Abstract
Notch and transforming growth factor-β (TGFβ) play pivotal roles during vascular development and the pathogenesis of vascular disease. The interaction of these two pathways is not fully understood. The present study utilized primary human smooth muscle cells (SMC) to examine molecular cross-talk between TGFβ1 and Notch signaling on contractile gene expression. Activation of Notch signaling using Notch intracellular domain or Jagged1 ligand induced smooth muscle α-actin (SM actin), smooth muscle myosin heavy chain, and calponin1, and the expression of Notch downstream effectors hairy-related transcription factors. Similarly, TGFβ1 treatment of human aortic smooth muscle cells induced SM actin, calponin1, and smooth muscle protein 22-α (SM22α) in a dose- and time-dependent manner. Hairy-related transcription factor proteins, which antagonize Notch activity, also suppressed the TGFβ1-induced increase in SMC markers, suggesting a general mechanism of inhibition. We found that Notch and TGFβ1 cooperatively activate SMC marker transcripts and protein through parallel signaling axes. Although the intracellular domain of Notch4 interacted with phosphoSmad2/3 in SMC, this interaction was not observed with Notch1 or Notch2. However, we found that CBF1 co-immunoprecipitated with phosphoSmad2/3, suggesting a mechanism to link canonical Notch signaling to phosphoSmad activity. Indeed, the combination of Notch activation and TGFβ1 treatment led to synergistic activation of a TGFβ-responsive promoter. This increase corresponded to increased levels of phosphoSmad2/3 interaction at Smad consensus binding sites within the SM actin, calponin1, and SM22α promoters. Thus, Notch and TGFβ coordinately induce a molecular and functional contractile phenotype by co-regulation of Smad activity at SMC promoters.
Keywords: Cell/Differentiation, Cytokines/TGFbeta, Development Differentiation/Muscle, DNA/Transcription, Receptors, Signal Transduction
Introduction
Smooth muscle cells (SMC)5 undergo significant phenotypic modulation during embryonic differentiation and following vascular injury. The differentiated SMC is associated with high expression of several specific contractile proteins including smooth muscle α-actin (SM actin), smooth muscle myosin heavy chain (SM-MHC), SM22α, and calponin1 (1). Multiple factors regulate SMC phenotype, particularly myocardin and serum-response factor, which function in a CArG-dependent pathway (1, 2). We previously reported that Notch activation strongly induces SM actin transcription and protein accumulation, and this process is antagonized by HRT disruption of the Notch-CBF1 complex at the SM actin promoter (3). In addition, other laboratories described Notch in SMC differentiation in vitro (4–9) and identified SM actin and SM-MHC as direct transcriptional targets of Notch-CBF1 (4, 5). Notch regulates differentiation through multiple mechanisms including direct transcription regulation, post-transcriptional regulation of mRNA (10), and regulation of protein turnover (11, 12).
Members of the transforming growth factor-β (TGFβ) family also induce SMC marker gene expression in multiple cell types (13, 14), although this has not been characterized in primary human SMC. Therefore, although signals mediated by TGFβ receptor and Notch receptors activate a similar phenotype in SMC, there is increasing appreciation for cross-talk of these pathways. TGFβ and Notch signaling interact in multiple cell types (15–18). Mechanisms of cooperation include regulation of expression of the other signaling pathway (ligands, receptors, effector molecules), co-regulation of target genes, and direct binding of Notch intracellular domain (NICD) to Smad. The relationship of Notch and TGFβ signaling in the regulation of SMC gene expression is unknown. Our goal was to address mechanisms of cross-talk between Notch and TGFβ in the regulation of SMC contractile marker genes at the molecular and functional level.
We utilized primary human aortic SMC, which express low but highly inducible levels of SMC contractile proteins. We extended our previous findings of Notch regulation of SM actin and demonstrate an overall activation of the SMC differentiation phenotype by both Notch and TGFβ signaling. This activation corresponds to a functional increase in SMC contractility. Our data support a model by which TGFβ-induced Smad transcriptional activity is synergistically increased by Notch activation via CBF1 interaction with phosphoSmad (pSmad) and increased pSmad binding to target SMC marker promoters. This provides an important mechanism by which SMC phenotype can be amplified rapidly following the activation of both Notch and TGFβ signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
Human aortic SMC from Cambrex (Walkersville, MD) were maintained in SmGM2 medium and used between passages 4 and 7. For TGFβ1 treatment, cells were grown for 24 h in serum-free medium before TGFβ1 (PeproTech) addition. For transduction with adenoviral expression constructs, we optimized conditions and tested for dose responsiveness of SMC marker expression to SMC marker expression (supplemental Fig. S1). The adenoviral concentration chosen (100 TCID50 viral particles/cell) in all experiments is indicated by 1x in supplemental Fig. S1. Cells at 50% confluence were transduced with adenoviral constructs for 12 h followed by change of medium as described earlier (3). For Jagged1 induction of Notch signaling, human IgG specific for the Fc region (1 μg/ml in phosphate-buffered saline) was coated on plates for 2 h at room temperature followed by a gentle wash with phosphate-buffered saline. 1 μg of Fc or recombinant rat Jagged1-Fc (1 μg/ml in phosphate-buffered saline) was applied and incubated for 16 h at 4 °C prior to plating cells. For collagen gel contraction assays, a bottom layer of 1.2 mg/ml collagen (Advanced BioMatrix) was polymerized in a 24-well plate (0.3 ml of gel/well). An SMC-embedded layer of 1.2 mg/ml collagen (1 × 106cells/ml) was plated above the lower gel (0.3 ml of gel/well). After polymerization, the gels were released from the sides of the well and cultured in medium with or without additions, and contraction was documented. The gel area was quantified using Scion Image and expressed as the percentage of the area of the entire well. Gene expression constructs and viral purification were previously described (19).
Immunoblotting and Immunohistochemistry
Cell lysates were prepared and an immunoblot was performed as described (20). Antibodies were: anti-V5 (Invitrogen), anti-β-actin (Sigma), anti-SM actin (Sigma), anti-HA (Sigma), anti-calponin1 (Sigma), anti-SM22α (Abcam), pSmad2/3 (21) (provided by V. Lindner), anti-FLAG (Sigma), anti-fibronectin (Sigma), and anti-procollagen (provided by V. Lindner). For immunofluorescence, cells were fixed in 4% paraformaldehyde followed by permeabilization in 0.2% Triton X-100/phosphate-buffered saline. Primary antibodies were used at a 1:400 dilution followed by detection using fluorescein isothiocyanate-conjugated secondary antibody.
RT-PCR and Quantitative RT-PCR
Total RNA was extracted using TRI Reagent (Sigma), treated with RNase-free DNase I (Promega), and reverse transcribed using 20 pmol/liter oligo(dT) with avian myeloblastosis virus reverse transcriptase (Promega). Quantitative RT-PCR was performed using the iCycler (Bio-Rad) using SYBR Green (Bio-Rad) with 20 ng of cDNA as template. Each sample was amplified in triplicate. Threshold cycle numbers were calculated at the log phase of amplification and normalized to cyclophilin as described previously (3). Primers to detect Notch receptors were: Notch1, 5′-TCCACCAGTTTGAATGGTCA-3′, 5′-AGCTCATCATCTGGGACAGG-3′; Notch2, 5′-CCCACCATGTACCAGATTCC-3′, 5′-AGCAGCATTTGAGGAAGCAT-3′; Notch3, 5′-GATGAGCTTGGGAAATCAGC-3′, 5′-GATCTCACGGTTGGCAAAGT-3′; Notch4, 5′-AAAGATGCCCAGGACAACAG-3′, 5′-GTCAGCAGATCCCAGTGGTT-3′.
Promoter Reporter Luciferase Assay
SMC were plated at 20,000 cells/well and transfected 24 h later using 100 TCID50 viral particles/cell adenovirus, 0.25 μg of reporter plasmid, 0.75 μl of GeneJuice (Invitrogen), and 25 ng of Renilla luciferase plasmid/well. 24 h after transfection, cells were starved in serum-free medium for 24 h and then treated with TGFβ1 for 24 h before collection for luciferase assay as described (20). Experiments were repeated at least four times, and the results from a representative experiment are shown with standard deviations.
Co-immunoprecipitation Assay
Cell lysates and immunoprecipitations were performed as described (20, 22). GFP- and NICD-transduced cells were placed in serum-free medium for 24 h before stimulation with TGFβ1 for 1 h. Cell lysates were immunoprecipitated with anti-pSmad2/3 antibodies followed by the addition of protein A/G plus agarose beads. Proteins were eluted and subjected to SDS-PAGE. Immunoblots were probed with anti-V5 or anti-HA antibodies.
Statistical Analysis
All experiments were performed independently a minimum of four times to assure reproducibility of results. Statistical analyses were performed using Student's t test with significance at p < 0.05. Data are presented as means ± S.D.
RESULTS
Notch Signaling Promotes, whereas HRT Suppresses, an SMC Contractile Phenotype
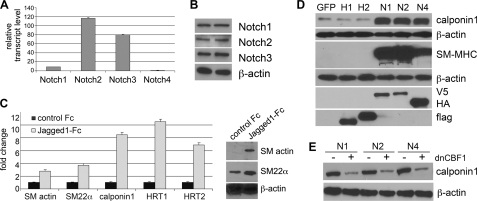
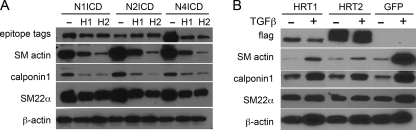
In vivo, Notch expression varies during development and vascular remodeling. We evaluated endogenous expression of Notch signaling components in primary human aortic SMC (Fig. 1, A and B). Under normal growth conditions, transcripts for all four Notch receptors were detectable, although Notch4 was significantly lower than the rest (8-fold lower than Notch1 transcript). At the protein level, we detected significant levels of Notch1, Notch2, and Notch3 protein by immunoblot, whereas Notch4 protein was undetectable. Because Jagged1 ligand has been implicated in regulation of SMC differentiation in vitro and in vivo (4, 23), we verified activation of endogenous Notch signaling using Jagged1-Fc to induce a contractile phenotype characterized by induction of SMC markers SM actin, SM22α, calponin1, and the Notch targets HRT1 and HRT2. Transcripts for SMC markers were induced as early as 8 h after ligand stimulation (Fig. 1C, left) followed later with protein accumulation (Fig. 1C, right). Because Jagged1 can activate multiple Notch receptors, we used constitutively active NICD forms of the Notch receptor in the study to enable better understanding of the individual receptors themselves. We have shown previously that Notch activation significantly increased SM actin and SM-MHC, whereas HRT1 or HRT2 expression did not mimic the effects of NICD (3). By immunoblot analysis (Fig. 1D), we confirmed the strong induction of contractile proteins following Notch activation. Although the CBF1 dependence of SM actin and SM-MHC activation by Notch has been documented (5, 6), we also found that Notch induction of calponin1 was dependent on CBF1 activity (Fig. 1E).
FIGURE 1.
Notch signaling promotes, whereas HRT suppresses, an SMC differentiated contractile phenotype. Primary human aortic SMC were studied. A, total RNA was collected and reverse transcribed, and mRNA for Notch1, Notch2, Notch3, and Notch4 was detected by quantitative RT-PCR. B, cell lysates were collected in duplicate and used for immunoblot analysis to detect Notch proteins. Notch4 protein was not detectable. C, left, the Jagged1-Fc ligand or control Fc was immobilized on anti-IgG antibodies, and SMC was plated on top for 8 h. Total RNA was collected for quantitative RT-PCR for the transcripts indicated. Data were normalized to -fold change when compared with expression on control Fc. Right, total cell lysates were collected from SMC activated by plating on Jagged1-Fc when compared with control Fc for 48 h. Immunoblots detected increased SM actin and SM22α in response to Jagged1-Fc. In A and C, data are presented as means ± S.D. D, SMC were transduced with GFP, HRT1 (H1), HRT2 (H2), Notch1ICD (N1), Notch2ICD (N2), or Notch4ICD (N4), and cell lysates were collected for immunoblot. Expression was verified by detection of epitope tags for N1/N2 (V5), N4 (HA), and H1/H2 (flag). Most lysates were analyzed 3 days after transduction, except for SM-MHC, which takes longer to accumulate and is shown 7 days after transduction. E, cells were stimulated with NICD in the presence (+) or absence (−) of dominant negative CBF1, and cell lysates were collected after 3 days for analysis of calponin1 protein by immunoblot. β-actin was used as a loading control in immunoblots.
TGFβ Activates a Similar Contractile Phenotype in Human SMC
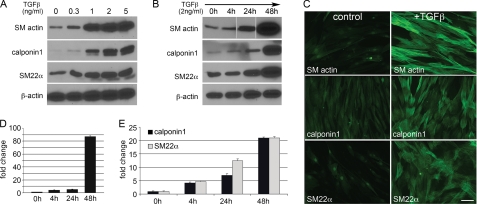
TGFβ plays an important role in SMC proliferation and differentiation, but few studies have addressed TGFβ1 in primary human SMC differentiation. We performed a dose-response study of increasing TGFβ1 on SMC contractile markers. Serum-starved SMC were stimulated for 48 h with TGFβ1, which caused a dose-dependent increase in SM actin, calponin1, and SM22α (Fig. 2A). In subsequent studies, we used 2 ng/ml TGFβ1 to induce SMC marker expression. Kinetic analysis showed major contractile protein accumulation between 24 and 48 h after TGFβ treatment (Fig. 2B). These changes were confirmed by immunofluorescence, which reflected induction of an organized cytoskeletal network in the presence of TGFβ1 (Fig. 2C). To determine whether marker gene transcription is regulated by TGFβ, quantitative RT-PCR was performed (Fig. 2, D and E). TGFβ1 induced an ∼80-fold increase in SM actin transcript by 48 h and a time-dependent accumulation of ∼20-fold calponin1 and SM22α transcripts after 48 h.
FIGURE 2.
TGFβ1 increases SMC differentiated contractile marker expression in a dosage- and time-dependent manner. A, primary human aortic SMC were serum-starved for 24 h and then treated with TGFβ1 at various concentrations for 2 days. Lysates were collected for immunoblot analysis for proteins indicated. B, SMC were serum-starved for 24 h and then treated with 2 ng/ml TGFβ1. Cell lysates were collected at the indicated times after TGFβ addition and used for immunoblot analysis. C, immunofluorescence staining was performed on control- or TGFβ1-treated SMC (2 ng/ml for 4 days) to localize SM actin, calponin1, and SM22α. Scale bar = 20 μm. D and E, serum-starved SMC were treated with 2 ng/ml TGFβ1 for the indicated times, and total RNA was collected for expression analysis by quantitative RT-PCR for SM actin (D), calponin, or SM22α (E). Data are presented as -fold change when compared with control SMC without TGFβ1. Data are presented as means ± S.D.
Notch and TGFβ Interact to Increase a Functional SMC Contractile Phenotype
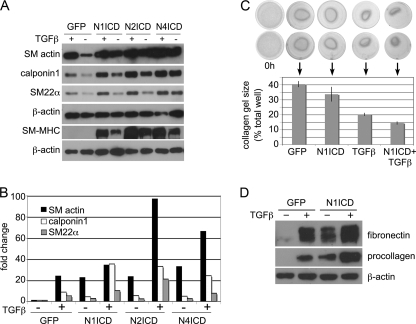
The interaction of Notch and TGFβ signaling has been reported in various cells (24–26). Although both induce an SMC contractile phenotype, the interaction or potential synergy of the two pathways has not been addressed in SMC. We measured SM actin, calponin1, SM22α, and SM-MHC with the combination of activated Notch signaling and TGFβ1 treatment (Fig. 3A). Activation of both pathways leads to increased accumulation of all SMC markers greater than either one alone. In addition, Notch activation and TGFβ1 stimulation displayed corresponding activation of SM actin, calponin1, and SM22α transcripts (Fig. 3B). To test whether this increased molecular contractile phenotype translated to increased functional contractile force, we used a collagen gel contraction assay. SMC embedded within a three-dimensional collagen matrix exert contractile force over time, leading to decreased gel size. We tested the contractile ability of control (GFP) cells, Notch-activated (N1ICD) cells, TGFβ1-treated cells, and cells with Notch activation plus TGFβ1 treatment (Fig. 3C). Although both NICD and TGFβ1 treatment led to increased gel contraction, SMC with both treatments had the greatest contractile response. These functional data correspond to our molecular observations of the SMC phenotype with increased procollagen and fibronectin (Fig. 3D), suggesting a cooperative effect on SMC differentiation.
FIGURE 3.
Notch and TGFβ1 function cooperatively to regulate SMC differentiation marker proteins and SMC contraction. A, control (GFP)- or NotchICD-transduced serum-starved SMC were treated with 2 ng/ml TGFβ1 for 2 days (for SM actin, calponin1, and SM22α analysis) or 4 days (for SM-MHC), and cell lysates were collected for immunoblot analysis for SMC marker proteins. B, SMC were transduced with GFP or NICD for 24 h, serum-starved for 24 h, and treated with 2 ng/ml TGFβ1 (+) for 24 h. Total RNA was collected for quantitative RT-PCR for SMC markers. Data are expressed as -fold change in transcripts when compared with GFP-transduced cells without TGFβ1. C, collagen gel contraction assays were used to determine the contractile ability of SMC expressing GFP or Notch1ICD (N1ICD) in the presence or absence of 2 ng/ml TGFβ1. Data are presented as means ± S.D. D, control (GFP) serum-starved or NotchICD-transduced serum-starved SMC were treated with 2 ng/ml TGFβ1 for 2 days, and cell lysates were analyzed for fibronectin and procollagen.
Notch and TGFβ1 Activate Parallel Pathways
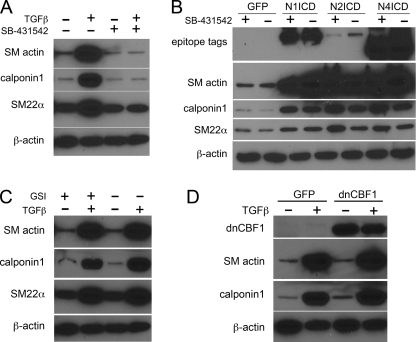
To explore the relationship between Notch and TGFβ signaling, we first asked whether the Notch pathway acts upstream of TGFβ or vice versa. To address this, GFP- or NICD-transduced SMC were treated with the TGFβ receptor inhibitor 4-(5-benzol[1,3]dioxol-5-yl-4-pyrldin-2-yl-1H-imidazol-2-yl)-benzamide hydrate (SB-431542) (27), which completely suppresses TGFβ1-induced accumulation of SM actin, calponin1, and SM22α protein (Fig. 4A). SMC were transduced with GFP or NICD and treated with SB-431542 or vehicle for 48 h, after which the cells were collected for immunoblot (Fig. 4B). Inhibition of TGFβ receptor with SB-431542 did not affect the ability of Notch to induce SMC contractile markers, showing that the Notch signal is independent of TGFβ signaling.
FIGURE 4.
Notch and TGFβ1 regulate SMC differentiation markers by parallel signal pathways. A, serum-starved human primary SMC were stimulated with 2 ng/ml TGFβ1 (+) in the presence or absence of SB-431542 (10 μm) for 2 days. Cell lysates were collected for immunoblot analysis. B, SMC were transduced with ICD forms of Notch1 (N1ICD), Notch2 (N2ICD), or Notch4 (N4ICD) or control GFP and grown in the absence (−) or presence (+) of (10 μm) of SB-431542 for 2 days before cell lysate collection for immunoblot. C, serum-starved SMC were stimulated with 2 ng/ml TGFβ1 (+) for 48 h in the presence or absence of 10 μm γ-secretase inhibitor (GSI +) or dimethyl sulfoxide (DMSO) control (−) before cell lysis for immunoblotting. D, SMC were transduced with GFP or dominant negative CBF1 (dnCBF1) and treated with 2 ng/ml TGFβ1 (+) for 2 days before collection for immunoblot.
Likewise, we examined whether TGFβ-mediated SMC marker induction occurs via the Notch signaling pathway. SMC were serum-starved and stimulated with TGFβ1 in the presence or absence of the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine-t-butylester (28) or vehicle for 48 h followed by analysis of SMC contractile protein levels. Although γ-secretase inhibitor suppresses Notch ligand-induced SM actin production in SMC (3), it had no effect on TGFβ1-induced SM actin, calponin1, or SM22α proteins (Fig. 4C).
We also tested a dominant negative CBF1 construct, which inhibits Notch-induced SM actin expression in SMC (3). Inhibition of CBF1 did not affect the ability of TGFβ1 to increase SM actin or calponin1 protein (Fig. 4D). TGFβ1 also does not affect endogenous expression of Notch receptors (not shown). These data suggest that Notch and TGFβ1 do not regulate SMC phenotype by controlling the other signaling pathway.
HRT Is a General Suppressor of SMC Contractile Protein Expression
Members of the HRT family of transcription factors are typically considered Notch effector proteins in selected cell types including SMC. However, HRT proteins also have negative regulatory activity in both Notch-induced and myocardin-induced SMC differentiation (3, 29). Therefore, we characterized HRT activity in the context of Notch- and TGFβ-induced SMC marker expression. We previously reported that HRT1 or HRT2 effectively inhibit Notch-induced SM actin accumulation (3), and in comparison, HRT had the same effect on calponin1 and SM22α protein (Fig. 5A). Similarly, the strong induction of all three markers by TGFβ1 was inhibited by HRT1 or HRT2 (Fig. 5B). These data further expand the activity of HRT proteins as antagonists of several pathways that drive the SMC differentiated contractile phenotype.
FIGURE 5.
HRTs antagonize Notch and TGFβ1 activity in SMC differentiation marker expression. A, primary human aortic SMC were transduced with NotchICD alone (−) or with HRT1 (H1) or HRT2 (H2) co-expression for 3 days before collection of cells for immunoblot analysis. Expression of NICD was verified by detection of epitope tags for N1ICD/N2ICD (V5) or N4 (HA). B, SMC transduced with GFP or HRT1 or HRT2 were grown in the absence (−) or presence (+) of 2 ng/ml TGFβ1 for 48 h before collection for immunoblot analysis. HRT expression was confirmed by detection of the FLAG epitope tag.
Notch-CBF1 Pathway Is Involved in Protein Interactions with Smad in SMC
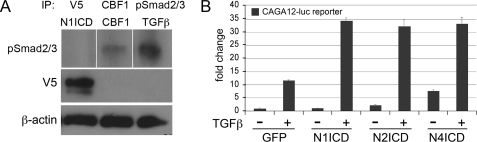
A potential mechanism of Notch/TGFβ cross-talk has been suggested via direct binding of NotchICD and Smad (17–19). To address this possibility, pSmad2/3 was immunoprecipitated from GFP- or NICD-transduced SMC that had been stimulated for 1 h with TGFβ1 before collection and immunoprecipitation. When the pSmad2/3 immunopellets were analyzed for NICD, we consistently detected Notch4ICD, but not Notch1ICD or Notch2ICD (data not shown). Although our findings are consistent with previous reports (24–26), it is unlikely that the interaction of Notch4ICD with pSmad2/3 explains the co-regulation of SMC markers. Cooperation with TGFβ1 signaling is common to activation of multiple Notch receptors, although neither Notch1ICD nor Notch2ICD could be immunoprecipitated with pSmad2/3 under comparable conditions. However, when the common downstream mediator CBF1 was expressed in SMC (3), we detected interaction with pSmad2/3 in immunoprecipitates (Fig. 6A), suggesting a novel mechanism of Smad regulation. If this interaction has functional consequences, we would expect that Notch activation would regulate Smad2/3 transcriptional activity. This was tested using the TGFβ-responsive CAGA12 construct (30) in the presence or absence of Notch activation. As expected, TGFβ1 treatment alone induced reporter activity ∼10-fold; however, concurrent activation of Notch significantly increased the activity of the Smad2/3 reporter >30-fold when compared with basal activity (Fig. 6B). We also tested the impact of TGFβ1 signaling on basal and Notch-induced CBF1 reporter transactivation, and no changes were observed (data not shown). Our results suggest that the interaction of Notch/TGFβ selectively modulates pSmad2/3 promoter binding activity.
FIGURE 6.
Molecular and functional interactions of Notch and TGFβ1 signaling networks. A, human primary SMC expressing Notch1ICD (left) or CBF1 (middle) or treated with TGFβ1 (right) were lysed and immunoprecipitated (IP) with antibodies against V5 (N1ICD epitope tag), CBF1, or pSmad2/3. Immunoprecipitates were separated and immunoblotted with anti-pSmad2/3. B, luciferase promoter transactivation assays were performed with the TGFβ1-responsive CAGA12-luc reporter construct. SMC were transduced as indicated and stimulated with 2 ng/ml TGFβ1 for 24 h before quantification of luciferase activity. Data are expressed as -fold change when compared with GFP-transduced cells without the addition of TGFβ1. Data are presented as means ± S.D.
Notch Activation Increases TGFβ1-induced Binding of pSmad2/3 to Promoters of SMC Markers
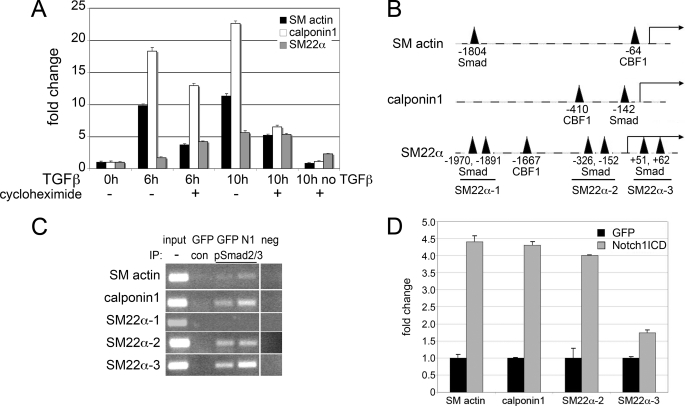
Regulation of SMC marker genes by TGFβ1 could be via Smad-mediated transcription by interaction with consensus binding regions in target promoters or via an indirect mechanism. To test whether protein synthesis was required for the changes in SMC marker expression in response to TGFβ1, we used cycloheximide to block translation (Fig. 7A). Although there were reduced SM actin and calponin1 transcripts in the presence of cycloheximide + TGFβ1 when compared with TGFβ1 alone, there was still 5–10-fold increase, suggesting that induction can still occur without new protein translation. Analysis of the 2-kb upstream promoter regions of these SMC genes identified Smad and CBF1 consensus binding sites in all genes (Fig. 7B), with regions upstream of the SM22α coding sequence having three potential Smad binding regions. Primers were designed to span the Smad consensus regions within each promoter, and chromatin immunoprecipitation assays were performed to detect pSmad2/3 binding to these regions (Fig. 7C). SMC were transduced with GFP or N1ICD and treated with TGFβ1 for 1 h, and cross-linked protein-DNA complexes were immunoprecipitated with either control IgG or anti-pSmad2/3. Following TGFβ1 treatment alone and immunoprecipitation with anti-pSmad2/3 (GFP pSmad2/3 lane), amplification of product spanning each of the predicted Smad binding sites was detected, with the exception of the SM22α-1 region encompassing the −1970/−1891 sites (Fig. 7B). In the absence of TGFβ1 treatment, we were unable to detect pSmad2/3 binding to the SM actin, calponin1, and SM22α promoters in the ChIP assay (data not shown). In addition, no product amplification was observed under any condition when immunoprecipitated with control IgG (GFP con lane). In the presence of Notch1ICD (N1 lane), we observed an apparent increase in product representing increased immunoprecipitation of specific DNA bound pSmad2/3. Using quantitative PCR, we verified that NotchICD in combination with TGFβ1 elevated pSmad2/3 binding as detected by consistently increased PCR product amplification in immunoprecipitates with NICD and TGFβ1 (Fig. 7D).
FIGURE 7.
Activated Notch signaling enhances Smad2/3 binding to SMC promoters. A, human aortic SMC were serum-starved and then stimulated with 2 ng/ml TGFβ1 for 6 or 10 h in the presence or absence of (10 μg/ml) cycloheximide. Cells were collected for quantitative RT-PCR. Data are expressed as -fold change when compared with cells with no TGFβ1 treatment and no cycloheximide (0h). B, promoter sequences were evaluated 2 kb upstream of the transcriptional start site. Indicated are consensus binding sites for Smad and CBF1. C, SMC were transduced with GFP or N1ICD (N1) and stimulated with 2 ng/ml TGFβ1 for 1 h. Cells were collected for chromatin immunoprecipitation (IP) assays using control antibody (con) or anti-pSmad2/3. Input shows material before immunoprecipitation. PCR amplification was performed to amplify the regions including the Smad binding sites of SM actin, calponin1, and the three regions in the SM22α promoter that contain Smad sites. neg, negative control. D, immunoprecipitated samples from C were used for quantitative RT-PCR to compare product with Notch activation. Values were normalized to amplification from GFP transfectants. Data are presented as means ± S.D.
DISCUSSION
Regulation of SMC phenotype is a complex, multifactoral process involving the myocardin-SRF complex and other pathways, including Notch and TGFβ signaling. We extend our previous characterization of Notch regulation of SM actin transcription (3) to show that Notch activation induces a functional contractile phenotype, as does TGFβ1, in primary human SMC. Further, HRT factors function as general inhibitors of the contractile phenotype and can effectively block SMC differentiation induced by multiple stimuli, including myocardin, Notch, and TGFβ. This negative feedback pathway is an adaptable mechanism that could account for initial vascular response to injury that includes suppression of the contractile phenotype. Although there is basal expression of Notch in the adult vasculature, injury leads to strong up-regulation of all Notch receptors in vascular cells (31). We predict that increased Notch signaling in SMC elevates HRT levels to an active threshold that antagonizes the differentiated phenotype, allowing for active SMC remodeling. As Notch signaling decreases, decreased HRT levels would allow re-establishment of the contractile phenotype.
The function of HRTs as transcriptional repressors is documented (3, 7, 22, 32), but this represents the first demonstration that HRT opposes TGFβ1. The potential mechanism needs further investigation, but there are several possibilities. HRTs may inhibit pSmad2/3 binding to SMC gene promoters directly or indirectly, similar to their inhibition of NICD/CBF1 binding to the CBF1 site in SM actin (3). Alternatively, HRTs may repress downstream TGFβ1 signaling via regulation of SRF and myocardin binding to SMC promoters. HRT2 has been shown to repress myocardin-induced SMC differentiation (29), and TGFβ up-regulates SRF expression in hepatic stellate cells (33). Therefore, interaction of HRTs with myocardin-SRF should be considered. Finally, analysis of SMC marker promoter sequences identified several HRT consensus sites within the SM actin and calponin1 promoters. Thus, direct DNA binding activity may mediate transcriptional repression.
Although TGFβ regulates SMC differentiation, recent studies highlight the importance of understanding cross-talk between Notch and the TGFβ/BMP superfamily. NICD blocks TGFβ-mediated growth arrest by indirectly deregulating c-Myc expression in epithelial cells (24). In C2C12 cells, the cooperative role of Notch and TGFβ in mediating Hes1 expression is through direct protein-protein interactions between the signal-transducing intracellular elements from both pathways (25). In neuroepithelial cells, BMP2 enhances Notch-induced Hes5 expression by NICD/Smad1 interaction in the presence of P/CAF and p300 (26). There are multiple mechanisms of Notch and TGFβ interaction that may be cell type-dependent. In the case of SMC, both Notch signaling and TGFβ signaling individually promote SMC differentiation and apparently intersect at the level of transcriptional regulation of specific target genes. We tested whether a potential Smad/NICD or Smad/CBF1 interaction would affect Notch target or TGFβ/Smad target genes. Although we utilized CBF1 reporter constructs and NICD to test Notch activation in the presence or absence of TGFβ1, we found that TGFβ1 addition had no effect on CBF1 promoter transactivation (not shown). TGFβ1 stimulation also did not enhance NICD transactivation of a 157-bp SM actin promoter construct containing only a CBF1 binding site (3) (not shown).
We therefore focused on the cooperative impact of the two signaling pathways on Smad activity. Two lines of evidence suggest a mechanism by which Notch activation with TGFβ1 signaling promotes Smad transcriptional activity. First, the combined interaction and activation of NICD and TGFβ1 on CAGA12 reporter transactivation suggests that Smad transcriptional activity is altered. One possibility is that NICD/CBF1 may stabilize Smad transcriptional complexes similar to the interaction of BMP2 and Notch in inducing Hes5 expression (26). Alternatively, CBF1 binds to pSmad2/3, and this interaction may promote DNA binding or transcriptional activation.
Second, transcript levels for SM actin, calponin1, and SM22α were increased by activation of both pathways, and Smad interaction with promoter binding sites of each of these genes was increased by Notch activation. Each of these promoters contains both CBF1 and Smad consensus binding sites; thus, it is possible that NICD/CBF1 complex binding to adjacent promoter regions provides a cis regulatory signal to promote Smad binding. However, the fact that an amplified transcriptional effect was observed in the artificial CAGA12 construct that does not bind NICD/CBF1 suggests that DNA binding to CBF1 sites is not required for regulation of Smad DNA binding or transcriptional activity. Rather, the binding of CBF1 to Smad may be sufficient to regulate its function. Similar regulatory mechanisms were defined for BMP to either repress or promote Smad activity. For example, binding of cGMP-dependent kinase I to Smad promotes BMP target activation (34), and Tbx20 binding to Smad1 and Smad5 inhibits BMP/Smad-dependent activation of target promoters by sequestration from Smad4 (35). Future mutation studies are necessary to determine the relationship of DNA binding of Notch-CBF1 complexes to regulation of Smad activity.
Another future consideration is the impact of Notch signaling on alternative TGFβ signaling pathways. TGFβ1 can phosphorylate Smad2/3 and Smad1/5/8 through ALK5 and ALK1, respectively, in endothelial cells, neurons, and human chondrocytes (36–39). We found that TGFβ1 can induce phosphorylation of Smad2/3 and Smad1/5/8 in SMC, and ALK5 is required for TGFβ1 regulation of SMC gene expression (not shown). Subsequent downstream signaling is complex, not only involving Smads but also kinases such as p38 mitogen-activated protein kinase, ERK1/2, and JNK (40). TGFβ1 activates Smad-independent pathways such as ERK/mitogen-activated protein (MAP) kinase signaling through direct phosphorylation of ShcA (41). Consistent with this, inhibition of ERK dramatically repressed TGFβ1-induced SMC gene expression in our system (data not shown). Thus, further clarification of TGFβ1-mediated pathways in SMC and the impact of Notch signaling on these alternative pathways will better define cooperative mechanisms between these important regulators of SMC phenotype.
In conclusion, we identified novel activities of HRTs as general inhibitors of SMC contractile phenotype as they counter both Notch and TGFβ1 pathways. Notch and TGFβ signaling regulates SMC gene expression cooperatively via parallel axes, which interact at the level of signal-transducing intracellular elements that regulate Smad activity. These studies provide novel evidence of cross-talk of Notch and TGFβ signaling in regulating SMC gene expression, which is important to understand SMC phenotypic transitions.
Supplementary Material
Acknowledgments
We thank our Viral Vector Core Facility for the amplification of adenoviral vectors and Drs. Jeong Yoon (Maine Medical Center Research Institute) and Howard Crawford (The State University of New York, Stony Brook, NY) for critical feedback on research strategy. We thank Dr. Volkhard Lindner (Maine Medical Center Research Institute) for the phosphoSmad and procollagen antibodies and helpful discussions. The Viral Vector Core Facility is supported by National Institutes of Health Grant P20RR15555 from the National Center for Research Resources.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL070865 (to L. L.) and P20RR15555 from the National Center for Research Resources (to R. E. Friesel and L. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- SMC
- smooth muscle cell(s)
- SM actin
- smooth muscle α-actin
- SM-MHC
- smooth muscle myosin heavy chain
- TGFβ
- transforming growth factor-β
- HRT
- hairy-related transcription factor
- NICD
- Notch intracellular domain
- RT-PCR
- reverse transcription-PCR
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- BMP
- bone morphogenetic protein
- SRF
- serum-response factor
- pSmad
- phosphoSmad
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida T., Kawai-Kowase K., Owens G. K. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1596–1601 [DOI] [PubMed] [Google Scholar]
- 3.Tang Y., Urs S., Liaw L. (2008) Circ. Res. 102, 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi H., Iso T., Sato H., Yamazaki M., Matsui H., Tanaka T., Manabe I., Arai M., Nagai R., Kurabayashi M. (2006) J. Biol. Chem. 281, 28555–28564 [DOI] [PubMed] [Google Scholar]
- 5.Noseda M., Fu Y., Niessen K., Wong F., Chang L., McLean G., Karsan A. (2006) Circ. Res. 98, 1468–1470 [DOI] [PubMed] [Google Scholar]
- 6.Morrow D., Scheller A., Birney Y. A., Sweeney C., Guha S., Cummins P. M., Murphy R., Walls D., Redmond E. M., Cahill P. A. (2005) Am. J. Physiol. Cell Physiol. 289, C1188–C1196 [DOI] [PubMed] [Google Scholar]
- 7.Proweller A., Pear W. S., Parmacek M. S. (2005) J. Biol. Chem. 280, 8994–9004 [DOI] [PubMed] [Google Scholar]
- 8.Wang W., Prince C. Z., Hu X., Pollman M. J. (2003) Biochem. Biophys. Res. Commun. 308, 596–601 [DOI] [PubMed] [Google Scholar]
- 9.Sweeney C., Morrow D., Birney Y. A., Coyle S., Hennessy C., Scheller A., Cummins P. M., Walls D., Redmond E. M., Cahill P. A. (2004) FASEB J. 18, 1421–1423 [DOI] [PubMed] [Google Scholar]
- 10.Thatcher E. J., Flynt A. S., Li N., Patton J. R., Patton J. G. (2007) Dev. Dyn. 236, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 11.Nie L., Wu H., Sun X. H. (2008) J. Biol. Chem. 283, 684–692 [DOI] [PubMed] [Google Scholar]
- 12.Wang C., Qi R., Li N., Wang Z., An H., Zhang Q., Yu Y., Cao X. (2009) J. Biol. Chem. 284, 16183–16190 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ono Y., Sensui H., Okutsu S., Nagatomi R. (2007) J. Cell. Physiol 210, 358–369 [DOI] [PubMed] [Google Scholar]
- 14.Hung S. C., Kuo P. Y., Chang C. F., Chen T. H., Ho L. L. (2006) Cell Tissue Res. 324, 457–466 [DOI] [PubMed] [Google Scholar]
- 15.Kennard S., Liu H., Lilly B. (2008) J. Biol. Chem. 283, 1324–1333 [DOI] [PubMed] [Google Scholar]
- 16.Niimi H., Pardali K., Vanlandewijck M., Heldin C. H., Moustakas A. (2007) J. Cell Biol. 176, 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zavadil J., Cermak L., Soto-Nieves N., Bottinger E. P. (2004) EMBO J. 23, 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y., Lowther W., Kato K., Bianco C., Kenney N., Strizzi L., Raafat D., Hirota M., Khan N. I., Bargo S., Jones B., Salomon D., Callahan R. (2005) Oncogene 24, 5365–5374 [DOI] [PubMed] [Google Scholar]
- 19.Aoki K., Barker C., Danthinne X., Imperiale M. J., Nabel G. J. (1999) Mol. Med. 5, 224–231 [PMC free article] [PubMed] [Google Scholar]
- 20.Havrda M. C., Johnson M. J., O'Neill C. F., Liaw L. (2006) Thromb. Haemost. 96, 361–370 [DOI] [PubMed] [Google Scholar]
- 21.LeClair R. J., Durmus T., Wang Q., Pyagay P., Terzic A., Lindner V. (2007) Circ. Res. 100, 826–833 [DOI] [PubMed] [Google Scholar]
- 22.King I. N., Kathiriya I. S., Murakami M., Nakagawa M., Gardner K. A., Srivastava D., Nakagawa O. (2006) Biochem. Biophys. Res. Commun. 345, 446–452 [DOI] [PubMed] [Google Scholar]
- 23.High F. A., Lu M. M., Pear W. S., Loomes K. M., Kaestner K. H., Epstein J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1955–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao P., Kadesch T. (2003) Mol. Cell. Biol. 23, 6694–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blokzijl A., Dahlqvist C., Reissmann E., Falk A., Moliner A., Lendahl U., Ibanez C. F. (2003) J. Cell Biol. 163, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takizawa T., Ochiai W., Nakashima K., Taga T. (2003) Nucleic Acids Res. 31, 5723–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watabe T., Nishihara A., Mishima K., Yamashita J., Shimizu K., Miyazawa K., Nishikawa S., Miyazono K. (2003) J. Cell Biol. 163, 1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comery T. A., Martone R. L., Aschmies S., Atchison K. P., Diamantidis G., Gong X., Zhou H., Kreft A. F., Pangalos M. N., Sonnenberg-Reines J., Jacobsen J. S., Marquis K. L. (2005) J. Neurosci. 25, 8898–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doi H., Iso T., Yamazaki M., Akiyama H., Kanai H., Sato H., Kawai-Kowase K., Tanaka T., Maeno T., Okamoto E., Arai M., Kedes L., Kurabayashi M. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2328–2334 [DOI] [PubMed] [Google Scholar]
- 30.Ozbun L. L., Martinez A., Jakowlew S. B. (2005) Biochim. Biophys. Acta 1728, 163–180 [DOI] [PubMed] [Google Scholar]
- 31.Lindner V., Booth C., Prudovsky I., Small D., Maciag T., Liaw L. (2001) Am. J. Pathol. 159, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa O., McFadden D. G., Nakagawa M., Yanagisawa H., Hu T., Srivastava D., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13655–13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann J., Haas U., Gressner A. M., Weiskirchen R. (2007) Biochim. Biophys. Acta 1772, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 34.Schwappacher R., Weiske J., Heining E., Ezerski V., Marom B., Henis Y. I., Huber O., Knaus P. (2009) EMBO J. 28, 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R., Horsthuis T., Farin H. F., Grieskamp T., Norden J., Petry M., Wakker V., Moorman A. F., Christoffels V. M., Kispert A. (2009) Circ. Res. 105, 442–452 [DOI] [PubMed] [Google Scholar]
- 36.Goumans M. J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., ten Dijke P. (2003) Mol. Cell 12, 817–828 [DOI] [PubMed] [Google Scholar]
- 37.Goumans M. J., Valdimarsdottir G., Itoh S., Rosendahl A., Sideras P., ten Dijke P. (2002) EMBO J. 21, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konig H. G., Kogel D., Rami A., Prehn J. H. (2005) J. Cell Biol. 168, 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finnson K. W., Parker W. L., ten Dijke P., Thorikay M., Philip A. (2008) J. Bone Miner. Res. 23, 896–906 [DOI] [PubMed] [Google Scholar]
- 40.Bobik A. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1712–1720 [DOI] [PubMed] [Google Scholar]
- 41.Lee M. K., Pardoux C., Hall M. C., Lee P. S., Warburton D., Qing J., Smith S. M., Derynck R. (2007) EMBO J. 26, 3957–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.