Abstract
When unfolded proteins accumulate in the endoplasmic reticulum (ER) causing ER stress, the unfolded protein response (UPR) responds rapidly to induce a transcriptional program that functions to alleviate the stress. However, under extreme conditions, when UPR activation is not sufficient to alleviate ER stress, the stress may persist long term. Very little is known about how the cell responds to persistent ER stress that is not resolved by the immediate activation of the UPR. We show that Hog1 MAP kinase becomes phosphorylated during the late stage of ER stress and helps the ER regain homeostasis. Although Hog1 is well known to function in osmotic stress and cell wall integrity pathways, we show that the activation mechanism for Hog1 during ER stress is distinct from both of these pathways. During late stage ER stress, upon phosphorylation, Hog1 translocates into the nucleus and regulates gene expression. Subsequently, Hog1 returns to the cytoplasm, where its phosphorylation levels remain high. From its cytoplasmic location, Hog1 contributes to the activation of autophagy by enhancing the stability of Atg8, a critical autophagy protein. Thus, Hog1 coordinates a multifaceted response to persistent ER stress.
Keywords: Autophagy, Endoplasmic Reticulum (ER), ER Stress, MAP Kinases (MAPKs), Yeast, Atg8, HSP12, Hog1, S. cerevisiae, Unfolded Protein Response (UPR)
Introduction
In eukaryotic cells, plasma membrane proteins, proteins that are secreted, and proteins that reside within the secretory pathway all begin their maturation within the endoplasmic reticulum (ER).3 The ER contains chaperones, glycosylation enzymes, and other protein-folding enzymes, and therefore provides an ideal environment for the folding of nascent proteins. Secretory proteins leave the ER for their targeted locations only after they have achieved their appropriate conformation, making the ER a gateway to the secretory pathway. In response to environmental or developmental changes, the demand for protein folding can increase dramatically, and overwhelm the protein-folding machinery of the ER, thus leading to an accumulation of unfolded proteins within the ER. This condition, known as ER stress, is deleterious to cells and has been implicated in many human pathologies, including Alzheimer, Parkinson, and Huntington diseases (1). Therefore, when ER stress is detected, cells react in multiple coordinated manners to alleviate the stress.
At the center of this stress response is the unfolded protein response (UPR), a conserved pathway that activates a transcriptional program, which helps expand the protein-folding capacity of the ER (2–5). In budding yeast the UPR is initiated when Ire1, an ER transmembrane protein, detects stress within the ER lumen and activates its own cytoplasmic ribonuclease domain (6). The ribonuclease removes a UPR-specific intron from HAC1 mRNA, thus initiating its splicing (7). HAC1 encodes a transcription factor that activates UPR target genes (8), but only the spliced form of HAC1 can be translated (9, 10). Thus, IRE1-dependent HAC1 mRNA splicing is a key regulatory step for the UPR pathway. Previous work has focused on characterizing the 381 Hac1 target genes that are transcribed rapidly, within 15 min of induction of ER stress. Many of these transcripts encode ER chaperones and other proteins that augment the functional capacity of the ER. However, if this initial wave of transcription is insufficient to alleviate the stress, and ER stress therefore persists after UPR activation, more drastic cellular events may be necessary. Precisely how the cell copes with persistent ER stress is not yet well understood.
One event that has recently been shown to be induced during late phase ER stress is autophagy (11, 12). Autophagy is a mechanism of enclosing intracellular components in a double membrane vesicle called an autophagosome, and then delivering these components to the vacuole for degradation. Autophagy is induced during starvation in both yeast and higher eukaryotes as a way of breaking down macromolecules to recycle their components (13, 14). In mammalian cells, autophagy additionally functions in regulating the cytoplasmic rearrangements that are necessary for many cell differentiation programs (15), in breaking down pathogens in the innate immune response (16), and in activating cell death pathways (15, 17, 18).
During ER stress, autophagy has several proposed functions. In mammalian cells, there is evidence that autophagy allows removal of toxic misfolded proteins to allow survival of the stress (19–21) but there is also evidence that autophagy is invoked as a means of killing cells when ER stress is too severe (22, 23). In yeast, it has been proposed that autophagy functions to remove the excess lipids that are generated by UPR-dependent activation of lipid biosynthetic genes (11).
The mechanism for inducing autophagy during ER stress is still unclear both in yeast and mammalian systems. One potential mediator of ER stress-induced autophagy that has not yet been examined is the MAP kinase p38/Hog1. p38, the mammalian homologue of Hog1 is involved in the induction of autophagy during the innate immune response in macrophages, and in human cancer cells following chemotherapeutic treatment (24–27). In yeast, when cells are simultaneously subjected to osmotic stress and nutrient deprivation, Hog1, appears to play a role in inducing autophagy, although it does not perform this function when each stress is encountered separately (28).
Although Hog1 has no previously known role in the ER stress response pathway, it does play an important role in other stress responses. Specifically, Hog1 is an important mediator of both the osmotic stress response pathway (29) and the cell wall integrity pathway (30, 31). Both of these pathways activate Hog1 by activating its mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK), Pbs2 (29). During osmotic stress, two pathways converge to activate Pbs2: one that is mediated by the plasma membrane protein Sho1, and one that is mediated by the cytoplasmic regulator Ssk1 (32–36). Upon activation, Hog1 is imported into the nucleus (37) where it regulates a transcriptional program that helps cells adapt to the stress (38–45). Following adaptation, Hog1 is dephosphorylated and returns to the cytoplasm where it is presumed to be inactive (46–49). By contrast, cell wall stress promotes Pbs2/Hog1 activation solely through the Sho1-mediated pathway. Hog1 is not imported into the nucleus during cell wall stress but does regulate the expression of genes that help enhance the integrity of the cell wall (30, 31, 50).
In this study, we have identified a new role for Hog1. We show that Hog1 becomes phosphorylated specifically during the later stages of ER stress, and facilitates recovery within the ER lumen. The mechanism of Hog1 phosphorylation during ER stress is distinct from the Hog1 phosphorylation mechanisms of osmotic stress or cell wall stress. During ER stress, phosphorylation requires SSK1, but not SHO1, and additionally requires both IRE1 and HAC1. Following its phosphorylation, Hog1 translocates into the nucleus and regulates the expression of genes that are specifically activated during late stage ER stress. In addition, Hog1 plays a critical role in the induction of key autophagy components during late stage ER stress. Thus, Hog1 regulates multiple aspects of the cellular response to long term ER stress.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
Strains are listed in Table 1. Deletion strains were generated using a one-step recombination-mediated technique (51). To monitor Atg8 levels, the plasmid pRS316 GFP-AUT7 (52), which encodes N-terminal GFP-tagged Atg8 from its native promoter, was used. Cells were grown in YPD medium at 30 °C, except for strains bearing the GFP-Atg8 reporter, which were grown in selective medium. During dithiothreitol (DTT) treatments, YPD was pH 5.4. To induce nitrogen starvation, cells were collected by centrifugation, washed twice, and resuspended in starvation medium (1× YNB without amino acids and ammonium sulfate (Difco), 2% dextrose, 0.5 mg/ml inositol). To induce osmotic stress, NaCl was added at a final concentration of 0.4 m. Cell wall stress was induced by treatment with 0.8 units/ml of zymolyase. To induce ER stress, tunicamycin (Tm, Calbiochem) was added at a final concentration of 1 μg/ml, or DTT (Fisher) was added at final concentrations of 2, 3, or 4 mm as indicated. Tm was stored as a 10 mg/ml stock in dimethyl sulfoxide, and DTT was stored as 1 m stock in H2O. Cycloheximide (CHX) was stored as a 50 mg/ml stock in dimethyl sulfoxide and added to a final concentration of 50 μg/ml.
TABLE 1.
Strains used in this study
| Strain number | Relevant genotype | Source |
|---|---|---|
| MNY1004 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3 | Ref. 2 |
| MNY1100 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, hog1Δ::KanMX | This study |
| MNY1101 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, his3–11,15::UPRE-lacZ:HIS3, HOG1-GFP::KanMX | This study |
| MNY1102 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, pbs2Δ::KanMX | This study |
| MNY1106 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, ssk1Δ::KanMX | This study |
| MNY1108 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, sho1Δ::KanMX | This study |
| MNY1111 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, ire1Δ::KanMX | This study |
| MNY1112 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, hac1Δ::KanMX | This study |
| MNY1116 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, ire1Δ::KanMX, ssk1Δ::NatMX | This study |
| MNY1117 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, pRS316 GFP-AUT7 | This study |
| MNY1118 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, hog1Δ::KanMX, pRS316 GFP-AUT7 | This study |
| MNY1119 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, pbs2Δ::KanMX, pRS316 GFP-AUT7 | This study |
| MNY1121 | MATa, leu2–3,112, trp1, ura3–52, pho8::pho8Δ60, pho13::URA3 | Ref. 54 |
| MNY1122 | MATa, leu2–3,112, trp1, ura3–52, pho8::pho8Δ60, pho13::URA3, hog1Δ::KanMX | This study |
| MNY1123 | MATa, leu2–3,112, trp1, ura3–52, pho8::pho8Δ60, pho13::URA3, atg8Δ::KanMX | This study |
| MNY1124 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, sho1Δ::KanMX, ire1Δ::URA3 | This study |
| MNY1125 | MATa, leu2–3,112, trp1-1, can1–100, ura3-1, ade2-1, his3–11,15::UPRE-lacZ:HIS3, hsp12Δ::KanMX | This study |
| MNY1126 | MATa, FUS1::lacZ::LEU2, hog1Δ::URA3 | Ref. 66 |
| MNY1127 | MATa, FUS1::lacZ::LEU2, hog1::URA3::HOG1-GFP-CCAAX | Ref. 66 |
| MNY1128 | MATa, FUS1::lacZ::LEU2, HOG1-GFP::kanMX | Ref. 66 |
| MNY1129 | MATa, FUS1::lacZ::LEU2, hog1Δ::URA3, pRS314 GFP-AUT7 | This study |
| MNY1130 | MATa, FUS1::lacZ::LEU2, hog1::URA3::HOG1-GFP-CCAAX, pRS314 GFP-AUT7 | This study |
| MNY1131 | MATa, FUS1::lacZ::LEU2, HOG1-GFP::kanMX, pRS314 GFP-AUT7 | This study |
Cell Extracts, Northern Blotting, and Immunoblotting
For Northern blotting, total RNA was isolated from cells as described previously (8). 10 (HAC1 Northern) or 25 μg (HSP12, YMR103C, and YPL088W) of RNA were loaded on a 1.5% agarose gel with 6.7% formaldehyde, and transferred to Zeta probe membrane (Bio-Rad) in 10× SSC by capillary action overnight. Following UV cross-linking, membranes were probed with a DNA probe generated by random primed DNA labeling.
For immunoblot analysis, protein was extracted as described previously (53). 20 μg of protein were separated on a 10% SDS-PAGE gel and transferred to nitrocellulose. Membranes were probed with primary antibodies against dually phosphorylated p38 (Cell Signaling) 1:1,000 overnight in 5% bovine serum albumin, total Hog1 (Santa Cruz Biotechnology) 1:2,000 in 5% milk for 1 h, PGK (Molecular Probes) 1:10,000 in 5% milk for 1 h, or GFP (Clontech) 1:10,000 in 5% milk for 1 h. Membranes were then exposed to horseradish peroxidase-conjugated secondary mouse (Bio-Rad) or rabbit (GE Healthcare) antibodies at a 1:10,000 dilution for 1 h, and developed with ECL Plus Western blotting detection reagent (GE Healthcare). Northern blots and immunoblots were scanned on a typhoon phosphorimager (GE Healthcare), and quantified using ImageQuant software (GE Healthcare).
Microscopy
To visualize Hog1 localization, a C-terminal GFP tag was integrated at the HOG1 genomic locus. This tag has been reported previously not to affect Hog1 function (37). Live GFP-expressing cells were stained with 0.2 μg/ml of 4′,6-diamidino-2-phenylindole for 5 min and imaged using an Axiovert 200M microscope (Carl Zeiss MicroImaging, Inc.) with a 100 × 1.3 NA objective. Images were captured using a monochrome digital camera (Axiocam; Carl Zeiss MicroImaging, Inc.) and analyzed using Axiovision software (Carl Zeiss MicroImaging, Inc.). To quantify GFP-Atg8 fluorescence, images were captured on a DeltaVision microscope and fluorescence intensity of 300 separate puncta for each cell type was quantified using Softworks software.
Alkaline Phosphatase Activity Assay
Strains bearing the pho8Δ60 mutation and deleted for PHO13, the other known alkaline phosphatase in yeast, were obtained from Y. Ohsumi. Alkaline phosphatase activity was determined by providing para-nitrophenyl phosphate (Sigma) as a substrate to extracts, and measuring its conversion to para-nitrophenol by reading the absorbance at 400 nm, as described previously (54, 55).
RESULTS
During ER Stress Hog1 Is Phosphorylated and Promotes Stress Recovery
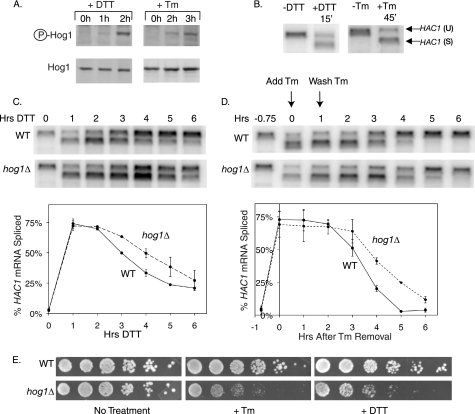
To investigate a possible involvement of Hog1 MAPK in the ER stress response, wild type cells were treated with DTT or Tm for 3 h. DTT disrupts intramolecular and intermolecular disulfide bonds and Tm is an inhibitor of N-linked glycosylation in the ER, thus both treatments lead to an accumulation of unfolded proteins in the ER, but by different mechanisms. Both treatments resulted in increased levels of phosphorylated Hog1 protein (Fig. 1A). Compared with the kinetics of UPR activation, the kinetics of Hog1 phosphorylation were significantly delayed. As measured by HAC1 mRNA splicing, UPR activation occurred after a 15-min DTT treatment or 45-min Tm treatment (Fig. 1B), whereas Hog1 phosphorylation was not induced until 2 h of treatment with either drug (Fig. 1A), suggesting that Hog1 might play a specific role in responding to long term ER stress.
FIGURE 1.
During ER stress Hog1 is phosphorylated and promotes stress recovery. A, phospho-Hog1 immunoblots of wild type cells treated with 2 mm DTT or 1 μg/ml of Tm. B, Northern analysis with HAC1-specific probe shows conversion of unspliced HAC1 (HAC1(U)) to spliced HAC1 (HAC1(S)) within 15 min of 2 mm DTT treatment, or 45 min of 1 μg/ml of Tm treatment. C and D, HAC1 Northern analysis during sustained 2 mm DTT treatment (C) or 45 min of 1 μg/ml of Tm treatment followed by removal of the drug from the medium (D) in wild type (WT) and hog1Δ cells. Graphs depict HAC1(S)/[HAC1(U) + HAC1(S)]. Error bars represent S.D. of three repeats. E, 5-fold serial dilutions of wild type and hog1Δ cells grown on medium alone, or medium containing 0.1 μg/ml of Tm or 8 mm DTT.
To investigate a potential role for HOG1 in alleviating ER stress, we measured HAC1 mRNA splicing in wild type and hog1Δ cells following induction of ER stress. Because the UPR pathway is activated at a level commensurate with the amount of stress in the ER, HAC1 mRNA splicing can be used to assay the extent of ER stress. When DTT was added to wild type and hog1Δ cells, HAC1 mRNA became 75% spliced within the first hour of treatment for both cell types (Fig. 1C). However, hog1Δ cells retained a high level of splicing longer than wild type cells, suggesting that hog1Δ cells are impaired in regaining ER homeostasis. To confirm that hog1Δ cells are impaired in ER stress recovery, we also performed UPR recovery experiments during Tm treatment. In wild type cells, the continued presence of Tm induces prolonged HAC1 mRNA splicing, whereas removal of Tm from the medium allows recovery of the ER (data not shown and Fig. 1D, top panel). Therefore, UPR recovery experiments were performed by removing Tm from the medium of both wild type and hog1Δ cells following 45 min of treatment. Again, hog1Δ cells were significantly delayed in their recovery from ER stress (Fig. 1D).
In addition to promoting recovery of the ER during prolonged ER stress, HOG1 also promoted survival of the stress. Compared with wild type cells, hog1Δ cells were hypersensitive to growth on both DTT and Tm plates (Fig. 1E). This result validated the heretofore unconfirmed findings of a genomewide genetic screen, which identified HOG1 among the many genes that confer DTT resistance (56).
ER Stress Activates Hog1 through a Unique Mechanism Mediated by SSK1 and the UPR
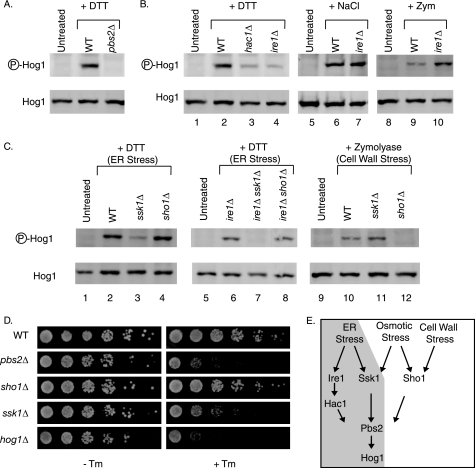
Pbs2 is the MEK that is known to phosphorylate Hog1 (29). Therefore, we examined the possibility that Pbs2 phosphorylates Hog1 during ER stress. In cells lacking PBS2, no Hog1 phosphorylation was observed during DTT treatment (Fig. 2A). In addition, pbs2Δ cells exhibited the same sensitivity to growth on Tm medium as did hog1Δ cells (Fig. 2D), indicating that Pbs2 phosphorylates Hog1 during ER stress.
FIGURE 2.
ER stress activates Hog1 through a unique mechanism mediated by SSK1 and the UPR. A–C, phospho-Hog1 immunoblots of cells treated with 4 mm DTT for 3 h, 0.4 m NaCl for 5 min, or 0.8 units/ml of zymolyase for 1 h. D, 5-fold serial dilutions of cells grown with or without 0.1 μg/ml of Tm. E, schematic depicting the overlapping but non-identical pathways of Hog1 activation during ER stress (shaded gray), osmotic stress, and cell wall stress. WT, wild type.
Because Pbs2 is a cytosolic protein and cannot directly detect ER stress, we tested the possibility that the canonical UPR signaling pathway acts upstream of Pbs2 to activate Hog1 phosphorylation. In the absence of IRE1 and HAC1, Hog1 phosphorylation was significantly reduced (Fig. 2B, lanes 2–4), indicating that the canonical UPR components contribute to Hog1 activation during ER stress.
Because Hog1 is also known to be activated by osmotic stress and cell wall stress (29, 30), and because ER stress is known to cause alterations in the cell wall composition (57), the possibility exists that ER stress actually activates Hog1 indirectly, by causing either osmotic stress or cell wall stress. In this case, ER stress should induce Hog1 activation through the same mechanism as either osmotic stress or cell wall stress. However, the mechanisms were not the same, as the UPR did not promote Hog1 phosphorylation during either osmotic stress or cell wall stress. During osmotic stress, Hog1 phosphorylation was unaffected by the deletion of IRE1 (Fig. 2B, lanes 6 and 7). During cell wall stress, Hog1 phosphorylation was enhanced by the deletion of IRE1 (Fig. 2B, lanes 9 and 10), which is consistent with previous reports showing that cell wall stress is exacerbated by the absence of the UPR (57). This contrasted sharply with ER stress, during which Hog1 phosphorylation was diminished in the absence of IRE1 (Fig. 2B). Thus, ER stress utilizes a unique mechanism to induce Hog1 phosphorylation.
In ire1Δ and hac1Δ cells, Hog1 phosphorylation was reduced, but not eliminated, suggesting that a parallel non-redundant pathway exists to phosphorylate Hog1 during ER stress. To identify mediators of this parallel pathway, we examined DTT-induced Hog1 phosphorylation upon deletion of SSK1 or SHO1, mediators of two known upstream activation pathways for Hog1 (32–36). Whereas deletion of SHO1 had no impact on Hog1 phosphorylation, deletion of SSK1 significantly reduced phosphorylation (Fig. 2C, lanes 2–4). Furthermore, ssk1Δ cells were hypersensitive to growth on medium containing Tm (Fig. 2D). The SSK1-dependent Hog1 activation pathway was found to be parallel to the IRE1-dependent activation pathway. Deletion of each gene separately only reduced phosphorylation levels (Fig. 2, B, lane 4, and C, lane 3), whereas deletion of both IRE1 and SSK1 completely eliminated Hog1 phosphorylation (Fig. 2C, lane 7). In contrast to ER stress, phosphorylation of Hog1 by cell wall stress was dependent upon SHO1, rather than SSK1 (Fig. 2C, lanes 10–12) as reported previously (30, 31). Taken together, these data show that during ER stress, Hog1 becomes phosphorylated through an IRE1- and SSK1-dependent mechanism, which is unique from the Hog1 phosphorylation mechanism of both osmotic stress and cell wall stress (Fig. 2E), and is therefore not the result of indirect activation of osmotic stress or cell wall stress pathways.
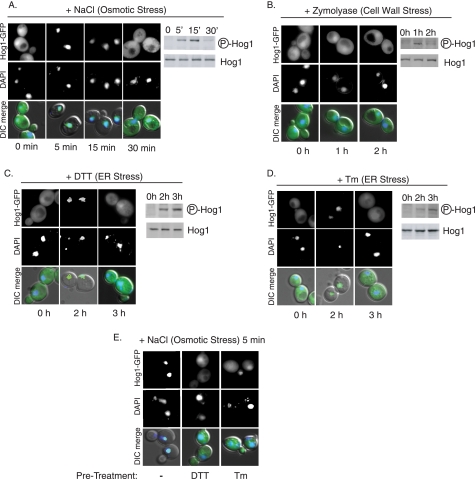
Hog1 Localization Is Regulated Uniquely during ER Stress
Hog1 localization is regulated differently during different types of stress, and plays an important role in Hog1 function. During osmotic stress, Hog1 phosphorylation results in its immediate import into the nucleus, where it interacts with various transcription factors to mediate a transcriptional response. Subsequently, Hog1 is exported from the nucleus with kinetics that correlate with its dephosphorylation (Fig. 3A and Ref. 37). During cell wall stress, although Hog1 is phosphorylated, it is not imported into the nucleus (Fig. 3B and Ref. 30). During ER stress, Hog1 localization was regulated differently from both osmotic stress and cell wall stress. After 2 h of DTT or Tm treatments, when phosphorylation was initially detected, Hog1-GFP was imported into the nucleus (Fig. 3, C and D). Subsequently, after 3 h of treatment, Hog1 remained phosphorylated, but returned to the cytoplasm, hinting that Hog1 might have a cytoplasmic function during ER stress. To investigate the mechanism of this cytoplasmic localization, ER-stressed cells with cytoplasmically localized Hog1 (3 h DTT or Tm treatments) were subjected to osmotic stress (0.4 m NaCl, 5 min). In the absence of prior ER stress treatment, osmotic stress caused Hog1 to be imported into the nucleus (Fig. 3E). In contrast, cells subjected to ER stress for 3 h did not import Hog1 into the nucleus, suggesting that ER stress causes active cytoplasmic retention or nuclear exclusion of Hog1. Together, these data suggest that Hog1 may have functions both in the nucleus and cytoplasm during ER stress.
FIGURE 3.
Hog1 localization is regulated uniquely during ER stress. A–D, cells expressing HOG1-GFP were treated with 0.4 m NaCl, 0.8 units/ml of zymolyase, 2 mm DTT, or 1 μg/ml of Tm, then collected for microscopy and phospho-Hog1 immunoblot analysis. E, cells expressing HOG1-GFP were treated with 0.4 m NaCl alone (left panel) or following 3 h of preincubation with 2 mm DTT (middle panel) or 1 μg/ml of Tm (right panel). DAPI, 4′,6-diamidino-2-phenylindole.
Hog1 Regulates mRNA Expression during ER Stress
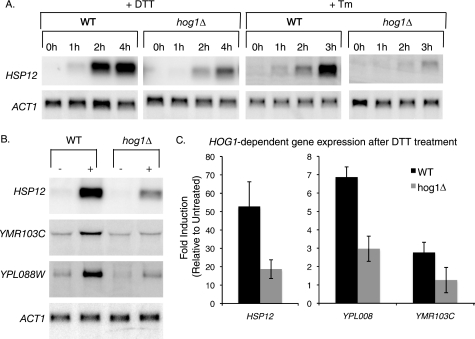
During osmotic stress, one function of Hog1 nuclear import is to regulate gene expression (38–44). Because Hog1 is also imported into the nucleus during ER stress, we examined the possibility that Hog1 regulates gene expression during the later stages of ER stress. Previously, a microarray study was conducted to define the IRE1/HAC1-dependent transcriptional response during ER stress (58). This study focused on the changes in gene expression that occur within the first hour of ER stress, but included gene expression data from wild type cells treated with DTT for 2 h. To identify potential Hog1-regulated genes, we searched for transcripts within the data set whose induction correlated with the kinetics of Hog1 phosphorylation. Candidate targets would remain uninduced during the first hour of ER stress, and become induced after 2 h of stress. Based on these criteria, HSP12 was identified as a candidate Hog1 target. HSP12 seemed an especially likely target because it is known to be induced by Hog1 during other types of stress (31, 39).
Northern analysis confirmed that during both Tm and DTT treatments, HSP12 is induced during late phase ER stress, but not during the earlier phase of the stress response. Furthermore, HSP12 induction was dependent upon HOG1 (Fig. 4A). Eight other candidate targets of Hog1-mediated gene regulation were also examined after 2 h of DTT treatment. Of the eight genes examined, YMR103C and YPL088W were induced in a HOG1-dependent manner during ER stress (Fig. 4B and quantified in Fig. 4C). Thus, regulation of gene expression is one function of Hog1 during the later stages of ER stress.
FIGURE 4.
Hog1 regulates mRNA expression during ER stress. A, HSP12 Northern blots of WT and hog1Δ cells treated with 2 mm DTT or 1 μg/ml of Tm. B and C, following 2 h of DTT treatment, the levels of HSP12, YMR103C, and YPL088W in wild type (WT) and hog1Δ cells were measured by Northern blot, normalized to actin, and presented as fold-increase compared with untreated cells. Error bars represent S.D. of three repeats.
Induction of Atg8 Protein during ER Stress Requires HOG1
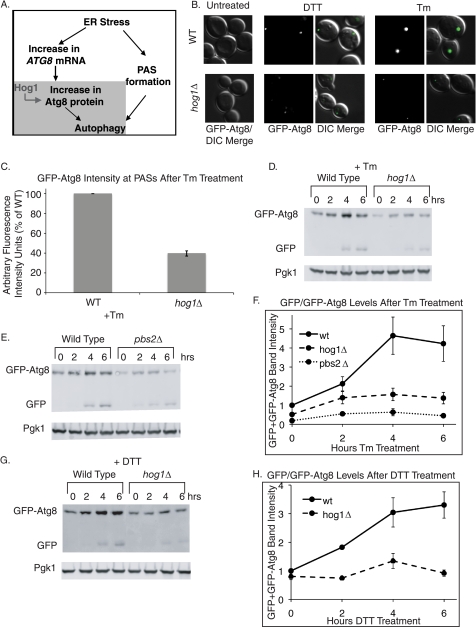
During persistent ER stress, autophagy is induced with kinetics that correlate with the kinetics of Hog1 phosphorylation (11, 12). Therefore, we examined the possibility that Hog1 regulates autophagy during ER stress. The induction of autophagy requires the convergence of two parallel pathways (Fig. 5A). One pathway induces the formation of pre-autophagosomal structures (PASs, also called phagophore assembly sites), which are necessary for nucleation of the autophagosome (52). The second pathway induces an increased abundance of the small ubiquitin-like protein Atg8 (59, 60). This increase is necessary for the formation of appropriately sized autophagosomes (61). Both PAS formation and Atg8 accumulation have been shown to occur during persistent ER stress (11, 12) although the mechanism for inducing these events is not known.
FIGURE 5.
Induction of Atg8 protein during ER stress requires HOG1. A, the schematic shows parallel pathways that converge to induce autophagy. PAS formation and increases in Atg8 protein levels are both necessary for appropriate levels of autophagy. Hog1 specifically regulates the increase in Atg8 protein levels and induction of autophagy downstream. B, cells expressing the GFP-ATG8 reporter were subjected to 4 h of 2 mm DTT or 1 μg/ml of Tm treatment before microscopic visualization to assess PAS formation. C, following 6 h of 1 μg/ml of Tm treatment, fluorescence intensity of Atg8-GFP puncta was quantified (n = 300). D, E, and G, GFP immunoblots of cells bearing the GFP-ATG8 reporter, treated with 1 μg/ml of Tm or 3 mm DTT. The top GFP band represents the GFP-Atg8 fusion protein, and the bottom GFP band represents free GFP. F and H, quantitation of GFP immunoblots. GFP-Atg8 + free GFP signal was normalized to Pgk1 (loading control). Error bars represent S.E. of at least three replicates.
Because Atg8 is targeted to PASs once they form, a GFP-ATG8 fusion gene expressed from the ATG8 promoter is widely used to monitor both PAS formation fluorescence and Atg8 protein accumulation by GFP immunoblot (52, 62, 63). Using this reporter, we found that both DTT and Tm treatments induced the formation of PASs, which are observed as discrete puncta throughout the cell (Fig. 5B), as well as an increase in GFP-Atg8 protein levels (Fig. 5, D–G). The formation of PASs and induction of Atg8p suggests that autophagy was induced during ER stress, confirming previous reports (11, 12). Furthermore, when autophagy is induced, some GFP-Atg8 becomes trapped in the autophagosome and delivered to the vacuole (60) where the Atg8 portion of the chimera is degraded and the GFP portion is resistant to degradation and remains intact (62). Thus, the presence of a free GFP band after DTT or Tm treatment (Fig. 5, D, E, and G, top panel, bottom band) also indicates that autophagy was induced.
During DTT and Tm treatment in hog1Δ cells, GFP-Atg8 became localized at discrete puncta, indicating that HOG1 is not necessary for PAS formation (Fig. 5B). However, the fluorescence intensity of the puncta was much lower in hog1Δ cells than in wild type cells (Fig. 5, B and quantified in C), suggesting that Hog1 is involved in a parallel pathway of increasing the production of GFP-Atg8. To confirm this, GFP-Atg8 levels were examined by GFP immunoblot during both Tm and DTT treatments. For both treatments, Atg8 protein accumulation (measured as the sum of GFP-Atg8 plus free GFP) was considerably inhibited by the absence of HOG1 (Fig. 5, D and G, and quantified in F and H). Therefore, Hog1 acts specifically to induce Atg8 protein accumulation during ER stress. This role of Hog1 in regulating Atg8 accumulation was dependent on the ability of Hog1 to become phosphorylated, as pbs2Δ cells, which cannot phosphorylate Hog1 (Fig. 2A), were also defective in Tm-induced accumulation of GFP-Atg8 (Fig. 5, E and quantified in F).
Hog1 Acts from the Cytoplasm and Regulates Atg8 Protein Stability during ER Stress
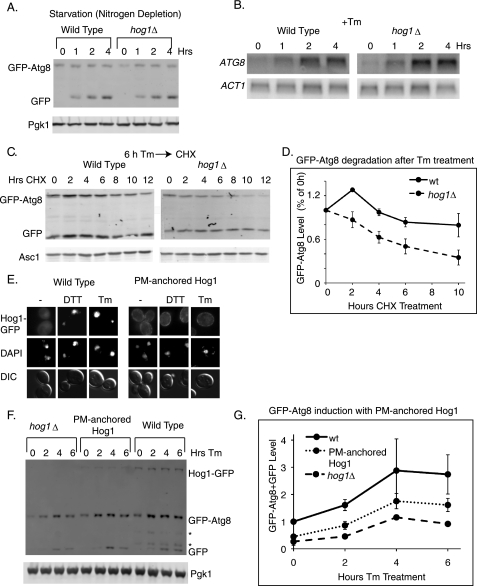
To determine the specificity of Hog1 in regulating Atg8 during ER stress, GFP-Atg8 induction in wild type and hog1Δ cells was examined during starvation. Autophagy is strongly induced by starvation, during which cytoplasmic degradation allows cells to recycle essential nutrients (64, 65). Upon removal of nitrogen from the medium, GFP-Atg8 was induced, as expected. However, this induction did not depend upon HOG1 (Fig. 6A). Thus, Hog1 induces Atg8 accumulation through a mechanism that is unique to ER stress.
FIGURE 6.
Hog1 acts from the cytoplasm and regulates Atg8 protein stability during ER stress. A, GFP immunoblots of cells bearing the GFP-ATG8 reporter following nitrogen deprivation. B, ATG8 Northern blots of wild type and hog1Δ cells treated with 1 μg/ml of Tm. C and D, following 6 h of 1 μg/ml of Tm treatment, 50 μg/ml of CHX was added to cells for 12 h. The GFP-Atg8 band was quantified and normalized to Asc1 levels. The protein level at each time point was divided by the level at the time of CHX addition to determine the % protein remaining. E, strains expressing Hog1-GFP or Hog1-GFP-CCAAX (PM-anchored) were visualized following 2 h of 2 mm DTT treatment or 1 μg/ml of Tm treatment. F, GFP immunoblots of a hog1Δ strain, a strain expressing Hog1-GFP (WT), and a strain expressing Hog1-GFP-CCAAX (PM-anchored) following treatment with 1 μg/ml of Tm. Each strain expressed the GFP-Atg8 marker. Note that in this strain background, GFP-Atg8 cleavage occurs less efficiently than in the strain background used for Fig. 5, and that additional GFP degradation products are observed (*). G, quantitation of F. GFP-Atg8 + free GFP bands were normalized to Pgk1. Error bars represent S.E. of three replicates.
During ER stress, in the absence of HOG1 or PBS2, GFP-Atg8 accumulation was reduced, but not eliminated (Fig. 5, D–H). This suggests that two separate signals collaborate to induce Atg8 expression during ER stress, and that only one of these signals is HOG1-dependent. To distinguish these two signals, ATG8 transcription and protein stability were examined during Tm treatment in both wild type and hog1Δ cells. As expected, Tm treatment resulted in increased levels of the ATG8 transcript. However, transcript induction did not depend upon HOG1 (Fig. 6B), suggesting that the HOG1-independent signal for Atg8 induction occurs at the transcriptional level. By contrast, the stability of the GFP-Atg8 protein during ER stress was found to be HOG1-dependent. Following 6 h of Tm treatment, CHX was added to wild type and hog1Δ cells to allow an examination of GFP-Atg8 degradation rates in the absence of new protein synthesis. In wild type cells, GFP-Atg8 was degraded slowly, with a half-life of 11 h, whereas in hog1Δ cells, GFP-Atg8 was degraded with a half-life of 6 h. This indicates that during ER stress, Hog1 facilitates the accumulation of Atg8 by stabilizing the protein (Fig. 6, C and D).
Because Hog1 is localized to the cytoplasm after 3 h of ER stress (Fig. 3, C and D) and controls the stability of GFP-Atg8 protein rather than its transcription, we examined the possibility that cytoplasmic localization of Hog1 is important for its role in autophagy. We took advantage of a strain designed by Westfall et al. (66) in which the HOG1 gene is replaced by HOG1-GFP-CCAAX fusion. The fusion protein contains the nine C-terminal residues of Ras2, and is thereby constitutively tethered to the plasma membrane. As expected, Hog1 was not imported into the nucleus during ER stress in this strain (Fig. 6E). We therefore examined GFP-Atg8 induction in this strain to determine whether cytoplasmically localized Hog1 could confer GFP-Atg8 regulation.
The plasma membrane-tethered Hog1-GFP protein was expressed at lower levels than the wild type Hog1-GFP (Fig. 6F, top band). Despite this reduced expression (quantified as 26% of wild type), cells with Hog1 tethered to the plasma membrane induced more GFP-Atg8 accumulation than hog1Δ cells of the same strain background, suggesting that Hog1 can act from the cytoplasm to induce Atg8 (Fig. 6F and quantified in Fig. 6G). However, GFP-Atg8 accumulation was less in the HOG1-GFP-CCAAX fusion strain than in cells expressing wild type HOG1-GFP. It is not yet clear whether this is the result of reduced Hog1 levels in the HOG1-GFP-CCAAX fusion, or reflects an additional nuclear function for Hog1 in regulating Atg8 stability.
Cellular Effects of the Role of Hog1 in Atg8 Induction
Modulating Atg8 protein levels has previously been shown to directly affect the amount of autophagy induced by the cell (61). Therefore, we anticipated that the regulation of Hog1 Atg8 levels would influence the rate of autophagy during ER stress. To test this, we took advantage of the Pho8 alkaline phosphatase, a protein that can only become activated when delivered to the vacuole where its C-terminal peptide is cleaved. The wild type protein is delivered to the vacuole constitutively, but the pho8Δ60 mutant form cannot be delivered to the vacuole unless autophagy is induced. Therefore, in strains expressing pho8Δ60 as their only alkaline phosphatase, alkaline phosphatase activity has been widely used as a way to directly measure autophagy (54).
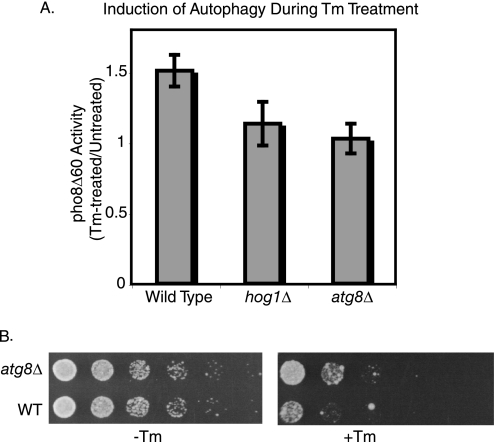
As measured by pho8Δ60 activity, a 6-h Tm treatment caused a 1.5-fold increase in autophagy (Fig. 7A), whereas atg8Δ cells showed no induction. In the absence of HOG1, autophagy was only marginally increased (1.1-fold) compared with untreated cells. Thus, Hog1-mediated increases in Atg8 levels are necessary for the induction of autophagy during the later stages of ER stress.
FIGURE 7.
Cellular effects of the role of Hog1 in Atg8 induction. A, alkaline phosphatase activity of cells bearing the pho8Δ60 mutation following a 6-h Tm treatment (compared with untreated cells). Error bars represent S.D. of three replicates. B, 5-fold serial dilutions of wild type and atg8Δ cells grown on medium alone, or medium containing 0.4 μg/ml of Tm.
To gain insight into the function of autophagy during ER stress, the growth of atg8Δ cells on medium containing Tm was examined. In the absence of ATG8, cells were more resistant than wild type cells to ER stress (Fig. 7B), suggesting that one function of autophagy may be to promote cell death when ER stress is particularly persistent or severe. Thus, during ER stress, Hog1 plays roles in both cell survival (Fig. 1E) and cell death (i.e. induction of Atg8 and autophagy).
In a previous study (11), autophagy-deficient strains were shown to be sensitive to growth on medium containing Tm, rather than resistant. The reasons for this inconsistency are currently unclear, but they might result from differences in growth conditions, the level of stress imposed, or the duration of the stress. Thus, under the specific conditions tested here, we have identified a novel role of Atg8 and the autophagy pathway in promoting cell death during ER stress.
DISCUSSION
A Late Phase ER Stress Response Pathway in Yeast
Recent studies in mammalian cells indicate that during ER stress, activation of apoptotic pathways only occurs during the later stages of stress, whereas pro-survival pathways dominate the early stress response (67). This highlights the cellular need to distinguish between early and persistent stress, and tailor its response accordingly. Our data shed light on the specific nature of the late phase ER stress response in yeast, as we have discovered that two processes, transcriptional regulation and induction of autophagy, are regulated in a coordinated manner by MAPK during persistent stress.
In this study, we have shown that Hog1 is activated during the late phase of the ER stress response and plays a role in helping the ER regain homeostasis, as hog1Δ cells recover more slowly than wild type cells during ER stress and exhibit increased sensitivity to the stress. Part of the function of Hog1 is to regulate late phase gene expression. We have identified three genes that are regulated by Hog1 during ER stress: HSP12, YMR103C, and YPL088W. Each of these genes is activated with similar kinetics, remaining uninduced during the early stages of ER stress, but specifically becoming activated during late stage ER stress (Fig. 4A and Ref. 58). Therefore, two distinct waves of gene transcription seem to occur: one that is activated rapidly by Hac1, and one that is activated much later by Hog1. In the future, genomewide studies will determine what other transcripts are regulated by Hog1 during this second wave of gene expression.
The functional roles of HSP12, YMR103C, and YPL088W during ER stress are not yet clear. HSP12 is known to be activated during various types of stress (31, 68–70), but its biochemical function is not known, and YMR103C, and YPL088W are uncharacterized. However, in other cases where a single stimulus activates sequential waves of gene expression, it has been noted that genes with similar biological function tend to cluster within specific expression groups. For example, when B cells are activated to secrete antibody, five waves of gene expression are sequentially induced and each wave is comprised of genes with a common biological function that is distinct from the other waves (71). Regulation of gene expression during ER stress may occur similarly, in which case genes that are specifically regulated during late phase ER stress might comprise one or more functional groups that are uniquely suited to help cells withstand persistent ER stress.
We have also shown that Hog1 functions in the regulation of autophagy during late stage ER stress. The function of autophagy during ER stress is not known, but several interesting propositions have been put forth. In one report, although Atg8 was clearly induced, it did not appear to be degraded in the vacuole (11), leading to the proposition that autophagosomes function not to degrade stressed ER, but to sequester it and prevent toxicity while the ER recovers. In the current study, we observed conversion of GFP-Atg8 into free GFP during both DTT and Tm treatments, as well as activation of Pho8Δ60 phosphatase, suggesting that some cellular components were delivered to the vacuole for degradation. Therefore, degradation is part of autophagy function during the late phase ER stress response. However, it is possible that ER sequestering is an additional important function of autophagy.
It has also been proposed that autophagy-induced ER degradation serves to reverse the UPR-stimulated expansion of the ER, thus helping cells return to a resting state after UPR induction (11). This model implies that proliferated ER is no longer necessary by the time autophagy is induced because the stress has been alleviated. However, the induction of a second wave of stress-responsive gene expression suggests that the stress has not been alleviated. Therefore, we believe that rather than helping to wind down the ER stress response, autophagy may help extend or intensify the response.
In mammalian cells, autophagy has been shown to assist in the degradation of misfolded ER proteins (19–21), a function that could help cells cope with ER stress. Autophagy has also been shown to promote non-apoptotic cell death during ER stress in mammalian cells (22, 23). During long term ER stress in yeast, both functions of autophagy may be important. Initially, autophagy may protect cells from the toxicity of ER stress by promoting degradation of misfolded proteins. However, after several days of growth under conditions of ER stress, autophagy-deficient cells survived better than wild type cells, suggesting that if activated long enough, autophagy may induce cell death during ER stress.
Specificity of Hog1-mediated Cellular Changes during Late Phase ER Stress
Although osmotic stress, cell wall stress, and ER stress each activate the phosphorylation of Hog1, the downstream effects of Hog1 activation are different among these different types of stress. First, during each condition, Hog1 stimulates unique changes in gene expression, with overlapping but non-identical targets among the different types of stress (Fig. 4, A and C, and Refs. 31 and 44). Second, persistent ER stress induces autophagy, whereas osmotic stress and cell wall stress do not. These differences highlight the capacity of Hog1 to customize its downstream response to its specific activation conditions.
This type of customization is a common feature of MAPKs, but how it is achieved remains an open question. In the case of Hog1, the specificity of the downstream response might be conferred by differences in the upstream activation mechanism, and/or by differences in the localization pattern of Hog1 during the different types of stress. During osmotic stress two upstream activation modules, mediated by SHO1 and SSK1, stimulate Hog1 phosphorylation (32, 35); during cell wall stress only the SHO1 activation module activates Hog1 (30, 31); and during ER stress the SSK1 activation module collaborates with the canonical UPR pathway to activate Hog1. Furthermore, osmotic stress induces nuclear localization that is tightly correlated with phosphorylation; cell wall stress does not induce changes in Hog1 localization; and ER stress induces both nuclear and cytoplasmic localizations that correlate with high levels of Hog1 phosphorylation. It is not yet clear whether the differences in the activation mechanism are responsible for the different localization patterns, and what role the activation mechanism and localization patterns play in specifying the downstream response of Hog1 activation.
Because the UPR pathway regulates transcription, and MAPKs are primarily regulated at the level of phosphorylation, the involvement of the UPR in Hog1 activation was surprising. Possible mechanisms for IRE1-dependent activation of Hog1 include down-regulation of transcripts encoding phosphatases that normally function to keep Hog1 phosphorylation levels low (72, 73), or a transcriptional up-regulation of proteins that lead to Hog1 phosphorylation. Future genomewide studies of the late phase ER stress response pathway will shed light on this mechanism.
HOG1-dependent Atg8 Regulation
Little is known about how Atg8 levels are regulated during autophagy, but most models presume that transcriptional activation of ATG8 accounts entirely for the increase in protein. Here, we show that during ER stress Atg8 is regulated at the level of protein stability. The regulation of Atg8 correlates with cytoplasmic localization of Hog1, and does not require nuclear translocation, suggesting that it might be conferred through one of the known cytoplasmic targets of Hog1: Rck1, Rck2, Hsl1, Sic1, or through a yet undiscovered novel or ER stress-specific cytoplasmic Hog1 target.
In this study, we have identified a late phase ER stress response in which Hog1 regulates downstream events in response to long term ER stress. Gene activation and autophagy induction are clearly part of this response. However, Hog1 might also regulate other physiological processes during persistent ER stress. Because past studies have focused primarily on the initial response of the cell to ER stress, aspects of cell physiology that are affected by persistent ER stress have gone previously unnoticed. Therefore, future studies will aim to identify additional downstream events that are regulated during persistent ER stress.
Acknowledgments
We thank Yoshinori Ohusmi for Atg8 antiserum and the pho8Δ60 strain, Daniel Klionsky for Atg8 antiserum and plasmids, and Jeremy Thorner for yeast strains. We also thank Randy Hampton for plasmids, yeast strains, and comments on this manuscript, Lorraine Pillus and Jenny B. DuRose for comments on this manuscript, and Peter Novick for technical advice.
This work was supported by American Cancer Society (Grant ACS RSG-05-01 GMC) and Searle (Grant 03–G107) (to M. N.).
- ER
- endoplasmic reticulum
- Tm
- tunicamycin
- DTT
- dithiothreitol
- UPR
- unfolded protein response
- PAS
- pre-autophagosomal structure
- CHX
- cycloheximide
- MAPK
- mitogen-activated protein kinase
- GFP
- green fluorescent protein.
REFERENCES
- 1.Lindholm D., Wootz H., Korhonen L. (2006) Cell Death Differ. 13, 385–392 [DOI] [PubMed] [Google Scholar]
- 2.Cox J. S., Shamu C. E., Walter P. (1993) Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 3.Mori K., Ma W., Gething M. J., Sambrook J. (1993) Cell 74, 743–756 [DOI] [PubMed] [Google Scholar]
- 4.Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 6.Sidrauski C., Walter P. (1997) Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 7.Kawahara T., Yanagi H., Yura T., Mori K. (1997) Mol. Biol. Cell 8, 1845–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J. S., Walter P. (1996) Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 9.Chapman R. E., Walter P. (1997) Curr. Biol. 7, 850–859 [DOI] [PubMed] [Google Scholar]
- 10.Rüegsegger U., Leber J. H., Walter P. (2001) Cell 107, 103–114 [DOI] [PubMed] [Google Scholar]
- 11.Bernales S., McDonald K. L., Walter P. (2006) PLoS Biol 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky D. J., Ohsumi Y. (1999) Annu. Rev. Cell Dev. Biol. 15, 1–32 [DOI] [PubMed] [Google Scholar]
- 14.He C., Klionsky D. J. (2009) Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cecconi F., Levine B. (2008) Dev Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid D., Dengjel J., Schoor O., Stevanovic S., Münz C. (2006) J. Mol. Med. 84, 194–202 [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. (2009) Curr. Top. Microbiol. Immunol. 335, 71–84 [DOI] [PubMed] [Google Scholar]
- 18.Kourtis N., Tavernarakis N. (2009) Cell Death Differ. 16, 21–30 [DOI] [PubMed] [Google Scholar]
- 19.Ding W. X., Ni H. M., Gao W., Yoshimori T., Stolz D. B., Ron D., Yin X. M. (2007) Am. J. Pathol. 171, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamimoto T., Shoji S., Hidvegi T., Mizushima N., Umebayashi K., Perlmutter D. H., Yoshimori T. (2006) J. Biol. Chem. 281, 4467–4476 [DOI] [PubMed] [Google Scholar]
- 21.Kruse K. B., Brodsky J. L., McCracken A. A. (2006) Mol. Biol. Cell 17, 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. (2007) J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 23.Ullman E., Fan Y., Stawowczyk M., Chen H. M., Yue Z., Zong W. X. (2008) Cell Death Differ. 15, 422–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y., Jagannath C., Liu X. D., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. (2007) Immunity 27, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Q., Tashiro S., Onodera S., Minami M., Ikejima T. (2007) J. Pharmacol. Sci. 105, 317–325 [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y., Qiu F., Ye Y. C., Guo Z. M., Tashiro S., Onodera S., Ikejima T. (2009) FEBS J. 276, 1291–1306 [DOI] [PubMed] [Google Scholar]
- 27.Liu B., Cheng Y., Zhang B., Bian H. J., Bao J. K. (2009) Cancer Lett. 275, 54–60 [DOI] [PubMed] [Google Scholar]
- 28.Prick T., Thumm M., Köhrer K., Häussinger D., Vom Dahl S. (2006) Biochem. J. 394, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C. (1993) Science 259, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 30.Bermejo C., Rodríguez E., García R., Rodríguez-Peña J. M., Rodríguez de la Concepción M. L., Rivas C., Arias P., Nombela C., Posas F., Arroyo J. (2008) Mol. Biol. Cell 19, 1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García R., Rodríguez-Peña J. M., Bermejo C., Nombela C., Arroyo J. (2009) J. Biol. Chem. 284, 10901–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda T., Wurgler-Murphy S. M., Saito H. (1994) Nature 369, 242–245 [DOI] [PubMed] [Google Scholar]
- 33.Posas F., Wurgler-Murphy S. M., Maeda T., Witten E. A., Thai T. C., Saito H. (1996) Cell 86, 865–875 [DOI] [PubMed] [Google Scholar]
- 34.Posas F., Saito H. (1997) Science 276, 1702–1705 [DOI] [PubMed] [Google Scholar]
- 35.Maeda T., Takekawa M., Saito H. (1995) Science 269, 554–558 [DOI] [PubMed] [Google Scholar]
- 36.Posas F., Saito H. (1998) EMBO J. 17, 1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiser V., Ruis H., Ammerer G. (1999) Mol. Biol. Cell 10, 1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Márquez J. A., Pascual-Ahuir A., Proft M., Serrano R. (1998) EMBO J. 17, 2543–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rep M., Reiser V., Gartner U., Thevelein J. M., Hohmann S., Ammerer G., Ruis H. (1999) Mol. Cell. Biol. 19, 5474–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proft M., Pascual-Ahuir A., de Nadal E., Ariño J., Serrano R., Posas F. (2001) EMBO J. 20, 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Nadal E., Casadomé L., Posas F. (2003) Mol. Cell. Biol. 23, 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., Posas F. (2004) Nature 427, 370–374 [DOI] [PubMed] [Google Scholar]
- 43.Proft M., Mas G., de Nadal E., Vendrell A., Noriega N., Struhl K., Posas F. (2006) Mol. Cell 23, 241–250 [DOI] [PubMed] [Google Scholar]
- 44.O'Rourke S. M., Herskowitz I. (2004) Mol. Biol. Cell 15, 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alepuz P. M., Jovanovic A., Reiser V., Ammerer G. (2001) Mol. Cell 7, 767–777 [DOI] [PubMed] [Google Scholar]
- 46.Jacoby T., Flanagan H., Faykin A., Seto A. G., Mattison C., Ota I. (1997) J. Biol. Chem. 272, 17749–17755 [DOI] [PubMed] [Google Scholar]
- 47.Mattison C. P., Ota I. M. (2000) Genes Dev. 14, 1229–1235 [PMC free article] [PubMed] [Google Scholar]
- 48.Warmka J., Hanneman J., Lee J., Amin D., Ota I. (2001) Mol. Cell. Biol. 21, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wurgler-Murphy S. M., Maeda T., Witten E. A., Saito H. (1997) Mol. Cell. Biol. 17, 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arroyo J., Bermejo C., Garcia R., Rodriguez-Pena J. M. (2009) Clin. Microbiol. Infect. 15, Suppl. 1, 44–46 [DOI] [PubMed] [Google Scholar]
- 51.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bicknell A. A., Babour A., Federovitch C. M., Niwa M. (2007) J. Cell Biol. 177, 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 55.Klionsky D. J. (2007) Methods Mol. Biol. 390, 363–371 [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., Feldman D. E., Deng C., Brown J. A., De Giacomo A. F., Gaw A. F., Shi G., Le Q. T., Brown J. M., Koong A. C. (2005) Mol. Cancer Res. 3, 669–677 [DOI] [PubMed] [Google Scholar]
- 57.Scrimale T., Didone L., de Mesy Bentley K. L., Krysan D. J. (2009) Mol. Biol. Cell 20, 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 59.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang W. P., Scott S. V., Kim J., Klionsky D. J. (2000) J. Biol. Chem. 275, 5845–5851 [DOI] [PubMed] [Google Scholar]
- 61.Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J., Huang W. P., Klionsky D. J. (2001) J. Cell Biol. 152, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shintani T., Klionsky D. J. (2004) J. Biol. Chem. 279, 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992) J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. (1994) FEBS Lett. 349, 275–280 [DOI] [PubMed] [Google Scholar]
- 66.Westfall P. J., Patterson J. C., Chen R. E., Thorner J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12212–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varela J. C., Praekelt U. M., Meacock P. A., Planta R. J., Mager W. H. (1995) Mol. Cell. Biol. 15, 6232–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Praekelt U. M., Meacock P. A. (1990) Mol. Gen. Genet. 223, 97–106 [DOI] [PubMed] [Google Scholar]
- 70.Sales K., Brandt W., Rumbak E., Lindsey G. (2000) Biochim. Biophys. Acta 1463, 267–278 [DOI] [PubMed] [Google Scholar]
- 71.van Anken E., Romijn E. P., Maggioni C., Mezghrani A., Sitia R., Braakman I., Heck A. J. (2003) Immunity 18, 243–253 [DOI] [PubMed] [Google Scholar]
- 72.Mettetal J. T., Muzzey D., Gómez-Uribe C., van Oudenaarden A. (2008) Science 319, 482–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macia J., Regot S., Peeters T., Conde N., Solé R., Posas F. (2009) Sci. Signal. 2, ra13. [DOI] [PubMed] [Google Scholar]