Abstract
The existence of circulating microRNAs (miRNAs) in the blood of cancer patients has raised the possibility that miRNAs may serve as a novel diagnostic marker. However, the secretory mechanism and biological function of extracellular miRNAs remain unclear. Here, we show that miRNAs are released through a ceramide-dependent secretory machinery and that the secretory miRNAs are transferable and functional in the recipient cells. Ceramide, whose biosynthesis is regulated by neutral sphingomyelinase 2 (nSMase2), triggers secretion of small membrane vesicles called exosomes. The decreased activity of nSMase2 with a chemical inhibitor, GW4869, and a specific small interfering RNA resulted in the reduced secretion of miRNAs. Complementarily, overexpression of nSMase2 increased extracellular amounts of miRNAs. We also revealed that the endosomal sorting complex required for transport system is unnecessary for the release of miRNAs. Furthermore, a tumor-suppressive miRNA secreted via this pathway was transported between cells and exerted gene silencing in the recipient cells, thereby leading to cell growth inhibition. Our findings shed a ray of light on the physiological relevance of secretory miRNAs.
Keywords: MicroRNA, RNA Silencing, RNA Interference (RNAi), Secretion, Vesicles, Ceramide, Exosome, Intercellular Transfer, Neutral Sphingomyelinase, Secretory microRNA
Introduction
It has been well known that extracellular RNAs circulate in the blood of healthy people and diseased patients with sufficient integrity, although ribonuclease is present in both plasma and serum (1). The spectrum of RNAs whose presence was demonstrated in plasma and other body fluids, such as urine and breast milk, extends from housekeeping genes to fetal genes detected in pregnant women and genes overexpressed in a variety of different tumors (2). To explain the stability of circulating RNAs, it was suggested that extracellular RNAs are included within lipoprotein vesicles. Indeed, exogenous RNAs added to plasma or blood are immediately degraded, whereas endogenous plasma RNAs are stable for hours under the same conditions (3). Moreover, the treatment of some detergents results in immediate degradation of plasma extracellular RNAs, apparently due to disruption of the lipid vesicles. These findings clearly indicate that extracellular RNAs are packaged in some kinds of secretory particles including apoptotic bodies and exosomes, and thus, they are protected from dominantly existing ribonucleases.
Apoptotic bodies are small membranous particles released during programmed cell death (4), and exosomes are small intraluminal vesicles (50–100 nm in diameter) of multivesicular bodies (MVB)3 released on exocytic fusion of MVB with plasma membranes (5). Currently, accumulating evidence suggests that these secretory vesicles can function as intercellular transmitters to convey their contents, in particular, microRNA (miRNA) (6–8). Recent studies reported that extracellular exosomal miRNAs were transferred into other cells and that apoptotic bodies delivered miR-126 into endothelial cells (9). Despite these advances, however, the underlying mechanism of the secretory process and the biological function of circulating miRNAs are not yet fully understood.
miRNAs, small 20–22-nucleotide-long members of the non-protein-coding RNA family, are expressed in the vast majority of eukaryotes, including humans (10). Not only do they inhibit translation of their target genes, they also degrade the target mRNAs through recognition of imperfect complementary sites, usually located in the 3′-untranslated regions of the target mRNAs, endowing miRNAs with the capacity to regulate numerous biological processes. Over the past several years, it was evident that dysregulations of many kinds of miRNAs have been linked to the initiation and progression of human cancer (11). Interestingly, the amounts of secretory miRNAs are up-regulated in the plasma of patients bearing tumors, including B cell lymphoma, prostate cancer, lung cancer, and ovarian cancer (12–15). Thus, detection and monitoring of tumors are now becoming possible by the evaluation of tumor-derived secretory miRNAs.
In this study, we have shown that secretion of miRNAs is controlled by neutral sphingomyelinase 2 (nSMase2), which is known as a rate-limiting enzyme of ceramide biosynthesis. Furthermore, we provide evidences that miRNAs secreted from donor cells can be taken up and function in the recipient cells. These findings propose a novel mechanism of intercellular communication mediated by secretory miRNAs.
EXPERIMENTAL PROCEDURES
Reagents
Rabbit polyclonal anti-nSMase2 (H-195) (sc-67305), goat polyclonal anti-Alix (Q-19) (sc-49268), and donkey anti-goat IgG (horseradish peroxidase) (sc-2020) were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-ROCK1 (ab36746) was from Abcam. Mouse monoclonal anti-actin, clone C4 (MAB1501), was from Millipore. Anti-CD63 monoclonal antibody was purchased from BD Pharmingen. Peroxidase-labeled anti-mouse and anti-rabbit antibodies were included in the Amersham Biosciences ECL PLUS Western blotting reagents pack (RPN2124) (GE HealthCare). Synthetic Caenorhabditis elegans miRNA cel-miR-39 was synthesized by Qiagen (Valencia, CA). Synthetic hsa-miR-146a (pre-miR-146a) was purchased from Ambion (Austin, TX). The duplexes of each small interfering RNA (siRNA), targeting human nSMase2 mRNA (s30925; target sequences of 5′-GGAGGUGUUUGACAAGCGAdTdT-3′ and 5′-UCGCUUGUCAAACACCUCCtg-3′), and negative control 1 (NC1) were purchased from Applied Biosystems, and an siRNA specific for human ALG-2 interacting protein (Alix) mRNA (target sequences of 5′-GAACCUGGAUAAUGAUGAAdTdT-3′ and 5′-UUCAUCAUUAUCCAGGUUCdTdT-3′) was purchased from Sigma-Genosys. GW4869 was purchased from Calbiochem (Darmstadt, Germany). Cisplatin was obtained from Alexis (Lausen, Switzerland). Geneticin was purchased from Invitrogen.
Cell Culture
HEK293 cells, a human embryonic kidney cell line (CRL-1573), and COS-7 cells, an African green monkey kidney fibroblast-like cell line (CRL-1651), were obtained from the American Type Culture Collection (Manassas, VA). These cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum and antibiotic-antimycotic (Invitrogen) at 37 °C in 5% CO2. PC-3M-luc cells (Xenogen) were cultured in RPMI containing 10% heat-inactivated fetal bovine serum and antibiotic-antimycotic at 37 °C in 5% CO2.
Preparation of Conditioned Medium and Exosome
Prior to collection of culture medium, HEK293 and COS-7 cells were washed three times with Advanced RPMI containing antibiotic-antimycotic and 2 mm l-glutamine (medium A), and the medium was switched to fresh medium A. After incubation during 3 days, medium A was collected and centrifuged at 2,000 × g for 15 min at room temperature. To thoroughly remove cellular debris, the supernatant was centrifuged again at 12,000 × g for 35 min at room temperature. Then the conditioned medium was used for miRNA extraction and functional assays as well as exosome isolation.
For exosome preparation, the conditioned medium was ultracentrifuged at 110,000 × g for 70 min at 4 °C. The pellets were washed with 11 ml of phosphate-buffered saline, and after ultracentrifugation, they were resuspended in phosphate-buffered saline. The exosome fraction was measured for its protein content using the Micro BCA protein assay kit (Thermo Scientific).
Isolation of MicroRNAs
Isolation of extracellular and cellular miRNAs was performed using the mirVana isolation kit (Ambion). One hundred μl of conditioned medium or cell lysate was diluted with 200 μl of lysis/binding solution. After a 5-min incubation, 20 μl of miRNA homogenate additive and 1 μl of 1 nm cel-miR-39 were added to each aliquot, followed by vortex for 30 s and incubation on ice for 10 min. Subsequent phenol extraction and filter cartridge work were carried out according to the manufacturer's protocol.
RNA Detection
Detection of RNAs was performed by using the Agilent Bioanalyzer 2100 (Agilent Technologies). Prior to the analysis, total RNAs were prepared with the Agilent RNA 6000 Pico kit (Agilent Technologies) according to the manufacturer's protocol.
RNase Treatment
To evaluate whether small RNAs were present inside the exosomes, RNase mixture (Ambion) was added to conditioned medium at a final concentration of 5 units/ml RNase A and 200 units/ml RNase T1 and then incubated at 37 °C for 30 min. Small RNAs were purified using the mirVana miRNA isolation kit (Ambion) as described above.
Quantitative Real Time PCR (QRT-PCR)
The method for QRT-PCR has been previously described (13). PCR was carried out in 96-well plates using the 7300 Real Time PCR System (Applied Biosystems). All reactions were done in triplicate. All of the TaqMan microRNA assays were purchased from Applied Biosystems. cel-miR-39 and hRNU6 were used as invariant control for conditioned medium and cell, respectively. The concentrations of extracellular miRNAs were calculated based on their Ct values normalized by those of cel-miR-39, which was spiked in each aliquot of QRT-PCR reaction at 1 nm.
Immunoprecipitation
Conditioned medium was incubated with magnetic beads (Invitrogen) coated with purified anti-CD63 antibody or mouse IgG1 control antibody (Millipore) at 4 °C overnight. After washing with phosphate-buffered saline, miRNAs were extracted and applied to QRT-PCR.
Immunoblot Analysis
SDS-PAGE gels (SuperSep Ace 5–20%, 194-15021, Wako) were calibrated with Precision Plus protein standards (161-0375) (Bio-Rad), and anti-CD63 (1:100), anti-nSMase2 (1:200), anti-Alix (1:100), anti-ROCK1 (1:200), and anti-actin (1:1,000) were used as primary antibodies. The dilution ratio of each antibody is indicated in parentheses. Two secondary antibodies (peroxidase-labeled anti-mouse and anti-rabbit antibodies) were each used at a dilution of 1:10,000. Bound antibodies were visualized by chemiluminescence using the ECL Plus Western blotting detection system (RPN2132) (GE HealthCare), and luminescent images were analyzed by a LuminoImager (LAS-3000; Fuji Film Inc.).
Plasmids
psiRNA-LucGL3 was purchased from InvivoGen. Primary-miR-143 and primary-miR-146a expression vectors were purchased from TaKaRa BIO. A full-length human nSMase2 cDNA was cloned into pIRES2-EGFP vector (Clontech). Primary miRNAs (pri-miR-155, pri-miR-16, and pri-miR-21) were PCR-amplified from human genomic DNA and cloned into the downstream of cytomegalovirus promoter in pIRES2-EGFP. For luciferase-based reporter gene assays, pLucNeo was constructed by inserting a firefly luciferase gene derived from pGL3-control (Promega) into pEYFP-1 vector (Clontech) at the BglII and AflII sites. Sensor vector for miR-146a was constructed by introducing tandem binding sites with perfect complementarity to miR-146a, separated by a four-nucleotide spacer into the NotI site of psiCHECK2 (Promega). The sequences of the binding site are as follows: 5′-AAACCTAGAGCGGCCGCAACCCATGGAATTCAGTTCTCAAAGAATTCTTAACCCATGGAATTCAGTTCTCAGCGGCCGCTGGCCGCAA-3′ (sense) and 5′-TTGCGGCCAGCGGCCGCTGAGAACTGAATTCCATGGGTTAAGAATTCTTTGAGAACTGAATTCCATGGGTTGCGGCCGCTCTAGGTTT-3′ (antisense). The “seed” sequence of miR-146a is indicated in bold and italics. In a mutated miR-146a sensor vector, the seed sequence, AGTTCTCA, was displaced with CTGGAGAC. All the plasmids were verified by DNA sequencing.
Transient Transfection Assays
Plasmid transfections to HEK293 and COS-7 cells were performed using Lipofectamine LTX (Invitrogen). Cell numbers and amounts of plasmids for each transfection were determined according to the manufacturer's protocol.
Transfections of siRNA and miRNA were accomplished with DharmaFECT transfection reagent (Thermo Scientific) according to the manufacturer's protocol. The total amounts of small RNAs for each transfection were equally adjusted by the addition of NC1.
Establishment of Stable Cell Lines
Stable HEK293 cell lines expressing firefly luciferase or miR-146a were generated by selection with 300 μg/ml Geneticin. HEK293 cells were transfected with 0.5 μg of pLucNeo vector or pri-miR-146a expression vector at 90% confluency in 24-well dishes using a Lipofectamine LTX reagent in accordance with the manufacturer's instructions. Twelve h after the transfection, the cells were replated in a 10-cm dish followed by a 3-week selection with the antibiotic. Ten surviving single colonies were picked up from each transfectant and were then cultured for another 2 weeks. The cells expressing the largest amount of firefly luciferase or miR-146a among transfectants were used as luciferase stably expressing cells and miR-146a stably expressing cells, respectively.
Luciferase Reporter Assay
HEK293 and COS-7 cells were cultured at a density of 5 × 104 and 1 × 104 cells/well, respectively, in 96-well tissue culture plates overnight, and miRNA transfections or the addition of conditioned medium were performed as described under the legend for Figs. 5–7. The cells were harvested, and Renilla luciferase activity was measured and normalized by firefly luciferase activity(16). All assays were performed in triplicate and repeated at least three times, and the most representative results are shown.
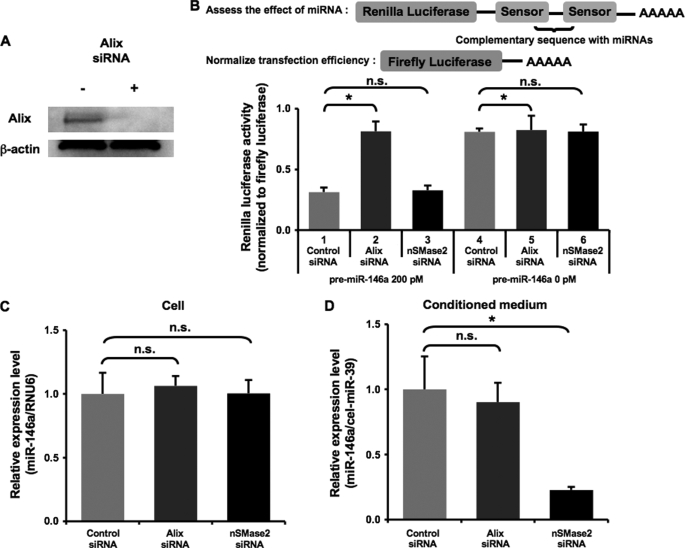
FIGURE 5.
The secretion of miRNAs does not require ESCRT system. A, knockdown of human Alix protein. HEK293 cells transfected with negative control siRNA or Alix siRNA were harvested and applied to immunoblot for Alix and β-actin proteins. B, the depletion of Alix impaired miRNA activity. Upper schematic, our sensor vector possesses Renilla luciferase and firefly luciferase for assessing the miRNA activity and for normalizing transfection efficiency, respectively. Lower graph, HEK293 cells were transfected with 0.1 μg of miR-146a sensor vector and the indicated siRNAs together with 200 or 0 pm pre-miR-146a in a 96-well plate. The following day, the cells were applied to a Dual luciferase reporter assay. The values on the y axis are normalized Renilla luciferase activity. C and D, cellular and extracellular miR-146a were not affected by Alix siRNA. MiR-146a stably overexpressing HEK293 cells were transfected with the indicated siRNAs. After the medium switch, the cells were cultured for another 24 h, and the cell lysate (C) and conditioned medium (D) were then applied to miRNA QRT-PCR analysis. The values on the y axis are depicted relative to control siRNA, which is arbitrarily defined as 1. B–D, each bar is presented as mean S.E. (n = 3). (*, p < 0.05, **, p < 0.005, as compared with control siRNAs; Student's t test); n.s. represents not significant.
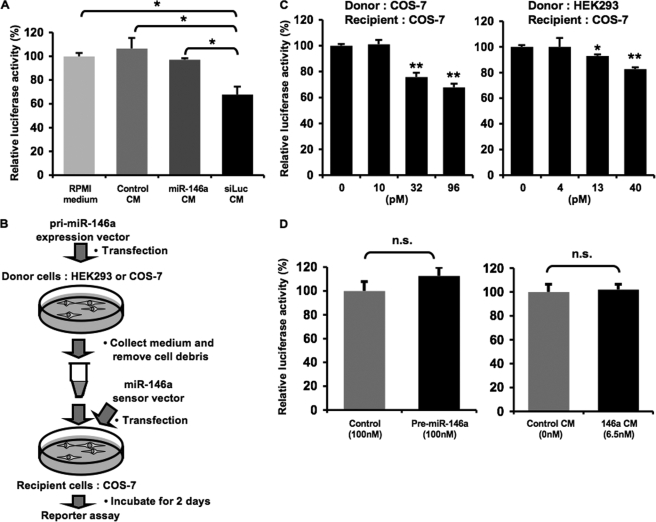
FIGURE 6.
Secretory small RNAs can be transferred to recipient cells. A, treatment with conditioned medium enriched in luciferase siRNA down-regulated luciferase activity. HEK293 cells stably transduced with firefly luciferase were incubated with the indicated medium for 3 days. The conditioned medium was prepared from HEK293 cells transfected with psiRNA-LucGL3 or empty vector. The luciferase siRNA was not detected in RPMI medium and control CM as well as miR-146a CM using QRT-PCR, whereas luciferase siRNA (siLuc) CM contained 200 pm luciferase siRNA as calculated under “Experimental Procedures.” The concentration of miR-146a in miR-146a CM was 123 pm. The cells were harvested and applied to luciferase reporter assay. The values on the y axis are depicted relative to firefly luciferase activity of RPMI medium-treated cells, which is defined as 100%. B, the diagram shows a reporter assay for testing the direct regulation of target gene expression by extracellular miRNAs. C, extracellular miR-146a derived from COS-7 and HEK293 cells suppressed luciferase activity of the sensor vector. COS-7 cells transfected with miR-146a sensor vector were used as recipient cells. The recipient cells were incubated in the conditioned medium containing extracellular miRNAs of the indicated concentrations. After a 2-day incubation, luciferase reporter assay was performed as described under “Experimental Procedures.” D, miR-146a did not reduce luciferase activity from the mutated sensor vector. COS-7 cells transfected with the mutated miR-146a sensor vector were used as recipient cells. The recipient cells were transfected with synthetic miRNAs (left graph) or incubated in the conditioned medium containing extracellular miRNAs at the indicated concentrations (right graph). Luciferase assay was carried out as described above. C and D, the values on the y axis are depicted relative to normalized Renilla luciferase activity of control cells, which is defined as 100%. A, C, and D, each bar is presented as mean S.E. (n = 3). (*, p < 0.05, **, p < 0.005, as compared with control recipient cells; Student's t test); n.s. represents not significant.
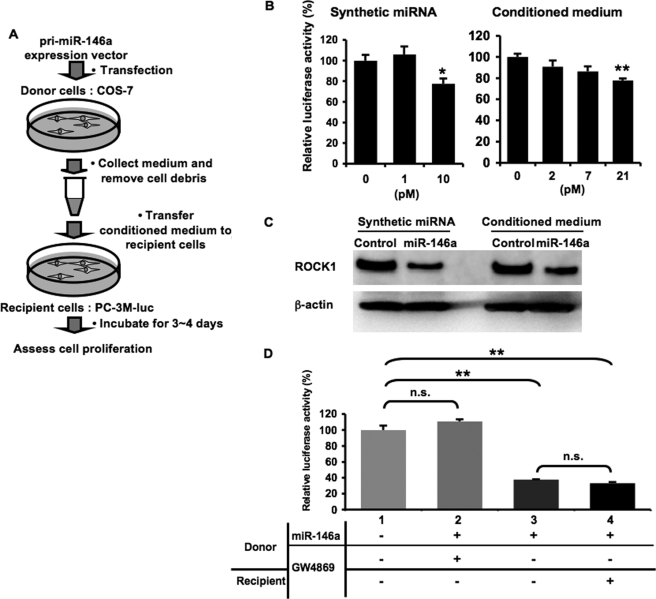
FIGURE 7.
Secretory tumor-suppressive miRNAs attenuated PC-3M cell proliferation. A, schematic representation of a cell proliferation assay. B, cell growth inhibition by synthetic miR-146a and extracellular miR-146a. PC-3M-luc cells were transfected with synthetic miRNAs (left graph) or incubated in the conditioned medium containing extracellular miRNAs at the indicated concentrations (right graph) followed by cell growth assay as described under “Experimental Procedures.” C, miR-146a-mediated ROCK1 suppression in PC-3M-luc cells. The final concentrations of synthetic and extracellular miR-146a are 10 and 21 pm, respectively. D, the treatment with GW4869 to donor cells restored the reduced cell growth by the exosomal miR-146a. Donor COS-7 cells transfected with pri-miR-146a expression vector were incubated in the presence (lane 2) or absence of 10 μm GW4869 for 3 days. The conditioned medium from COS-7 cells transfected with empty vector was used as a control (lane 1). GW4869-untreated conditioned medium enriched in miR-146a was divided into two aliquots, one of which was treated with 10 μm GW4869 (lane 3) and the other of which was not treated (lane 4) before the transfer to recipient cells. The following assay was conducted as described above. B and D, the values on the y axis are depicted relative to normalized Renilla luciferase activity of control cells, which is defined as 100%. Each bar is presented as mean S.E. (n = 3). (*, p < 0.05, **, p < 0.005, as compared with untreated PC-3M-luc cells; Student's t test); n.s. represents not significant.
Cell Growth Assay
PC-3M-luc cells were seeded at a density of 2 × 103 cells/well in a 96-well plate. The following day, the cells were transfected with mature miRNAs or were incubated with a conditioned medium. Twenty-four h later, the culture medium of the transfected cells was switched to medium A, whereas the conditioned medium was not changed. After a 3-day culture, cells were harvested for the measurement of firefly luciferase activity.
Measurement of Caspase-3/7 Activities
On day 0, HEK293 cells were seeded at a density of 2.5 × 104 cells/well in a 96-well tissue culture plate. The following day, the cells were transfected with nSMase2 expression vector or were treated with 10 μm cisplatin. On day 2, the cells were applied to an Apo-ONE homogeneous caspase-3/7 assay (Promega). After a 12-h incubation, the fluorescence of each well was measured at an excitation wavelength of 480 nm and an emission wavelength of 520 nm using Envision (Wallac).
RESULTS
Overexpression of miRNA in Cells Leads to an Increased Secretion
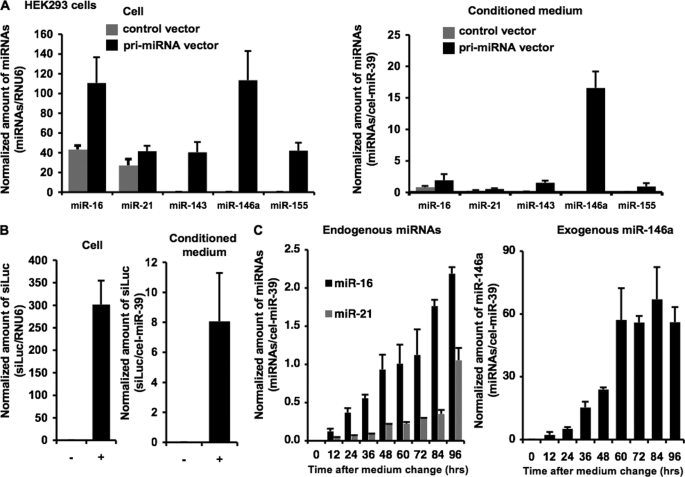
To reveal the physiological roles of extracellular miRNAs, we investigated the mechanism of their secretion and whether or not secretory miRNAs can function in cells beyond their own cell origin. Based on the observation that expression patterns of miRNAs in a cell and culture medium are well correlated (7, 17), we hypothesized that up-regulation of cellular miRNAs leads to an increase of extracellular miRNAs in a conditioned medium. To test this hypothesis, we quantified the amount of extracellular and cellular miRNAs (miR-16, -21, -143, -146a, and -155) in HEK293 and COS-7 cells transfected with each primary miRNA expression vector or empty vector. We avoided the use of synthetic analogues of mature miRNAs for this overexpression experiment because they might persist in the medium and interfere with accurate quantification of extracellular miRNAs. The transfected cells were thoroughly washed prior to the medium change to remove any surplus liposome left by the original transfection. QRT-PCR analysis showed that secretion of the miRNAs was enhanced in proportion to their cellular amounts induced by the transfections of pri-miRNA vectors (Fig. 1A; supplemental Fig. 1A). To exactly estimate what percentages of the miRNAs were excreted, we re-evaluated cellular amounts of the miRNAs by the same normalization method as the extracellular miRNAs (supplemental Fig. 1B). As a result of the calculations, the amounts of secreted miRNAs were quite lower in conditioned medium than in parental cells. The ratios of secreted miRNAs/cellular miRNAs are shown as follows: endogenous miR-16, 1.21 ± 0.12%; endogenous miR-21, 1.62 ± 0.63%; exogenous miR-143, 2.57 ± 1.03%; exogenous miR-146a, 15.6 ± 1.62%; exogenous miR-155, 1.38 ± 0.84%. Additionally, siRNAs targeting luciferase were also released from the siRNA-overexpressing cells (Fig. 1B). We further observed that endogenous miR-16 and -21 as well as overexpressed miR-146a time-dependently accumulated in the conditioned medium during 96 h (Fig. 1C). We observed by microscopic examination that these cells did not suffer from any damage and that they maintained the original cell shape. Moreover, the treatment of an apoptotic inducer, cisplatin, did not increase the miRNA secretion (data not shown). These findings strongly suggest that living cells can actively secrete endogenous and exogenous miRNAs as well as artificial small RNAs.
FIGURE 1.
Characterization of extracellular microRNAs. A, miRNAs were released in proportion to the cellular expression level. HEK293 cells were set up at a density of 2.5 × 105 cells/well in a 24-well plate. The following day, the cells were transfected with 0.5 μg of the indicated primary miRNA expression vectors or pCMV empty vector as a control as described under “Experimental Procedures.” Twenty-four h later, after washing three times by Advanced RPMI containing antibiotic-antimycotic and 2 mm l-glutamine (medium A), culture medium was switched to medium A. After a 24-h incubation, preparation of conditioned medium and isolation of total RNAs were performed as described under “Experimental Procedures.” Expression levels of miRNAs were analyzed using quantitative real-time PCP (QRT-PCR). B, siRNAs targeting luciferase gene (siLuc) were secreted into culture medium. HEK293 cells were set up at a density of 2.5 × 105 cells/well in a 24-well plate. Transfection with psiRNA-LucGL3 vector (+) or control vector (−) and preparation of conditioned medium were conducted as described above. The amount of luciferase siRNAs generated from the expression vector was measured by QRT-PCR with a custom-designed TaqMan small RNA Assay (Applied Biosystems) specific for the luciferase siRNAs. C, time course analysis of extracellular miRNAs. Conditioned medium was collected at the indicated time and applied to quantitative miRNA RT-PCR. A–C, cel-miR-39 and hRNU6 were used as invariant control for conditioned medium and cell, respectively. Each bar is presented as mean S.E. (n = 3).
Secreted miRNAs Are Contained in Exosomes
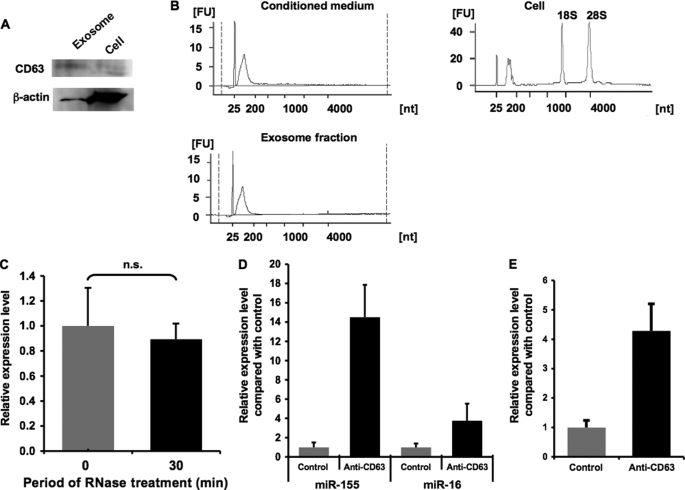
It was revealed that exosomes can transfer some of their contents to other cell types, and importantly, that miRNAs exist in exosomes and are protected from RNases (6). First, we isolated exosome fractions from HEK293 cells by the standard ultracentrifugation method. Consistent with the previous investigations, these exosome fractions were positive for a surface marker of exosome, CD63 (Fig. 2A). To characterize the exosomal RNAs, we conducted an electrophoresis of total RNA extracted from conditioned medium, exosomes, and their donor cells. Conditioned medium and exosomes applied to this analysis were prepared from the supernatant of the same numbers of donor cells. The Bioanalyzer 2100 profiles shown in Fig. 2B revealed that conditioned medium and exosome fractions share a very similar size-distribution pattern and expression intensity, which shows the enrichment of small sized RNAs, whereas the cellular RNA profile indicates two conspicuous peaks of 18 S and 28 S ribosomal RNAs and a broad peak of small RNAs. These data suggest that small RNAs are preferentially released from the cells and that most of the secreted RNAs are contained in exosome fractions. To evaluate whether extracellular miRNAs are contained inside exosomes, the conditioned medium from HEK293 cells was exposed to RNases before RNA extraction. As a result, endogenous miR-21 still existed even after 30 min of treatment with RNase A and T1 (Fig. 2C). Contrarily, exogenously added synthetic cel-miR-39 was completely decomposed by the same treatment (supplemental Fig. 2). Moreover, immunoprecipitant obtained with an anti-CD63 antibody was enriched in endogenous miR-16 and -155 as well as overexpressed luciferase siRNA (Fig. 2, D and E). These results collectively indicate that extracellular miRNAs are contained within the CD63-positive exosomes and therefore are protected from external RNases by the surrounding membrane.
FIGURE 2.
Purification and characterization of secretory exosomes. A, purified exosomes secreted from HEK293 cells are enriched in CD63 protein. The exosome fractions and whole cell lysate were analyzed by immunoblotting with an anti-CD63 antibody or anti-β-actin antibody. B, total RNAs isolated from conditioned medium, exosome fractions, and their donor HEK293 cells were detected using a Bioanalyzer 2100. The obtained data show the amount and the size distribution of total RNAs. The peak migrating around 25 nucleotides (nt) represents an internal standard. FU, fluorescence units. C, effect of RNase treatment on extracellular miR-21. Conditioned medium from HEK293 cells was exposed to RNase A and RNase T1 at 37 °C followed by the isolation of miRNAs at the indicated times. The amount of miR-21 was determined using quantitative miRNA RT-PCR. The values on the y axis are depicted relative to the amount of miR-21 at 0 min, which is arbitrarily defined as 1. D and E, CD63+ exosome fraction is enriched in miR-155 and miR-16 (D) and luciferase siRNA (E). Conditioned medium of HEK293 cells was immunoprecipitated with anti-CD63 antibody or isotype control. The values on the y axis are depicted relative to control, which is arbitrarily defined as 1. C–E, each bar is presented as mean S.E. (n = 3). n.s. represents not significant.
The Secretion of miRNAs Is Regulated by a Ceramide-dependent Pathway
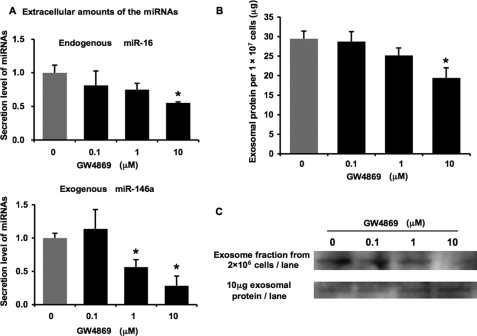
Exosomes are produced in MVB and released from a variety of cells. They are originally thought to function to dispose of cellular garbage, including degraded protein, but now they draw much attention as a cell-cell communication tool (5). The biogenesis of exosomes has appeared to associate with the endosomal sorting complex required for transport (ESCRT) machinery because the machinery is highly involved in the exocytosis of some cellular contents and various viruses. On the other hand, Trajkovic et al. (18) reported that exosomes are released independently from ESCRT machinery but triggered by sphingolipid ceramide. Thus, it is still unclear whether many types of exosomal contents, such as proteins, lipids, and nucleic acids, are released by the same mechanism. To address the question as to whether miRNA secretion is regulated by the ceramide-dependent pathway or the ESCRT pathway, we first treated HEK293 cells with an nSMase inhibitor, GW4869, which is known to inhibit ceramide biosynthesis (18). As a result of this treatment, extracellular endogenous miR-16 and exogenous miR-146a were dose-dependently reduced, whereas their cellular expression levels remained unchanged (Fig. 3A; supplemental Fig. 3). Consistently, exosomal protein was also decreased by the treatment of GW4869 in a dose-dependent manner (Fig. 3B). Furthermore, a reduction in the signals of an exosome marker, CD63, was observed by immunoblotting of exosomes purified from the supernatant of the same number of GW4869-treated HEK293 cells when compared with control cells (Fig. 3C, upper panel). Nevertheless, the amounts of CD63 protein were unchanged in exosomes from GW4869-treated cells when the same amounts of exosomal proteins were analyzed (Fig. 3C, lower panel). These data show that the treatment of GW4869 reduces the amount of exosomes released into the conditioned medium but does not modify exosomal protein composition.
FIGURE 3.
Secretion of miRNA was attenuated by GW4869. A, secretion of miRNAs was suppressed by the treatment with GW4869. HEK293 cells were transfected with pri-miR-146a vector in a 6-well plate. The cells were reseeded and cultured in a 24-well plate for 48 h in the indicated concentrations of GW4869. After the incubation, the medium was subjected to QRT-PCR for miR-16 and -146a. The values on the y axis are depicted relative to the amount of each miRNA at 0 μm GW4869, which is arbitrarily defined as 1. B, after the treatment with the indicated concentrations of GW4869 for 24 h, the total amounts of proteins in the exosomal pellet purified from large scale cultures of stably miR-146a-transduced HEK293 cells were quantified by a BCA assay and are presented as the values per 10 million secreting cells. A and B, each bar is presented as mean S.E. (n = 3). C, purified exosomes secreted by equal numbers of control or GW4869-treated HEK293 cells and equal amounts of total exosomal proteins (quantified by BCA assay) secreted by either control or GW4869-treated HEK293 cells were analyzed by immunoblotting for the presence of CD63 protein. (*, p < 0.05, **, p < 0.005, as compared with untreated cells; Student's t test).
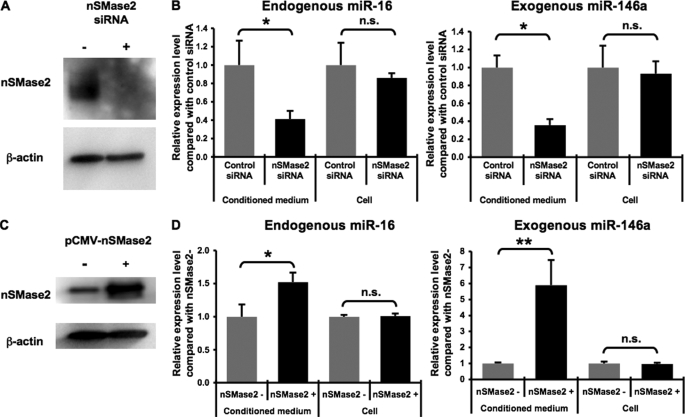
We next employed an RNA interference approach to knock down the expression of endogenous nSMase2 in HEK293 cells. A transfection of an siRNA duplex targeting human nSMase2 mRNAs resulted in a robust reduction of nSMase2 protein levels in HEK293 cells (Fig. 4A). Under these conditions, secretion of endogenous miR-16 as well as exogenous miR-146a was attenuated by the nSMase2 siRNA as compared with control cells where negative control siRNAs were transfected, whereas cellular expression of miRNAs was not changed (Fig. 4B). Furthermore, to complement the results of this knockdown study, we observed that overexpressed human nSMase2 induced the secretion of endogenous miR-16 as well as exogenous miR-146a and luciferase siRNA from HEK293 cells without affecting cellular amounts of the miRNAs (Fig. 4, C and D, and supplemental Fig. 4A). Although it was already known that nSMase2 overexpression increases the amount of ceramide in the raft fraction and subsequently induces an apoptotic cell death (19), we did not observe any elevated caspase-3/7 activities in nSMase2-overexpressing cells (supplemental Fig. 4B). These results suggest that a ceramide-dependent secretory pathway is involved in miRNA secretion.
FIGURE 4.
miRNAs were secreted through nSMase2-dependent pathway. A and B, siRNA-mediated knockdown of nSMase2. HEK293 cells were transfected with pri-miR-146a vector in a 6-well plate. One day later, these cells were reseeded in a 24-well plate and transfected with either negative control or nSMase2 siRNA. The following day, conditioned medium and cell extract were applied to QRT-PCR for miR-16 as well as miR-146a (B). In parallel, cell extract was applied to immunoblot for nSMase2 and β-actin proteins (A). The values on the y axis are depicted relative to the amount of miRNAs of control, which is arbitrarily defined as 1. C and D, augmentation of miRNA secretion by overexpressed human nSMase2. HEK293 cells were transfected with human nSMase2 expression vector or control vector, along with pri-miR-146a vector. After a 24-h incubation, conditioned medium and cell extract were applied to QRT-PCR (D), and cell extract was applied to immunoblot (C). The values on the y axis are depicted relative to the amount of miRNAs of nSMase2−, which is arbitrarily defined as 1. B and D, each bar is presented as mean S.E. (n = 3). (*, p < 0.05, **, p < 0.005, as compared with control cells; Student's t test); n.s. represents not significant.
The Secretion of miRNAs Does Not Require ESCRT System
Gibbings et al. (17) showed that a knockdown of some ESCRT-associated proteins, including Alix, vps36, and hrs, deteriorated ESCRT function, resulting in an inhibition of the activity of cellular miRNAs. To examine whether Alix is involved in the secretory process of miRNAs, the knockdown experiment was performed in HEK293 cells carrying a sensor vector. As shown in Fig. 5B, upper schematic, our sensor vector is available for assessing miR-146a specific activity, which expresses firefly luciferase and Renilla luciferase containing complementary miR-146a sequences at its 3′-untranslated region, allowing us to simultaneously monitor transfection efficiency and miR-146a activity. Consistent with the previous finding, the depletion of Alix by an siRNA transfection fully restored the miR-146a-suppressed Renilla luciferase activity (Fig. 5, A and B; compare lanes 1, 2, and 4). In other words, ESCRT integrity was compromised by the Alix knockdown. On the contrary, nSMase2 siRNA had no effect on the gene silencing by miR-146a, suggesting that ceramide synthesis is irrelevant to ESCRT function (Fig. 5B; compare lanes 1 and 3). The result in the setting of non-transfection of miR-146a ruled out the possibility that the application of these siRNAs changed the basal Renilla luciferase activities (Fig. 5B; compare lanes 4, 5, and 6). QRT-PCR analysis showed that the blocked RNA silencing by the Alix knockdown was not because of the decreased amount of cellular miR-146a (Fig. 5C). Intriguingly, in the HEK293 cells where the ESCRT system was impaired by the Alix siRNA, we did not detect any significant difference of the amount of extracellular miR-146a as compared with a control transfectant (Fig. 5D). Taken together with the results of Figs. 3 and 4, it is unequivocally demonstrated that secretion of miRNAs depends on a ceramide-triggered secretory mechanism but not ESCRT machinery.
Secretory Small RNAs Can Be Transferred to Recipient Cells
Our finding that miRNA secretion is tightly regulated by ceramide biosynthesis inspires us to envisage that extracellular miRNAs are not just discards from the originating cells but biologically active molecules. In fact, many reports suggest that small RNAs migrate beyond their cell origin and function in the recipient cells (6, 9). To test whether or not artificial small RNA can be transported into recipient cells under our experimental conditions, we treated firefly luciferase stably expressing HEK293 cells with a conditioned medium enriched in the luciferase siRNAs. The amounts of small RNAs in the conditioned medium were evaluated by QRT-PCR as described under “Experimental Procedures.” As a result, the luciferase activities were negatively regulated by the treatment of conditioned medium containing 200 pm luciferase siRNAs as compared with control conditioned medium and fresh RPMI medium as well as miR-146a-enriched conditioned medium (Fig. 6A). In addition to artificial siRNAs, we next examined a transfer of a class of naturally occurring small RNAs, miRNAs. To measure a direct regulation of gene expression by extracellular miRNAs, we performed a reporter assay based on the sensor vector according to an experimental scheme illustrated in Fig. 6B. As shown in Fig. 6C, the normalized Renilla luciferase activities were dose-dependently reduced by the treatment of miR-146a-enriched conditioned medium derived from COS-7 and HEK293 cells transfected with pri-miR-146a expression vectors. This effect was not due to a transfer of free miRNAs remaining in cellular debris and conditioned medium because a direct addition of 100 nm naked miR-146a analogues did not change the luciferase activity (supplemental Fig. 5). Furthermore, to exclude the possibility that the inhibitory effect is independent of the specific interaction between the transferred miR-146a and the complementary binding sites in the sensor vector, we used a mutated miR-146a sensor vector constructed as described under “Experimental Procedures.” As shown in Fig. 6D, the normalized Renilla luciferase activities from the mutated sensor vector were not changed by the addition of the conditioned medium from miR-146a-overexpressing HEK293 cells. These results show that extracellular miRNAs packaged in secretory exosome vesicles can be delivered into recipient cells and act as physiologically functional molecules to exert gene silencing through the same mechanism as cellular miRNAs.
Secretory Tumor-suppressive miRNAs Attenuate PC-3M Cell Proliferation
To gain further insight into the biological activity of secretory miRNAs, we investigated whether they can make an impact on a cellular phenotype. miR-146a is known to be down-regulated in prostate cancer (20), and reconstitution of its expression results in growth inhibition of prostate cancer cells (21). As schematically depicted in Fig. 7A, supernatants containing transduced miR-146a derived from COS-7 cells were added to the culture medium of metastatic prostate cancer cell line PC-3M-luc cells. This cell line stably harbors a firefly luciferase gene, enabling us to assess cell numbers with the luciferase activity. After a 3-day incubation, PC-3M-luc cells showed an ∼20% decrease in proliferation in the presence of 21 pm miR-146a in the conditioned medium, whereas liposomally transfected synthetic miRNAs at 10 pm suppressed cell growth to a similar extent (Fig. 7B). To determine whether extracellular miRNA exerts regulatory action on its target gene, we examined the expression of ROCK1, a target gene for miR-146a, in the treatment with extracellular miR-146a (21). The addition of the conditioned medium of miR-146a-transduced COS-7 cells significantly knocked down ROCK1 protein expression in PC-3M-luc cells to the same degree as the miR-146a analogue molecule under conditions where the amount of β-actin was not affected (Fig. 7C). To confirm that the cell growth inhibition resulted from the transferred exosomal miR-146a, we examined whether the suppressive effect can be cancelled by a reduced exosome secretion. Conditioned medium from GW4869-treated donor cells had no inhibitory activity on cell proliferation after a 4-day incubation (Fig. 7D; compare lanes 1 and 2). We also did not observe any direct effect of GW4869 on the restoration of miR-146a-suppressed cell growth (Fig. 7D; compare lanes 3 and 4). These findings suggest that itinerant exosomal miRNAs can induce some phenotypic changes in the recipient cells.
DISCUSSION
Our data provide a fresh insight into the secretory machinery of miRNAs. Through our mechanistic studies with a chemical inhibitor as well as nSMase2 overexpression and knockdown, we contend that miRNAs can be incorporated into exosomes and released via a ceramide-dependent pathway independently of ESCRT machinery. It remains elusive, however, how miRNAs are sorted into exosomes for their secretion. Two recent studies provide evidences that ESCRT complex associates with components of miRNA effector complexes and that RNA silencing takes place on MVB (22). Taken together, MVB could be a crossroads of miRNAs bound for the secretory pathway and gene-silencing cycle. Further investigations are needed to unveil a sorting mechanism by which miRNAs are determined to be secreted or to stay and function in their originating cells.
There is growing evidence that secretory miRNAs play a role in a variety of physiological phenomena. Zernecke et al. (9) showed that the transfer of miR-126 loaded into endothelial apoptotic bodies induced the expression of CXCL12 and the recruitment of progenitor cells, thereby alleviating atherosclerosis. Additionally, Rechavi et al. (8) proposed that viruses could tamper with the host immune response by transportation of viral non-autonomously encoded small RNAs. Both vascular protection and manipulation of the immune system by viruses occur for a long term. These findings prompt us to conceive of the idea that secretory miRNAs could mainly serve in chronic biological events, such as the formation of a tumor microenvironment.
Many tumors have a remarkable ability to mold their stromal environment to their own advantage (23, 24). Recent studies show the importance of communication between cancer cells and their surroundings through shedding of membrane exosomes, which can fuse to cells in the vicinity (7, 25, 26). For instance, epidermal growth factor receptor vIII proteins were transferred into glioma cells lacking epidermal growth factor receptor vIII via secretory membrane microvesicles (26). Together, membrane microvesicles or exosomes of cancer cells can contribute to a horizontal propagation of oncogenes and their associated transforming phenotype among subsets of cancer cells. Here, we show that re-expression of miR-146a through intercellular transfer leads to cell growth inhibition of PC-3M cells, where the expression of the miRNA is at a very low level. It is well known that many kinds of miRNAs are down-regulated in cancer cells, resulting in tumorigenesis, tumor progression, and metastasis (27). We therefore hypothesize that down-regulated miRNAs in cancer cells are compensated during the initial stage of tumorigenesis by the surrounding cells that supply exosomes containing the decreased miRNAs. However, once the surrounding cells cannot meet the demand, cancer cells end up entering an advanced stage. Secretory miRNAs could be conducive to the maintenance and surveillance system against cancer progression.
Small RNAs including antisense, siRNA, and miRNA are emerging as promising therapeutic agents against a wide array of diseases (28). Effective delivery of these molecules is crucial to their successful clinical application. Now, exosomes have entered the limelight in delivery of therapeutic nucleic acids with their ability to transfer exogenous miRNAs into tumor cells. Exosomes are naturally produced membrane vesicles that circulate in the bloodstream and are taken up by various types of cells. These aspects might be beneficial for the improvement of durability, stability, and safety for in vivo delivery. In fact, tumor exosomes generated tumor regression in vivo, acting as a cancer vaccine through their capacity of antigen presentation (29). It is essential to examine whether exosomes can deliver their cargoes in an in vivo animal model.
In conclusion, we revealed the secretory machinery of miRNAs and their intercellular transfer. Our results raise the possibility that in the animal kingdom, circulating miRNAs could play a pivotal and general role as a signaling molecule in physiological and pathological events. We anticipate that our study could pave the way for explorations of signal network mediated by secretory miRNAs.
Acknowledgment
We thank Ayano Matsumoto for excellent technical assistance.
This work was supported in part by a grant-in-aid for the third-term comprehensive 10-year strategy for cancer control, a grant-in-aid for scientific research on priority areas cancer from the Ministry of Education, Culture, Sports, Science and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- MVB
- multivesicular bodies
- Alix
- ALG-2 interacting protein
- ESCRT
- endosomal sorting complex required for transport
- miRNA
- microRNA
- nSMase2
- neutral sphingomyelinase 2
- siRNA
- small interfering RNA
- QRT-PCR
- quantitative real time PCR
- CM
- conditioned medium.
REFERENCES
- 1.Fleischhacker M., Schmidt B. (2007) Biochim. Biophys. Acta 1775, 181–232 [DOI] [PubMed] [Google Scholar]
- 2.El-Hefnawy T., Raja S., Kelly L., Bigbee W. L., Kirkwood J. M., Luketich J. D., Godfrey T. E. (2004) Clin. Chem. 50, 564–573 [DOI] [PubMed] [Google Scholar]
- 3.Tsui N. B., Ng E. K., Lo Y. M. (2002) Clin. Chem. 48, 1647–1653 [PubMed] [Google Scholar]
- 4.Hasselmann D. O., Rappl G., Tilgen W., Reinhold U. (2001) Clin. Chem. 47, 1488–1489 [PubMed] [Google Scholar]
- 5.Cocucci E., Racchetti G., Meldolesi J. (2009) Trends Cell Biol. 19, 43–51 [DOI] [PubMed] [Google Scholar]
- 6.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 7.Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rechavi O., Erlich Y., Amram H., Flomenblit L., Karginov F. V., Goldstein I., Hannon G. J., Kloog Y. (2009) Genes Dev. 23, 1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zernecke A., Bidzhekov K., Noels H., Shagdarsuren E., Gan L., Denecke B., Hristov M., Köppel T., Jahantigh M. N., Lutgens E., Wang S., Olson E. N., Schober A., Weber C. (2009) Sci. Signal. 2, ra81. [DOI] [PubMed] [Google Scholar]
- 10.Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 11.Croce C. M. (2009) Nat. Rev. Genet. 10, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrie C. H., Gal S., Dunlop H. M., Pushkaran B., Liggins A. P., Pulford K., Banham A. H., Pezzella F., Boultwood J., Wainscoat J. S., Hatton C. S., Harris A. L. (2008) Br. J. Haematol. 141, 672–675 [DOI] [PubMed] [Google Scholar]
- 13.Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O'Briant K. C., Allen A., Lin D. W., Urban N., Drescher C. W., Knudsen B. S., Stirewalt D. L., Gentleman R., Vessella R. L., Nelson P. S., Martin D. B., Tewari M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Zhang Y., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Wang J., Zen K., Zhang J., Zhang C. Y. (2008) Cell Res. 18, 997–1006 [DOI] [PubMed] [Google Scholar]
- 15.Taylor D. D., Gercel-Taylor C. (2008) Gynecol. Oncol. 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 16.Takeshita F., Patrawala L., Osaki M., Takahashi R. U., Yamamoto Y., Kosaka N., Kawamata M., Kelnar K., Bader A. G., Brown D., Ochiya T. (2010) Mol. Ther. 18, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbings D. J., Ciaudo C., Erhardt M., Voinnet O. (2009) Nat. Cell Biol. 11, 1143–1149 [DOI] [PubMed] [Google Scholar]
- 18.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 19.Hannun Y. A., Obeid L. M. (2002) J. Biol. Chem. 277, 25847–25850 [DOI] [PubMed] [Google Scholar]
- 20.Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S. L., Chiang A., Chang D., Ying S. Y. (2008) RNA 14, 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y. S., Pressman S., Andress A. P., Kim K., White J. L., Cassidy J. J., Li X., Lubell K., Lim do H., Cho I. S., Nakahara K., Preall J. B., Bellare P., Sontheimer E. J., Carthew R. W. (2009) Nat. Cell Biol. 11, 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhowmick N. A., Neilson E. G., Moses H. L. (2004) Nature 432, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 25.Wolfers J., Lozier A., Raposo G., Regnault A., Théry C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T., Angevin E., Amigorena S., Zitvogel L. (2001) Nat. Med. 7, 297–303 [DOI] [PubMed] [Google Scholar]
- 26.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. (2008) Nat. Cell Biol. 10, 619–624 [DOI] [PubMed] [Google Scholar]
- 27.Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D'Urso L., Pagliuca A., Biffoni M., Labbaye C., Bartucci M., Muto G., Peschle C., De Maria R. (2008) Nat. Med. 14, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 28.Castanotto D., Rossi J. J. (2009) Nature 457, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignot G., Roux S., Thery C., Ségura E., Zitvogel L. (2006) J. Cell Mol. Med. 10, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]