmonella.24 In contrast, SPI-1-deficient Salmonella was absolutely dependent on DCs to cross the intestinal epithelium.24 Consistent with these findings, it was observed that infection with virulent Salmonella is more severe in mice in which DCs are unable to form transepithelial dendrites, because of a deficiency on CXCR3.23 Our observation that WT Salmonella can inhibit phagocytosis in DCs in vitro supports the notion that interference with DC function might be beneficial for bacterial survival and dissemination. It is likely that capture by transepithelial DCs could promote an inflammatory response that contributes to restricting Salmonella replication and dissemination.
Our data suggest that impairment of SPI-1 function can promote Salmonella entrance via DC uptake, which would reduce the capacity of this pathogen to prevent an inflammatory response and disseminate systemically. However, the scarce frequency of DCs able to translocate ΔInvC strain may explain the reduced dissemination and organ colonization observed for this strain. Further research is needed to evaluate the effect of the lack of SPI-1 function on the intestinal inflammatory response in vivo.
Acknowledgments
The authors are supported by grants FONDECYT no. 1070352, FONDECYT no. 1050979, FONDECYT no. 3060041, FONDECYT no. 11075060, FONDECYT no. 1040349, SavinMuco-Path-INCO-CT-2006-032296; IFS#B/3764-1, VRAID-INICIO 20/2007, Red-15 PBCT, and Millennium Nucleus on Immunology and Immunotherapy (P04/030-F).
Supporting Information
Additional Supporting information may be found in the online version of this article:
Figure S1. ΔInvC Salmonella does not secrete SPI-1 effector proteins and shows a reduced capacity to induce cell death on DCs. (A) Genetic characterization of ΔInvC Salmonella strain. PCR amplification produced a 1311 bp long fragment corresponding to normal invC gene in WT Salmonella. In contrast, the same PCR reaction produced a longer fragment (2000 bp long) ΔInvC Salmonella, due to insertion of the kanamycin resistance cassette. As a positive control, PCR amplification of avrA (SPI-1 effector) was included and produced the expected 1057 bp long fragment for both WT and ΔInvC Salmonella. (B) Functional characterization of ΔInvC Salmonella. WT and ΔInvC Salmonella were grown in LB medium and the presence of SopE (SPI-1 effector) was evaluated in pellets (P) and supernatants (S) by Western blot. (C) and (D) Percentage of cell death induced by Salmonella strains. DCs were infected either with WT or ΔInvC Salmonella (MOI = 25) and the percentage of Annexin-V+/PI+ (C) and TUNEL+/PI+ (D) positive cells were quantified by flow cytometry at 2, 4, 18 and 24 hr after infection. As a positive control, DCs treated with 4.2 μM camptothecin (an apoptosis inducer) were included. Data shown are means of at least three independent experiments. Bars represent SE (Student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001).
Figure S2. ΔSPI-1 Salmonella shows increased entry into DCs. DCs were infected with WT, ΔInvC or ΔSPI-1 S.Typhimurium strains (MOI = 25). At the indicated times, intracellular bacteria were released from DCs and seeded on LB-agar plates. After 12 hr of incubation at 37°, colonies were quantified. CFUs were expressed as a percentage of the maximum value for each experiment. Data are means of three independent experiments and bars represent SE.
Figure S3. ΔInvC Salmonella strain shows a reduced capacity to enter J774.3 macrophages. J774.3 cells were infected with either WT or ΔInvC S.Typhimurium strains (MOI = 25). At the indicated times, intracellular bacteria were released from the cells and seeded on LB-agar plates. After 12 hr of incubation at 37°, colonies were quatified. CFUs were expressed as a percentage of the maximum value for each experiment. Data are means of two independent experiments and bars represent SE. (Student's t-test: *P < 0.05; ***P < 0.001).
Figure S4. Infection with either WT or ΔInvC strains of Salmonella does not affect Dextran-FITC uptake by DCs. DCs were infected with live or heat killed Salmonella strains in the presence of 100 μg/ml Dextran-FITC, incubated at 37° for 4 hr, and then analyzed by flow cytometry to detect CD11c+/Dextran-FITC+ cells. As controls, uninfected DCs and DCs treated for 12 hr with 2 μg/ml LPS were included. In addition, uninfected DCs incubated at 4° were included as a background control. (A) Histograms showing representative FACS profiles of Dextran- FITC fluorescence for CD11c+ population (CD11c+ gated). Marker establishes the background level of FITC fluorescence, according to the uninfected cells incubated at 4° controls. (B) Graph showing the percentage of CD11c+/Dextran-FITC+ cells obtained in four independent experiments. Bars represent mean ± SE.
Figure S5. WT or ΔInvC Salmonella do not prevent the processing and presentation of exogenously added OVA protein. DCs were infected with WT or ΔInvC S.Typhimurium and pulsed with 10 μg/ml of OVA protein for 1 hr. Then, cells were treated with 50 μg/ml gentamicin. After 20 hr, DCs were co-cultured with purified OT-I cells and 24 hr later CD69 expression (A) and IL-2 secretion (B) were determined by flow cytometry and ELISA, respectively. As a positive control, uninfected DCs were pulsed with 10 μg/ml of OVA. Data shown are means of two independent experiments and bars represent SE.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52:259–74. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 2.Clark MA, Jepson MA, Simmons NL, Hirst BH. Preferential interaction of Salmonella typhimurium with mouse Peyer’s patch M cells. Res Microbiol. 1994;145:543–52. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 3.Jepson MA, Clark MA. The role of M cells in Salmonella infection. Microbes Infect. 2001;3:1183–90. doi: 10.1016/s1286-4579(01)01478-2. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins SA, Niedergang F, Corthesy-Theulaz IE, Kraehenbuhl JP. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer’s patch dendritic cells. Cell Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-5822.2000.00035.x. [DOI] [PubMed] [Google Scholar]
- 5.Jepson MA, Collares-Buzato CB, Clark MA, Hirst BH, Simmons NL. Rapid disruption of epithelial barrier function by Salmonella typhimurium is associated with structural modification of intercellular junctions. Infect Immun. 1995;63:356–9. doi: 10.1128/iai.63.1.356-359.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–91. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhou D, Galan J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001;3:1293–8. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 9.Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int J Med Microbiol. 2002;291:479–85. doi: 10.1078/1438-4221-00156. [DOI] [PubMed] [Google Scholar]
- 10.Monack DM, Raupach B, Hromockyj AE, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci U S A. 1996;93:9833–8. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiama SG, Dreher D, Cochand L, Kok M, Obregon C, Nicod L, Gehr P. Host cell responses of Salmonella typhimurium infected human dendritic cells. Immunol Cell Biol. 2006;84:475–81. doi: 10.1111/j.1440-1711.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 12.Grant AJ, Sheppard M, Deardon R, Brown SP, Foster G, Bryant CE, Maskell DJ, Mastroeni P. Caspase-3-dependent phagocyte death during systemic Salmonella enterica serovar Typhimurium infection of mice. Immunology. 2008;125:28–37. doi: 10.1111/j.1365-2567.2008.02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 14.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–73. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 15.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–9. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A. 2000;97:10225–30. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akeda Y, Galan JE. Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J Bacteriol. 2004;186:2402–12. doi: 10.1128/JB.186.8.2402-2412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichelberg K, Ginocchio CC, Galan JE. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–10. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–5. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 20.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–8. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 21.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–92. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 22.Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immun. 2001;2:361–7. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 23.Niess JH, Brand S, Gu X, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–8. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 24.Hapfelmeier S, Muller AJ, Stecher B, et al. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med. 2008;205:437–50. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rescigno M. Dendritic cells and the complexity of microbial infection. Trends Microbiol. 2002;10:425–61. doi: 10.1016/s0966-842x(02)02425-3. [DOI] [PubMed] [Google Scholar]
- 26.Hapfelmeier S, Stecher B, Barthel M, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–85. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 27.Bueno SM, Tobar JA, Iruretagoyena MI, Kalergis AM. Molecular interactions between dendritic cells and Salmonella: escape from adaptive immunity and implications on pathogenesis. Crit Rev Immunol. 2005;25:389–403. doi: 10.1615/critrevimmunol.v25.i5.40. [DOI] [PubMed] [Google Scholar]
- 28.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–84. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Galan JE. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 30.Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol. 2005;43(Spec No):85–92. [PubMed] [Google Scholar]
- 31.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 32.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 33.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 34.Tobar JA, Gonzalez PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fcγ receptors on dendritic cells. J Immunol. 2004;173:4058–65. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez PA, Carreno LJ, Figueroa CA, Kalergis AM. Modulation of immunological synapse by membrane-bound and soluble ligands. Cytokine Growth Factor Rev. 2007;18:19–31. doi: 10.1016/j.cytogfr.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, Kalergis AM. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–48. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bueno SM, Gonzalez PA, Schwebach JR, Kalergis AM. T cell immunity evasion by virulent Salmonella enterica. Immunol Lett. 2007;111:14–20. doi: 10.1016/j.imlet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Bueno SM, Gonzalez PA, Kalergis AM. Use of genetically modified bacteria to modulate adaptive immunity. Current gene therapy. 2009;9:171–84. doi: 10.2174/156652309788488587. [DOI] [PubMed] [Google Scholar]
- 39.Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–11. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 40.Kuhle V, Hensel M. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci. 2004;61:2812–26. doi: 10.1007/s00018-004-4248-z. [DOI] [PubMed] [Google Scholar]
- 41.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun. 2005;73:7161–9. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun. 2005;73:3219–27. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halici S, Zenk SF, Jantsch J, Hensel M. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect Immun. 2008;76:4924–33. doi: 10.1128/IAI.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yrlid U, Wick MJ. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon Salmonella encounter. J Immunol. 2002;169:108–16. doi: 10.4049/jimmunol.169.1.108. [DOI] [PubMed] [Google Scholar]
- 45.Kuhle V, Jackel D, Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–70. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 46.Herrada AA, Contreras FJ, Tobar JA, Pacheco R, Kalergis AM. Immune complex-induced enhancement of bacterial antigen presentation requires Fcγ receptor III expression on dendritic cells. Proc Natl Acad Sci U S A. 2007;104:13402–7. doi: 10.1073/pnas.0700999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wick MJ. Monocyte and dendritic cell recruitment and activation during oral Salmonella infection. Immunol Lett. 2007;112:68–74. doi: 10.1016/j.imlet.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Cheminay C, Mohlenbrink A, Hensel M. Intracellular Salmonella inhibit antigen presentation by dendritic cells. J Immunol. 2005;174:2892–9. doi: 10.4049/jimmunol.174.5.2892. [DOI] [PubMed] [Google Scholar]
- 49.Alaniz RC, Cummings LA, Bergman MA, Rassoulian-Barrett SL, Cookson BT. Salmonella typhimurium coordinately regulates FliC location and reduces dendritic cell activation and antigen presentation to CD4+ T cells. J Immunol. 2006;177:3983–93. doi: 10.4049/jimmunol.177.6.3983. [DOI] [PubMed] [Google Scholar]
- 50.Brumme S, Arnold T, Sigmarsson H, et al. Impact of Salmonella Typhimurium DT104 virulence factors invC and sseD on the onset, clinical course, colonization patterns and immune response of porcine salmonellosis. Vet Microbiol. 2007;124:274–85. doi: 10.1016/j.vetmic.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 51.Finlay BB, Falkow S. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie. 1988;70:1089–99. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
- 52.Kalergis AM, Goyarts EC, Palmieri E, Honda S, Zhang W, Nathenson SG. A simplified procedure for the preparation of MHC/peptide tetramers: chemical biotinylation of an unpaired cysteine engineered at the C-terminus of MHC-I. J Immunol Methods. 2000;234:61–70. doi: 10.1016/s0022-1759(99)00215-x. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez PA, Prado CE, Leiva ED, Carreno LJ, Bueno SM, Riedel CA, Kalergis AM. Respiratory syncytial virus impairs T cell activation by preventing synapse assembly with dendritic cells. Proc Natl Acad Sci U S A. 2008;105:14999–5004. doi: 10.1073/pnas.0802555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A. 1999;96:2396–401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol. 1999;181:3433–7. doi: 10.1128/jb.181.11.3433-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Penheiter KL, Mathur N, Giles D, Fahlen T, Jones BD. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer’s patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 57.Boddicker JD, Jones BD. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect Immun. 2004;72:2002–13. doi: 10.1128/IAI.72.4.2002-2013.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng H, Wu H, Sloane V, Jones R, Yu Y, Lin P, Gewirtz AT, Neish AS. Flagellin/TLR5 responses in epithelia reveal intertwined activation of inflammatory and apoptotic pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G96–108. doi: 10.1152/ajpgi.00273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivasan A, McSorley SJ. Activation of Salmonella-specific immune responses in the intestinal mucosa. Arch Immunol Ther Exp (Warsz) 2006;54:25–31. doi: 10.1007/s00005-006-0003-5. [DOI] [PubMed] [Google Scholar]
- 60.Salazar-Gonzalez RM, Niess JH, Zammit DJ, et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity. 2006;24:623–32. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell EK, Mastroeni P, Kelly AP, Trowsdale J. Inhibition of cell surface MHC class II expression by Salmonella. Eur J Immunol. 2004;34:2559–67. doi: 10.1002/eji.200425314. [DOI] [PubMed] [Google Scholar]
- 62.McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:3344–8. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–13. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galan JE, Curtiss R., III Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–8. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galan JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–49. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stender S, Friebel A, Linder S, Rohde M, Mirold S, Hardt WD. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol Microbiol. 2000;36:1206–21. doi: 10.1046/j.1365-2958.2000.01933.x. [DOI] [PubMed] [Google Scholar]
- 67.Flentie KN, Qi M, Gammon ST, et al. Stably integrated luxCDABE for assessment of Salmonella invasion kinetics. Mol Imaging. 2008;7:222–33. [PMC free article] [PubMed] [Google Scholar]
- 68.Celli J, Olivier M, Finlay BB. Enteropathogenic Escherichia coli mediates antiphagocytosis through the inhibition of PI 3-kinase-dependent pathways. EMBO J. 2001;20:1245–58. doi: 10.1093/emboj/20.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Persson C, Carballeira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–18. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quitard S, Dean P, Maresca M, Kenny B. The enteropathogenic Escherichia coli EspF effector molecule inhibits PI-3 kinase-mediated uptake independently of mitochondrial targeting. Cell Microbiol. 2006;8:972–81. doi: 10.1111/j.1462-5822.2005.00680.x. [DOI] [PubMed] [Google Scholar]
- 71.Knodler LA, Celli J, Hardt WD, Vallance BA, Yip C, Finlay BB. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol Microbiol. 2002;43:1089–103. doi: 10.1046/j.1365-2958.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 72.Jang MH, Kweon MN, Iwatani K, et al. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc Natl Acad Sci U S A. 2004;101:6110–5. doi: 10.1073/pnas.0400969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kops SK, Lowe DK, Bement WM, West AB. Migration of Salmonella typhi through intestinal epithelial monolayers: an in vitro study. Microbiol Immunol. 1996;40:799–811. doi: 10.1111/j.1348-0421.1996.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 74.Jepson MA, Lang TF, Reed KA, Simmons NL. Evidence for a rapid, direct effect on epithelial monolayer integrity and transepithelial transport in response to Salmonella invasion. Pflugers Arch. 1996;432:225–33. doi: 10.1007/s004240050128. [DOI] [PubMed] [Google Scholar]
- 75.Arques JL, Hautefort I, Ivory K, Bertelli E, Regoli M, Clare S, Hinton JC, Nicoletti C. Salmonella induces flagellin- and MyD88-dependent migration of bacteria-capturing dendritic cells into the gut lumen. Gastroenterology. 2009;37:579–87. doi: 10.1053/j.gastro.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 76.Alaniz RC, Sandall S, Thomas EK, Wilson CB. Increased dendritic cell numbers impair protective immunity to intracellular bacteria despite augmenting antigen-specific CD8+ T lymphocyte responses. J Immunol. 2004;172:3725–35. doi: 10.4049/jimmunol.172.6.3725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.