Abstract
Cellular inhibitor of apoptosis (cIAP) proteins, cIAP1 and cIAP2, are important regulators of tumor necrosis factor (TNF) superfamily (SF) signaling and are amplified in a number of tumor types. They are targeted by IAP antagonist compounds that are undergoing clinical trials. IAP antagonist compounds trigger cIAP autoubiquitylation and degradation. The TNFSF member TWEAK induces lysosomal degradation of TRAF2 and cIAPs, leading to elevated NIK levels and activation of non-canonical NF-κB. To investigate the role of the ubiquitin ligase RING domain of cIAP1 in these pathways, we used cIAP-deleted cells reconstituted with cIAP1 point mutants designed to interfere with the ability of the RING to dimerize or to interact with E2 enzymes. We show that RING dimerization and E2 binding are required for IAP antagonists to induce cIAP1 degradation and protect cells from TNF-induced cell death. The RING functions of cIAP1 are required for full TNF-induced activation of NF-κB, however, delayed activation of NF-κB still occurs in cIAP1 and -2 double knock-out cells. The RING functions of cIAP1 are also required to prevent constitutive activation of non-canonical NF-κB by targeting NIK for proteasomal degradation. However, in cIAP double knock-out cells TWEAK was still able to increase NIK levels demonstrating that NIK can be regulated by cIAP-independent pathways. Finally we show that, unlike IAP antagonists, TWEAK was able to induce degradation of cIAP1 RING mutants. These results emphasize the critical importance of the RING of cIAP1 in many signaling scenarios, but also demonstrate that in some pathways RING functions are not required.
Keywords: Apoptosis, Cancer Therapy, NF-κB Transcription Factor, Protein-Protein Interactions, Tumor Necrosis Factor (TNF), Dimerization, Smac Mimetic, TRAF2, TWEAK, cIAP1
Introduction
Cellular inhibitors of apoptosis (cIAPs)5 were identified as proteins that bound directly to the adaptor protein TRAF2, which in turn binds to tumor necrosis factor receptor 2 (TNF-R2) (1). cIAPs have three baculoviral IAP repeat domains and interaction of cIAP1 with TRAF2 is dependent on residues within the N-terminal, baculoviral IAP repeat 1 (BIR1) (2, 3). cIAP1 and cIAP2 also bear a C-terminal RING E3 ligase domain, and it is this domain that is required for IAP antagonist compound (IAC)-induced autoubiquitylation and proteasomal degradation (4–7). cIAPs control activation of the NF-κB family of transcription factors in both unstimulated and cytokine-treated cells. In the absence of cytokines, together with TRAF2 and TRAF3, cIAPs ubiquitylate and promote proteasomal degradation of NF-κB inducing kinase (NIK), thereby preventing NIK from activating NF-κB2. Consistent with a central role for cIAPs in these processes, antagonism of IAPs by IACs is sufficient to activate the non-canonical NF-κB pathway through NIK stabilization and processing of p100 to p52 (6–9).
TNF-like weak inducer of apoptosis (TWEAK) binding to its receptor Fn14 is a physiological activator of NF-κB. Receptor engagement results in lysosomal mediated degradation of TRAF2 and cIAP1 and activation of non-canonical NF-κB resulting from NIK stabilization and subsequent processing of p100 to the active NF-κB subunit p52 (7, 10, 11). In support of this idea, TRAF2 or cIAP1 knock-out MEFs were shown to have high constitutive levels of p52 and low levels of p100 compared with wild-type cells (6, 12). cIAPs also play an important role at the TNF-R1 signaling complex in TNF-induced activation of canonical NF-κB. RIPK1 ubiquitylation can contribute to activation of canonical NF-κB by recruiting NEMO-IKK1-IKK2 and TAB2-TAK1, initiating a signaling cascade leading to translocation of p65-RelA NF-κB subunits to the nucleus, however, RIPK1 is not essential for TNF-induced NF-κB (13). The E3 ligase responsible for RIPK1 ubiquitylation is still not clear; IAPs are an attractive candidate because loss of cIAPs by either IAC treatment, small interfering RNA knockdown or disruption of both genes significantly inhibits RIPK1 ubiquitylation (14–17). However, interestingly some minor ubiquitylation of RIPK1 in TNF-stimulated cells can still be seen even in the absence of cIAPs, suggesting other E3 ligases are involved (18). Several new E3 ligases have recently been reported to play a role in activating NF-κB. RNF11 was shown to play a role in the recruitment of A20 to RIPK1 and thereby negatively regulate NF-κB and c-Jun N-terminal kinase (JNK) signaling (19). The RING E3 ligases HOIL-1 and HOIP are also required for a sustained TNF-induced NF-κB signal (17, 20). Subtle differences in response from the E3 ligases following TNF might also be cell type dependent, a result from the different forms of available TNF (membrane bound or soluble), or even due to their effects on other signaling pathways. Nevertheless, it is clear that removal of cIAPs sensitizes many different cell types to TNF-induced death.
Although the mechanism is still unclear, it appears that RING dimerization is important for E3 ligase function. Structural studies have revealed RING domain homodimers for many E3 ligases, including MDM2, MDMX, cIAP2, and RAG1 (5, 21, 22) and several RINGs, such as MDM2-MDMX, BRCA1-BARD1, and XIAP-cIAP1 can heterodimerize (22–27). Consistent with an important role for dimerization in RING function, cIAP RING dimerization is required for IAC-induced degradation of cIAP1 (5, 27). RING domains are required to recruit UBC E2 enzymes to substrates and cIAP1 has been shown by several groups to recruit UbcH5 (16, 27–29).
We generated point mutations in the baculoviral IAP repeat 1 (BIR1) of cIAP1 that specifically affect binding by TRAFs, as well as mutations in the RING that prevent dimerization and E2 binding. These mutations abolish the ability of cIAP1 to protect cells from TNF-induced apoptosis. These mutations also interfere with the ability of cIAP1 to activate canonical NF-κB in response to TNF, and destroy the regulation of non-canonical NF-κB by cIAP1. Interestingly, TWEAK-induced degradation of cIAP1 was not affected by these RING mutations indicating a significantly different role for cIAP1 at different TNFSF receptors.
EXPERIMENTAL PROCEDURES
Transient Transfections, Antibodies, and Reagents
Transient transfections, typically using 1 μg of plasmid DNA/10-cm plate of cells, were performed with EffecteneTM according to the manufacturer's instructions (Qiagen). Antibodies were used as follows: monoclonal anti-β actin (Sigma), polyclonal mouse anti-TRAF2, 1:1000 (Santa Cruz); monoclonal anti-cIAP1, 1:500 (Alexis); polyclonal anti-p65, 1:1000 (Western blotting) and 1:100 (immunostaining) (Santa Cruz); polyclonal anti-p100-p52, 1:1000 (Cell Signaling); polyclonal anti-IκBα, 1:1000 (Santa Cruz); polyclonal anti-NIK, 1:1000 (Cell Signaling); anti-FLAG, 1:2000 (Sigma); anti-ubiquitin, 1:1000 (Cell Signaling); fluorescein isothiocyanate-conjugated anti-rabbit, 1:500 (Invitrogen). 4-Hydroxytamoxifen (4HT, Sigma) was used at concentrations ranging from 5 to 100 nm. The IAP antagonist, compound A, has previously been described (6) and was used at 500 nm.
Cell Culture and Lentivirus Production
All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 8–10% fetal bovine serum, 2 mm l-glutamine and penicillin/streptomycin, and grown at 37 °C, 10% CO2. To generate lentiviral particles, 293T cells were transfected with packaging constructs pCMV ΔR8.2, pVSVg, and at a relevant lentiviral plasmid ratio of 1:0.4:0.6. After 24 h the virus containing supernatants were harvested and filtered (0.8 μm). Target cells were infected with virus supernatant for 24 h. The medium was subsequently changed and successful infection was selected with puromycin (5 μg/ml, pF 5× UAS selection) or hygromycin B (300 μg/ml, GEV16 selection). pF 5× UAS inducible constructs were induced with 4HT for 16 h prior to lysate harvesting for Western blotting or 72 h (5–10 nm) prior to death assays.
Constructs
Inducible mouse cIAP1 wild-type and cIAP1 F610A cloned into pF 5× UAS SV40 Puro have been previously described (5). Mouse cIAP1 E2 binding mutants V567A/D570A, L593A/I598A, L593A, and V567A were generated by spliced overlap PCR mutagenesis and cloned into the pF 5× UAS SV40 Puro vector using standard techniques and confirmed by sequencing. The Ubc13 shRNA and GFP shRNA lentiviral vectors were a kind gift from Dr. M. A. Kelliher (University or Massachusetts). Complete sequence of all constructs can be obtained upon request.
Generation of MEFs
Generation of MEFs has been described in detail elsewhere (6, 10). Briefly, primary MEFs were generated from embryos using standard protocols and then infected with SV40 large T antigen expressing lentivirus to generate immortal cell lines. Double knock-out cIAP1 and cIAP2 MEFs (DKO) were obtained from a cIAP1LoxP/LoxP and cIAP2FRT/FRT mouse crossed with a Cre transgenic and the resulting progeny were backcrossed to generate cIAP1−/−cIAP2FRT/FRT mice. These mice were subsequently crossed with Flp transgenic mice to generate cIAP1−/−cIAP2−/− (DKO) mice in the same manner. DKO MEFs were generated using E10 embryos rather than E15 embryos. These MEFs were also immortalized with SV40 large T expressing lentivirus. Immortal cIAP1−/− MEFs from E15 embryos obtained from a cIAP1LoxP/LoxPcIAP2FRT/FRT, Cre transgenic cross were generated in the same manner.
Death Assays
Cells were seeded on 12-well tissue culture plates at ∼40% confluence and allowed to adhere for 16–20 h. Cells were incubated with or without 4HT for 72 (MEFs) or 24 h (D645s and Kym1s) and then human Fc-TNF (100 ng/ml) or human Fc-TWEAK (100 ng/ml) were added to cells for 24 h and cell death was measured by propidium iodide staining and flow cytometry. In each sample, 5000 events were measured and cell death (% propidium iodide positive cells) was quantified.
Western Blotting
Samples were lysed in DISC lysis buffer containing 1% Triton X-100 supplemented with protease inhibitor mixture (Roche Applied Science) and N-ethylmaleimide (NEM) on ice for 30 min and clarified by centrifugation. Alternatively, for whole cell lysates, samples were boiled in the presence of 1% SDS and sheared through a 19-gauge needle. Samples were separated on precast 4–20% polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes for antibody detection. All membrane blocking steps and antibody dilutions were performed with 5% skim milk in PBS containing 0.1% Tween 20 (PTBS), and washing steps performed with PTBS. Proteins on Western blots were visualized by ECL (Amersham Biosciences) following incubation of membranes with horseradish peroxidase-coupled secondary antibodies.
Immunostaining
Cells were grown on 25-mm coverslips overnight. Cells were treated as indicated and fixed immediately with 3% paraformaldehyde for 30 min and washed three times with PBS. Cells were incubated with 150 mm glycine/PBS for 15 min to block amine groups, and washed three times with PBS. Cells were permeablized with 0.5% Triton X-100/PBS for 5 min and washed three times with PBS. Cells were blocked in 1% bovine serum albumin/PBS for 30 min subsequent to incubation with polyclonal p65 antibody (1:100) (Santa Cruz) for 30 min. Cells were washed four times and incubated with fluorescein isothiocyanate-conjugated secondary anti-rabbit antibody (1:500) (Invitrogen) for 30 min. Cells were washed three times with PBS. The coverslips were then mounted to slides with Polymount (anti-fade) and sealed with nail polish. Slides were viewed under a ×40 fluorescence microscope objective.
RESULTS
IAP Antagonist-induced Degradation of cIAP1 Requires E2 Binding and RING Dimerization
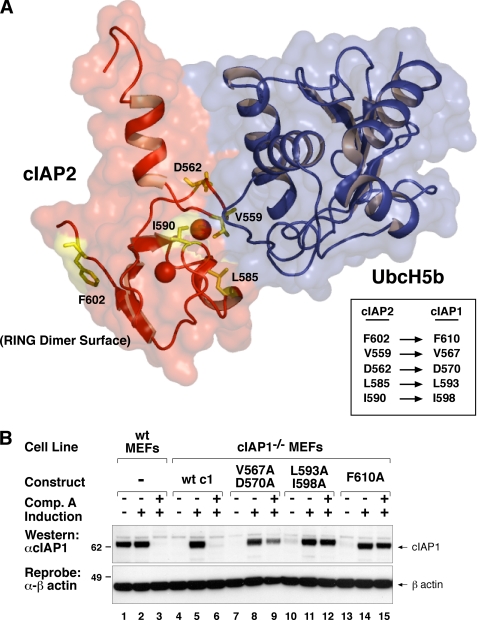
Using the crystal structure of the RING domain of cIAP2 bound to the E2 UbcH5 (5), we identified residues that were likely to be important for RING dimerization and interaction with the E2 enzyme. The F610A mutant prevents cIAP1 RING dimer formation and has been previously described (5). Point mutations at positions Val567, Asp570, Leu593, and Ile598 in mouse cIAP1 were introduced to disrupt E2 enzyme binding.
To investigate the features of the RING domain required for IAP antagonist-induced degradation we reintroduced these cIAP1 RING mutants into cIAP1−/− MEFs using an inducible lentiviral system, achieving close to endogenous levels (Fig. 1B, cf. lane 2 with lanes 5, 8, 11, 14, and 17). Single mutations such as Val567 or Leu593 alone were able to partially protect cIAP1 from IAC degradation (5). Double mutations at either Val567 and Asp570 or Leu593 and Leu598 were, however, more effective at protecting cIAP1 from IAC-induced degradation, with V567A/D570A being slightly less effective than L593A/I598A (Fig. 1B, lanes 9 and 12). As previously described, the F610A mutant that disrupts dimer formation was resistant to IAP antagonist (compound A (6)) induced degradation (Fig. 1B, lane 15 (5). IAP antagonist compounds also trigger the autoubiquitylation of IAPs, which provides the signal for their degradation via the proteasomal pathway (7). Consistent with our RING mutants being resistant to IAC-induced degradation, they were also not ubiquitylated following IAC treatment (supplemental Fig. S1A, cf. lanes 4, 8, and 12). These results show that targeted mutations that inhibit either E2 binding or RING dimerization prevent IAP antagonist-induced ubiquitylation and degradation of IAPs.
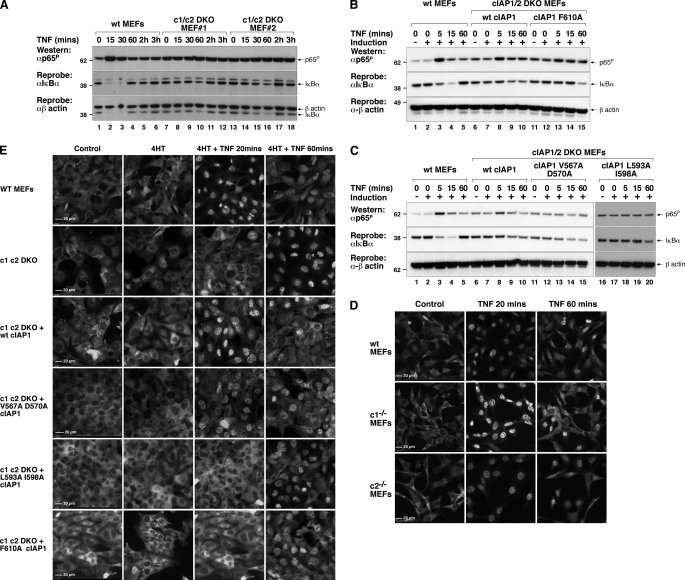
FIGURE 1.
RING mutants of cIAP1 are resistant to compound A-induced degradation. A, a schematic representation of the crystal structure of the cIAP2 RING domain (red) bound to UbcH5b (blue) overlaid on a surface representation of the structure with E2 interacting residues Val559 and Asp562 and the dimer interface residues highlighted in yellow and stick form. The equivalent cIAP1 residues are indicated in the box. B, E2 binding and dimer formation are required for IAP antagonist-induced degradation cIAP1. cIAP1−/− MEFs were immortalized with SV40 large T and infected with a lentivirus expressing inducible cIAP1 constructs. Single clones expressing wild-type or point mutants in the dimerization (F610A) or E2 binding (V567A, D570A, L593A, I598A) interface were generated. Clones were simultaneously induced with 5 nm 4HT and treated with or without 500 nm compound A (6) for 16 h. Cells were lysed in DISC lysis buffer supplemented with protease inhibitors and NEM. Lysates were separated using SDS-PAGE and blotted with a cIAP1 antibody.
TWEAK Induces Degradation of cIAP1 RING Mutants
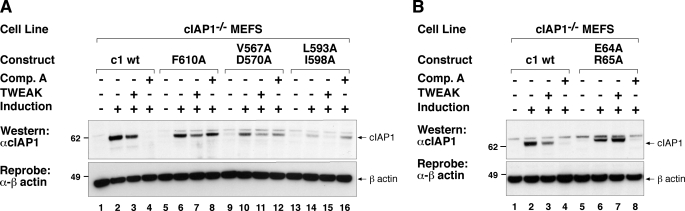
The TNFSF ligand TWEAK triggers lysosomal degradation of cIAP1 and TRAF2, whereas IAP antagonists induce proteasomal degradation of cIAP1 (4, 6, 7, 10, 15, 30). It was therefore interesting to ask whether the RING activity of cIAP1 was also required for TWEAK-induced lysosomal degradation of cIAP1. As previously reported TWEAK-induced cIAP1 degradation was less complete compared with degradation induced by our IAP antagonist compound (e.g. Fig. 2A, cf. lanes 3 and 4 (10)). IAP antagonist-treated cells were included as an internal control, and consistent with our observations in Fig. 1A our dimerization mutant (F610A) and E2 binding mutants (V567A/D570A, L593A/I598A) were unaffected by addition of compound A (Fig. 2A, cf. lanes 4, 8, 12, and 16). TWEAK, on the other hand, effectively reduced the levels of wild-type cIAP1 and that of the RING mutants (Fig. 2A, cf. lanes 3, 7, 11, and 15). This result shows, surprisingly, that neither cIAP1 E2 binding nor cIAP1 RING dimerization are required for TWEAK Fn14-mediated cIAP1 loss.
FIGURE 2.
RING mutants of cIAP1 distinguish between TWEAK- and compound A-triggered loss of cIAP1. A, inducible cIAP1−/− MEFs used in Fig. 1B were induced with 5 nm 4HT for 16 h and incubated with or without 100 ng/ml of TWEAK or 500 nm compound A. DISC lysates were separated using SDS-PAGE and analyzed by Western blot for cIAP1 levels. B, cIAP1 TRAF2 interaction is required for TWEAK-induced degradation of cIAP1. cIAP1 knock-out MEFs were reconstituted with inducible lentivirus expressing a TRAF2 interacting mutant (E64A/R65A (2)) as described in the legend to Fig. 1B. Cells were induced with 5 nm 4HT for 16 h and incubated in the presence or absence of 100 ng/ml of TWEAK or 500 nm compound A for 16 h and analyzed as in A.
Degradation of TRAF2 following TWEAK stimulation depends upon whether a soluble or membrane-bound mimic form (Fc TWEAK) of the ligand is used to stimulate cells (31). Both forms of the ligand, however, drive cIAP1 and TRAF2 into a Triton X-100-insoluble fraction (10, 31). We therefore further examined whether recruitment of TRAF2 to the insoluble fraction was dependent on cIAP1 RING dimerization. The F610A mutant also supported loss of TRAF2 from the supernatant (supplemental Fig. S2A, lane 12) and prevented accumulation in the pellet fraction (supplemental Fig. S2A, lane 16) in a similar manner to the wild-type cIAP1. This is evident even in the uninduced samples where there is, nevertheless, significant expression of cIAP1 (supplemental Fig. S2A, lanes 9 and 11, long exposure).
To further test the importance of the cIAP1-TRAF2 interaction in TWEAK signaling we reconstituted cIAP1−/− MEFs with a TRAF2 binding mutant of cIAP1, E64A/R65A. If the cIAP-TRAF2 interaction is required for TWEAK-induced degradation of cIAP1, such a mutant would be expected to be resistant to TWEAK-induced degradation but not to IAC-induced degradation. Consistent with our previous observations in TRAF2 knock-out cells (10), the TRAF2 binding mutant of cIAP1 was resistant to TWEAK-induced degradation, yet still able to undergo compound A-triggered degradation (Fig. 2B, cf. lanes 7 and 8). Together these results demonstrate that cIAPs can be degraded by two distinct processes and that only IAC-induced degradation depends on the integrity of the RING domain of cIAPs.
TNF has been reported to induce RING-mediated translocation of TRAF2 to a detergent-insoluble compartment through a Ubc13-dependent mechanism (32). Ubc13 mediates the attachment of Lys63-linked polyubiquitin chains to substrate proteins (33). We therefore tested whether Ubc13 was required for degradation of cIAP1 and TRAF2 following TWEAK and/or compound A. shRNA knockdown of Ubc13 levels by ∼70% did not, however, affect degradation of either cIAP1 or TRAF2 following TWEAK treatment (supplemental Fig. S2B). Consistent with a role for UbcH5 in compound A-mediated cIAP degradation, knock-down of Ubc13 also had no effect on the degradation of cIAP1 by compound A (supplemental Fig. S2B).
RING Functions Are Required for cIAP1 to Inhibit Activation of Non-canonical NF-κB
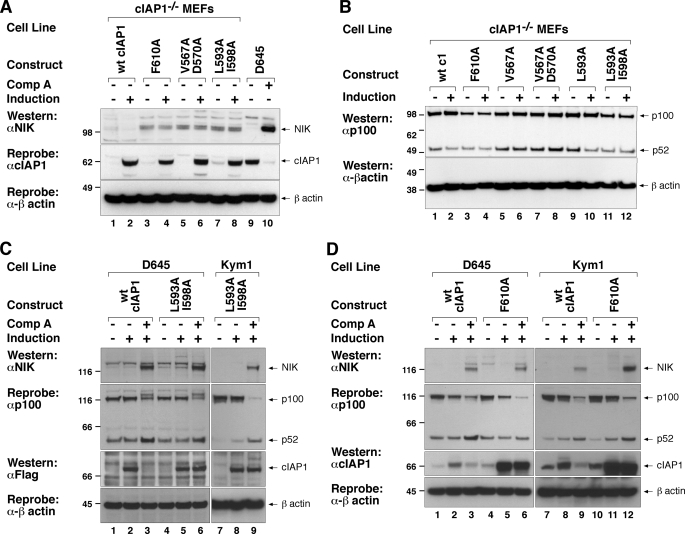
Following TWEAK stimulation or treatment with compound A, the non-canonical NF-κB pathway is activated through loss of cIAP1 and stabilization of NIK (a Lys48 ubiquitin substrate of cIAP1 (6, 7)). In cIAP1 or TRAF2 knock-out cells NIK levels are therefore constitutively high (6, 12) and can be reduced by re-introduction of wild-type cIAP1 (Fig. 3A) or TRAF2 (18), respectively. To test the importance of E2 binding and RING dimerization in this particular function of cIAP1 we therefore induced cIAP1 mutants in cIAP1−/− cells and Western blotted for NIK levels. As a positive control for detection of NIK, we treated D645 cells with compound A, which promotes cIAP1 degradation and NIK accumulation (Fig. 3A, lanes 9 and 10). cIAP1−/− MEFs have constitutively high levels of NIK that are reduced by induction of wild-type cIAP1 (Fig. 3A, lanes 1 and 2). Induction of either a dimerization or E2 binding mutant of cIAP1 had no effect on the endogenous levels of NIK (Fig. 3A, cf. lane 2 with lanes 4, 6, and 8).
FIGURE 3.
cIAP1 RING functions are required for cIAP1 to inhibit constitutive non-canonical NF-κB and for full activation following TWEAK. A, cells used in Figs. 1 and 2 were incubated with or without 5 nm 4HT for 16 h, lysed, and the lysates separated by SDS-PAGE. NIK levels were assayed by Western blot using a NIK antibody. B, RING mutants of cIAP1 cannot prevent the constitutive processing of p100 to p52 observed in cIAP1−/− MEFs. Cells were induced with 5 nm 4HT for 72 h, lysed in DISC lysis buffer, and analyzed by Western blotting using a p100 antibody that recognizes both p100 and p52. C, D645 and Kym1 cells were infected with a lentivirus expressing inducible mouse wild-type cIAP1 or a construct containing point mutations in the E2 binding interface (L593A/I598A). Polyclonal populations were induced with 10 nm 4HT for 16 h and then treated with or without 500 nm compound A for 4 h. Cells were lysed in DISC lysis buffer supplemented with protease inhibitors and NEM. Lysates were separated using SDS-PAGE and analyzed by Western for NIK and p100/p52 and cIAP1. D, D645 and Kym1 cells were infected with a lentivirus expressing inducible mouse wild-type cIAP1 or a construct containing point mutations in the dimerization interface (F610A). Polyclonal populations were induced with 10 nm 4HT for 16 h and then treated with or without 500 nm compound A for 4 h. Cells were lysed in DISC lysis buffer supplemented with protease inhibitors and NEM. Lysates were separated using SDS-PAGE and analyzed by Western blots for NIK, p100/p52, and cIAP1.
Stabilization of NIK leads to phosphorylation and proteasomal processing of NF-κB2 p100 to the p52 subunit resulting in translocation of the p52-RelB heterodimer to the nucleus. To further investigate the ability of cIAP1 mutants to regulate non-canonical NF-κB we assayed for the processing of p100 to p52. Induction of wild-type cIAP1 for 72 h reduced the levels of p52 and increased p100 levels (Fig. 3B, cf. lanes 1 and 2). The weak E2 binding mutant (L593A) was also able to reset the p100-p52 ratio in a similar way to wild-type cIAP1, but neither the dimer mutant nor the other E2 binding mutants of cIAP1 were effective in resetting the p100:p52 ratio (Fig. 3B, lanes 4, 6, 8, and 12). Taken together, these results indicate that the basal activation of the non-canonical NF-κB pathway requires cIAP1 RING function.
These results demonstrate that RING mutants expressed in cIAP1−/−cIAP2−/− MEFs cannot regulate the non-canonical NF-κB pathway. We therefore wondered whether they might inhibit wild-type cIAP function. To address this question, and extend our observations to other cell types, we inducibly expressed cIAP1 and mutants thereof in Kym1 and D645 cell lines. Both D645 and Kym1 cells express detectable levels of endogenous cIAP1 and therefore NIK levels are constitutively low. Induction of wild-type cIAP1 or any of our RING mutants in D645 and Kym1 cells had no discernible effect on the regulation of NIK by endogenous cIAP1 or the basal processing of NF-κB2 to p52 (E2 mutants, Fig. 3C, lanes 2, 5, and 8; dimer mutants, Fig. 3D, lanes 5 and 11). This indicates that overexpressed RING mutants did not act as strong dominant negatives to inhibit wild-type cIAP1 function over the time course of this assay. We further tested our mutants by depleting endogenous cIAP1 with an IAC. As described previously, IAC treatment of Kym1 and D645 cells resulted in degradation of wild-type cIAP1, NIK levels rose and p100 was processed to p52 (6) (Fig. 3, C and D). Consistent with our observations in MEFs, none of our RING mutants were able to prevent the activation of non-canonical NF-κB following IAC treatment, further demonstrating their lack of activity in different experimental settings.
TWEAK Further Increases NF-κB Activity in cIAP DKO and TRAF2 KO Cells
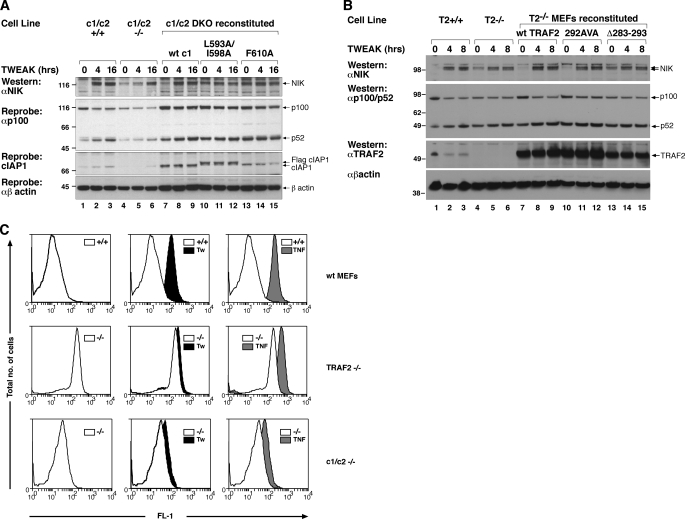
It has been proposed that several members of the TNFSF, including TWEAK, have the ability to activate non-canonical NF-κB simply by promoting degradation of TRAF2 and cIAP1 (10, 34). This model of TWEAK signaling would therefore predict that TWEAK would be unable to promote further activation of non-canonical NF-κB in either TRAF2 or cIAP1 and -2 double knock-out cells. To test this prediction we incubated cIAP1−/−cIAP2−/− MEFs with TWEAK and monitored NIK levels and p100 processing at 4 and 16 h post-TWEAK stimulation. Surprisingly, NIK levels increased, as did p100 processing, even in cIAP DKO MEFs, although this was clearly less than in wild-type MEFs and also delayed. Reconstitution of the DKO MEFs with wild-type cIAP1 reduced the basal level of NIK and rescued the ability of the knock-out cells to respond to TWEAK at the early 4-h time point (Fig. 4A, lanes 7–9). Neither the E2 binding mutant nor the dimer mutant were able to rescue and, if anything, even responded less to TWEAK than the DKO MEFs (Fig. 4A, lanes 10–15).
FIGURE 4.
TWEAK further increases NF-κB activity in cIAP DKO and TRAF2 KO cells. A, cIAP1 and cIAP2 DKO MEFs were immortalized with SV40 large T and reconstituted with inducible cIAP1 constructs as previously described for cIAP1−/− cells. Cells were induced for 72 h with 10 nm 4HT and treated with or without 100 ng/ml of TWEAK for the indicated times. Cells lysates were analyzed as described in the legend to Fig. 3, A and B. B, binding to TRAF2 is required for cIAP1 to inhibit non-canonical NF-κB activation. TRAF2−/− MEFs were immortalized with SV40 large T and infected with inducible TRAF2 constructs that contained either point mutations or a deletion in the CIM (13). Cells were induced for 72 h with 5 nm 4HT and treated with or without 100 ng/ml of TWEAK for the indicated times. Cell lysates were analyzed by Western blot for p100, p52, NIK, and TRAF2 levels. C, cIAP DKO MEFs and TRAF2−/− MEFs constitutively activate NF-κB that can be further increased by addition of TWEAK. Wild-type MEFs, cIAP DKO MEFs, or TRAF2 KO MEFs immortalized with SV40 large T containing lentivirus were infected with a lentivirus containing a enhanced green fluorescent protein NF-κB reporter. Cells were treated with 100 ng/ml of TWEAK or TNF for 16 h. Cells were analyzed by flow cytometry for fluorescence in the FL-1 (green) channel.
To further explore the finding of increased NIK stabilization and p100 to p52 processing in cIAP DKO MEFs we looked at TWEAK signaling in TRAF2 knock-out MEFs. We recently identified a cIAP interacting motif (CIM) in TRAF2 that is required for cIAPs to interact with TRAF2. In particular, deletion of amino acids 283–293 from TRAF2 prevented the binding of cIAP to TRAF2 and point mutation of both Glu292 and Glu294 showed reduced TRAF2-cIAP interaction (18). Consistent with the cIAP DKO MEFs, TRAF2−/− MEFs were also still able to respond to TWEAK by elevating NIK levels and increasing processing of p100, albeit significantly less well than wild-type cells. Reconstitution of the knock-out MEFs with wild-type TRAF2 restored signaling similar to wild-type cells, as did reconstitution with the weak cIAP binding mutant (Fig. 4B, cf. lanes 1–3, 7–9, and 10–12). TRAF2−/− MEFs reconstituted with the TRAF2 CIM mutant, however, behaved almost identically to the knock-out cells (Fig. 4B, cf. lanes 4–6 and 13–15). To further analyze the increase following TWEAK, we infected the cIAP DKO and TRAF2−/− cells with an NF-κB-GFP reporter. Both the cIAP DKO and TRAF2−/− cells displayed constitutively high basal NF-κB activity compared with the wild-type MEFS (Fig. 4C). A clear shift in GFP fluorescence can be seen following TWEAK (black) or TNF (gray) stimulation in wild-type MEFs, with a slightly larger shift post-TNF treatment. TNF is a strong activator of canonical NF-κB and this most likely explains the different response compared with the weaker shift seen with TWEAK. Consistent with the Western results, both cIAP DKO and TRAF2−/− MEFs showed an increase in NF-κB activity, above the high basal activity, following TWEAK or TNF stimulation (Fig. 4C). This assay is, however, unable to distinguish between canonical and non-canonical NF-κB activity, and although it is consistent with the Western analysis must be interpreted carefully. Taken together these results demonstrated that although cIAP and TRAF2 are important regulators of NF-κB, TWEAK can enhance the non-canonical pathway in cIAP and TRAF2 independent manners.
RING Mutants of cIAP1 Severely Impair the Activation of Canonical NF-κB following TNF Treatment
TNF is a well studied activator of canonical NF-κB. TNF binding to TNF-R1 in MEFs results in p65-RelA phosphorylation within 5 min and almost complete IκBα degradation within 15 min, thereby allowing p50-p65 translocation into the nucleus (35). Loss of IκBα is only transient because IκBα is a transcriptional target of NF-κB, and its levels are restored within 60 min of TNF addition. cIAPs are thought to be important regulators of this pathway, because loss of both cIAP1 and cIAP2 inhibits TNF-induced activation of NF-κB (14, 16). Using cIAP1 and cIAP2 DKO MEFs reconstituted with inducible cIAP1 mutants we therefore decided to test the contribution of the RING domain to TNF activation of NF-κB. Consistent with previous reports that used different methods to deplete both cIAPs (14, 16), TNF-induced degradation of IκBα was significantly reduced and delayed in two independent cIAP1 and cIAP2 DKO MEF lines compared with wild-type MEFs (Fig. 5A, cf. lanes 1–6 with lanes 7–12, and lanes 13–18 (14, 16)). It is noteworthy that TNF-induced IκBα degradation still occurs in cIAP DKO MEFs, so activation of NF-κB is not abolished, but rather reduced and delayed, and that both cIAP DKO MEF lines had high basal levels of phosphorylated p65. Wild-type cIAP1 was able to partially restore IκBα degradation in response to TNF in cIAP DKO MEFs, with degradation occurring within 15 min and not rebounding within 60 min as in wild-type MEFs (Fig. 5B, compare lanes 1–5 with lanes 6–10). Wild-type cIAP1 also reduced basal p65 phosphorylation and allowed TNF to promote increased p65 phosphorylation in these cells. The dimerization mutant was ineffective at promoting IκBα degradation, and degradation of IκBα occurred with similar kinetics to the parental DKO MEF line (Fig. 5B, lanes 11-15). Similarly, reconstitution of cIAP1 E2 binding mutants into cIAP DKO MEFs resulted in partial restoration of IκBα degradation with the weak E2 binding mutant (Fig. 5C, lanes 11–15) and no restoration with the strong E2 mutant (Fig. 5C, lanes 16–20). Furthermore, expression of our strong E2 binding mutant in D645 cells did not interfere with TNF-induced IκBα degradation, measured at 15 and 30 min (supplemental Fig. 3B, cf. lanes 3 and 4 and 7 and 8). This lack of activity is consistent with the inability of our RING mutants to change the kinetics of IκBα degradation induced by TNF in cIAP DKO MEFs.
FIGURE 5.
RING mutants of cIAP1 inhibit TNF-induced canonical NF-κB activation. A, wild-type MEFs and MEFs derived from two cIAP1/2 double knock-out embryos immortalized with SV40 large T antigen were incubated with or without 100 ng/ml of Fc-TNF for the indicated times. Lysates were made and separated using SDS-PAGE and blotted for phosphorylated p65 and IκBα. B, a RING dimer mutant of cIAP1 is unable to restore normal TNF-induced p65 phosphorylation and IκBα degradation in cIAP1 cIAP2 DKO MEFs. Polyclonal populations described in the legend to Fig. 3 were incubated with or without 5 nm 4HT for 16 h prior to incubation with 100 ng/ml of Fc-TNF for the indicated times. C, E2-binding mutants of cIAP1 are unable to restore normal TNF-induced p65 phosphorylation and IκBα degradation in cIAP1 cIAP2 DKO MEFs. Polyclonal populations described in the legend to Fig. 3 were incubated with or without 5 nm 4HT for 16 h prior to incubation with 100 ng/ml of Fc-TNF for the indicated times. D, p65 translocates normally in cIAP1−/− and cIAP2−/− MEFs following TNF. Wild-type, cIAP1−/−, or cIAP2−/− MEFs transformed with SV40T were grown on coverslips overnight and treated with TNF for the indicated times. Cells were fixed immediately and stained with p65 antibody as described under ”Experimental Procedures“ and viewed under a ×40 fluorescence microscope objective. E, RING functions of cIAP1 are critical for normal TNF-induced translocation of p65 to the nucleus. Cells used in Fig. 4, B and C, were grown on coverslips overnight and treated as in D.
Differences in the protein level in Western blots, even if performed multiple times, are not completely reliable measures of signaling responses. Therefore to further examine the effect of wild-type cIAP1 and cIAP1 mutants on TNF-induced NF-κB we looked at p65 translocation to the nucleus by immunofluorescence microscopy. p65 translocates to the nucleus within 20 min after addition of TNF and begins to exit by 60 min in wild-type MEFs, and neither translocation is affected by single deletion of either cIAP1 or cIAP2 (Fig. 5D). We considered the possibility that the RING mutants reintroduced into the single cIAP1−/− MEFs might function as dominant negatives that could interfere with the activity of the endogenous cIAP2. However, these mutants did not affect p65 translocation in cIAP1−/− cells (supplemental Fig. 3A). Therefore we restricted our analysis to cIAP DKO cells that show a marked delay in p65 translocation. Consistent with the Western analysis (Fig. 5, A–C), deletion of both cIAP1 and cIAP2 significantly delayed translocation of p65 to the nucleus, which occurred at 60 min instead of the usual 20 min (Fig. 5E). Reconstitution of cIAP1 and cIAP2 DKO MEFs with wild-type cIAP1 restored normal p65 nuclear translocation following TNF treatment (Fig. 5E), whereas reconstitution with a cIAP1 mutant that could no longer form RING dimers or with mutants that are defective in recruiting E2 did not rescue the DKO phenotype except in the case of the weaker, V567A/D570A E2 binding mutant (Fig. 5E). Therefore, both dimerization and E2 binding are essential for normal canonical NF-κB activation following TNF stimulation.
Dimerization and E2 Recruitment Are Required for cIAP1 to Protect MEFs from TNF-induced Cell Death
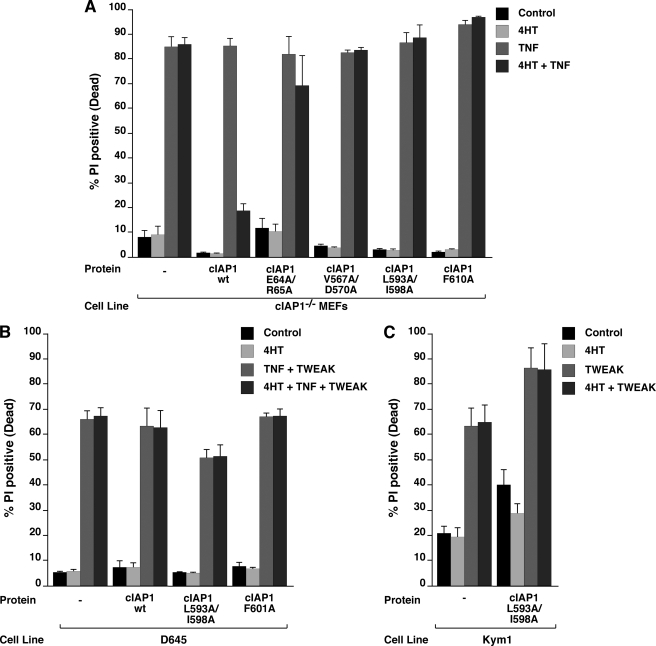
MEFs depleted of cIAP1 by knock-out or IAC are sensitive to exogenous TNF, and re-introduction of wild-type cIAP1 is able to provide substantial protection (6). The mechanism for cIAP1-mediated protection against TNF is not completely clear, particularly if the fact that p65 translocation occurs normally in these cells, is taken into account. To try and dissect the functions of cIAP1 that might be required to protect cells from TNF we tested TRAF2 binding, the dimer and E2 binding mutants for their ability to protect against TNF. Consistent with our previous results (6) addition of TNF to cIAP1−/− cells resulted in ∼80% cell death and this high percentage of cell death was reduced to ∼20% upon induction of wild-type cIAP1 (Fig. 6A). None of the mutants tested were capable of providing protection against TNF. Together, these results demonstrate that cIAP1 RING functions are critical for protection from TNF killing.
FIGURE 6.
cIAP1-mediated protection against TNF-induced apoptosis in MEFs is dependent on dimerization and E2 association. A, cells from Fig. 1B were left induced or un-induced with 5 nm 4HT for 72 h and treated with 100 ng/ml of TNF for 24 h. Cells were stained with propidium iodide (PI) and analyzed by flow cytometry. B, cells from Fig. 3, C and D, were induced with 10 nm 4HT for 24 h and treated with 100 ng/ml of TWEAK or TNF for 24 h. Cells were stained with PI and analyzed by flow cytometry. C, cells were analyzed as in B. Error bars represent S.E., n = 3.
cIAP1 RING dimerization and E2 binding are, however, not required for loss of cIAP1 following TWEAK. To analyze involvement of the cIAP1 RING domain in TWEAK or TWEAK/TNF-mediated cell death, we tested whether our mutants interfered with TWEAK signaling in cells with endogenous wild-type cIAP expression. Neither the dimerization nor the E2 binding mutants interfered with TWEAK/TNF-induced cell death in D645 cells or TWEAK-triggered apoptosis in Kym1 cells (Fig. 6, B and C). However, because TWEAK-induced death requires the production of autocrine TNF (6, 36) it is difficult to distinguish between the effects of cIAPs at the TNF and TWEAK signaling receptors in TWEAK-mediated cell death. In conclusion, although the RING domain of cIAP1 is required to protect cells from the cytotoxic actions of TNF, the cIAP1 RING is, however, not required for cell death induced by the TNFSF ligand TWEAK.
DISCUSSION
cIAP1 and its less well characterized paralog cIAP2 are important E3 ligases whose ubiquitylation activities are still incompletely understood. Our recent experiments have focused on the function of cIAP1 and show that it is important in regulating both TWEAK-Fn14 and TNF-TNF-R1 signaling (6, 10). Using the cIAP2 RING UbcH5 crystal structure we designed point mutations in the predicted RING dimer and E2 binding interfaces of cIAP1 (5) and tested the ability of these mutants to rescue signaling in cIAP1−/− and cIAP1−/−cIAP2−/− (DKO) MEFs.
IAP antagonist compounds target cIAP1, cIAP2, and XIAP are presumed to activate the E3 ligase activity of the cIAPs. As predicted from this model, using the experimental system described above, we show that loss of cIAP1 E2 binding prevents IAP antagonist-induced ubiquitylation and degradation. Although a precise function for RING dimerization has not been discovered a RING dimer mutant of cIAP1 is also protected from IAP antagonist-induced degradation, as previously reported (18).
Several groups have shown that treatment of cells with IACs or small interfering RNAs that deplete cIAPs prevents TNF-induced RIPK1 modification (14–17), suggesting that cIAPs are the E3 ligases that ubiquitylate RIPK1. Consistent with the hypothesis that cIAPs are key E3 ligases for RIPK1, cIAP DKO MEFs do not ubiquitylate RIPK1 following TNF treatment (17). Our cIAP1 F610A RING dimer mutant was also incapable of rescuing RIPK1 ubiquitylation following TNF signaling in cIAP1−/−cIAP2−/− MEFs (17) supporting the idea that this mutant affects not only autoubiquitylation, but also substrate ubiquitylation.
Using these mutants we then tested their ability to regulate TWEAK and TNF signaling. Following TWEAK stimulation, TRAF2 and cIAP1 are recruited to Fn14 and then relocate to a detergent-insoluble compartment (10). Translocation probably precedes degradation in the lysosome because translocation, but not degradation, occurs in some cell lines if a soluble form of TWEAK is used (31). Lysosomal degradation and translocation may also be dependent upon A20 because TWEAK-induced TRAF2 degradation was abolished in A20−/− cells (11). Other reports have also observed nonproteasomal degradation of TRAF2 and cIAP1 downstream of TNFSF receptors such as TNF-R2 and CD30 (37, 38). Such results suggest that the requirements for cIAP RING function might be different between IAP antagonist-induced degradation and in TWEAK signaling. However, we were surprised to find that neither E2 binding nor RING dimer formation were required for TWEAK-induced cIAP and TRAF2 translocation. Consistent with our previous results in TRAF2−/− cells (18) a cIAP1 mutant that cannot bind TRAF2 is not degraded following TWEAK stimulation. These results strongly support a model for TWEAK signaling that requires cIAP recruitment by TRAF2 to Fn14 but that does not require cIAP RING function for TRAF2 and cIAP degradation. We considered the possibility that TRAF2 functions via the Lys63 E2, Ubc13, to promote cIAP1 and TRAF2 degradation, but knock-down of UBC13 had no discernible affect on this process. These results are in line with a recent paper (39) that demonstrates that TRAF2 is unable to interact with UBC13. Indirectly our data also provides support for our earlier finding and other more recent ones that TRAF2 and cIAP1 degradation following TWEAK signaling does not occur via a proteasomal pathway (10, 11). Several groups have shown that signaling through TNFSF receptors such as CD40, TNF-R2, and BAFFR induce cIAP-dependent proteasome degradation of TRAF2 (9, 40, 41) and/or TRAF3 (9, 41). The differences between these results might reflect either the different ligand/receptor systems, or even the type of ligand used (34). We have used a mimic of the membrane-bound form of TWEAK, Fc-TWEAK, and it is known that soluble and membrane-bound forms of TNFSF ligands have different signaling capabilities (31, 42). The key issue to address with TWEAK will therefore be which form of ligand is used in which physiological signaling situation.
TWEAK is a strong activator of non-canonical NF-κB (43). IAP antagonist compounds also activate the non-canonical NF-κB pathway by depleting IAPs, leading to stabilization of NIK and processing of NF-κB2 to p52 (6–9). Furthermore, several recent reports have shown that loss of cIAPs, TRAF2, or TRAF3 is sufficient to result in constitutive activation of non-canonical NF-κB (6–9, 12, 44–46). These results have led to the proposal that cIAPs, TRAF2, and TRAF3 together form a complex that targets NIK to the proteasome and that cIAPs are the Lys48 E3 ligases that ubiquitylate NIK to induce its degradation (7–9, 16). Our data are consistent with this model because cIAP1−/−cIAP2−/− MEFs have high basal levels of NIK that are reduced if these cells are reconstituted with wild-type cIAP1 but not a RING dimer or E2 binding mutant. The RING-dependent regulation of NIK also seems to be conserved across cell types, as both D645 and Kym1 cell lines exhibited the same RING-dependent regulation of NIK by cIAP1.
Based on this model of cIAP-TRAF2-TRAF3 regulation of NIK, we have proposed that TWEAK activates non-canonical NF-κB simply by promoting the translocation and degradation of TRAF2 and cIAPs thereby allowing NIK levels to accumulate (10). If this proposal is correct, however, TWEAK would be unable to promote enhanced non-canonical activation in cIAP DKO cells or TRAF2 KO cells. However, whereas cIAP and TRAF2 KO cells were clearly significantly impaired in their ability to activate non-canonical NF-κB above the increased basal levels in these knock-out cells, some further stabilization of NIK occurred following TWEAK stimulation. This shows that NIK levels are not solely regulated by cIAPs and TRAF2 and that other mechanisms exist to regulate NIK levels. This cIAP-TRAF2 independent pathway can be regulated by TWEAK/FN14 signaling. It is possible that as these cells were generated from cells that lost cIAPs or TRAF2 during embryonic development that some adaptation has occurred, and the extent to which this alternative pathway of regulating NIK levels exists in normal cells is therefore still an open question.
There is also substantial data showing that cIAPs regulate TNF-induced activation of the canonical NF-κB pathway. For example, reduction in the levels of both cIAP1 and cIAP2 was shown to prevent TNF-induced IκBα degradation (14, 16). Loss of cIAPs induced by IAP antagonist treatment also severely impairs RIPK1 ubiquitylation. Thus cIAPs might contribute to TNF-induced NF-κB by providing a platform for recruitment of TAK1 and IKK complexes. Consistent with this idea, we previously published (18) that a TRAF2 CIM mutant that cannot bind cIAPs could not restore normal NF-κB or ubiquitylation of RIPK1 in response to TNF in TRAF2 and TRAF5 DKO MEFs (18). In this work we confirmed and extended these earlier results by demonstrating that cIAP1 and cIAP2 DKO MEFs display delayed and reduced IκBα degradation compared with wild-type cells in response to TNF as well as a significant delay in p65 translocation to the nucleus. This is consistent with ours and others (18, 47) recently published data demonstrating that TRAF2 and TRAF5 DKO MEFs have impaired canonical NF-κB activity in response to TNF. Possibly, in the absence of the major E3 ligases a backup signaling mechanism is initiated. This sort of flexibility seems inherent to the TNF-R1 signaling complex because, in addition to the data presented here it has recently been shown that neither TRADD nor RIPK1 are required for TNF-induced NF-κB (13, 48, 49).
Reintroduction of wild-type cIAP1, but not dimerization or E2 binding mutants, restored the normal NF-κB response in cIAP DKO MEFs compared with wild-type MEFs. Therefore consistent with previous observations we show that cIAP1 is a critical regulator of TNF-induced NF-κB, and that RING dimerization and E2 binding are required to control this activation.
cIAP1, in addition to regulating TNF-induced NF-κB, also provides protection that is independent of its NF-κB function. This protective signal is dependent on cIAP1 RING dimerization and E2 recruitment, and it is possible therefore that the protective signal involves limiting the death inducing activity of RIPK1 that has recently been demonstrated (50–52).
In summary, by reconstituting cIAP1 knock-out and cIAP1 and cIAP2 double knock-out cells with point mutants of cIAP1 we have tested the requirements for E2 binding and RING dimer formation in several cIAP1-regulated signaling pathways. Our data provide further support for models proposing that the E3 ligase function of cIAP1 is required to limit basal activation of NF-κB2 by promoting proteasomal degradation of NIK. However, data also demonstrates that other cIAP-TRAF2-independent pathways exist to regulate NIK levels and that TWEAK can increase NIK levels even in cIAP1 and cIAP2 double knock-out cells. Our data supports the proposal that the E3 ligase activity of cIAPs is important in regulating TNF-induced NF-κB, but also show that TNF can induce a delayed NF-κB response even in cIAP1 and cIAP2 double knock-out cells. Surprisingly we find that both E2 binding and RING dimerization are dispensable in proximal TWEAK signaling events. These data therefore provide further support for the proposal that TWEAK induces a non-proteasomal degradation of TRAF2 and cIAP1. It will now be interesting to see how this pathway is regulated, how prevalent this signaling mechanism is in TNFSF signaling, and finally how the form of ligand affects these proximal signaling events.
Supplementary Material
Acknowledgments
We thank Dr. M. A. Kelliher for the Ubc13 shRNA and GFP shRNA lentiviral vectors, TetraLogic Pharmaceuticals for the gift of compound A, and Hiroyasu Nakano for the gift of TRAF2−/−TRAF5−/− cells.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- cIAP
- cellular inhibitor of apoptosis
- IAP
- inhibitor of apoptosis
- NF-κB
- nuclear factor κ light chain enhancer of activated B cells
- TNF
- tumor necrosis factor
- TNF-R1
- tumor necrosis factor receptor 1
- TRAF
- TNF receptor-associated factor
- RIPK1
- receptor interacting protein kinase 1
- TNFSF
- TNF superfamily
- IκBα
- inhibitor NF-κB
- IKK
- IκB kinase
- RING
- really interesting new gene
- TAK1
- transforming growth factor β and 1113088 activated kinase-1
- TAB
- TAK1-binding protein
- NEMO
- NF-κB essential modulator
- NEM
- N-ethylmaleimide
- CIM
- cIAP interacting motif
- IAC
- IAP antagonist compound
- NIK
- NF-κB inducing kinase
- MEF
- mouse embryonic fibroblast
- 4HT
- 4-hydroxytamoxifen
- shRNA
- short hairpin RNA
- GFP
- green fluorecent protein
- DKO
- double knock-out
- PBS
- phosphate-buffered saline
- TWEAK
- TNF-like weak inducer of apoptosis
- UAS
- upstream activation sequence.
REFERENCES
- 1.Rothe M., Pan M., Henzel W., Ayres T., Goeddel D. (1995) Cell 83, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 2.Samuel T., Welsh K., Lober T., Togo S. H., Zapata J. M., Reed J. C. (2006) J. Biol. Chem. 281, 1080–1090 [DOI] [PubMed] [Google Scholar]
- 3.Varfolomeev E., Wayson S. M., Dixit V. M., Fairbrother W. J., Vucic D. (2006) J. Biol. Chem. 281, 29022–29029 [DOI] [PubMed] [Google Scholar]
- 4.Gaither A., Porter D., Yao Y., Borawski J., Yang G., Donovan J., Sage D., Slisz J., Tran M., Straub C., Ramsey T., Iourgenko V., Huang A., Chen Y., Schlegel R., Labow M., Fawell S., Sellers W. R., Zawel L. (2007) Cancer Res. 67, 11493–11498 [DOI] [PubMed] [Google Scholar]
- 5.Mace P. D., Linke K., Feltham R., Schumacher F. R., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) J. Biol. Chem. 283, 31633–31640 [DOI] [PubMed] [Google Scholar]
- 6.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 7.Varfolomeev E., Blankenship J., Wayson S., Fedorova A., Kayagaki N., Garg P., Zobel K., Dynek J., Elliott L., Wallweber H., Flygare J., Fairbrother W., Deshayes K., Dixit V., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 8.Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P. H., Keats J. J., Wang H., Vignali D. A., Bergsagel P. L., Karin M. (2008) Nat. Immunol. 9, 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., Soetandyo N., Wang Q., Ye Y. (2009) Biochim. Biophys. Acta 1793, 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grech A. P., Amesbury M., Chan T., Gardam S., Basten A., Brink R. (2004) Immunity 21, 629–642 [DOI] [PubMed] [Google Scholar]
- 13.Wong W. W., Gentle I. E., Nachbur U., Anderton H., Vaux D. L., Silke J. (2010) Cell Death Differ. 17, 482–487 [DOI] [PubMed] [Google Scholar]
- 14.Mahoney D. J., Cheung H. H., Mrad R. L., Plenchette S., Simard C., Enwere E., Arora V., Mak T. W., Lacasse E. C., Waring J., Korneluk R. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 16.Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., Walczak H. (2009) Mol. Cell 36, 831–844 [DOI] [PubMed] [Google Scholar]
- 18.Vince J. E., Pantaki D., Feltham R., Mace P. D., Cordier S. M., Schmukle A. C., Davidson A. J., Callus B. A., Wong W. W., Gentle I. E., Carter H., Lee E. F., Walczak H., Day C. L., Vaux D. L., Silke J. (2009) J. Biol. Chem. 284, 35906–35915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shembade N., Parvatiyar K., Harhaj N. S., Harhaj E. W. (2009) EMBO J. 28, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 21.Bellon S. F., Rodgers K. K., Schatz D. G., Coleman J. E., Steitz T. A. (1997) Nat. Struct. Biol. 4, 586–591 [DOI] [PubMed] [Google Scholar]
- 22.Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 23.Cheung H. H., Plenchette S., Kern C. J., Mahoney D. J., Korneluk R. G. (2008) Mol. Biol. Cell 19, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallery D. L., Vandenberg C. J., Hiom K. (2002) EMBO J. 21, 6755–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia Y., Pao G. M., Chen H. W., Verma I. M., Hunter T. (2003) J. Biol. Chem. 278, 5255–5263 [DOI] [PubMed] [Google Scholar]
- 27.Mace P. D., Shirley S., Day C. L. (2009) Cell Death Differ. 17, 46–53 [DOI] [PubMed] [Google Scholar]
- 28.Yang Q. H., Du C. (2004) J. Biol. Chem. 279, 16963–16970 [DOI] [PubMed] [Google Scholar]
- 29.Hu S., Yang X. (2003) J. Biol. Chem. 278, 10055–10060 [DOI] [PubMed] [Google Scholar]
- 30.Wang L., Du F., Wang X. (2008) Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- 31.Wicovsky A., Salzmann S., Roos C., Ehrenschwender M., Rosenthal T., Siegmund D., Henkler F., Gohlke F., Kneitz C., Wajant H. (2009) Cell Death Differ. 16, 1445–1459 [DOI] [PubMed] [Google Scholar]
- 32.Habelhah H., Takahashi S., Cho S. G., Kadoya T., Watanabe T., Ronai Z. (2004) EMBO J. 23, 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann R. M., Pickart C. M. (1999) Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 34.Silke J., Brink R. (2009) Cell Death Differ. 17, 35–45 [DOI] [PubMed] [Google Scholar]
- 35.Inoue J., Kerr L. D., Kakizuka A., Verma I. M. (1992) Cell 68, 1109–1120 [DOI] [PubMed] [Google Scholar]
- 36.Schneider P., Schwenzer R., Haas E., Mühlenbeck F., Schubert G., Scheurich P., Tschopp J., Wajant H. (1999) Eur. J. Immunol. 29, 1785–1792 [DOI] [PubMed] [Google Scholar]
- 37.Duckett C. S., Thompson C. B. (1997) Genes Dev. 11, 2810–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fotin-Mleczek M., Henkler F., Samel D., Reichwein M., Hausser A., Parmryd I., Scheurich P., Schmid J. A., Wajant H. (2002) J. Cell Sci. 115, 2757–2770 [DOI] [PubMed] [Google Scholar]
- 39.Yin Q., Lamothe B., Darnay B. G., Wu H. (2009) Biochemistry 48, 10558–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Yang Y., Ashwell J. D. (2002) Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 41.Matsuzawa A., Tseng P. H., Vallabhapurapu S., Luo J. L., Zhang W., Wang H., Vignali D. A., Gallagher E., Karin M. (2008) Science 321, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baccam M., Bishop G. A. (1999) Eur. J. Immunol. 29, 3855–3866 [DOI] [PubMed] [Google Scholar]
- 43.Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. (2003) J. Biol. Chem. 278, 36005–36012 [DOI] [PubMed] [Google Scholar]
- 44.Keats J. J., Fonseca R., Chesi M., Schop R., Baker A., Chng W. J., Van Wier S., Tiedemann R., Shi C. X., Sebag M., Braggio E., Henry T., Zhu Y. X., Fogle H., Price-Troska T., Ahmann G., Mancini C., Brents L. A., Kumar S., Greipp P., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M., Valdez R., Trent J., Stewart A. K., Carpten J., Bergsagel P. L. (2007) Cancer Cell 12, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardam S., Sierro F., Basten A., Mackay F., Brink R. (2008) Immunity 28, 391–401 [DOI] [PubMed] [Google Scholar]
- 46.Annunziata C. M., Davis R. E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., Dave S., Hurt E. M., Tan B., Zhao H., Stephens O., Santra M., Williams D. R., Dang L., Barlogie B., Shaughnessy J. D., Jr., Kuehl W. M., Staudt L. M. (2007) Cancer Cell 12, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Blackwell K., Thomas G. S., Sun S., Yeh W. C., Habelhah H. (2009) J. Mol. Biol. 389, 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pobezinskaya Y. L., Kim Y. S., Choksi S., Morgan M. J., Li T., Liu C., Liu Z. (2008) Nat. Immunol. 9, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ermolaeva M. A., Michallet M. C., Papadopoulou N., Utermöhlen O., Kranidioti K., Kollias G., Tschopp J., Pasparakis M. (2008) Nat. Immunol. 9, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 50.Cho Y. S., Challa S., Moquin D., Genga R., Ray T. D., Guildford M., Chan F. K. (2009) Cell 137, 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He S., Wang L., Miao L., Wang T., Du F., Zhao L., Wang X. (2009) Cell 137, 1100–1111 [DOI] [PubMed] [Google Scholar]
- 52.Zhang D. W., Shao J., Lin J., Zhang N., Lu B. J., Lin S. C., Dong M. Q., Han J. (2009) Science 325, 332–336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.