Abstract
In the Archaea only a handful of ribonucleases involved in RNA processing and degradation have been characterized. One potential group of archaeal ribonucleases are homologues of the bacterial RNase J family, which have a β-CASP metallo-β-lactamase fold. Here we show that β-CASP proteins encoded in the genomes of the hyperthermophilic Euryarchaeota Pyrococcus abyssi and Thermococcus kodakaraensis are processive exoribonucleases with a 5′ end dependence and a 5′ to 3′ directionality. We named these enzymes Pab-RNase J and Tk-RNase J, respectively. RNAs with 5′-monophosphate or 5′-hydroxyl ends are preferred substrates of Pab-RNase J, whereas circularized RNA is resistant to Pab-RNase J activity. Degradation of a 3′ end-labeled synthetic RNA in which an internal nucleoside is substituted by three ethylene glycol units generates intermediates demonstrating 5′ to 3′ directionality. The substitution of conserved residues in Pab-RNase J predicted to be involved in the coordination of metal ions demonstrates their importance for ribonuclease activity, although the detailed geometry of the catalytic site is likely to differ from bacterial RNase J. This is the first identification of a 5′-exoribonuclease encoded in the genomes of the Archaea. Phylogenetic analysis shows that euryarchaeal RNase J has been inherited vertically, suggesting an ancient origin predating the separation of the Bacteria and the Archaea.
Keywords: Archaebacteria, Enzymes, Ribonuclease, RNA, RNA Metabolism, β-CASP Protein
Introduction
The study of RNA metabolism in Archaea is in its infancy (for a recent review, see Ref. 1). The control of RNA processing and mRNA stability are important steps of gene expression. As in all domains of life, processing of tRNA in Archaea is performed by the ribonucleoprotein RNase P (2) and the endoribonuclease RNase Z (3, 4). The RNA-splicing endoribonuclease removes introns from archaeal pre-tRNA and participates in rRNA processing (5, 6). The coupling of transcription and translation in Archaea (7) suggests that mRNA degradation in the Archaea might be more closely related to processes in bacteria than in eukaryotes. To our knowledge, archaeal mRNA is not capped, nor have homologues of eukaryotic enzymes involved in mRNA capping, decapping, and 5′- to 3′-exoribonucleolytic degradation been identified in archaeal genomes (1, 8, 9). However, a eukaryotic-like exosome with a 3′- to 5′-exoribonucleolytic activity is present in most Archaea (10, 11) with the exception of the halophilic archaeon Haloferax where this activity is carried out by RNase R (12). Overall, our comprehension of mRNA degradation in Archaea is very limited, whereas it is quite advanced in Eukarya and bacteria.
In bacteria as well as in Eukarya, the nature of the 5′ end of mRNA is an important determinant for stability. In the model Gram-negative bacterium Escherichia coli, RNase E, a key enzyme in mRNA degradation, is a 5′ end-dependent endoribonuclease (13, 14). Indeed substrates with 5′-monophosphate ends stimulate RNase E activity (15–17). In the model Gram-positive bacterium Bacillus subtilis, the recently identified and characterized RNase J was discovered in part due to efforts to identify an enzyme with RNase E-like activity (18–20). Although the first work on RNase J demonstrated an endoribonuclease activity, subsequent work showed that RNase J also has an exoribonuclease activity with 5′ to 3′ directionality (19, 21, 22). In vitro and in vivo evidence suggest that primary transcripts with a 5′-triphosphate end are protected from degradation by RNase J, whereas transcripts with 5′-monophophosphate or 5′-hydroxyl ends are readily degraded (19, 21, 23, 24). Unlike eukaryotes, where decapping and 5′ end-dependent mRNA degradation by Xrn1 appear to belong to a pathway recently elaborated in evolution (9), studies with E. coli and B. subtilis, which are highly divergent species with truly ancient origins, suggest that at least two distinct pathways for 5′ end-dependent mRNA degradation have arisen in the prokaryotes.
Putative archaeal proteins with sequence similarity to part of RNase E have been reported (25). However, the only protein characterized to date, FAU-1 of the thermophilic archaeon Pyrococcus furiosus, has been shown to have RNA binding activity but not ribonuclease activity (26). More recently, it has been suggested that most euryarchaeal genomes encode a protein related to B. subtilis RNase J (20), which is a member of the β-CASP family of metallo-β-lactamases (27). The β-CASP protein family includes enzymes with DNase and RNase activity (28). Based on sequence comparisons and structural studies, bacterial RNase J has three distinct domains corresponding to a metallo-β-lactamase domain, a β-CASP domain, and a C-terminal domain that is conserved among the bacterial homologues (19). The structure of Thermus thermophilus RNase J with UMP bound shows a unique catalytic site formed by the metallo-β-lactamase and the β-CASP domains. These domains contribute contacts to the 5′-monophosphate of UMP in a structure reminiscent of the 5′ end-sensing pocket of RNase E (19). Although it seemed likely that the β-CASP proteins in the Euryarchaeota would have nucleolytic activity, until now their specificity and functionality were unknown.
Here, we have biochemically characterized homologues of B. subtilis RNase J designated Pab-RNase J and TK-RNase J from the Thermococcale Pyrococcus abyssi and Thermococcus kodakaraensis Archaea, respectively. The corresponding His-tagged proteins expressed in E. coli possess a 5′- to 3′-exoribonucleolytic activity that is 5′ end-dependent. We have determined the importance of the conserved β-CASP key residues in this activity. Furthermore, our phylogenetic study provides evidence for an ancient evolutionary origin of the euryarchaeal RNase J.
EXPERIMENTAL PROCEDURES
Multiple Sequence Alignment and Phylogenetic Tree Constructions
The archaeal phylogenetic tree was inferred from a concatenated dataset of 70 protein families. Each family included proteins encoded by genes conserved as single copy in all taxa (orthologous genes). A large majority of the families were composed of ribosomal proteins or tRNA synthetases. For each family, the sequences were aligned using Muscle (29), and all gaps and ambiguously aligned regions were removed with Gblocks (30). The 70 multiple alignments obtained were concatenated into a single large file of 10,855 aligned amino acid positions. The phylogenetic inferences were made with the maximum likelihood method using PHYML (31). We used the JTT model of amino acid evolution with a gamma distribution fitted to the data by likelihood maximization (32). The RNase J tree was computed with the same approach, and both trees were edited to highlight their congruence. The trees were arbitrarily rooted.
Construction of Plasmid Vectors for the Expression of Pab-RNase J and Variants
Supplemental Table S1 summarizes all the mentioned oligonucleotides. The plasmid used for expression of His-tagged Pab-RNase J was pET15b. The coding sequence (PAB1751) was cloned as an XhoI-BamHI PCR fragment using the PAJ5XhO and PAJ3 oligonucleotides to give the plasmid pEJ9. The H86A, E209A, H388A, and H410A variants were generated by site-directed mutagenesis of pEJ9 with the appropriate oligonucleotides and a QuikChange kit (Stratagene). Deletion of sequences corresponding to loops 1 and 2 were introduced into pEJ9 by reverse PCR with the appropriate oligonucleotides.
Overexpression and Purification of Wild Type Pab-RNase J and Variants
The BL21-CodonPlus (DE3) E. coli strain carrying pEJ9 or plasmids bearing mutations was induced at an A600 of 0.6 by the addition of 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and incubated 3 h at 30 °C. A cell extract was heated to 70 °C for 10 min and clarified by low speed centrifugation. TALON Metal resin (Clontech) was used for immobilized metal ion affinity chromatography purification of the His-tagged protein. Protein (∼50 ng/μl) was dialyzed against 20 mm Hepes, pH 7.5, 300 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and 10% glycerol and stored at 4 °C after the addition of bovine serum albumin (0.2 mg/ml). The enzyme, analyzed by Coomassie-stained 12% SDS-PAGE, migrated at 50 kDa, in agreement with the predicted molecular mass of 50.5 kDa, and was judged greater than 95% pure by visual inspection.
RNA Synthesis and Labeling
In vitro transcription with T7 RNA polymerase was performed as described by the manufacturer (Promega) using PCR fragments as templates. The leader mRNA ThrS, sR47, pre-tRNATrp, and tRNALeu templates were prepared as described (20, 33, 34). The 21-RNA and 21#-RNA were synthesized by Dharmacon. RNA transcripts were treated with calf intestinal phosphatase to obtain dephosphorylated RNAs and subsequently treated with T4 polynucleotide kinase to obtain monophosphorylated RNAs. [α-32P]CTP was used to synthesize uniformly labeled transcripts. 5′ end-labeling with T4 polynucleotide kinase was performed on dephosphorylated RNA or synthetic RNA in the presence of [γ-32P]ATP. 3′ end labeling was carried out with T4 RNA ligase in presence of 5′-[32P]pCp and DMSO. Circular RNA substrates were obtained by incubating 5′ end-labeled RNA with T4 RNA ligase. All labeled RNAs were purified on denaturing 8 or 10% PAGE. RNA circularization was verified by RNase T1 digestion (supplemental Fig. S3).
Degradation Assays
Activity was assayed using labeled in vitro-transcribed RNAs or synthetic RNA in single turnover conditions with a 2–10-fold Pab-RNase J excess or in multiple turnover condition with a 20-fold excess of substrate. A typical enzyme excess reaction in a final volume of 20 μl contained 32P-RNA (5 fmol/μl), 10 or 50 nm wild type Pab-RNase J or variants, 20 mm Hepes, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 0.2 mg/ml bovine serum albumin, and 10% glycerol. In a substrate excess reaction, 20 pmol of nonradioactive RNA was added to the mixture. Reactions were started by the addition of the enzyme and incubated at 65 °C. Samples were withdrawn at the indicated times, and the reaction was stopped by adding formamide-containing dye supplemented with 10 mm EDTA or spotted directly on TLC plates (PEI-cellulose, Nagel). Reaction products in formamide dye mix were separated on denaturing (7 m urea) 8, 10, or 20% PAGE. TLC plates were developed with 0.25 m KH2PO4. All results were analyzed by autoradiography and quantified by phosphorimaging. Initial rates were determined by measuring the slope representing the percentage of substrate degraded during the linear phase of the reaction in at least three independent experiments. The migration positions of nonradioactive CMP and GMP (Fluka) were determined by UV shadowing on TLC plates; pCp was determined by autoradiography of 5′-[32P]pCp (PerkinElmer Life Sciences).
RESULTS
To identify RNase J homologues in the Archaea, we used the NCBI genome blast tool with the B. subtilis RNase J1 sequence (YkqC) as the query. A family of β-CASP proteins related to RNase J was detected throughout the Euryarchaeota except in the Archaeoglobales and the Thermoplasmatales. Protein sequence alignments of the bacterial and euryarchaeal proteins revealed a high degree of similarity, including all the signature motifs characteristic of the β-CASP proteins. The sequence alignment in Fig. 1, which is based on the comparison of the bacterial and euryarchaeal homologues available at the time of the analysis (supplemental Fig. S1), is an overview in which only three sequences are shown. YkqC is the RNase J1 of B. subtilis; PAB1751 and TK1469 are euryarchaeal homologues from P. abyssi and T. kodakaraensis, respectively. Motifs 1–4 correspond to conserved elements in the metallo-β-lactamase domain; Motifs A–C correspond to conserved elements in the β-CASP domain. The major characteristic feature of the euryarchaeal homologues is the lack of the region corresponding to the C-terminal domain of the bacterial homologues. Additional features consist of two short insertions labeled as Loop 1 and Loop 2 and a short deletion just upstream of motif B (Fig. 1 and supplemental Fig. S1). The euryarchaeal homologues, which are on average about 100 residues shorter than their bacterial counterparts, due mainly to the absence of the C-terminal domain, are thus predicted to be canonical β-CASP proteins composed of a metallo-β-lactamase and a β-CASP domain, with no additional domains.
FIGURE 1.
Sequence alignment of RNase J1 of B. subtilis (YkqC) with RNase J homologues from P. abyssi (PAB1751) and T. kodakaraensis (TK1469). This alignment was extracted from a large multiple sequence alignment of ∼25 bacterial and 25 euryarchaeal protein sequences (supplemental Fig. S1). The graph at the bottom of the alignment indicates the extent of sequence conservation at each position. Conserved sequence motifs characteristic of the β-CASP proteins are indicated in red (metallo-β-lactamase motifs) and blue (β-CASP motifs). The archaeal loops (green) indicate the position of two insertions that are conserved in the euryarchaeal homologues. The conserved bacterial C-terminal domain (black) is absent in the euryarchaeal homologues.
PAB1751 and TK1469 Encode an Exoribonuclease
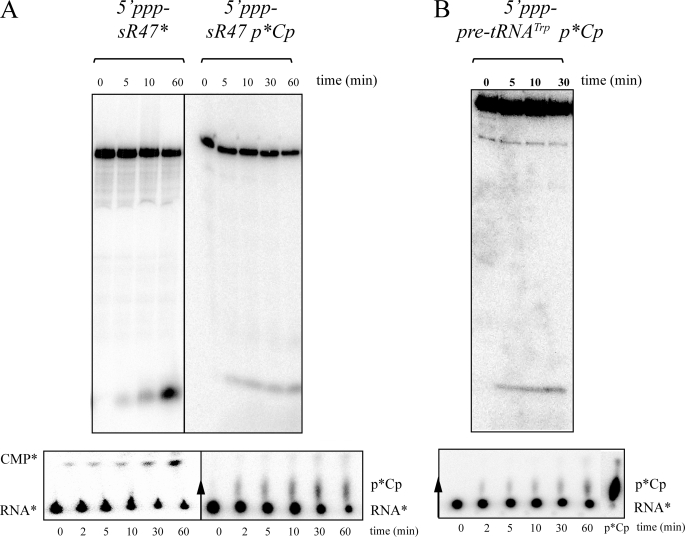
The protein encoded by PAB1751 with an N-terminal His tag was expressed in E. coli and purified. A 64-nt2 RNA substrate corresponding to the P. abyssi sR47 box C/D guide RNA, whose structure and function has been well characterized (33, 35), was either body-labeled with [α-32P]CTP or 3′ end-labeled with [32P]pCp. These substrates, which were synthesized by transcription with T7 RNA polymerase, have a 5′-triphosphate end. The optimal growth temperature of P. abyssi is 103 °C (36). However, the experiments presented here were carried out at 65 °C to avoid significant chemical (nonenzymatic) hydrolysis of RNA at a higher temperature. RNA degradation was analyzed by denaturing gel electrophoresis (Fig. 2A, upper panel), and the fast-migrating products were identified as [α-32P]CMP or [32P]pCp by thin layer chromatography (Fig. 2A, lower panel). These results demonstrate that the product of PAB1751 is an exoribonuclease as the RNA substrate is degraded to mononucleotides. We, therefore, named this enzyme Pab-RNase J. Pilot studies in which the His tag on Pab-RNase J was removed showed that the tag did not affect ribonuclease activity (data not shown). The tagged version was used throughout the work reported here. We also expressed and purified the protein from T. kodakaraensis encoded by TK1469 and named this enzyme Tk-RNase J because its activity is similar if not identical to the activity of the Pab-RNase J (data not shown). In the following sections, we simplify the presentation by only showing the results with Pab-RNase J, although most of the experiments were performed with both enzymes and comparable results were obtained with Tk-RNase J.
FIGURE 2.
Degradation of 5′-triphosphate RNA substrates by Pab-RNase J. A, degradation of uniformly labeled (5′ppp-sR47*) or 3′ end-labeled (5′ppp-sR47p*Cp) 64-nt sR47 RNA is shown. B, degradation of 3′ end-labeled (5′ppp-pre-tRNATrp p*Cp) 149-nt pre-tRNATrp RNA is shown. Uniformly labeled or 3′ end-labeled RNA (100 fmol) was incubated at 65 °C with 10 nm Pab-RNase J (200 fmol). The asterisk indicates the position of the labeled phosphate in each substrate. The products of the reaction were analyzed by denaturing 8% PAGE (upper panel) and by thin layer chromatography (lower panel). CMP* and GMP* were identified by comparison with the migration positions of nonradioactive CMP and GMP determined by UV shadowing on TLC plates (not shown). Radiolabeled 5′-[32P]pCp was included as a marker (B, lower panel).
Careful visual inspection of the denaturing gels in Fig. 2A did not reveal the appearance of intermediates in degradation. Three other 5′-triphosphorylated substrates, known to adopt specific structures, were tested; that is, the 350-nt thrS leader mRNA, previously employed in studies of B. subtilis RNase J1 (19, 20), P. abyssi pre-tRNATrp, and P. abyssi tRNALeu (35). Results comparable with those with the sR47 substrate were obtained, although the degradation of the pre-tRNATrp (Fig. 2B) and tRNALeu substrates (data not shown) was slower than the degradation of the thrS leader mRNA and sR47.
Throughout the experiments with 5′-triphosphorylated substrates, there was no indication of an endoribonuclease activity with either Pab-RNase J or Tk-RNase J. In particular, it has been demonstrated that highly purified RNase J1 of B. subtilis has endoribonuclease activity on 5′-triphosphorylated thrS leader mRNA (19). Although we cannot exclude the possibility that the higher temperature used in these experiments destabilizes secondary structure in the thrS leader mRNA, necessary for endoribonuclease cleavage, even at temperatures as low as 45 °C, the only detectable activity of the archaeal homologues on the thrS leader mRNA was exoribonucleolytic.
The Phosphorylation State of the RNA 5′ End Affects Pab-RNase J Activity
To test whether the phosphorylation state of the 5′ end of the RNA substrate affected Pab-RNase J activity, we synthesized several variants of sR47 and of pre-tRNATrp and measured their initial rates of degradation by Pab-RNase J. The sR47 substrate was labeled either at its 5′ end as a 5′-[32P]monophosphate or at its 3′ end by the addition of [32P]pCp. Two forms of the 3′ end-labeled substrate were prepared, 5′-hydroxyl and 5′-monophosphate. Two principal degradation products of the 3′ end-labeled substrates, [32P]pCp and [32P]CMP, were observed (Fig. 3A). Thus, pCp is converted to 5′-CMP in our assay. The overall analysis of our experimental results suggests that it is an activity of the Pab-RNase J itself and not due to a contaminating phosphatase. In particular, wild type Pab-RNase J can convert pCp to 5′-CMP, whereas variants that are inactive for exoribonuclease activity do not catalyze this reaction (data not shown). The principal product of the 5′ end-labeled RNA is [32P]GMP as guanosine is the 5′-nucleotide of sR47 (Fig. 3A).
FIGURE 3.
Degradation of 5′-monophosphate and 5′-hydroxyl RNA substrates by Pab-RNase J. A, degradation of the 5′ and 3′ end-labeled 64-nt sR47 RNA variants is shown. The nucleotide ladder was generated by alkaline hydrolysis of the 5′ end-labeled sR47 RNA. B, degradation of the 5′ and 3′ end-labeled 149 nt pre-tRNATrp RNA variants is shown. Lane M, markers, 5′ end-labeled digest of Φx174 DNA with HinfI. 5′ or 3′ end-labeled RNAs (100 fmol) were incubated at 65 °C with 10 nm Pab-RNase J (200 fmol). The products of the reaction were analyzed by denaturing 10% PAGE (upper panel) and by thin layer chromatography (lower panel). 5′p* indicates 5′ end-labeled RNA; 5′p-3′p*Cp indicates 5′-monophosphorylated RNA labeled at its 3′ end with pCp; 5′OH-3′p*Cp indicates 5′-hydroxyl RNA labeled at its 3′ end with pCp. Radiolabeled CMP* and GMP* were identified by comparison with the migration positions of nonradioactive CMP and GMP on TLC plates as determined by UV shadowing (A). Radiolabeled 5′-[32P]pCp was loaded on the gel and spotted on TLC plates as a marker (B).
There are significant differences in the kinetics of degradation between a substrate with a 5′-triphosphate and the substrates with a 5′-hydroxyl or 5′-monophosphate (Fig. 4). The observed initial rate of degradation of the substrate with the 5′-triphosphate end is 5-fold slower than the substrate with the 5′-hydroxyl end or with the 5′-monophosphate end (Table 1). This is the lower limit of the difference as chemical hydrolysis of the 5′-triphosphorylated substrate contributes to its degradation. Thus, sR47 RNA with a 5′-triphosphate end is a poor substrate for Pab-RNase J (Fig. 2A). We observed a comparable Pab-RNase J preference for a 5′-hydroxyl or 5′-monophosphate end with the thrS leader mRNA (data not shown).
FIGURE 4.
Kinetics of degradation of the substrate variants. The symbols correspond to the following variants of the sR47 substrate: ▴, 5′p*-sR47; ○, 5′ppp-sR47p*Cp; ◇, 5′OH-sR47p*Cp; □, 5′p-sR47p*Cp. 100 fmol of sR47 RNA was incubated at 65 °C with 200 fmol of Pab-RNase J. The fraction of substrate remaining was calculated from quantification of TLC results. The experiment was performed at least in triplicate. The error bars represent S.D.
TABLE 1.
Initial degradation rates of sR47 5′ and 3′ end-labeled variants
Rates are based on initial velocities as determined from kinetic analyses as described in Fig. 4. The rates and S.D. were calculated from at least three independent experiments.
| RNA substrates | fmol of substrate degraded/min |
|---|---|
| 5′ppp-sR47p*Cp | 0.6 ± 0.2 |
| 5′p*-sR47 | 3 ± 1.5 |
| 5′p-sR47p*Cp | 2 ± 1.5 |
| 5′OH-sR47p*Cp | 3 ± 1 |
The pre-tRNATrp substrate is poorly degraded by Pab-RNase J independently of the nature of its 5′ end (Figs. 2B and 3B). The higher content of RNA secondary structure and the base pairing of the 5′ end in an RNA stem may account for the resistance of pre-tRNATrp to Pab-RNase J degradation. In the case of poor substrates such as pre-tRNATrp, the pCp is not significantly converted to 5′-CMP (Fig. 2B), possibly because the concentration of pCp produced in these reaction is significantly below the Kd for binding to the enzyme. In conclusion, the protective effects of the 5′-triphosphate and/or a 5′-stem suggest that the degradation reaction starts by binding to a 5′-single-stranded end and proceeds with an overall 5′ to 3′ directionality. It is important to note that the position of the labeled [32P]phosphorus (5′ or 3′) in the RNA substrate does not affect the initial rate of RNA degradation (Figs. 3A and 4).
We further characterized the enzymatic properties of Pab-RNase J using 5′ end-labeled sR47 RNA. The enzymatic activity is highest between pH 6.5 and 8 (supplemental Fig. S2A). The addition of divalent ions to the reaction buffer did not stimulate the reaction, nor did the addition of EDTA inhibit the reaction. However, the activity was strongly reduced by the addition of 1,10-phenanthroline (20 mm), suggesting a requirement for tightly bound zinc ions. The Pab-RNase J activity increased with temperature up to 95 °C, the maximum temperature tested (supplemental Fig. S2B). We also tested if DNA was a substrate. Single-stranded DNA was degraded by Pab-RNase J (supplemental Fig. S2C), suggesting that the euryarchaeal RNase J homologues do not discriminate between ribose and 2′-deoxyribose nucleosides. Moreover, in line with the pre-tRNATrp results (Fig. 3B), where the 5′ end is embedded into a duplex structure, double-stranded DNA is resistant to Pab-RNase J degradation (data not shown).
Circular RNA Substrates Are Highly Resistant to Degradation by Pab-RNase J
Further evidence for 5′ end-dependent exoribonuclease activity comes from experiments with circular RNA substrates. The 5′-[32P]monophosphorylated sR47 and 21-nt synthetic RNA (corresponding to the first 21 nucleotides of sR47) were treated with RNA ligase to obtain circular RNAs. Pab-RNase J degradation assays were performed on these substrates (Fig. 5). With the linear substrates, the reaction was complete after 20 min. In contrast, the circular substrates were highly resistant to Pab-RNase J. The trace amount of product detected after 20 min is likely due to the exonucleolytic degradation of circles nicked either by chemical hydrolysis or by a very weak Pab-RNase J endoribonuclease activity. That the gel-purified circular RNAs were digested with the endoribonuclease RNase T1 to confirm their topology (supplemental Fig. S3) argues against the hypothesis that circularization “collapses” the RNA into structures that are inherently nuclease-resistant. These results strongly support the conclusion that Pab-RNase J requires a free 5′ end to initiate degradation.
FIGURE 5.
Degradation of circularized RNAs. Circular RNA was prepared as described under “Experimental Procedures.” 250 fmol of linear (L) or circular (C) 5′ end-labeled 21-nt synthetic RNA (5′p*21-RNA) or sR47 RNA (5′p*sR47) was incubated at 65 °C with 50 nm Pab-RNase J (500 fmol). The products of the reaction were analyzed on denaturing 20% PAGE. The nucleotide ladder was generated by alkaline hydrolysis of linear 5′ end-labeled sR47.
Pab-RNase J Has a 5′ to 3′ Mode of Degradation
The observation that Pab-RNase J degrades 5′-triphosphorylated RNA poorly (Fig. 3) suggests a 5′ to 3′ mode of degradation. If so, the release of the radiolabeled nucleotide will be the first step of the reaction with a 5′ end-labeled substrate and the last step with a 3′ end-labeled substrate. Therefore, degradation reactions with a 3′ end-labeled substrate would be expected to generate radiolabeled intermediates if the enzyme is distributive or has a slow progression on the substrate. However, we failed to detect degradation intermediates in a variety of experiments including limiting amounts of enzyme with 3′ end-labeled sR47, degradation of highly structured substrates such as pre-tRNATrp, analysis of products on high resolution sequencing gels, and reaction temperatures as low as 10 °C. Additional experiments in which we tried to block Pab-RNase J by the addition of an RNA binding protein (L7Ae, see Ref. 33) failed to reveal degradation intermediates.3 Therefore, RNA binding proteins and RNA secondary structure, which might be expected to block the progression of Pab-RNase J, have little effect under the conditions employed here.
To obtain direct evidence for a 5′-3′ direction of hydrolysis, we blocked the progression of the enzyme artificially. We analyzed on high resolution polyacrylamide gels the products of degradation of a modified synthetic RNA corresponding to the first 21 nucleotides of sR47 RNA, which is interrupted by a spacer formed by three ethylene glycol units at position 9 in the sequence. This synthetic RNA oligonucleotide (referred to as 21#-RNA) was labeled either at its 5′ end as a 32P-5′-monophosphate or at its 3′ end by the addition of [32P]pCp. In parallel, a 21-nt synthetic RNA oligonucleotide (referred to as 21-RNA) with the same sequence but without the interruption was used as a control. This experiment was conducted in two different conditions, in enzyme excess (4-fold, data not shown) and in substrate excess (20-fold, Fig. 6). Both conditions generated the same degradation pattern. Quantification of the digestion in Fig. 6 showed that up to 15 pmol of each substrate was converted into product after 1 h of incubation in a reaction that contained 20 pmol of substrate and 1 pmol of enzyme. Both synthetic RNAs containing a 5′-32P yielded GMP as principal product as guanosine is the 5′-nucleotide. No oligonucleotide intermediates were detected with the 5′ end-labeled substrate. As expected, the 21-RNA labeled at its 3′ end was degraded to pCp and 5′-CMP (as shown in Fig. 3 with the sR47 RNA substrate). However, degradation of the 3′ end-labeled 21#-RNA produced 3′ end-labeled degradation intermediates in addition to pCp and 5′-CMP (Fig. 6). These 3′ end-labeled intermediates correspond in length to the position of the spacer in the RNA oligonucleotide, demonstrating that Pab-RNase J pauses at the spacer. Thus, because labeled intermediates were only observed with the 3′ end-labeled substrate, these results provide very strong evidence that Pab-RNase J has a 5′ to 3′ directionality.
FIGURE 6.
Degradation of the synthetic 21-RNA and 21#-RNA oligonucleotides. The sequence of the synthetic 21-RNA corresponds to the first 21 nucleotides of sR47 (5′-GAUGAAGAUGAUGAGCUCGGC-3′). The synthetic 21#-RNA is interrupted by a nine-atom spacer (9S) formed by three ethylene glycol units replacing the ninth nucleotide from the 5′ end. The RNAs were either 5′ end-labeled (5′p*) or 3′ end-labeled (3′p*Cp). Excess RNA substrate (20 pmol) was incubated at 65 °C with 50 nm Pab-RNase J (1 pmol). The products of the reaction were analyzed by denaturing 20% PAGE. The nucleotide ladder was generated by alkaline hydrolysis of 3′ end-labeled 21#-RNA. The arrow indicates an intermediate in degradation of the 3′ end-labeled 21#-RNA whose size corresponds to the position of the modified linkage.
The lack of intermediates during degradation of the various unmodified RNAs in enzyme or substrate excess and in particular with the 3′ end-labeled pre-tRNATrp (Fig. 3B) suggests that once Pab-RNase J has initiated the reaction, it continues RNA degradation until the last nucleotide without dissociating from the RNA. Moreover, as already mentioned above, the position of the labeled [32P]phosphorus (5′ or 3′) does not affect the observed rate of degradation (Figs. 3A, 4, and 6 and Table 1). Thus, the rate of the removal of the first nucleotide is as fast as the release of the last nucleotide, meaning that the enzyme progresses rapidly through the substrate and that this progression is not the limiting step of the reaction. Thus, the initial rates reported in Table 1 are likely to reflect a slow step of the reaction, which could be the initial binding of Pab-RNase J to the 5′ end of the substrate. Overall, our results suggest that Pab-RNase J is a highly processive 5′- to 3′-exoribonuclease.
Enzymatic Activity of Pab-RNase J Variants
To characterize the catalytic site of Pab-RNase J, we started with the assumption that it was structurally homologous with the bacterial enzyme. A high resolution structure of bacterial RNase J was obtained from crystals of the enzyme from T. thermophilus (19). The catalytic site is formed by a pair of zinc ions that are coordinated by a network of five histidine and two aspartic acid residues. This network corresponds to highly conserved residues in the signature motifs of the β-CASP proteins. We made individual substitutions of His-86, His-388, His-410, and Glu-209 of Pab-RNase J with alanine. His-86 (Motif 2) and His-410 (Motif C) are predicted to directly coordinate zinc ions (see Fig. 1). His-388 (Motif B) is predicted to interact indirectly with a zinc ion by a bridging water molecule. Glu-209 (Motif A) is a glutamic acid in Pab-RNase J that corresponds to a strictly conserved aspartic acid in the bacterial enzyme. Glu-209 is predicted to lock His-388 into the optimal orientation for interacting with a zinc ion via a bridging water molecule. The exoribonuclease activity of each purified recombinant protein variant was tested in excess of enzyme. The H86A, E209A, and H388A variants have significantly reduced activity compared with the wild type protein (Table 2), which is consistent with a role for these residues in the coordination of zinc ions. However, the H410A variant has full activity, making it very unlikely that this histidine is involved in the coordination of a zinc ion. The same results were obtained with Tk-RNase J (data not shown). The substitution of His-410 with valine, obtained only in the Tk-RNase J context (Table 2), results in a significant loss of activity suggesting that the larger valine residue perturbs the structure of the catalytic site. We also constructed variants of Pab-RNase J in which the archaeal specific loops were deleted. Pab-RNase J(Δ1), deletion of Loop 1, has reduced but detectable ribonuclease activity. Pab-RNase J(Δ2), deletion of Loop 2, has significantly reduced ribonuclease activity. Loop 1 and Loop 2 are, thus, required for a fully active enzyme.
TABLE 2.
Activity of Pab-RNase J variants
5′-End-labeled sR47 (5′p*sR47) (100 fmol) was incubated with 1 pmol of protein. The initial velocity of degradation was determined from at least three independent experiments. Relative activity represents the rates normalized to wild-type Pab-RNase J, which was taken as 100%. The H410V1 variant is a derivative of Tk-RNase J as we were unable to construct the equivalent variant in Pab-RNase J.
| Motif | Variant | Relative activity |
|---|---|---|
| Motif 2 | H86A | ∼5% |
| Motif A | E209A | ∼5% |
| Motif B | H388A | <1% |
| Motif C | H410A | 100% |
| H410V1 | <1% | |
| Loop 1 | Δ1 | 25% |
| Loop 2 | Δ2 | ∼8% |
Archaeal RNase J Has Been Inherited Vertically
To investigate the evolutionary origin of the euryarchaeal homologues, we constructed a phylogenetic tree based on a set of 70 concatenated ubiquitous protein sequences in the available euryarchaeal genomes. It is assumed that this tree reflects evolution of the species. This tree is compared with a tree obtained with the RNase J sequences from the Euryarchaeota (Fig. 7). The congruence between the two trees, except for two poorly bootstrap supported branches (<60%) in the RNase J homologues, shows that the euryarchaeal homologues have evolved through vertical inheritance. This phylogenetic analysis suggests that the ancestral protein of the bacterial and euryarchaeal homologues of RNase J was an ancient β-CASP protein that existed before the separation of the Archaea and the bacteria.
FIGURE 7.
Phylogenetic trees of the Euryarchaeota and euryarchaeal RNase J. The tree of the Euryarchaeota was constructed from a concatenated sequence of 70 ubiquitous proteins (see “Experimental Procedures”). The euryarchaeal RNase J tree was constructed from sequences that were available at the time of the analysis (supplemental Fig. S1). The trees are arbitrarily rooted. Bootstrap values are indicated at the branch points.
DISCUSSION
In the Euryarchaeota most of the genomes that have been sequenced encode a β-CASP protein that is related to the B. subtilis RNase J. Here, we have characterized PAB1751 from P. abyssi and TK1469 from T. kodakaraensis and named them Pab-RNase J and Tk-RNase J, respectively. These enzymes are 5′ end-dependent exoribonucleases with a 5′ to 3′ directionality. Phylogenetic analysis of euryarchaeal homologues shows that they have been inherited vertically and suggests that they have an ancient origin predating the separation of the Bacteria and the Archaea. Taken together, our results suggest that the ancestral protein of the bacterial and euryarchaeal homologues of RNase J was a primordial β-CASP protein that had 5′-exonuclease activity.
A search for putative RNase J homologues in the Crenarchaeota revealed the presence of a family of β-CASP proteins.3 In sequence comparisons with bacterial RNase J, the conservation in the crenarchaeal homologues was below 20% as compared with 32 and 34% identity for Pab-RNase J and Tk-RNase J, respectively. The crenarchaeal homologues have been annotated as the small subunit of a hypothetical cleavage and polyadenylation specificity factor (CPSF). CPSF is a nuclear endoribonuclease involved in the cleavage and polyadenylation of eukaryotic pre-mRNAs. To our knowledge the Archaea are not known to perform this type of mRNA maturation (1). In Sulfolobus solfataricus, a crenarchaeon, the translation initiation factor a/eIF2(-γ) binds to RNA 5′-triphosphorylated ends and protects transcripts from a 5′ end-dependent degradation pathway (37). The ribonuclease(s) in this pathway remain to be identified. In Methanococcus jannaschii, another crenarchaeon, mRNA processing at a site 12–16 nt upstream of the AUG translation start codon appears to be performed by an endoribonuclease (38). The β-CASP proteins in the Crenarchaea are potential ribonucleases for activities recently described in S. solfataricus and M. jannaschii, but their specificity (DNA or RNA) and functionality (exo- or endonuclease) have not yet been determined.
The products of Pab-RNase J are nucleotide 5′-monophosphates, which are characteristic of an exoribonuclease. Our results show that RNA substrates are completely degraded to monophosphates without detectable oligonucleotide intermediates or end products except for the substrate interrupted by three ethylene glycol units, suggesting that Pab-RNase J is a processive exoribonuclease. The analysis of the degradation of substrates with 5′-triphosphate, 5′-monophosphate, and 5′-hydroxyl ends shows that Pab-RNase J is a 5′ end-dependent exoribonuclease with a preference for substrates with 5′-monophosphate or 5′-hydroxyl ends. A substrate with a 5′-triphosphate end is degraded at least 5-fold slower than a substrate with a 5′-monophosphate end, and this value is likely to be an underestimation. Under the conditions employed here, the slow degradation of the 5′-triphosphorylated substrate is at least partly due to chemical hydrolysis of RNA, which creates RNA fragments with 5′-hydroxyl ends that are then degraded by Pab-RNase J. Circularized RNA substrates and the structured pre-tRNATrp are resistant to degradation by Pab-RNase J, demonstrating the importance of a free end for the initiation of exoribonucleolytic degradation. Comparable results were obtained in experiments employing Tk-RNase J. Results with the modified synthetic RNA oligoribonucleotide containing the substitution of an internal nucleoside by three ethylene glycol units demonstrated the 5′ to 3′ directionality of Pab-RNase J. It is surprising that the three ethylene glycol units did not create a stronger barrier to Pab-RNase J degradation. The reported broad substrate specificity of β-lactamase enzymes with active sites accessible to a number of different molecules (28, 39, 40) or an inherent Pab-RNase J endonucleolytic activity could explain this result. Finally, the degradation of substrates with labeled [32P]phosphorus at the 3′ end position in limiting enzyme conditions suggests that Pab-RNase J rapidly degrades the RNA in a processive reaction and that the slow step of the reaction appears to be the initial binding at the 5′ end of the substrate. Additional biochemical and kinetic experiments will be required to elucidate the detailed mechanism of Pab-RNase J-mediated RNA degradation.
The euryarchaeal RNase J homologues have all the signature motifs characteristic of the β-CASP family of proteins. The catalytic core of bacterial RNase J is formed by a network of charged residues that coordinate a pair of zinc ions and that correspond to highly conserved residues in the signature motifs of the β-CASP proteins (19). By analogy to bacterial RNase J, we substituted four residues predicted to be involved in zinc ion coordination with alanine (Fig. 1). Pab-RNase J variants with the H86A (Motif 2), E209A (Motif A), and H388A (Motif B) substitutions have significantly reduced activity, confirming their importance for euryarchaeal RNase J activity. However, a Pab-RNase J variant with the H410A (Motif C) substitution retains ribonuclease activity. The result with the H410A variant is not consistent with the prediction that His-410 is involved in coordinating a zinc ion and suggests that the detailed architecture of the catalytic site of the archaeal enzyme differs from that of the bacterial enzyme.
In all organisms RNA degradation is a prevalent activity. Three major classes of RNases have been characterized; they are endonucleases that cut RNA internally, 3′-exonucleases, and 5′-exonucleases. In the thermococcal Archaea, all three activities have now been reported. Several endoribonucleases in P. furiosus have been shown to be required for the biogenesis of tRNA (41) and CRISPR RNA (42). The P. abyssi and P. horikoshii exosome has 3′-exoribonuclease activity as shown by structural and functional studies (43). Now, our biochemical characterization of Tk- and Pab-RNase J demonstrates that a 5′-exoribonuclease is encoded in the genomes of the Thermococcales. However, we have not been able to detect endoribonuclease activity with either Pab- or Tk-RNase J. The B. subtilis RNase J1 has previously been demonstrated to cleave 5′-triphosphorylated thrS leader mRNA in a single-stranded region upstream of an RNA stem-loop involved in transcription termination (19, 20). Using the identical substrate, we did not detect cleavage at this site even in experiments in which we lowered the temperature of the reaction to avoid the possibility that the secondary structure of the thrS leader mRNA was perturbed at 65 °C. It is possible that the Thermococcale RNase J homologues do not have an endoribonuclease activity. If this is not the case, then other explanations include altered substrate specificity of the archaeal enzyme. That is, thrS leader mRNA might not be a substrate for endoribonuclease cleavage, but such substrates, which remain to be identified, might exist in the Thermococcales.
Except for the difference in temperature optima, the characteristics of the exoribonuclease activity of Pab-RNase J are very similar to the exoribonuclease activity of RNase J1 from B. subtilis (19, 21), suggesting that Pab-RNase J might have a role in RNA processing and degradation. The fact that Pab-RNase J and Tk-RNase J degrade single-stranded DNA suggests potential multi-functional roles in vivo. Three other exonucleases from the β-CASP family are known to degrade both RNA and DNA. Two human exonucleases involved in DNA repair, hSNM1 and Artemis, degrade either DNA or RNA substrates in vitro with equal efficiency (44, 45). The CPSF-73 factor involved in pre-mRNA 3′ end processing has a 5′-exonuclease activity that is almost insensitive to deoxynucleotides (46). In addition, the 3′-5′-exoribonuclease RNase T from E. coli can also act as a DNA single-strand specific enzyme in vitro (47), and in yeast, the primary 5′-3′-exoribonuclease Xrn1 (48), which was first purified as an enzyme promoting DNA strand exchange, has modest DNase activity (49). The lack of discrimination between RNA and DNA among the exonucleases may be more common than appreciated as this specificity has not been examined systematically. What gives members of the β-CASP family their specificity in vivo is unknown, although clues to this puzzle might come from CSPF-73, which is part of a multiprotein complex targeted to pre-mRNA 3′ end processing.
Supplementary Material
Acknowledgments
We thank G. Rauffet for help with protein sequence alignments, I. Omrane and G. Petre for help with the characterization of Pab-RNase J and Tk-RNase J, H. Putzer for the gift of the plasmid encoding the thrS mRNA (pHMS25), C. Turlan and V. Khemici for helpful discussions, and B. Luisi for critical comments on the manuscript.
This work was supported by the CNRS with additional funding from the Agence Nationale de la Recherche (ANR) (Grant BLAN08–1_329396).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
B. Clouet-d'Orval, D. Rinaldi, Y. Quentin, and A. J. Carpousis, unpublished results.
- nt
- nucleotide(s)
- CPSF
- cleavage and polyadenylation specificity factor.
REFERENCES
- 1.Evguenieva-Hackenberg E., Klug G. (2009) Prog. Mol. Biol. Transl. Sci. 85, 275–317 [DOI] [PubMed] [Google Scholar]
- 2.Frank D. N., Pace N. R. (1998) Annu. Rev. Biochem. 67, 153–180 [DOI] [PubMed] [Google Scholar]
- 3.Schiffer S., Rösch S., Marchfelder A. (2002) EMBO J. 21, 2769–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redko Y., Li de Lasierra-Gallay I., Condon C. (2007) Nat. Rev. Microbiol. 5, 278–286 [DOI] [PubMed] [Google Scholar]
- 5.Li H., Trotta C. R., Abelson J. (1998) Science 280, 279–284 [DOI] [PubMed] [Google Scholar]
- 6.Tang T. H., Rozhdestvensky T. S., d'Orval B. C., Bortolin M. L., Huber H., Charpentier B., Branlant C., Bachellerie J. P., Brosius J., Hüttenhofer A. (2002) Nucleic Acids Res. 30, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French S. L., Santangelo T. J., Beyer A. L., Reeve J. N. (2007) Mol. Biol. Evol. 24, 893–895 [DOI] [PubMed] [Google Scholar]
- 8.Brown J. W., Reeve J. N. (1985) J. Bacteriol. 162, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantharaman V., Koonin E. V., Aravind L. (2002) Nucleic Acids Res. 30, 1427–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin E. V., Wolf Y. I., Aravind L. (2001) Genome Res 11, 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. (2005) Nat. Struct. Mol. Biol. 12, 575–581 [DOI] [PubMed] [Google Scholar]
- 12.Portnoy V., Schuster G. (2006) Nucleic Acids Res. 34, 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackie G. A. (1998) Nature 395, 720–723 [DOI] [PubMed] [Google Scholar]
- 14.Bouvet P., Belasco J. G. (1992) Nature 360, 488–491 [DOI] [PubMed] [Google Scholar]
- 15.Jiang X., Belasco J. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9211–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., Luisi B. F. (2005) Nature 437, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 17.Jourdan S. S., McDowall K. J. (2008) Mol. Microbiol. 67, 102–115 [DOI] [PubMed] [Google Scholar]
- 18.Condon C., Putzer H., Luo D., Grunberg-Manago M. (1997) J. Mol. Biol. 268, 235–242 [DOI] [PubMed] [Google Scholar]
- 19.de la Sierra-Gallay I. L., Zig L., Jamalli A., Putzer H. (2008) Nat. Struct. Mol. Biol. 15, 206–212 [DOI] [PubMed] [Google Scholar]
- 20.Even S., Pellegrini O., Zig L., Labas V., Vinh J., Bréchemmier-Baey D., Putzer H. (2005) Nucleic Acids Res. 33, 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathy N., Bénard L., Pellegrini O., Daou R., Wen T., Condon C. (2007) Cell 129, 681–692 [DOI] [PubMed] [Google Scholar]
- 22.Britton R. A., Wen T., Schaefer L., Pellegrini O., Uicker W. C., Mathy N., Tobin C., Daou R., Szyk J., Condon C. (2007) Mol. Microbiol. 63, 127–138 [DOI] [PubMed] [Google Scholar]
- 23.Collins J. A., Irnov I., Baker S., Winkler W. C. (2007) Genes Dev. 21, 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daou-Chabo R., Mathy N., Bénard L., Condon C. (2009) Mol. Microbiol. 71, 1538–1550 [DOI] [PubMed] [Google Scholar]
- 25.Lee K., Cohen S. N. (2003) Mol. Microbiol. 48, 349–360 [DOI] [PubMed] [Google Scholar]
- 26.Kanai A., Oida H., Matsuura N., Doi H. (2003) Biochem. J. 372, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callebaut I., Moshous D., Mornon J. P., de Villartay J. P. (2002) Nucleic Acids Res. 30, 3592–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominski Z. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 67–93 [DOI] [PubMed] [Google Scholar]
- 29.Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castresana J. (2000) Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 31.Guindon S., Gascuel O. (2003) Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 32.Jones D. T., Taylor W. R., Thornton J. M. (1992) Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 33.Nolivos S., Carpousis A. J., Clouet-d'Orval B. (2005) Nucleic Acids Res. 33, 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bortolin M. L., Bachellerie J. P., Clouet-d'Orval B. (2003) Nucleic Acids Res. 31, 6524–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clouet d'Orval B., Bortolin M. L., Gaspin C., Bachellerie J. P. (2001) Nucleic Acids Res. 29, 4518–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005) Genome Res. 15, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasenöhrl D., Lombo T., Kaberdin V., Londei P., Bläsi U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2146–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Olsen G. J. (2009) RNA 15, 1909–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel A., Schilling O., Niecke M., Bettmer J., Meyer-Klaucke W. (2002) J. Biol. Chem. 277, 29078–29085 [DOI] [PubMed] [Google Scholar]
- 40.Daiyasu H., Osaka K., Ishino Y., Toh H. (2001) FEBS Lett. 503, 1–6 [DOI] [PubMed] [Google Scholar]
- 41.Tsai H. Y., Pulukkunat D. K., Woznick W. K., Gopalan V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16147–16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carte J., Wang R., Li H., Terns R. M., Terns M. P. (2008) Genes Dev. 22, 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos C. R., Oliveira C. L., Torriani I. L., Oliveira C. C. (2006) J. Biol. Chem. 281, 6751–6759 [DOI] [PubMed] [Google Scholar]
- 44.Hejna J., Philip S., Ott J., Faulkner C., Moses R. (2007) Nucleic Acids Res. 35, 6115–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y., Pannicke U., Schwarz K., Lieber M. R. (2002) Cell 108, 781–794 [DOI] [PubMed] [Google Scholar]
- 46.Yang X. C., Sullivan K. D., Marzluff W. F., Dominski Z. (2009) Mol. Cell. Biol. 29, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viswanathan M., Dower K. W., Lovett S. T. (1998) J. Biol. Chem. 273, 35126–35131 [DOI] [PubMed] [Google Scholar]
- 48.Stevens A., Poole T. L. (1995) J. Biol. Chem. 270, 16063–16069 [DOI] [PubMed] [Google Scholar]
- 49.Johnson A. W., Kolodner R. D. (1991) J. Biol. Chem. 266, 14046–14054 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.