Abstract
Mycobacterial species, like other microbes, spontaneously form multicellular drug-tolerant biofilms when grown in vitro in detergent-free liquid media. The structure of Mycobacterium tuberculosis biofilms is formed through genetically programmed pathways and is built upon a large abundance of novel extracellular free mycolic acids (FM), although the mechanism of FM synthesis remained unclear. Here we show that the FM in Mycobacterium smegmatis biofilms is produced through the enzymatic release from constitutively present mycolyl derivatives. One of the precursors for FM is newly synthesized trehalose dimycolate (TDM), which is cleaved by a novel TDM-specific serine esterase, Msmeg_1529. Disruption of Msmeg_1529 leads to undetectable hydrolytic activity, reduced levels of FM in the mutant, and retarded biofilm growth. Furthermore, enzymatic hydrolysis of TDM remains conserved in M. tuberculosis, suggesting the presence of a TDM-specific esterase in this pathogen. Overall, this study provides the first evidence for an enzymatic release of free mycolic acids from cell envelope mycolates during mycobacterial growth.
Keywords: Bacteria, Bacterial Genetics, Bacterial Metabolism, Enzymes, Lipid, Biofilms, Free Mycolic Acids, Mycobacteria, Serine Esterase, Trehalose Dimycolate
Introduction
Many microbial species grow as organized communities on surfaces and interfaces, forming a range of multicellular structures from colonies on submerged biotic and abiotic surfaces to pellicles at the air-medium interfaces (1–6). Individual cells within these biofilms are typically embedded in a complex architecture of extracellular matrices primarily composed of secreted polymeric substances including polysaccharides, lipids, proteins, and DNA (1, 3, 7–9). Biofilm development involves coordinated communication between cells and programs of gene expression needed for individual cells to respond to varying microenvironments within the biofilm structure (10–12).
The architectural complexities of microbial biofilms, built on the scaffold of a extracellular matrix, lead to a widespread heterogeneity in the interior microenvironments to which the resident cells presumably adapt by adjusting their physiological and metabolic activities (13, 14). Therefore, heterogeneous microenvironments of biofilms foster a range of phenotypic diversity in the population and may even trigger cellular differentiation in the community (15, 16). Taken together, mature biofilms provide suitable microenvironments for the development of a subpopulation of stress-tolerant cells, which can persist against a wide range of environmental challenges including antibiotics (17, 18). The extraordinary drug tolerance of microbes in biofilms is particularly relevant in clinical settings where pathogens responsible for chronic infectious diseases such as cystic fibrosis, otitis media, infective endocarditis, and urinary tract infections colonize the host surfaces to form persistent and drug tolerant communities (19–22).
Mycobacterium tuberculosis, the causative agent of human tuberculosis, kills about 2 million people annually, and treatment of its infection requires 6–9 months of multidrug chemotherapy (23). These extended periods of chemotherapy are required to minimize relapse and reoccurrence of disease, presumably by eradicating the subpopulation of cells that escape the early phase of treatment (24). Although the basis for the tolerance of these persisters is not clear, the duration of treatment as well as the rate of relapse correlates closely with the bacterial burden in lung lesions such that the large number of bacilli associated with caseating granulomas and cavities may be especially difficult to eliminate by chemotherapy (25, 26). The microbiological nature of M. tuberculosis in these high burden infections remains unclear, although we note that in vitro M. tuberculosis bacilli form parallel rope-like cords in detergent-free growth medium as well as pellicles at the air-medium interface (27). These M. tuberculosis cords are rich in an extracellular glycolipid, trehalose dimycolate (TDM),3 that has been shown to induce granulomatous responses in animal models (28–30), and mutants defective in cording show loss of lethal chronic persistence in mice (31).
Recent studies show that development of multicellular structures of mycobacteria has distinct physiological and genetic requirements and promote drug tolerance in the population (32–37); these structures can, therefore, be broadly categorized as biofilms. Mutants of Mycobacterium smegmatis and Mycobacterium marinum with defective biosynthesis of either cell wall or cell surface lipids like glycopeptidolipids, mycolyldiacylglycerol, and lipooligosaccharides fail to form biofilms (33, 38, 39). However, the composition of mycobacterial biofilms is significantly different from that of other bacteria, containing an extracellular matrix rich in lipids rather than polysaccharides (8, 34, 37). Previously, we observed that mycolic acid synthesis during biofilm maturation of M. smegmatis was regulated by groEL1 through its direct interaction with KasA (a β-keto-acyl carrier protein synthase), a component of the multienzyme complex type II fatty acid synthase involved in mycolic acid synthesis (34). We further found that M. tuberculosis biofilms also have substantial amounts of extracellular free mycolic acids (FM) (37). The abundance of FM in biofilms was novel in a way that mycolic acids have generally been understood to be present as esters of sugars, although the processes whereby FM is synthesized and released during biofilm formation remain unclear.
Here we show that FM-rich M. smegmatis biofilms contain enzymatic activity that hydrolyzes trehalose dimycolate to release FM. We also identify a gene, Msmeg_1529, that encodes a cutinase-like serine carboxyesterase responsible for TDM hydrolysis. Disruption of Msmeg_1529 leads to loss of hydrolytic activity in the mutant. We further show that disrupted TDM synthesis through mutation in mycolyltransferases or TDM hydrolysis through mutation in Msmeg_1529 impairs biofilm development of M. smegmatis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
All M. smegmatis strains were maintained in 7H9ADCTween liquid medium or on 7H10ADC agar. Mutants were grown in 150 μg/ml hygromycin and the complemented strains in 150 μg/ml hygromycin and 40 μg/ml kanamycin. For experiments, the strains were grown either planktonically or as biofilms in biofilm media as described previously (34). An attenuated strain of M. tuberculosis mc27000 (37) was used as a parent wild type in this study. The strain was maintained in 7H9OADC with 100 μg/ml pantothenic acid in liquid culture and on 7H11OADC + pantothenic acid on plates. For experiments, the strain was cultured in Sauton's medium with pantothenic acid before processing.
Genetic Manipulations of M. smegmatis
All the genetic manipulations in M. smegmatis were carried using a recently developed recombineering technique in mycobacteria (53). The allelic exchange substrate for every deletion was prepared by amplifying the left and right arms of the gene using primers listed in supplemental Table S1. The kanr, hygr recombinant colonies were screened for correct gene replacement by PCR. For complementation, the mutants were rescued of the recombineering plasmid, pJV53, and then transformed with complementing plasmids. The transformants were selected on 7H10ADC/hyg/kan. The complementing plasmids, pMs6398 and pMs1529, were constructed by cloning the PCR- amplified full-length ORF downstream of Phsp60 of pJL37 (54). The primers used for amplification of Msmeg_6398 and Msmeg_1529 ORF are listed in supplemental Table S2.
Lipid Purification and Analysis
The total organic extractable apolar and polar lipids of mc2155, cultured either as biofilms or as a planktonic suspension, were extracted in petroleum ether as described previously (37). For analysis of radiolabeled lipids, the cultures were exposed to [14C]glucose for 6 h before extraction in petroleum ether. The extracted lipids were developed on one- or two-dimensional TLC in the solvent systems as indicated for every experiment. The radiolabeled lipids were analyzed by autoradiography. The FM were developed in chloroform:methanol (96:4) and visualized by spraying the TLC plate with 5% molybdophosphoric acid in ethanol and charring the plate at 120 °C for 10 min. The TDM was developed in chloroform:methanol:water (90:10:1) and visualized by charring the plate with 10% H2SO4 in ethanol at 120 °C for 10 min. Methyl esters of FM and total cellular mycolic acids were prepared as described previously and resolved in hexane:diethyl ether (95:5) before visualizing the plate with 5% molybdophosphoric acids.
Structural Analysis of TDM and FM
The lipid spot migrating similar to M. tuberculosis FM in solvent C (37) was extracted from the silica plate in chloroform:methanol (2:1) and analyzed by NMR and mass spectrometry. Similarly, lipid comigrating with a standard Mtb TDM (Sigma) was extracted from silica plate in chloroform:methanol (2:1) and analyzed by mass spectrometry. Mass spectrometry analyses of purified mycolates and TDM were done on a Voyager Elite reflectron matrix-assisted laser desorption ionization time-of-flight mass spectrometer (PerSeptive Biosystems, Framingham, MA) equipped with a 337-nm UV laser. Samples were solubilized in 1 ml of chloroform/methanol (2:1) and mixed on target with 1 ml of a 2,5-dihydroxybenzoic acid matrix solution (10 mg/ml dissolved in chloroform/methanol 2:1). For NMR analyses the same molecules were dissolved in deuterated chloroform containing 0.01% of tetramethylsilane and transferred into Shigemi tubes matched for D2O. Then 0.1 ml of deuterium oxide was added to avoid solvent evaporation during long acquisition. One-dimensional proton NMR spectra were recorded at 299 K on an 800 MHz Avance II Bruker spectrometers equipped with a 1H/13C/15N/2H probe. After structural confirmation of TDM and FM, the purified lipids were used as reference for all subsequent TLCs.
In Vitro Assay for the Synthesis of FM
For all the in vitro assays described, radiolabeled substrate, either total cellular lipids, TDM, or cell wall, equivalent to 100,000 cpm were homogeneously resuspended in the assay buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 25 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, and 0.2% Triton N101) by mild sonication. The homogenate was then mixed to 100 μg of proteins from M. smegmatis lysates prepared from either planktonic or biofilm cells lysed by sonication in the assay buffer. The mixture was incubated at 37 °C for 2 h. The lipids in the reaction mixture were extracted in an equal volume of petroleum ether, dried, and analyzed on TLC developed in chloroform:methanol 96:4. For testing the heat sensitivity of the active component, the biofilm lysate was incubated at 95 °C for 15 min before the assay. For assays with either purified TDM or cell wall, the radiolabeled homogenate in the assay was substituted with equivalent counts of TDM or cell wall, isolated using the previously described methods. For assay with purified proteins, 5 μg of Msmeg_1529 or D29 LysB were used.
Cloning and Characterization of Serine Esterases from M. smegmatis
A search of serine esterase from M. smegmatis genome webpage of the J. Craig Venter Institute (Comprehensive Microbial Resource website of the J. Craig Venter Institute) revealed six ORFs. All six ORFs along with Msmeg_6398 were amplified using the primers listed in supplemental Table S3. The amplicons were cloned in the NdeI and XhoI sites of pET21b, and the recombinant plasmid expressing the ORFs was sequenced to confirm the clone identity. Each recombinant clone was then transformed into Escherichia coli strain BL-21 DE3. The transformants were grown in LB-ampicillin until A0.6 before being induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. The induced cells were harvested, washed, and sonicated in assay buffer. 100 μg of total culture lysate protein was mixed with 100,000 cpm of purified [14C]TDM, and the esterase activity in the lysates were assayed as described above.
Drug Tolerance Assay of M. smegmatis Biofilms
A drug tolerance assay of M. smegmatis mc2155 in biofilms cultures was done as described previously (37). Cells were grown in biofilms for 5 days before exposing to 400 μg/ml rifampicin for 7 days. Before harvesting cells, the biofilms plates were incubated with 0.2% Tween 80 to solubilize the biofilms. The solubilized biofilms were then rocked in the presence of detergent and glass beads for 24 h at 4 °C. The homogeneous suspension of cells was then diluted and plated on 7H10 agar to determine their colony-forming units.
RESULTS
Regulated Synthesis of FM in M. smegmatis Biofilms
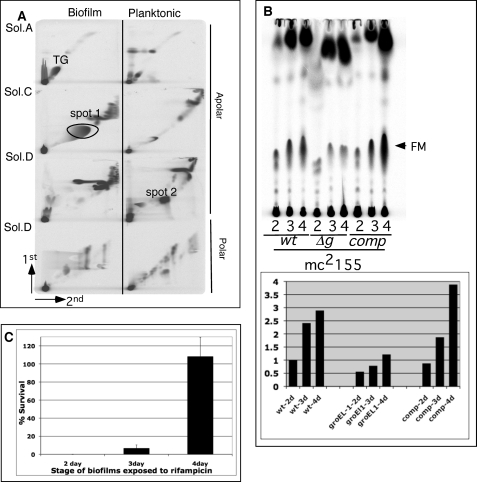
We previously described the presence of FM in the biofilms of M. tuberculosis (37), although the mechanisms underlying FM synthesis remained unclear. To further study the mechanism of FM synthesis, we sought to exploit the fast growing and non-pathogenic species M. smegmatis, which was preliminarily observed to make a lipid in biofilms that co-migrated with Mtb FM (37). Before undertaking a detailed study on FM synthesis in this species, we extended those preliminary studies and performed a complete lipid analysis using the same technique that we described for Mtb (37). We examined total polar and apolar lipid synthesis in planktonic and biofilm cultures of M. smegmatis by 14C-labeling and two-dimensional thin layer chromatography (Fig. 1A). Apart from the large abundance of a lipid (marked as spot 1) corresponding to the position of Mtb FM (37), we also noted two other major changes in biofilm cells; that is, increased levels of triacylglyceride (TG) and decreased levels of an unknown lipid (spot 2). There are other minor changes in the total lipids of the two cultures that we have not pursued further.
FIGURE 1.
Regulated synthesis of FM in M. smegmatis biofilms. A, two-dimensional radio-thin layer chromatography of 14C apolar and polar lipids equivalent to 50,000 cpm extracted from planktonic and biofilm cultures of M. smegmatis is shown. The TLCs were developed in solvent (Sol.) systems A, C, and D as described previously (37). The lipids with significantly different level of 14C incorporation in the two cultures are annotated as Spot 1, Spot 2, and TG. The direction of migration in each dimension is marked at the left bottom. B, radio-TLC of 14C apolar lipids equivalent to 50,000 cpm extracted from 2-, 3-, and 4-day (d) biofilms of M. smegmatis, mc2155 wild-type (wt), ΔgroEL1 (Δg), and ΔgroEL1 complemented with pMsgroEL1 (comp) is shown. The arrow denotes FM. The quantity of FM is normalized to the 2-day biofilms of wild-type M. smegmatis. C, shown is exposure of 2-, 3-, and 4-day biofilms to 400 μg/ml rifampicin. Percentage survival was calculated using untreated culture at the respective stage as a reference.
Spot 1 was purified and structurally characterized by 1H NMR and mass spectrometry to be a mixture of free alpha and epoxy mycolic acids (supplemental Fig. S1). Further detailed analysis of this lipid by TLC is described in supplemental Fig. S1. The identity of spot 2 is described below.
Because the nonessential M. smegmatis chaperone GroEL1 associates directly with the type II fatty acid synthase enzyme KasA during biofilm formation and is, therefore, directly implicated in M. smegmatis mycolic acid synthesis (34), we asked whether the defect in the later stages of biofilm development of ΔgroEL-1 mutant correlate with decreased accumulation of FM. We determined the stage-specific synthesis of FM by radiolabeling the newly synthesized lipid for 6 h with [14C]acetate. We observed an ∼2.5-fold increase in radiolabeled mycolic acids from day 2 to day 4 of biofilm growth of wild type, and the induction was largely abrogated in the ΔgroEL1 mutant and could be restored in the complemented strain (Fig. 1B).
Because a matured structure is important for drug tolerance behavior of M. tuberculosis biofilms (37), we predicted that the drug tolerance in the wild-type M. smegmatis biofilms would also correlate with the induced synthesis of FM (Fig. 1B) and would progressively increase from the 2- to 4-day stage. Upon exposure to rifampicin there was a very sharp increase in the number of drug tolerant bacilli in 4-day biofilms compared with 2- and 3-day biofilms (Fig. 1C). Overall, these data together established a tight correlation between FM synthesis, structural maturation, and drug tolerance properties of M. smegmatis biofilms.
FM Is Made through Hydrolysis of Trehalose Dimycolate
Because GroEL1 is known to associate with the fatty acid synthase II enzyme KasA (34), it seems unlikely that synthesis of FM in M. smegmatis biofilms involves an entirely new enzymatic pathway but, rather, derives from mycolic acid derivatives made by FAS-II. However, mycolic acids are typically esterified to other molecules with the major forms being mycolylarabinogalactan-peptidoglycan (mAGP), in which the mycolic acids are covalently linked to the cell wall components, and TDM, in which two mycolic acid chains are linked to the sugar trehalose (40). These two molecules are, therefore, the major candidates for substrates for the generation of FM, although cellular esterases that hydrolyze these have not been described. We note though that the mycobacteriophage lysin B enzyme has recently been shown to cleave mAGP (41), and there are several mycobacterial cutinase-like proteins for which lipolytic activity has been reported (42).
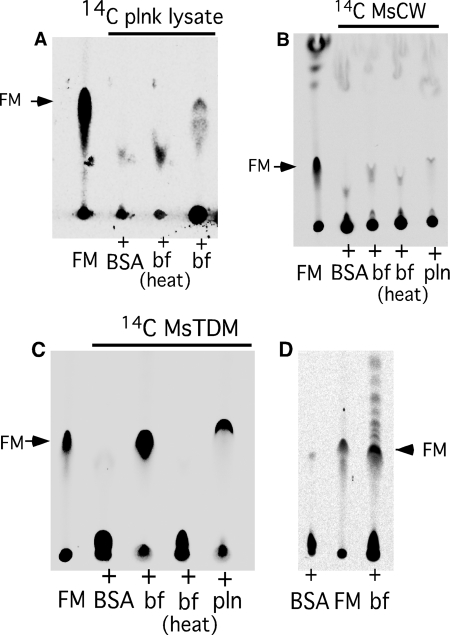
To test the hypothesis that biofilm-grown cells contain an enzymatic activity responsible for synthesizing FM from precursor mycolic acid-containing substrates, planktonically grown M. smegmatis cells were labeled with [14C]glucose, and a total lysate was prepared; we then incubated this with a total extract prepared from unlabeled biofilm-grown cells (Fig. 2A). As shown in Fig. 2A, the planktonically grown substrate did not contain FM, but it was generated upon incubation with the biofilm extract (Fig. 2A). The activity in the biofilm extract is likely to be enzyme-mediated as heat treatment abolished the activity. Having determined that the biofilm extract contains the enzymatic activity of interest, we examined whether it was capable of generating FM from purified mAGP. However, no activity was observed, indicating that mAGP is not the immediate precursor for FM synthesis (Fig. 2B). We then tested this with TDM as a substrate using 14C-radiolabeled TDM purified from planktonically grown M. smegmatis (Fig. 2C). Because TDM from M. smegmatis is relatively uncharacterized, we purified the apolar lipid co-migrating with M. tuberculosis TDM and confirmed its identity by NMR and mass spectrometry (supplemental Fig. S2). We observed that the biofilm lysate has potent activity that released FM from TDM. However, we also observed a detectable activity in the planktonically grown cells (Fig. 2C). Although the product from planktonic lysate in Fig. 2C (fifth lane) appears to migrate slightly faster than reference FM (first lane), the difference in mobility was not reproducible. An activity of TDM esterase was also observed in lysates prepared from M. tuberculosis biofilms (Fig. 2D), suggesting that both species contain enzymes capable of cleaving TDM to generate FM.
FIGURE 2.
FM is released from TDM in the presence of mycobacterial lysates. A, radio-TLC of the lipids in reaction mixtures of 14C-labeled M. smegmatis planktonic lysate with either lysates of M. smegmatis biofilms (bf), heat-inactivated biofilm lysates (heat bf), or with BSA is shown. B, radio-TLC of the lipids in the reaction mixture of purified 14C-labeled cell wall (MsCW) with buffer containing BSA, biofilm lysate (bf), heat-inactivated biofilms lysate (bf heat), and planktonic lysate (plnk) is shown. C, radio-TLC of the reaction mixture as described in panel B is shown except that 14C-MsTDM was used instead of MsCW as a substrate. D, radio-TLC shows lipids in the reaction mixture of purified 14C-MsTDM with M. tuberculosis biofilms lysate (bf) and buffer containing BSA. Purified FM was used as a control in panels A–D. TLCs in panel A–D were developed in chloroform:methanol 96:4.
Reduced Synthesis of TDM in Maturing Biofilms
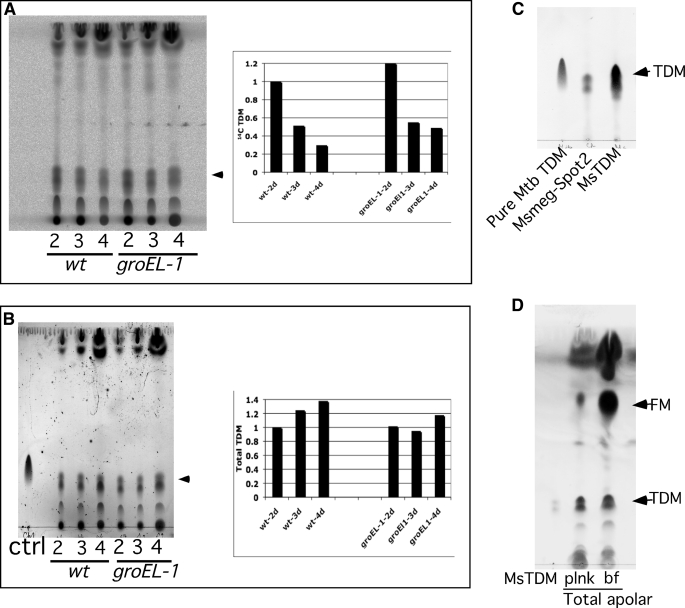
If TDM is a precursor for FM synthesis, then we predict that it is present at reduced levels as cells form mature biofilms. Indeed we observed that the synthesis of 14C-labeled TDM is reduced as cells mature from 2- to 4-day-grown M. smegmatis biofilms (Fig. 3A). We postulate that this likely arises from an increase in subsequent processing of synthesized TDM to yield FM. We note that a reduction also occurs in the ΔgroEL1 mutant, although this could arise from loss of synthesis (see Fig. 1B) as opposed to enhanced processing (Fig. 3A). Although the levels of newly synthesized TDM decreased during biofilm growth, the total amount of TDM in both the wild-type and mutant strains remained unchanged (Fig. 3B). It is likely that the site of TDM synthesis is spatially separated from its ultimate location on the outside of the cell envelope, which is then not accessible to the proposed biofilms esterase. We propose that when active synthesis of FM begins, the newly synthesized TDM that has yet to be delivered to its outer membrane location is captured and converted to FM by the esterase before translocation.
FIGURE 3.
Decreased synthesis of TDM in maturing biofilms. A, shown is radio-TLC of 14C apolar lipids corresponding to 50,000 cpm from 2-, 3-, and 4-day (d) biofilms of M. smegmatis, mc2155 wild-type (wt), and mc2155-ΔgroEL1 extracted in petroleum ether. The plot shows relative amount of spots corresponding to [14C]TDM using 2-day biofilms of wild type as reference. B, the total lipid content in each sample of panel A was visualized by charring the plate with 10%H2SO4 in ethanol. A cold purified TDM was used as a control (ctrl). The position of TDM is marked by arrow. The plot shows the relative amount of total TDM using 2-day biofilms of wild type as reference. C, TLC of Spot 2, originally identified with differential 14C incorporation in planktonic and biofilm cultures (Fig. 1 panel A), along with purified M. tuberculosis TDM (Mtb TDM) and M. smegmatis (MsTDM) is shown. D, shown is TLC of total apolar lipids equivalent to 50,000 cpm from 14C-labeled planktonic (plnk) and 4-day biofilm (bf) cultures seen after charring as described above. A cold purified MsTDM was used as a control. The TLCs were developed in chloroform:methanol:water (90:10:1).
Finally, we note that if newly synthesized TDM is the precursor for FM, then spot 2 in Fig. 1A, which is abundant in planktonically grown cells but barely detectable in biofilms (Fig. 1A), may correspond to TDM. To confirm this we purified spot 2 after two-dimensional TLC separation and showed that it has similar migration patterns as TDM isolated from both M. smegmatis and M. tuberculosis (Fig. 3C). Because spot 2 in Fig. 1A was identified by a radio-TLC, we predicted that the difference in TDM levels of planktonic and 4-day biofilm cultures, like the one observed for early and late stage biofilms (Figs. 3, A and B), will remain limited only to the newly made TDM. We looked at the total TDM in the apolar lipids of the two cultures by charring the TLC plate, and indeed we observed that the levels of total TDM in the cultures were very similar, although we note a clear accumulation of total FM in the biofilm culture (Fig. 3D). Taken together, these observations strongly support the hypothesis that biofilm formation occurs through a transition in growth phase in which FM is generated by esterase-dependent cleavage of newly synthesized TDM.
An Antigen 85 Homologue Is Required for Biofilm Formation
We reasoned that if TDM is a precursor for FM and that these are required for mature biofilm formation, then mutants defective in TDM synthesis would also be altered in biofilm development. TDM synthesis is dependent on mycolyltransferases, members of the serine carboxyesterases superfamily but originally identified as fibronectin-binding proteins (fbp), and members of the antigen-85 (Ag85) complex, which transfer mycolyl chains from 6-hydroxy group of trehalose monomycolate (TMM) to the 6-hydroxy group of a second TMM molecule to form TDM (43). They can also transfer mycolyl chains from TMM to arabinogalactan-peptidoglycan to form the mAGP complex (43). M. tuberculosis encodes four homologues of mycolyltransferases (fbpA:Rv3804c, fbpB:Rv1886c, fbpC:Rv0129c and fbpD:Rv3803c), of which FbpD is likely devoid of enzymatic activity (44). Although there is some functional redundancy in the remaining three, loss of FbpA activity leads to a strong depletion of TDM (45), suggesting that FbpA plays a major role in TDM synthesis. A similar role for FbpA has been suggested in M. smegmatis (46).
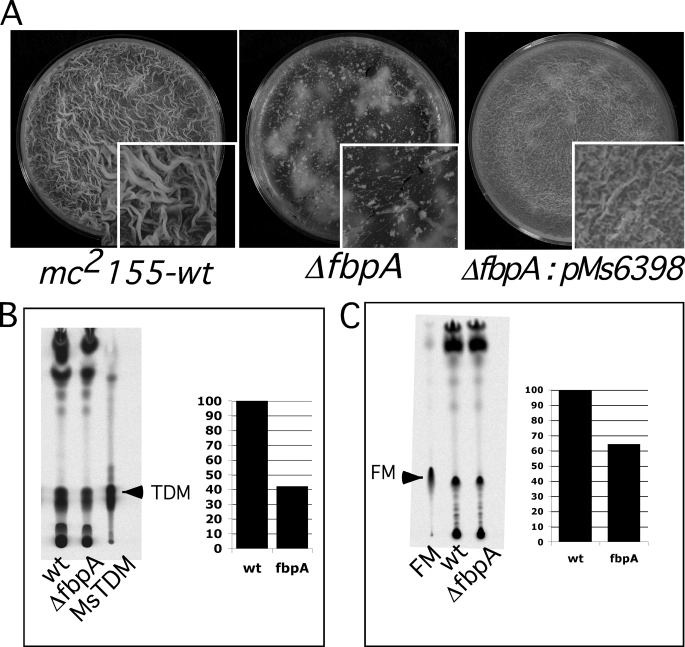
Apart from fbpA (Msmeg_6398), we noted four additional mycolyltransferases in M. smegmatis, fbpB (Msmeg_2078), fbpC (Msmeg_6396), and two genes closely related to fbpD (Msmeg_3580, and Msmeg_6399), and we constructed deletion mutants of each of these. Each of the mutants was examined for biofilm formation, and we observed that the ΔfbpA mutant showed a strong biofilm defect (Fig. 4A) that could be completely restored by complementation (Fig. 4A); the other four mutants formed normal biofilms (supplemental Fig. S3). TDM synthesis was depleted by about 43% in the ΔfbpA mutant, consistent with previous findings (46) (Fig. 4B). As predicted, a similar partial reduction in the synthesis of FM was also observed in the ΔfpbA mutant (Fig. 4C), consistent with the hypothesis that newly synthesized TDM is a substrate for FM synthesis.
FIGURE 4.
Defective biofilms of mc2155:fbpA mutant. A, 5-day biofilms of M. smegmatis, mc2155 wild-type (wt), mc2155:ΔfbpA, and mc2155:ΔfbpA:pMs6398 (complemented) strains are shown. B, shown is radio-TLC, developed in chloroform:methanol:water (90:10:1), of apolar lipids from M. smegmatis wild type (wt) and ΔfbpA mutant showing incorporation of 14C into MsTDM. The plot shows the relative amount of MsTDM from the two strains. C, shown is radio-TLC of the apolar lipids from the wild-type and mutant strains developed in chloroform:methanol (96:4) showing 14C incorporation into FM. The plot shows the relative amount of FM in the two strains. 5-Day biofilms of the strains were labeled for 6 h with [14C]glycerol, and total apolar lipids equivalent to 50,000 cpm from each sample were loaded and compared with either purified MsTDM in panel B (marked with an arrow) or purified FM in panel C.
Msmeg_1529 Encodes a TDM-specific Esterase
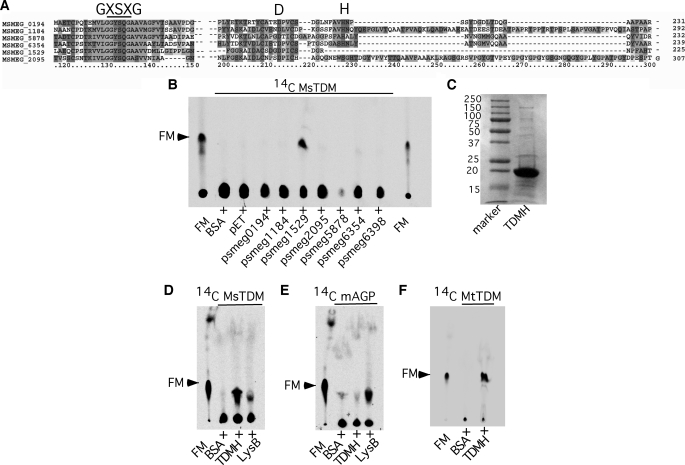
The release of FM from TDM by biofilm lysates of M. smegmatis and M. tuberculosis suggests the presence of an esterase that hydrolyzes the linkage between the sugar and lipid moieties in TDM. This is unlikely to be catalyzed by the mycolyltransferases discussed above as these have strong specificity for TMM (43). However, such enzymes may share common mechanisms with the mycolyltransferases, which use a catalytic serine (within a conserved GXSXG motif) and have conserved glutamine and histidine residues in the active site (47). We note that the recently described cutinase-like phage-encoded lysin B is also a serine esterase, with conserved serine, aspartic acid, and histidine residues in the active site, and cleaves mycolylarabinogalactan to release FM (41). In a search for related enzymes with potential serine-esterase activity, we looked for ORFs annotated as serine esterase in the M. smegmatis genome web page (Comprehensive Microbial Resource at the J. Craig Venter Institute) and identified six ORFs (Fig. 5A). All six candidate ORFs were cloned and expressed in E. coli, and total cell lysates from induced cells were tested for the ability to generate FM from radiolabeled M. smegmatis TDM. Five of the ORFs as well as FbpA (Msmeg_6398) had no activity, but one (Msmeg_1529) was evidently active (Fig. 5B). The Msmeg_1529 protein was purified to greater than 90% homogeneity from E. coli (Fig. 5C), and its activity was verified on purified M. smegmatis TDM (Fig. 5D). Interestingly this enzyme is specific for TDM, and no activity was observed on purified mAGP (Fig. 5E); it is, however, active on M. tuberculosis TDM (Fig. 5F). This activity is distinct from the phage lysin B protein, which acts preferentially on mAGP even though it has some activity on TDM (Fig. 5, D and E). We propose to call this enzyme a TDM hydrolase.
FIGURE 5.
Msmeg_1529 is a TDM-specific hydrolase. A, multiple sequence alignment of serine esterases from M. smegmatis by ClustalX is shown. The conserved catalytic triad of GXSXG, Asp, and His are annotated. B, shown is radio-TLC of apolar lipids extracted in petroleum ether from a reaction mixture containing purified 14C-MsTDM lysates of recombinant E. coli expressing putative M. smegmatis serine esterase identified in panel A. The recombinant strains expressed the ORFs (marked below each lane) under isopropyl 1-thio-β-d-galactopyranoside-inducible LacI-dependent expression system. Reactions with BSA and recombinant E. coli containing empty vector (pET21b) were used as negative control. C, SDS-PAGE shows TDM hydrolase (TDMH) (Msmeg_1529) purified from recombinant strain used in panel B. D, shown is radio-TLC of lipids extracted from the reaction mixture of pure 14C-MsTDM with either TDM hydrolase or LysB from mycobacteriophage D29. E, radio-TLC of lipids extracted from reaction mixture containing pure [14C]mAGP with either TDM hydrolase or LysB. F, shown is radio-TLC of lipids extracted from a reaction mixture containing pure 14C-MtTDM (TDM from M. tuberculosis) with TDM hydrolase. Buffer with BSA as a negative control and pure FM as a reference was used in panels D, E, and F.
A ΔM_smeg1529 Mutant Is Altered in Biofilm Formation
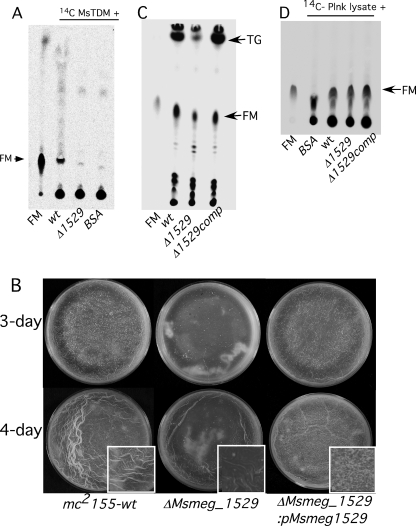
Given the in vitro activity of Msmeg_1529 and the proposed relationship between TDM and FM during biofilm formation, we constructed a mutant with a deletion in this ORF. Expectedly, the lysate of ΔMsmeg_1529 mutant had undetectable hydrolytic activity against purified TDM, indicating that Msmeg_1529 is the major enzyme in M. smegmatis responsible for TDM hydrolysis (Fig. 6A). However, ΔMsmeg_1529 mutant is capable of forming biofilms but with a retarded rate as compared with wild type (Fig. 6B). The difference was clearly noticeable at 3- and 4-day stages (Fig. 6B), but upon prolonged incubation the mutant biofilms were indistinguishable from the parent strain both in appearance as well as in rifampicin tolerance (data not shown). The delayed development in the mutant was complemented by expression of Msmeg_1529 by an episomal plasmid, pMs_1529 (Fig. 6B). The compromised TDM hydrolysis and partial defect in the biofilm growth of Msmeg_1529 suggested that the mutant biofilms would also be compromised in making FM. We indeed found nearly 50% reduction in the synthesis of FM in the mutant biofilms at the 4-day stage, and the synthesis was restored to the normal level in the complemented strain (Fig. 6C). Interestingly, we also observed a dramatic reduction in the synthesis of TG in the mutant, suggesting that the induction of FM and TG in M. smegmatis biofilms could be linked (Fig. 6C). The FM and TG in the mutant, however, were restored to wild-type levels upon extended incubation (data not shown).
FIGURE 6.
Phenotype of ΔMsmeg_1529 mutant. A, radio-TLC shows the petroleum ether-extracted 14C lipids from the reaction mixture containing [14C]TDM and lysates of either wild-type (wt) or ΔMsmeg_1529. BSA was used as a negative control. Purified FM (marked with an arrow) was used as a reference. B, 3- and 4-day biofilms of mc2155 (wild type), mc2155:ΔMsmeg_1529, and mc2155:ΔMsmeg_1529 complemented with pMsmeg_1529 are shown. C, shown is radio-TLC of 14C-labeled apolar lipids equivalent to 50,000 cpm made at the 4-day stage of the three strains described in panel B. Lipids were labeled with [14C]acetate for 6 h before petroleum ether extraction. FM and TG are annotated. D, radio-TLC shows the petroleum ether-extracted 14C lipids from the reaction mixture containing 14C-labeled planktonic (plnk) lysate and lysate from either wild-type or ΔMsmeg_1529-, or ΔMsmeg_1529-complemented strains. BSA in the buffer was used as a negative control, and purified FM from M. smegmatis (marked with an arrow) was used as a reference.
We further reasoned that if FM synthesis is only partially reduced in ΔMsmeg_1529 mutant, then there would be alternate esterase-dependent pathway to produce FM in M. smegmatis. We tested the presence of alternate pathways in ΔMsmeg_1529 using the assay described in Fig. 2A, and we found that the biofilm lysate of the mutant was able to generate FM when mixed with 14C-label planktonic lysate (Fig. 6D). However, identity of alternate esterase and its substrate remains unknown, although mAGP of the cell wall as a substrate is ruled out from the experiment described in Fig. 2B.
DISCUSSION
Although growth of mycobacterial surface pellicles has been reported for many years, it is only recently that these structures have been revisited from a biofilm perspective and found to involve a specific developmental genetic program and harbor drug-tolerant cells in the extracellular matrix presumably made of secreted FM (37). Although the role of mycobacterial biofilms in their pathogenicity remains unclear, the presence of drug tolerant persisters and altered lipid profiles suggests that this is an important question to address.
Here we show that FM is newly synthesized during growth of both M. smegmatis and M. tuberculosis biofilms. The data reported here also show that newly synthesized but not the previously made TDM is one of the precursors for FM and is processed by a TDM-specific esterase. It is perhaps not surprising that mAGP is not a precursor for FM synthesis, as the only previously described enzyme that cleaves mAGP to release FM (Lysin B) is also involved in cell lysis during phage growth. However, it is known from previous studies that although TDM is the major component of cord factor and plays a role in M. tuberculosis pathogenicity (29, 31), mutants with reduced levels of TDM are still viable.
We propose that the distinction between newly synthesized TDM and pre-existing TDM in the synthesis of FM is determined by cellular localization of the enzyme and its cognate substrate. Synthesis of mycolic acids leading to formation of TMM is thought to occur in the cytoplasm and that TMM is transported across the cytoplasmic membrane where it serves as a precursor for linkage of mycolic acids to arabinogalactan-peptidoglycan or for TDM synthesis (40); both of these reactions are catalyzed by the Ag85 complex (40). We propose that TDM is generated within the periplasmic space between the mycobacterial inner and outer membranes and that this is the site of its conversion into FM by esterases such as Msmeg_1529. The FM is then secreted to form the biofilm matrix. We further propose that the Msmeg_1529 enzyme is restricted to this location and, therefore, does not have access to pre-existing TDM that is an integrated component of the outer membrane. Two further lines of evidence support a specific role for Msmeg_1529. First, expression of this gene is induced about 4-fold in biofilm formation (48); second, Msmeg_1529 contains a strongly predicted signal peptide at its N terminus, consistent with its proposed site of action outside of the cytoplasmic membrane, where newly synthesized TDM is made.
There appears to be considerable redundancy in many of the enzymatic pathways involved in mycolic acid synthesis and processing. For example, there are at least four mycolyltransferases in both M. tuberculosis and in M. smegmatis, and although FbpA plays a major role in forming both mAGP and TDM, it is not an essential gene (49). Likewise, although only Msmeg_1529 from the six putative esterases we overexpressed has the ability to convert TDM to FM in vitro, the ΔMsmeg_1529 mutant showed only a partial reduction in FM synthesis, suggesting that there are other pathways for FM synthesis. This is further supported by the presence of mycolyl esterase activity in the ΔMsmeg_1529 mutant. Further identification of novel pathways involved in FM synthesis will be critical for understanding the role of this lipid in biofilm formation.
The specific role of GroEL1 in biofilm formation remains unclear. Because GroEL1 interacts directly with KasA during biofilm formation (34), it acts at a relatively early stage before TMM transport across the membrane and before the processing of TDM into FM. An intriguing possibility is that GroEL1 helps to deliver newly synthesized TMM to a specific ABC transporter, which is turn is associated on the cell wall side of the membrane with Msmeg_1529 and perhaps other esterases. This newly synthesized TMM would then be targeted for conversion to TDM by antigen-85 followed by closely coupled processing into FM by Msmeg_1529. The recent report of phosphorylation of GroEL1 provides a possible mechanism for activation of its role during biofilm growth (50).
The pathways for mycolic acid synthesis and processing are closely related in M. smegmatis and M. tuberculosis. In this study we show that M. tuberculosis biofilm-grown cells also possess an activity that generates FM from purified radiolabeled TDM from M. smegmatis (Fig. 2D). Seven cutinase-like proteins, with esterase activities in some of them, have been described in M. tuberculosis (42, 51), and we suggest that one or more of these behaves similarly to Msmeg_1529, although none has yet been shown to cleave either TDM or mAGP. Rv3452, designated Culp4, is the closest relative of Msmeg_1529, has a putative signal peptide, and has been reported to be cell wall-associated (52). We, therefore, suggest that Rv3452 is a likely candidate for TDM hydrolase in M. tuberculosis.
We demonstrated previously that M. tuberculosis biofilms contains a subset of cells that show high tolerance to antibiotics (37), and we have shown here (Fig. 1C) that the drug tolerance of M. smegmatis biofilm correlates with induced synthesis of FM. Thus, FM is very likely to play an important role in development of drug tolerant persisters. Interestingly, it has also been reported that a ΔfbpA mutant of M. smegmatis is hypersensitive to anti-mycobacterial drugs, consistent with a role of TDM in FM synthesis and consequent development of drug tolerant persisters. Intriguingly, although both ΔfbpA and ΔMsmeg_1529 mutants have comparable defects in FM synthesis, ΔfbpA is severely defective in biofilm growth. The severity of the ΔfbpA mutant could be due to possible defect in its cell wall biosynthesis as the gene has been previously implicated in the synthesis of mycolyl esters of cell wall (43, 45).
Although complete identification of pathways involved in FM synthesis remains to be identified, identification of a TDM hydrolase provides an important insight into the dynamic nature of mycobacterial cell wall during their growth in biofilms. Furthermore, given that Mtb TDM is a highly immunogenic lipid and can be hydrolyzed by Msmeg_1529, the esterase can also be exploited as a reagent to understand the pathogenesis of tuberculosis.
Supplementary Material
Acknowledgments
We acknowledge technical support provided by Amy Vogelsberger and Christina Ferreira.
This work was supported, in whole or in part, by National Institutes of Health Grants AI079288 (to A. K. O.) and AI064494 (to G. F. H.). This work was also supported by an award from the American Lung Association (to A. K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
- TDM
- trehalose dimycolate
- FM
- free mycolic acid
- TG
- triacylglyceride
- mAGP
- mycolylarabinogalactan-peptidoglycan
- TMM
- trehalose monomycolate
- ORF
- open reading frame
- Mtb
- M. tuberculosis
- BSA
- bovine serum albumin.
REFERENCES
- 1.Hall-Stoodley L., Stoodley P. (2002) Curr. Opin. Biotechnol. 13, 228–233 [DOI] [PubMed] [Google Scholar]
- 2.Costerton J. W., Irvin R. T., Cheng K. J. (1981) Crit. Rev. Microbiol. 8, 303–338 [DOI] [PubMed] [Google Scholar]
- 3.Blankenship J. R., Mitchell A. P. (2006) Curr. Opin. Microbiol. 9, 588–594 [DOI] [PubMed] [Google Scholar]
- 4.Kolter R., Greenberg E. P. (2006) Nature 441, 300–302 [DOI] [PubMed] [Google Scholar]
- 5.Kolter R., Losick R. (1998) Science 280, 226–227 [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L., Costerton J. W., Stoodley P. (2004) Nat. Rev. Microbiol. 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 7.Allison D. G. (2003) Biofouling 19, 139–150 [DOI] [PubMed] [Google Scholar]
- 8.Branda S. S., Vik S., Friedman L., Kolter R. (2005) Trends Microbiol. 13, 20–26 [DOI] [PubMed] [Google Scholar]
- 9.Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. (2002) Science 295, 1487. [DOI] [PubMed] [Google Scholar]
- 10.Henke J. M., Bassler B. L. (2004) Trends Cell Biol. 14, 648–656 [DOI] [PubMed] [Google Scholar]
- 11.Kearns D. B., Chu F., Branda S. S., Kolter R., Losick R. (2005) Mol. Microbiol. 55, 739–749 [DOI] [PubMed] [Google Scholar]
- 12.Sakuragi Y., Kolter R. (2007) J. Bacteriol. 189, 5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teal T. K., Lies D. P., Wold B. J., Newman D. K. (2006) Appl. Environ. Microbiol. 72, 7324–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart P. S., Franklin M. J. (2008) Nat. Rev. Microbiol. 6, 199–210 [DOI] [PubMed] [Google Scholar]
- 15.Stoodley P., Sauer K., Davies D. G., Costerton J. W. (2002) Annu. Rev. Microbiol. 56, 187–209 [DOI] [PubMed] [Google Scholar]
- 16.Vlamakis H., Aguilar C., Losick R., Kolter R. (2008) Genes Dev. 22, 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeill K., Hamilton I. R. (2003) FEMS Microbiol. Lett. 221, 25–30 [DOI] [PubMed] [Google Scholar]
- 18.Fux C. A., Costerton J. W., Stewart P. S., Stoodley P. (2005) Trends Microbiol. 13, 34–40 [DOI] [PubMed] [Google Scholar]
- 19.Post J. C., Stoodley P., Hall-Stoodley L., Ehrlich G. D. (2004) Curr. Opin. Otolaryngol. Head Neck Surg. 12, 185–190 [DOI] [PubMed] [Google Scholar]
- 20.Lam J., Chan R., Lam K., Costerton J. W. (1980) Infect. Immun. 28, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson G. G., Dodson K. W., Hooton T. M., Hultgren S. J. (2004) Trends Microbiol. 12, 424–430 [DOI] [PubMed] [Google Scholar]
- 22.Costerton J. W., Stewart P. S., Greenberg E. P. (1999) Science 284, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization (2009) WHO Report 2009: Global Tuberculosis Control-Epidemiology, Strategy, Financing, World Health Organization, Geneva [Google Scholar]
- 24.Jindani A., Doré C. J., Mitchison D. A. (2003) Am. J. Respir. Crit. Care Med. 167, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control (2003) in Morbidity and Mortality Weekly Report, Vol. 52, pp. 1–77, Center for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 26.Connolly L. E., Edelstein P. H., Ramakrishnan L. (2007) PLoS Med. 4, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middlebrook G., Dubos R. J., Pierce C. (1947) J. Exp. Med. 86, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noll H., Bloch H., Asselineau J., Lederer E. (1956) Biochim. Biophys. Acta 20, 299–309 [DOI] [PubMed] [Google Scholar]
- 29.Hunter R. L., Olsen M. R., Jagannath C., Actor J. K. (2006) Ann. Clin. Lab. Sci. 36, 371–386 [PubMed] [Google Scholar]
- 30.Geisel R. E., Sakamoto K., Russell D. G., Rhoades E. R. (2005) J. Immunol. 174, 5007–5015 [DOI] [PubMed] [Google Scholar]
- 31.Glickman M. S., Cox J. S., Jacobs W. R., Jr. (2000) Mol. Cell 5, 717–727 [DOI] [PubMed] [Google Scholar]
- 32.Marsollier L., Brodin P., Jackson M., Korduláková J., Tafelmeyer P., Carbonnelle E., Aubry J., Milon G., Legras P., André J. P., Leroy C., Cottin J., Guillou M. L., Reysset G., Cole S. T. (2007) PLoS Pathog. 3, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Recht J., Kolter R. (2001) J. Bacteriol. 183, 5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojha A., Anand M., Bhatt A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. (2005) Cell 123, 861–873 [DOI] [PubMed] [Google Scholar]
- 35.Teng R., Dick T. (2003) FEMS Microbiol. Lett. 227, 171–174 [DOI] [PubMed] [Google Scholar]
- 36.Carter G., Wu M., Drummond D. C., Bermudez L. E. (2003) J. Med. Microbiol. 52, 747–752 [DOI] [PubMed] [Google Scholar]
- 37.Ojha A. K., Baughn A. D., Sambandan D., Hsu T., Trivelli X., Guerardel Y., Alahari A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. (2008) Mol. Microbiol. 69, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J. M., German G. J., Alexander D. C., Ren H., Tan T., Liu J. (2006) J. Bacteriol. 188, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren H., Dover L. G., Islam S. T., Alexander D. C., Chen J. M., Besra G. S., Liu J. (2007) Mol. Microbiol. 63, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 40.Takayama K., Wang C., Besra G. S. (2005) Clin. Microbiol. Rev. 18, 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne K., Sun Q., Sacchettini J., Hatfull G. F. (2009) Mol. Microbiol. 73, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West N. P., Chow F. M., Randall E. J., Wu J., Chen J., Ribeiro J. M., Britton W. J. (2009) FASEB J. 23, 1694–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 44.Kremer L., Maughan W. N., Wilson R. A., Dover L. G., Besra G. S. (2002) Lett. Appl. Microbiol. 34, 233–237 [DOI] [PubMed] [Google Scholar]
- 45.Puech V., Guilhot C., Perez E., Tropis M., Armitige L. Y., Gicquel B., Daffé M. (2002) Mol. Microbiol. 44, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 46.Nguyen L., Chinnapapagari S., Thompson C. J. (2005) J. Bacteriol. 187, 6603–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronning D. R., Klabunde T., Besra G. S., Vissa V. D., Belisle J. T., Sacchettini J. C. (2000) Nat. Struct. Biol. 7, 141–146 [DOI] [PubMed] [Google Scholar]
- 48.Ojha A., Hatfull G. F. (2007) Mol. Microbiol. 66, 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armitige L. Y., Jagannath C., Wanger A. R., Norris S. J. (2000) Infect. Immun. 68, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canova M. J., Kremer L., Molle V. (2009) J. Bacteriol. 191, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schu M., Maurin D., Dhouib R., Bakala N′goma J. C., Delorme V., Lambeau G., Carriere F., Canaan S. (2010) FASEB J., in press [DOI] [PubMed] [Google Scholar]
- 52.Parker S. K., Curtin K. M., Vasil M. L. (2007) J. Bacteriol. 189, 4153–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kessel J. C., Hatfull G. F. (2007) Nat. Methods 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 54.Lewis J. A., Hatfull G. F. (2000) Mol. Microbiol. 35, 350–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.