Abstract
In tomato (Solanum lycopersicum), resistance to Pseudomonas syringae pv. tomato is elicited by the interaction of the host Pto kinase with the pathogen effector protein AvrPto, which leads to various immune responses including localized cell death termed the hypersensitive response. The AGC kinase Adi3 functions to suppress host cell death and interacts with Pto only in the presence of AvrPto. The cell death suppression (CDS) activity of Adi3 requires phosphorylation by 3-phosphoinositide-dependent protein kinase 1 (Pdk1) and loss of Adi3 function is associated with the hypersensitive response cell death initiated by the Pto/AvrPto interaction. Here we studied the relationship between Adi3 cellular localization and its CDS activity. Adi3 is a nuclear-localized protein, and this localization is dictated by a nuclear localization signal found in the Adi3 T-loop extension, an ∼80 amino acid insertion into the T-loop, or activation loop, which is phosphorylated for kinase activation. Nuclear localization of Adi3 is required for its CDS activity and loss of nuclear localization causes elimination of Adi3 CDS activity and induction of cell death. This nuclear localization of Adi3 is dependent on Ser-539 phosphorylation by Pdk1 and non-nuclear Adi3 is found in punctate structures throughout the cell. Our data support a model in which Pdk1 phosphorylation of Adi3 directs nuclear localization for CDS and that disruption of Adi3 nuclear localization may be a mechanism for induction of cell death such as that during the Pto/AvrPto interaction.
Keywords: Cell Death, Nuclear Translocation, Phosphatidylinositol-dependent kinase-1 (PDK1), Protein Kinases, Threonine-Serine Protein Kinase, AGC Kinase, Adi3, Programmed Cell Death
Introduction
During the resistance response of plants to pathogens, programmed cell death (PCD)2 occurs as part of the hypersensitive response (HR), which functions to limit pathogen spread (1, 2). It has been over 100 years since the first description of the HR and its associated cell death (3). However, not until relatively recent times has the search for genes and pathways regulating PCD during the HR received much attention. This search has been difficult, but has identified several kinases and transcription factors involved in PCD (1, 4, 5) as well as lipid biosynthetic genes that produce lipid second messengers regulating PCD (6–9).
In tomato (Solanum lycopersicum), the causative agent of bacterial speck disease is Pseudomonas syringae pv. tomato (Pst). Interaction of the tomato resistance protein kinase Pto with the Pst effector protein AvrPto brings about the HR and resistance to Pst (10). Studies have been undertaken to identify genes involved in PCD associated with Pto-mediated HR and revealed a downstream MAP kinase, MAPKKKα, that functions in the induction of cell death during both resistance and susceptibility (11).
Another gene that was identified as a Pto-interacting protein was the tomato protein kinase Adi3, which only interacts with Pto in the presence of AvrPto (12). Subsequently, we have shown Adi3 to function as a negative regulator of plant cell death (13) and thus, it may be the functional homologue of PKB (aka Akt), a major PCD suppressor in mammals (13–15). Adi3 is phosphorylated by 3-phosphoinositide-dependent protein kinase-1 (Pdk1) at Ser-539, which is required for full Adi3 kinase activity and cell death suppression (CDS) ability (13). Mutation of Ser-539 to Asp is capable of mimicking this phosphorylation event, creating a constitutively active Adi3 capable of CDS (13). Adi3 cell death control can also be associated with MAPKKKα that is involved in Pto-mediate HR cell death (11, 13).
Adi3 is a member of the AGC kinase family, which is a conserved family of eukaryotic Ser/Thr protein kinases that regulate many basic cellular processes such as transcription, translation, cell growth, apoptosis, and cytoskeletal remodeling (16). In mammalian systems, AGC kinases affect downstream signaling components through direct mechanisms, including regulation of nuclear shuttling, activities of transcription factors (17), phosphorylation-dependent trafficking of signaling proteins (18), and chromatin remodeling (19). The cell death (apoptosis) regulator PKB is also an AGC kinase family member.
Little is known about the functions of plant AGC kinases. However, there has been several recent studies reported. As with mammalian systems, many plant AGC kinases are activated by Pdk1 (13, 16, 20–23). Arabidopsis contains at least 39 AGC kinase family members (16, 21, 24) and some of their functions include blue-light signaling (25), root hair development (22, 26, 27), oxidative burst signaling (23, 27), and auxin signaling (24, 28). Group VIIIa AGC kinases (of which Adi3 is a member) are specific to plants and are mainly distinguished from mammalian kinases by a large 70–100 amino acid insertion in the activation loop, or T-loop, referred to as the T-loop extension (16). Similar, but much shorter (30–60 amino acids) T-loop extensions are also present in other AGC kinases such as the Ndr family of AGC kinases (29). In mammals and yeast, Ndr kinases regulate processes such as cell morphological changes, exit from mitosis, and apoptosis. The Ndr T-loop extension functions in cell localization and regulation of kinase activity (29–32). Very little is known about the function of the T-loop extension in plant AGC kinases. The T-loop extension of only two Arabidopsis group VIIIa AGC kinases have been studied and appear to contain cellular localization signals (21). However, the amino acid motifs within these T-loop extensions responsible for directing cellular localization have not been identified.
Here we show that the Adi3 T-loop extension is required for nuclear localization and that Adi3 nuclear localization is required for its CDS activity. Non-nuclear localization confines Adi3 to intracellular punctate membrane structures and a concomitant loss of CDS. These studies raise the possibility of restricting Adi3 nuclear localization as a means to induce plant PCD.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Mutagenesis
The Adi3ΔT-loop construct was created by producing an Adi3 PCR fragment lacking the T-loop extension (bp 1369–1608). First, a PCR product containing Adi3 bp 1336–1368 + 1609–2103 was synthesized and called fragment 1. Next another Adi3 PCR product, fragment 2, from bp 1–1368 was produced that overlapped fragment 1. Both fragment 1 and 2 were then used as templates for overlapping PCR (33) with a 30-bp forward primer originating at Adi3 bp 1 and a 27 bp reverse primer originating at Adi3 bp 2103 to produce a PCR product lacking the T-loop extension. For bacterial expression, full-length tomato Adi3 (13) and Adi3ΔT-loop were PCR amplified and cloned into pMal-c2 (New England Biolabs). Mutations were created using site directed mutagenesis with Pfu Turbo (Stratagene). For protoplast expression, transient leaf expression, and subcellular localization studies, Adi3, Adi3ΔT-loop, and other Adi3 mutants were cloned into the BamHI/XbaI restriction sites of pTEX-eGFP to create N-terminal GFP fusions. For Agrobacterium-mediated transient expression, 35Spro:eGFP-Adi3 was EcoRI/HindIII digested from pTEX and cloned into the same sites of pCambia1300. For GFP-NES/NLS constructs, eGFP was PCR amplified with a GFP-specific forward primer and a reverse primer containing the Adi3 NLS plus GFP (5′-CCGggattcCTAAGGCTTAGGTGTTTTTTTCTTGCTCTTTTGAGGTTTGTATAGTTCATC-3′; BamHI lowercase; linker bold; NLS underlined, bp 1509–1542; GFP italics) for 3′-end fusion or with a GFP-specific reverse primer and a forward primer containing the Adi3 NES plus GFP (5′-CCGggtaccATGCCTTTCTTGGACGATCTTTCAATCCGGATGCCAAGTAAAGGAGAACAA-3′; KpnI lowercase; linkers bold; NES underlined, bp 373–490; GFP italics) for 5′-end fusion and cloned into the KpnI/BamHI restriction sites of pTEX.
The tomato exportin-1 (Xpo1) cDNA was cloned by a combination of methods. First, the Arabidopsis Xpo1 cDNA sequence (34) (GenBankTM accession Y18469) was used to screen the tomato EST data base for contigs containing the tomato Xpo1 cDNA. From this search, clone cTOF-19-M20 was identified as containing a partial tomato Xpo1 cDNA lacking the first 1466 5′ base pairs. A full-length cDNA (GenBankTM accession GU126514) was identified by cloning the 1466 5′ base pairs by 5′-RACE and a full-length open reading frame amplified by RT/PCR using a forward primer based at the ATG start codon (5′-ATGGCGGCGGAGAAGCTGAGA-3′) and a reverse primer based at the TGA stop codon (5′-TCATGAATCCACCATTTCATC-3′). The resulting 3228-bp PCR product was cloned into the yeast two-hybrid bait vector pEG202, and protein interactions analyzed by yeast two-hybrid as previously described (13).
Protoplast Transient Expression and Cellular Fractionation
Protoplasts were isolated from leaves of 3-week-old PtoR or prf-3 tomato plants and transformed with 20 μg of DNA for each construct as previously reported (13). Samples of transformed protoplasts were taken 16 h after transformation for protein expression determination, subcellular fractionation, and confocal microscopy. This time point was selected to minimize amount of dead cells, which is maximal at 24 h, to visualize expressed protein. Protoplast transformation efficiency was determined to be 80% by counting the number of GFP-positive protoplasts out of a total of 100 cells. Subcellular fractions were isolated using the following protocol: Total cell extract was obtained by gently shaking transformed protoplasts in 100 μl of Buffer A (10 mm MES-HCl pH 5.7, 1 m sucrose, 5 mm MgCl2, 2 mm β-mercaptoethanol, 10 μl/ml phosphatase inhibitors (Sigma), 10 μl/ml proteinase inhibitors (Sigma), 10 μm MG-132, 0.2% Triton X-100). Nuclei were pelleted from total cell extract by centrifugation at 5,000 × g for 10 min at 4 °C followed by three washes with 100 μl of Buffer A and centrifugation at 5,000 × g for 10 min at 4 °C. Nuclei were lysed for SDS-PAGE analysis by vortexing in 30 μl of Buffer B (10 mm MES-HCl pH 5.7, 1 m sucrose, 5 mm MgCl2, 2 mm β-mercaptoethanol, phosphatase inhibitors (Sigma), 10 μl/ml plant proteinase inhibitors (Sigma), 10 μm MG-132, 1% Triton X-100). The supernatant from nuclei pelleting was centrifuged at 100,000 × g for 1 h at 4 °C and the pellet used for the membrane fraction and the supernatant for the soluble fraction. Membrane proteins were solubilized by sonication in 30 μl of Buffer B. All fractions were separated by SDS-PAGE and analyzed by α-GFP (Santa Cruz Biotechnology) immunoblotting. Membrane fraction purity was determined using the Golgi marker soybean α-1, 2-mannosidase (GmMan1) tagged with m-Cherry (35) as expressed in protoplasts and analyzed by α-GFP immunoblotting. Purity of the nuclear fraction was determined by immunoblotting using α-histone H3 antibody provided by Dr. Mary Bryk, Texas A& M University. Subcellular fractionation analysis was done three independent times and the presented Western blots are representative of all experiments.
Confocal Microscopy
Tomato protoplasts, after 16 h of transformation and leaf sections taken from PtoR tomato leaves that were Agrobacterium-infiltrated, were imaged using an Olympus FV1000 confocal microscope at the Texas A&M University Microscopy and Imaging Center. Images were collected using system software FV10-ASW 1.6 with a 60×/1.2 water immersion objective in the XYZ scan mode (1.14 μm/slice). Excitation and emission wavelengths were as follows: eGFP, 488 nm excitation, 507 nm emission; hoechst 33342 (Sigma), 343 nm excitation, 460 nm emission; and chlorophyll autofluorescence, 470 nm excitation, 680 nm emission. All cell localization analyses with confocal microscopy were carried out at least three independent times with a minimum of 30 protoplast cells and five leaf sections from three independently infiltrated leaves viewed each time. All confocal images are shown as Z-stacks and presented images are representative of typical cell localization for all proteins seen during independent experiments. For imaging protoplasts transformed with GFP-Adi3 and GFP-Adi3 mutants, the wavelength power was increased 2-fold in comparison to the power used for imaging protoplasts transformed with GFP alone.
Kinase Assays
In vitro kinase assays were done at least three independent times using purified Escherichia coli-expressed MBP-tagged and immunoprecipitated GFP-tagged proteins as previously described (13).
AvrPto and Other Treatments
PtoR and pfr-3 protoplasts expressing eGFP-Adi3 or other eGFP-Adi3 mutants for 14 h were transformed with pTEX-avrPto-FLAG. Samples were collected at 0.5, 1.5, 3, and 4 h for determination of cell viability. For nuclei staining and leptomycin B treatment, protoplasts expressing the indicated constructs for 14 h were treated with 10 μm Hoechst 33342 for 2 h or 20 nm leptomycin B (Sigma) for 90 min followed by confocal microscopy imaging. All experiments were carried out a minimum of three times, and images presented are representative of the experiments.
Agrobacterium-mediated Transient Expression
Agrobacterium tumefaciens strain GV2260 was used for Agrobacterium-mediated transient expression in Nicotiana benthamiana and PtoR tomato leaves as previously reported (13). Adi3 constructs were expressed from pCambia-1300 and avrPto-FLAG from pBTEX.
Cell Viability Measurements
Conductivity tests were carried out on 3 leaf discs (1-cm diameter) obtained from leaves transiently expressing Adi3 proteins. Disks were placed in 3 ml of dH2O for 4 h with gentle shaking at room temperature and conductivity measured with an Acorn Con 5 meter (Oakton Instruments, Vernon Hills, IL). Cell death in leaves transiently expressing Adi3 proteins was visualized by vacuum infiltrating 0.1% Evans blue into leaf disks followed by washing with dH2O and depigmentation with Carnoy's solution (60% ethanol, 30% CHCl3, 10% acetic acid). Protoplast cell viability was determined using Evans blue as previously reported (13). All cell viability assays were carried out a minimum of three independent times.
RESULTS
Adi3 Contains a Nuclear Localization Signal in the T-loop Extension and an N-terminal Nuclear Export Signal
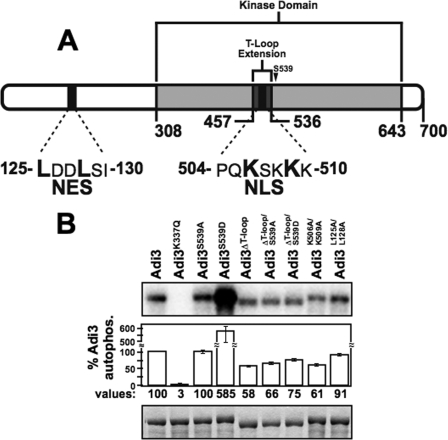
A search of the Adi3 protein sequence for cellular localization signals using the WoLF PSORT protein localization predictor (36) revealed a monopartite basic nuclear localization signal (NLS) in the Adi3 T-loop extension from amino acids 504–510 (PQKSKKK, Fig. 1A). Additionally, the NESnet nuclear export signal (NES) search engine, identified a leucine-rich NES in the N-terminal region of Adi3 from amino acids 125–130 (LDDLSI, Fig. 1A).
FIGURE 1.
T-loop extension deletion effects on autophosphorylation activity of Adi3. A, Adi3 protein domains showing location of the T-loop extension and NLS and NES signals. B, kinase activity of Adi3 mutants tested by in vitro autophosphorylation assays. MBP-tagged proteins were expressed en E. coli, purified, and 5 μg of each protein incubated with [γ-32P]ATP in an in vitro kinase assay. Quantification software was used to normalize the autophosphorylation level to the protein levels for each sample. Values are reported as a percentage of wild-type Adi3 autophosphorylation and are the average of three independent experiments. Error bars are standard error. Top panel: kinase assay (phosphorimage). Bottom panel: assay input (Coomassie Blue-stained gel).
Because the Adi3 NLS is located in the T-loop extension, two different mutants to analyze contributions of this sequence to Adi3 function were generated as well as a mutant of the NES sequence. The Adi3 T-loop extension was deleted by overlapping PCR to generate the Adi3ΔT-loop mutant, and the first (Lys-506) and third (Lys-509) Lys residue of the NLS were mutated to Ala creating the Adi3K506A/K509A mutant. Mutation of equivalent Lys residues in NLS sequences similar to that in Adi3 is sufficient to prevent protein nuclear localization (37, 38). Similarly, in NES sequences analogous to that in Adi3, mutation of one or more Leu residues is sufficient to show nuclear accumulation of the protein (39). Thus, Leu-125 and Leu-128 were mutated to Ala to generate the Adi3L125A/L128A mutant.
To determine if deletion of the Adi3 T-loop extension affects Adi3 kinase activity, autophosphorylation activity of the Adi3ΔT-loop protein was tested and compared with known Adi3 autophosphorylation mutants. As seen previously, the S539D mutation produces a constitutively active Adi3 with greatly enhanced kinase activity (13) (Fig. 1B). Deletion of the T-loop extension (Adi3ΔT-loop) reduced Adi3 autophosphorylation activity ∼42%, which may be a cause of deleting the 79 amino acids of the T-loop extension within the catalytic kinase domain. Introduction of the S539D mutation into Adi3ΔT-loop (Adi3ΔT-loop/S539D) increased Adi3ΔT-loop autophosphorylation by ∼19% (Fig. 1B) indicating that mimicking Pdk1 phosphorylation of Adi3ΔT-loop (Adi3ΔT-loop/S539D) does not increase Adi3ΔT-loop autophosphorylation activity to the extent of wild type. Interestingly, the NLS mutant (Adi3K506A/K509A) had autophosphorylation activity close to that of Adi3ΔT-loop (Fig. 1B) suggesting these Lys residues offer important structural features to Adi3 required for autophosphorylation activity. The NES mutant, Adi3L125A/L128A did not drastically affect Adi3 autophosphorylation activity (Fig. 1B). The K337Q mutation (Adi3K337Q) eliminates ATP binding and kinase activity in wild-type protein (13). Because the T-loop extension deletion and the NLS mutation affected Adi3 autophosphorylation activity, we next tested if these mutations had effects on Adi3 CDS activity and cellular localization.
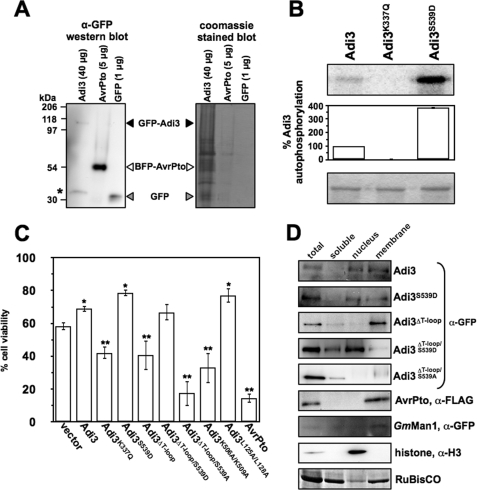
In Vivo Expression Levels of GFP-Adi3 Expressed from the 35S Promoter and Functionality of the GFP-Adi3 Protein
Our Adi3 functional and localization studies use a GFP-Adi3 protein expressed from the cauliflower mosaic virus 35S promoter (35Spro). Use of the 35Spro can lead to overexpression and accumulation of large amounts of protein, possibly leading to mislocalization. Thus, we analyzed the GFP-Adi3 protein levels in tomato protoplasts expressing 35Spro:GFP-Adi3 and compared this to the protein levels of two other proteins expressed from the 35Spro:AvrPto-BFP, a Pst effector protein fused to BFP, and GFP alone. Western blot analysis indicated that the detection levels of GFP-Adi3 were very low and required 8 times and 40 times as much total protein to be detected compared with AvrPto-BFP or GFP, respectively (Fig. 2A). This suggests that GFP-Adi3 proteins do not reach levels that may cause problems in cellular localization analysis. In addition, we found one nonspecific band in our Western blot that could be detected only when high concentrations of total protein were used (asterisk in Fig. 2A and supplemental Fig. S1). This protein is larger than that of GFP alone suggesting it is not GFP and indicating the presence of intact GFP-Adi3 as expressed in protoplasts. Additionally, the GFP-Adi3 protein used for subsequent localization and functional analyses was also fully kinase active as expected (Fig. 2B).
FIGURE 2.
Cellular localization of GFP-Adi3 proteins regulates cell death suppression (CDS) activity. A, GFP-Adi3 protein expression levels under the 35S promoter. Tomato protoplasts were transformed with the indicated DNA constructs and protein expressed for 16 h followed by total protein extraction and α-GFP Western blot analysis. B, GFP-Adi3 autophosphorylation activity. Recombinant GFP-Adi3 protein was immunoprecipitated using α-GFP-agarose and tested in an in vitro kinase assay as in Fig. 1B. Values are reported as a percentage of wild-type Adi3 autophosphorylation and are average of three independent experiments. Error bars are standard error. Top panel: kinase assay (phosphorimage). Bottom panel: assay input (Coomassie Blue-stained gel). C, cell viability of protoplasts expressing GFP-Adi3 plus NLS and NES mutants. Tomato protoplasts expressing the indicated proteins for 24 h were analyzed for cell viability by Evans Blue staining to identify dead cells. Data represent the average of five independent experiments. Error bars are standard error. One asterisk and two asterisks indicate significant increase or decrease, respectively, in cell viability compared with vector-only expression (Student's t test, p < 0.05). D, subcellular distribution of GFP-Adi3 proteins. The indicated proteins were expressed for 16 h in tomato protoplasts followed by subcellular fractionation and Western blot with the indicated antibodies. Markers used: membrane, AvrPto-FLAG (73) and soy bean α-1,2-mannosidase (GmMan1)-mCherry (35); nuclei, histone H3; soluble fraction and loading control, RuBisCo (Coomassie Blue stain of membrane). Western blots are representative of three independent experiments.
Ser-539 Phosphorylation and an Intact NLS Are Important for Adi3 CDS Activity
We used our Adi3 expressing protoplast system (13) to test different Adi3 kinase activity mutants for compromised cell death control (Fig. 2C). These assays make two premises. First, PCD is a process that is always “on” and there are proteins such as PKB and Adi3 that continually suppress PCD to prevent cell death (13–15). Loss of these proteins relieves suppression of PCD and initiates cell death as has been seen for PKB (40, 41) and we have seen for Adi3 (13). Second, overexpression of functionally compromised or constitutively active forms of PCD suppressor kinases will outcompete the endogenous protein, acting in a dominant negative or dominant manner, to induce or suppress cell death, respectively. This has been shown for PKB (42–46) and Adi3 (13). In our current assays, GFP-Adi3 proteins were expressed in protoplasts for 24 h and cell viability measured by Evans blue staining as we have previously done (13). These assays used protoplasts from PtoR tomato plants, which contain a functional Pto gene and show strong induction of HR cell death in response to the Pst effector protein AvrPto (10). For this reason, we used AvrPto expression as a positive control for strong cell death induction. Expression of wild-type Adi3 showed a moderate increase in cell viability over the vector alone (Fig. 2C) supporting its role in CDS (13). The kinase-inactive Adi3K337Q showed a reduction in CDS and the constitutively active Adi3S539D had increased CDS (Fig. 2C) further indicating that Adi3 kinase activity and Pdk1 phosphorylation (via the Pdk1 phosphorylation mimic) is required for Adi3 PCD control. Expression of Adi3ΔT-loop showed a reduction in CDS similar to Adi3K337Q (Fig. 2C). However, introduction of the phosphomimic S539D mutation (Adi3ΔT-loop/S539D) was able to restore CDS activity for Adi3ΔT-loop to near wild-type levels (Fig. 2C). Introduction of the nonphosphorylatable S539A mutation into Adi3ΔT-loop (Adi3ΔT-loop/S539A) showed a strong loss of CDS (Fig. 2C). The NLS mutant Adi3K506A/K509A also showed loss of CDS equal to that of Adi3ΔT-loop (Fig. 2C). The Adi3 NES mutant, Adi3L125A/L128A, had increased CDS comparable to Adi3S539D (Fig. 2C). These data would suggest that a functional Adi3 NLS is required for proper CDS and that mimicking the Pdk1 phosphorylation can restore CDS activity to nonfunctional NLS mutants.
Subcellular Distribution of Adi3ΔT-loop Proteins
To analyze the effect on Adi3 subcellular localization due to the loss of the T-loop extension, subcellular fractions from protoplasts expressing GFP-Adi3 proteins for 16 h were analyzed by Western blot (Fig. 2D). The Adi3 and Adi3S539D proteins showed similar localization in the nucleus and membrane fractions, while no protein was detected in the soluble fraction (Fig. 2D). This localization pattern of GFP-Adi3 was confirmed with Adi3 tagged with the small, 9-amino acid HA epitope (supplemental Fig. S2A) indicating the large GFP protein did not affect localization of the Adi3 protein. The Adi3ΔT-loop protein showed strong localization to the membrane fraction and drastically reduced detection in the nuclear fraction (Fig. 2D) indicating a lack of a functional NLS in the Adi3ΔT-loop protein. Introduction of the S539D mutation (Adi3ΔT-loop/S539D) caused a substantial shift of the Adi3ΔT-loop protein to the nuclear fraction and reduction in membrane localization (Fig. 2D). The Adi3ΔT-loop/S539A protein was not found in the nuclear fraction but was detectable in the membrane fraction only as a high molecular weight smear, suggesting post-translational modification (Fig. 2D). These data indicate that the Adi3 NLS in the T-loop extension directs nuclear localization and that mimicking Pdk1 phosphorylation of Adi3 without the T-loop extension (Adi3ΔT-loop/S539D) can redirect the protein to the nucleus. Taken together with the CDS assays (Fig. 2C), these data suggest that loss of Adi3 nuclear localization (deletion of T-loop extension or NLS mutant) impairs the ability of Adi3 to suppress cell death. Restoring nuclear localization (S539D introduction into Adi3ΔT-loop) is capable of restoring Adi3 CDS. Additionally, our data support our earlier findings that overexpression of non-functional mutant forms of Adi3 act in a dominant negative manner with regard to the endogenous Adi3 protein (13).
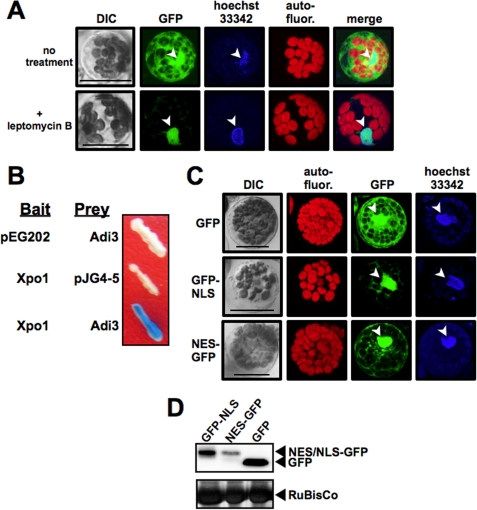
Confirmation of Adi3 Nuclear Localization
To confirm Adi3 nuclear localization, GFP-Adi3 expressing protoplasts were treated with the nuclear export inhibitor leptomycin B, which restricts nuclear/cytoplasmic shuttling proteins to the nucleus (47, 48). The protoplasts were also treated with the DNA stain Hoechst 33342 to identify nuclei (49) followed by confocal microscopy imaging of the GFP and Hoechst 33342 signals. The results showed that leptomycin B treatment reduced cytoplasmic GFP-Adi3 and restricted the protein to the nucleus (Fig. 3A). We also cloned the nuclear export protein Exportin-1 (Xpo1), which binds leucine-rich NESs (34) and showed that Adi3 interacts with Xpo1 in a yeast two-hybrid assay (Fig. 3B). The functionality of the Adi3 NLS was also confirmed by fusing the NLS to the C terminus of GFP (GFP-NLS) and analyzing for nuclear accumulation of GFP. The protein was expressed in tomato protoplasts treated with hoechst 33342 for nucleus identification and cellular localization viewed by confocal microscopy. Protein expression was confirmed by α-GFP Western blot (Fig. 3D). GFP-NLS protein localization was reduced in the cytoplasm, but retained in the nucleus suggesting the Adi3 NLS is a functional nuclear localization signal (Fig. 3C). A similar analysis was carried out with the Adi3 NES sequence, which was fused to the N terminus of GFP (NES-GFP). If the NES is functional, NES-GFP protein should accumulate in the cytoplasm. However, the Adi3 NES was incapable of reducing GFP nuclear localization (Fig. 3C) indicating the Adi3 NES may be a weak export signal unable to maintain nuclear exclusion of the small GFP protein, which can freely enter the nucleus (50, 51).
FIGURE 3.
Adi3 contains a functional NLS. A, leptomycin B treatment. Tomato protoplasts expressing GFP-Adi3 for 14 h were incubated with 20 nm leptomycin B for 90 min followed by incubation with 10 μm Hoechst 33342 for 2 h and visualized by confocal microscopy. Merge, overlay of GFP, Hoechst 33342, and autofluorescence images. Bar, 20 μm; arrowhead, nucleus. B, Adi3 interaction with Xpo1. Xpo1 from tomato was cloned and tested for yeast two-hybrid interaction with Adi3 in the LexA system. C, functional analysis of Adi3 NES and NLS. The indicated GFP fusion proteins were expressed in tomato protoplasts for 14 h, treated with 10 μm Hoechst 33342 for 2 h, and visualized by confocal microscopy. Bar, 20 μm; arrowhead, nucleus. D, expression of GFP NLS and NES fusion proteins in tomato protoplasts as detected by α-GFP Western blot.
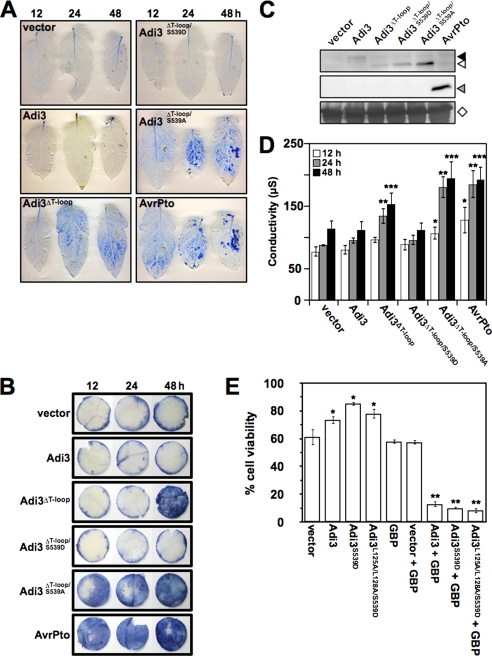
Adi3 Nuclear Localization Is Required for PCD Suppression in Tomato and Tobacco Leaves
Because Adi3 NLS loss, either by deletion of the T-loop extension or NLS mutation, causes Adi3 to have compromised PCD control in protoplasts, functionality of GFP-Adi3 and GFP-Adi3ΔT-loop proteins in leaf tissue was analyzed utilizing Agrobacterium-mediated transient expression in tomato (Fig. 4A) and N. benthamiana (Fig. 4B). As with our protoplast functionality assays (Fig. 2C), we used PtoR tomato plants, which produce strong HR cell death in response to the Pst effector AvrPto (10). N. benthamiana plants also produce HR cell death in response to AvrPto (10). Thus, expression of AvrPto-FLAG was used as a strong cell death inducer positive control (Fig. 4, A and B). In the assays, leaves (tomato) or leaf disks (N. benthamiana) were taken at 12, 24, and 48 h after Agrobacterium infiltration, stained with Evans blue to identify dead cells, depigmented, and photographed. Protein expression in leaf tissue was confirmed by α-GFP and α-FLAG Western blot (Fig. 4C). In both tomato and N. benthamiana, expression of Adi3 showed no loss of CDS (Fig. 4, A and B). The Adi3ΔT-loop protein showed a loss of CDS by 24 h after infiltration in tomato (Fig. 4A) and 48 h in N. benthamiana (Fig. 4B). Expression of Adi3ΔT-loop/S539D was capable of restoring CDS to wild-type levels in both tomato and N. benthamiana, while the expression of Adi3ΔT-loop/S539A caused a very strong loss of CDS nearly equivalent to that of AvrPto (Fig. 4, A and B). This would suggest that Adi3 nuclear localization driven by mimicking Pdk1 phosphorylation of Ser-539 is required for CDS.
FIGURE 4.
Nuclear-localized Adi3 is required for CDS. A, CDS loss in tomato leaves expressing Adi3ΔT-loop proteins. Agrobacterium tumefaciens-mediated transient expression of GFP-Adi3 and GFP-Adi3ΔT-loop proteins was carried out in PtoR tomato leaves. After indicated times, whole leaves were Evans Blue stained, depigmented, and photographed. AvrPto-FLAG was used as positive control for cell death. B, CDS loss in N. benthamiana leaves expressing Adi3ΔT-loop proteins. Agrobacterium-mediated transient expression of GFP-Adi3 and GFP-Adi3ΔT-loop proteins was carried out in N. benthamiana leaves. After indicated times, 1 cm in diameter leaf disks were treated as in A for visualization of induced cell death. C, Western blot of proteins expressed in tomato leaves. Total protein extract was obtained from leaves transiently expressing GFP-Adi3 and GFP-Adi3ΔT-loop proteins and Western analysis carried out using α-GFP (top panel) and α-FLAG (bottom panel) antibodies. Filled triangle, GFP-Adi3; open triangle, GFP-Adi3ΔT-loop proteins; gray triangle, AvrPto-Flag; open diamond, RuBisCo loading control. D, ion leakage as indication of cell death induction. Conductivity tests were carried out on three 1-cm diameter disks from tomato leaves transiently expressing GFP-Adi3 and GFP-Adi3ΔT-loop proteins at indicated times. Values are the average of three independent experiments. Error bars are standard error. One asterisk, two asterisks, and three asterisks indicate significant increase in ion leakage at 12, 24, and 48 h, respectively, compared with vector only expression at those time points (t test, p < 0.05). E, restriction of GFP-Adi3 to cytoplasm by the GFP-binding protein eliminates Adi3 CDS. The indicated combinations of GFP-Adi3 and GBP were expressed in tomato protoplasts for 16 h and observed for cell death using Evans blue staining. All data represent average of three independent experiments. Error bars are standard error. One asterisk and two asterisks indicate significant increase or decrease, respectively, in cell viability compared with vector-only expression (t test, p < 0.05).
A reliable assessment of plant cell death is to measure cell ion leakage from leaf disks by measuring conductivity of a surrounding solution (52). Cell death control in leaf disks from Fig. 4B was analyzed by conductivity (Fig. 4D). The results matched the visualization of cell death seen with Evans blue staining; Adi3ΔT-loop and Adi3ΔT-loop/S539A were unable to suppress cell death, with Adi3ΔT-loop/S539A showing the strongest loss (Fig. 4D). The Adi3ΔT-loop/S539D protein had restored ability to suppress cell death (Fig. 4D). These CDS assays using intact leaves confirm the results seen in our protoplast system (Fig. 1C) and confirms that the protoplast system is a reliable platform for assessing Adi3 function.
As a further confirmation that nuclear Adi3 is required for its CDS activity, we coexpressed GFP-Adi3 in PtoR tomato protoplasts with a GFP-binding protein (GBP) for 16 h and analyzed for loss of Adi3 CDS by Evans blue-stained dead cells. This GBP protein has been shown in plants to effectively bind GFP fusion proteins in vivo and disrupt the function of GFP fusions of nuclear-localized cell death-regulating proteins by restricting them to the cytoplasm (53). Expression of GBP alone did not alter cell viability (Fig. 4E). However, coexpression of GBP with Adi3, Adi3S539D, or Adi3L125A/L128A/S539D showed a strong loss of Adi3 CDS activity (Fig. 4E) further indicating a nuclear-localized Adi3 is required for full CDS. A reduction in the nuclear localization of the GFP-Adi3 proteins expressed with GBP was confirmed by α-GFP Western blot (supplemental Fig. S2B).
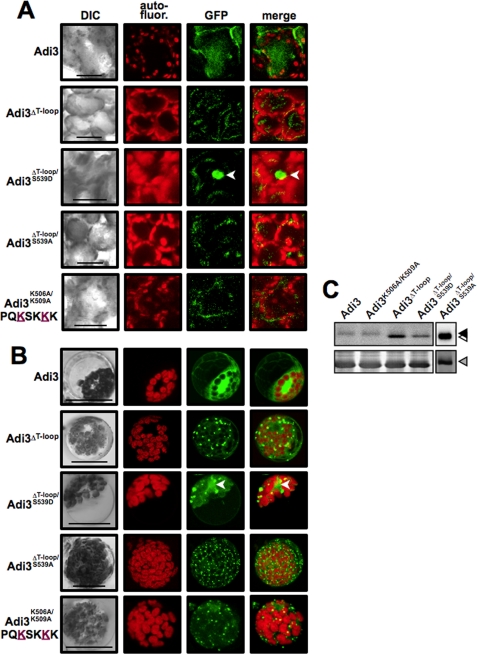
Deletion of the T-loop Extension Causes Localization of Adi3 to Punctate Cellular Structures
To confirm subcellular fractionation results (Fig. 2D), confocal microscopy was used to visualize the localization of GFP-Adi3 and GFP-Adi3 NLS mutants in Agrobacterium-infiltrated intact tomato leaf mesophyll cells (Fig. 5A) and tomato mesophyll protoplasts (Fig. 5B). Protein expression of GFP-Adi3 proteins were confirmed by α-GFP Western blot analysis (Fig. 5C). Both systems showed similar results. Adi3 wild type appeared to be localized throughout the entire cell, while GFP-Adi3ΔT-loop protein was localized to punctate structures throughout the cell (Fig. 5, A and B). Localization of the GFP-Adi3ΔT-loop protein appears to be regulated by the mimicking phosphorylation status of the Pdk1 phosphorylation site, Ser-539. Introduction of the S539D mutation into the Adi3ΔT-loop protein decreased punctate structure localization and directed some protein to the nucleus (Fig. 5, A and B), which was also seen in the subcellular fractionation (Fig. 2D). Introduction of the S539A mutation caused an increase in the number of Adi3ΔT-loop punctate structures with no nuclear localization (Fig. 5B). Expression of the Adi3K506A/K509A protein also showed localization to punctate structures similar to the Adi3ΔT-loop protein (Fig. 5, A and B). The number of GFP-Adi3ΔT-loop punctate structures seen in protoplasts were quantified for each mutant and showed that expression of GFP-Adi3ΔT-loop/S539A is located to the most punctate structures (supplemental Fig. S3). Because the Adi3ΔT-loop/S539A protein has very strong loss of CDS activity and a high amount of localization to punctate cellular structures, there appears to be a strong correlation between the number of punctate structures and induction of PCD. Several attempts to identify these punctate structures have not yielded positive results. However, the Adi3ΔT-loop proteins do appear to be enriched in the membrane fraction (Fig. 2D) suggesting the punctate structures are an intracellular membrane system.
FIGURE 5.
Adi3 NLS mutants are located in punctate cellular structures. A, localization of GFP-Adi3 proteins in intact PtoR tomato leaf mesophyll cells. After Agrobacterium-mediated transient expression for 48 h (GFP-Adi3), 24 h (GFP-Adi3ΔT-loop; GFP-Adi3ΔT-loop/S539D), or 12 h (GFP-Adi3ΔT-loop/S539A; GFP-Adi3K506A/K509A), leaf sections were cut from 3 different leaves and visualized by confocal microscopy. Images are representative of three independent experiments. Merge, overlay of GFP and autofluorescence images. Bar, 20 μm; arrowhead, nucleus. B, localization of GFP-Adi3 proteins in PtoR tomato protoplasts. The indicated GFP-Adi3 proteins were expressed in protoplasts for 16 h and visualized by confocal microscopy. Labeling as in A. C, Western blot using α-GFP antibody on total protein from expression of the indicated GFP-Adi3 proteins. Filled triangle, GFP-Adi3; open triangle, GFP-Adi3ΔT-loop proteins; gray triangle, RuBisCo loading control (Coomassie Blue-stained membrane).
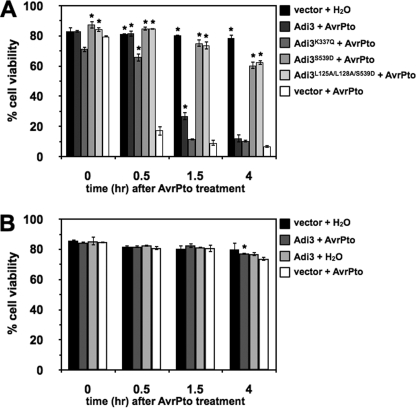
Nuclear-localized Adi3 Can Suppress AvrPto-induced Cell Death
To test our findings that nuclear-localized Adi3 can suppress cell death, we tested the ability of constitutively active and nuclear forms of Adi3 to suppress the induction of cell death caused by AvrPto. PtoR tomato protoplasts expressing GFP alone (vector) or wild-type and mutant GFP-Adi3 proteins for 14 h were transformed with BFP-AvrPto or a water control and cell death measured with Evans Blue over a 4-h time period. Expression of Adi3 and AvrPto were confirmed by α-GFP Western blot (supplemental Fig. S4). Cell death was strongly induced by expression of AvrPto alone as compared with GFP expression only (Fig. 6A, compare first and last column of each time point). Wild-type Adi3 was capable of suppressing this cell death induced by AvrPto at 0.5 h and to a small level at 1.5 h after AvrPto expression (Fig. 6A, compare second and last column of each time point). The constitutively active Adi3S539D and the constitutively nuclear-localized and active Adi3L125A/L128A/S539D were both able to very strongly suppress AvrPto-induced cell death up to 4 h after AvrPto expression (Fig. 6A, compare fourth and fifth column with last column of each time point). Interestingly, kinase-inactive Adi3K337Q was able to suppress AvrPto-induced cell death at 0.5 h after AvrPto expression (Fig. 6A, compare third and last column of each time point). These assays were also carried out in protoplasts from prf-3 tomato plants which have a mutation in the Prf gene (54, 55). This gene is required for Pto-mediated cell death in response to AvrPto, and cell death is not induced in prf-3 plants when treated with AvrPto (10, 54, 55). As expected, cell viability was maintained at or near the GFP alone control in all combinations of Adi3 and AvrPto in prf-3 protoplasts (Fig. 6B).
FIGURE 6.
Nuclear-localized Adi3 protects against AvrPto-induced cell death. A, PtoR tomato protoplasts or B, pfr-3 tomato protoplasts were transformed with indicated GFP-Adi3 constructs, proteins were expressed for 14 h and transformed with an avrPto construct, samples were taken at the indicated times, and cell was viability determined by Evans Blue staining. Graphs represent the average of three independent experiments. Error bars are standard error. One asterisk indicates significant increase in cell viability compared with AvrPto only expression within each time point (Student's t test, p < 0.05).
We also tested the ability of Adi3 to suppress non-pathogen induced forms of cell death in yeast, such as the cell death induced by H2O2 (56). Expression of GFP-Adi3 in yeast was capable of reducing the cell death induced by H2O2 (supplemental Fig. S5) suggesting the Adi3 CDS mechanism is highly conserved in eukaryotes. In these assays the Pst effector protein AvrPtoB was used as a positive control for H2O2 CDS (57).
Taken together, these data confirm that nuclear-localized Adi3 is capable of suppressing cell death and that the function of overexpressed, active forms of Adi3 can be analyzed in vivo because they outcompete endogenous Adi3 protein as we have seen previously (13). Our in vivo functional studies, which use GFP-Adi3 proteins, also suggest that the localization of the GFP-Adi3 proteins is correct since the CDS activity of these GFP-Adi3 proteins matches predicted Adi3 function and that previously seen for constitutively active Adi3S539D (13).
DISCUSSION
Identification of plant proteins that regulate PCD has been difficult and yielded few results as compared with mammalian systems (1, 4, 5, 58). However, several MAP kinases and transcription factors have been identified that regulate plant PCD (11, 59–64). Additionally, a few homologues of mammalian PCD regulators have been found in plant systems (65). Given the important roles of PCD in plant development and pathogen interactions, identification, and characterization of such proteins is critical for understanding how plant PCD functions. We identified the protein kinases Adi3 and Pdk1 as negative regulators of plant PCD (13). Our studies here firmly place Adi3 as a suppressor of PCD in tomato and provide insights into the role of cellular localization in controlling Adi3 CDS activity that are strikingly similar to the apoptosis suppressing function of PKB in mammalian systems. Because Adi3-like sequences are found in many plants (Arabidopsis, rice, corn), it is possible that PCD is regulated in a similar manner in most plants.
The NLS in the Adi3 T-loop Extension Directs Nuclear Localization
Our data show that the Adi3 T-loop extension contains an NLS that is required for nuclear localization. Other studies have suggested that this T-loop extension of plant AGC kinases also functions in determining cellular localization. The T-loop extension of the plant AGC kinases PID and AGC1–7 directs these proteins to the plasma membrane and cytoplasm, respectively (21). Nuclear localization of two other plant VIIIa AGC kinases, KIPK and WAG1, has been shown, but the requirement of the T-loop extension has not been reported (21). The Ndr family of mammalian AGC kinases also contains T-loop extensions similar to plant VIIIa AGC kinases. The extension encodes an NLS for Ndr1, but it appears to inhibit autophosphorylation in other Ndr kinases (29–32). The Adi3 T-loop extension does not appear to be an inhibitor of autophosphorylation. Sequence identity and amino acid length of both mammalian and plant AGC kinase T-loop extensions are quite variable. However, they all contain a stretch of basic amino acids near the C-terminal end of the extension that appears to give functionality to the extension (21, 29). In Ndr1 (31) and Adi3, these basic amino acids encode the NLS and in other Ndr kinases they are responsible for autophosphorylation inhibition (32). Examination of other plant VIIIa AGC kinase T-loop extensions is required to determine if these basic amino acids are required for cellular localization or kinase activity.
Pdk1 Phosphorylation of Adi3 Is Required for Nuclear Localization and CDS
The presented results indicate that in addition to phosphorylation of Adi3 at Ser-539 for activation of kinase activity, mimicking Pdk1-mediated phosphorylation at Ser-539 is sufficient for Adi3 nuclear localization (Figs. 2D and 5, A and B). Interestingly, the S539D mutation can alter localization of the Adi3ΔT-loop protein from punctate structures to the nucleus even though this protein lacks the identified NLS (Fig. 5, A and B), suggesting the S539D phosphorylation mimic may produce a conformational change sufficient for interaction with nuclear import machinery or exposure of a nonconsensus NLS. Pdk1-dependent nuclear localization of plant AGC kinases has not been reported. However, the mammalian AGC kinase PKB is phosphorylated at the plasma membrane on Thr-308 by Pdk1, which mobilizes PKB to the nucleus (14, 66, 67).
Mimicking Pdk1 phosphorylation of Adi3 Ser-539 is also required for CDS as indicated by restoration of CDS to the Adi3ΔT-loop/S539D protein and strong loss of CDS for Adi3ΔT-loop/S539A (Figs. 2C and 4, A, B, D) as well as the ability of Adi3S539D and Adi3L125A/L128A/S539D to suppress AvrPto-induced cell death (Fig. 6A). This is similar to Pdk1-phosphorylated, activated, and nuclear-localized PKB, which can phosphorylate nuclear substrates to suppress cell death (67–71). Interestingly, the S539D mutation does not increase Adi3ΔT-loop kinase activity to the level of wild-type Adi3 (Fig. 1B) even though Adi3ΔT-loop/S539D is capable of CDS, suggesting full autophosphorylation activity is not required for complete Adi3 CDS and that one mechanism by which Adi3 suppresses cell death may be protein interaction for functional inhibition independent of full kinase activity. This is supported by the finding that the kinase-inactive Adi3K337Q is capable of limited CDS (Fig. 6A). From our results a picture is evolving of a Pdk1/Adi3 kinase signaling cascade for suppressing cell death that is similar to mammalian Pdk1/PKB; Pdk1 phosphorylation of Ser-539 activates Adi3 kinase activity, which is sufficient to direct nuclear localization and is required for CDS activity.
A Model for Adi3 Function in Cell Death Control
Based on our results presented here and previous studies (13) we suggest a model of Adi3 function in PCD regulation. We have shown that the Pdk1-Adi3 interaction is constitutive (13) and most likely takes place at the plasma membrane because that is where Pdk1 is known to activate substrates (72). Mimicking phosphorylation of Adi3 Ser-539 by Pdk1 drives Adi3 nuclear localization where it functions to suppress PCD. Loss of Adi3 nuclear localization through deletion of the T-loop extension or NLS mutation directs Adi3 to intracellular punctate structures where it no longer has CDS activity. These punctate structures appear to be an intracellular membrane system. This model, and the fact that Adi3 interacts with Pto and AvrPto (12, 13), raises the possibility that the induction of HR cell death during the pathogen resistance response is mediated by redirecting Adi3 from the nucleus to the punctate membrane structures seen here. This would inactivate Adi3 and induce PCD. In the future, it will be important to determine exactly how Adi3 is being mobilized to the nucleus and how this localization is regulated under pathogen-challenged situations. Additionally, it will be essential to identify the punctate membrane structures to which nonnuclear Adi3 is located, as well as to identify nuclear Adi3 substrates for CDS.
Supplementary Material
Acknowledgments
We thank Dr. Stan Vitha of the Texas A&M Microscopy and Imaging Center for assistance and critical advice with confocal imaging and members of the Devarenne Laboratory for comments and constructive discussions. The eBFP2 construct was kindly provided by Michael W. Davidson, Florida State University. The GFP-binding protein construct was kindly provided by Sophien Kamoun, The Sainsbury Laboratory.
This work was supported by USDA-CSREES Grant 2007-35319-17832 (to G. B. M. and T. P. D.), USDA-AFRI Grant 2010-65108-20526 (to T. P. D.), and by Texas A&M University Dept. of Biochemistry and Biophysics start-up funds (to T. P. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PCD
- plant cell death
- Adi3
- AvrPto-dependent Pto-interacting protein 3
- AGC
- cAMP-dependent protein kinase (PKA)/protein kinase G/protein kinase C (PKC) protein kinase family
- CDS
- cell death suppression
- DIC
- differential interference contrast image
- eGFP
- enhanced green fluorescent protein
- HR
- hypersensitive response
- MBP
- maltose-binding protein
- Ndr kinase
- nuclear Dbf2-related kinase
- NES
- nuclear exportation signal
- NLS
- nuclear localization signal
- Pdk1
- 3-phosphoinositide-dependent protein kinase-1
- Pst
- P. syringae pv. tomato
- Xpo1
- Exportin-1 (yeast CRM1-homolog)
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Lam E. (2004) Nat. Rev. Mol. Cell Biol. 5, 305–315 [DOI] [PubMed] [Google Scholar]
- 2.Greenberg J. T., Yao N. (2004) Cell Microbiol. 6, 201–211 [DOI] [PubMed] [Google Scholar]
- 3.Mur L. A., Kenton P., Lloyd A. J., Ougham H., Prats E. (2008) J. Exp. Bot. 59, 501–520 [DOI] [PubMed] [Google Scholar]
- 4.Hoeberichts F. A., Woltering E. J. (2003) Bioessays 25, 47–57 [DOI] [PubMed] [Google Scholar]
- 5.Lam E. (2008) Crit. Rev. Plant Sci. 27, 413–423 [Google Scholar]
- 6.Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., Ausubel F. M., Glazebrook J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk A., Feys B. J., Frost L. N., Jones J. D. G., Daniels M. J., Parker J. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kachroo P., Shanklin J., Shah J., Whittle E. J., Klessig D. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang H., Yao N., Song J. T., Luo S., Lu H., Greenberg J. T. (2003) Genes Dev. 17, 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedley K. F., Martin G. B. (2003) Annu. Rev. Phytopathol. 41, 215–243 [DOI] [PubMed] [Google Scholar]
- 11.del Pozo O., Pedley K. F., Martin G. B. (2004) EMBO J. 23, 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogdanove A. J., Martin G. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devarenne T. P., Ekengren S. K., Pedley K. F., Martin G. B. (2006) EMBO J. 25, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vivanco I., Sawyers C. L. (2002) Nat. Rev. Cancer 2, 489–501 [DOI] [PubMed] [Google Scholar]
- 15.Vara J. A. F., Casado E., de Castro J., Cejas P., Belda-Iniesta C., Gonzales-Baron M. (2004) Cancer Trt. Reviews 30, 193–204 [DOI] [PubMed] [Google Scholar]
- 16.Bögre L., Okrész L., Henriques R., Anthony R. G. (2003) TRENDS Plant Sci. 8, 424–431 [DOI] [PubMed] [Google Scholar]
- 17.Sobko A. (2006) Sci. STKE 2006, re9. [DOI] [PubMed] [Google Scholar]
- 18.Bivona T. G., Quatela S. E., Bodemann B. O., Ahearn I. M., Soskis M. J., Mor A., Miura J., Wiener H. H., Wright L., Saba S. G., Yim D., Fein A., Pérez de Castro I., Li C., Thompson C. B., Cox A. D., Philips M. R. (2006) Mol. Cell 21, 481–493 [DOI] [PubMed] [Google Scholar]
- 19.Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., Hung M. C. (2005) Science 310, 306–310 [DOI] [PubMed] [Google Scholar]
- 20.Zegzouti H., Anthony R. G., Jahchan N., Bögre L., Christensen S. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6404–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zegzouti H., Li W., Lorenz T. C., Xie M., Payne C. T., Smith K., Glenny S., Payne G. S., Christensen S. K. (2006) J. Biol. Chem. 281, 35520–35530 [DOI] [PubMed] [Google Scholar]
- 22.Anthony R. G., Henriques R., Helfer A., Mészáros T., Rios G., Testerink C., Munnik T., Deák M., Koncz C., Bögre L. (2004) EMBO J. 23, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anthony R. G., Khan S., Costa J., Pais M. S., Bögre L. (2006) J. Biol. Chem. 281, 37536–37546 [DOI] [PubMed] [Google Scholar]
- 24.Galván-Ampudia C. S., Offringa R. (2007) Trends Plant Sci. 12, 541–547 [DOI] [PubMed] [Google Scholar]
- 25.Briggs W. R., Christie J. M. (2002) TRENDS Plant Sci. 7, 204–210 [DOI] [PubMed] [Google Scholar]
- 26.Oyama T., Shimura Y., Okada K. (2002) Plant J. 30, 289–299 [DOI] [PubMed] [Google Scholar]
- 27.Rentel M. C., Lecourieux D., Ouaked F., Usher S. L., Petersen L., Okamoto H., Knight H., Peck S. C., Grierson C. S., Hirt H., Knight M. R. (2004) Nature 427, 858–861 [DOI] [PubMed] [Google Scholar]
- 28.Robert H. S., Offringa R. (2008) Curr. Opin. Plant Biol. 11, 495–502 [DOI] [PubMed] [Google Scholar]
- 29.Hergovich A., Cornils H., Hemmings B. A. (2008) Biochim. Biophys. Acta 1784, 3–15 [DOI] [PubMed] [Google Scholar]
- 30.Tamaskovic R., Bichsel S. J., Hemmings B. A. (2003) FEBS Lett. 546, 73–80 [DOI] [PubMed] [Google Scholar]
- 31.Millward T., Cron P., Hemmings B. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5022–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A. (2004) J. Biol. Chem. 279, 35228–35235 [DOI] [PubMed] [Google Scholar]
- 33.Higuchi R., Krummel B., Saiki R. K. (1988) Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haasen D., Köhler C., Neuhaus G., Merkle T. (1999) Plant J. 20, 695–705 [DOI] [PubMed] [Google Scholar]
- 35.Nelson B. K., Cai X., Nebenführ A. (2007) Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 36.Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) Nucleic Acids Res. 35, W585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C., Hodgins D., Calvert J. G., Welch S. K., Jolie R., Yoo D. (2006) Virology 346, 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meares G. P., Jope R. S. (2007) J. Biol. Chem. 282, 16989–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. (1999) EMBO J. 18, 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., Hay N. (2001) Genes Dev. 15, 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawlor M. A., Alessi D. R. (2001) J. Cell Sci. 114, 2903–2910 [DOI] [PubMed] [Google Scholar]
- 42.Luo H. R., Hattori H., Hossain M. A., Hester L., Huang Y., Lee-Kwon W., Donowitz M., Nagata E., Snyder S. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11712–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudek H., Datta S. R., Franke T. F., Birnbaum M. J., Yao R., Cooper G. M., Segal R. A., Kaplan D. R., Greenberg M. E. (1997) Science 275, 661–665 [DOI] [PubMed] [Google Scholar]
- 44.Arico S., Pattingre S., Bauvy C., Gane P., Barbat A., Codogno P., Ogier-Denis E. (2002) J. Biol. Chem. 277, 27613–27621 [DOI] [PubMed] [Google Scholar]
- 45.Kulp S. K., Yang Y. T., Hung C. C., Chen K. F., Lai J. P., Tseng P. H., Fowble J. W., Ward P. J., Chen C. S. (2004) Cancer Res. 64, 1444–1451 [DOI] [PubMed] [Google Scholar]
- 46.Zhu J., Huang J. W., Tseng P. H., Yang Y. T., Fowble J., Shiau C. W., Shaw Y. J., Kulp S. K., Chen C. S. (2004) Cancer Res. 64, 4309–4318 [DOI] [PubMed] [Google Scholar]
- 47.Kudo N., Khochbin S., Nishi K., Kitano K., Yanagida M., Yoshida M., Horinouchi S. (1997) J. Biol. Chem. 272, 29742–29751 [DOI] [PubMed] [Google Scholar]
- 48.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodin S. A., Pis'menskii V. F., Nikitin A. M., Surovaya A. N., Gursky G. V. (2001) Dokl. Biochem. Biophys. 379, 235–238 [DOI] [PubMed] [Google Scholar]
- 50.Böhm C., Seibel N. M., Henkel B., Steiner H., Haass C., Hampe W. (2006) J. Biol. Chem. 281, 14547–14553 [DOI] [PubMed] [Google Scholar]
- 51.Seibel N. M., Eljouni J., Nalaskowski M. M., Hampe W. (2007) Anal. Biochem. 368, 95–99 [DOI] [PubMed] [Google Scholar]
- 52.Rizhsky L., Shulaev V., Mittler R. (2004) in Measuring Programmed Cell Death in Plants, Apoptosis Methods and Protocols (Brady H. J. M., ed) pp. 179–180, Humana Press, Totowa: [DOI] [PubMed] [Google Scholar]
- 53.Schornack S., Fuchs R., Huitema E., Rothbauer U., Lipka V., Kamoun S. (2009) Plant J. 60, 744–754 [DOI] [PubMed] [Google Scholar]
- 54.Salmeron J. M., Barker S. J., Carland F. M., Mehta A. Y., Staskawicz B. J. (1994) Plant Cell 6, 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salmeron J. M., Oldroyd G. E., Rommens C. M., Scofield S. R., Kim H. S., Lavelle D. T., Dahlbeck D., Staskawicz B. J. (1996) Cell 86, 123–133 [DOI] [PubMed] [Google Scholar]
- 56.Madeo F., Fröhlich E., Ligr M., Grey M., Sigrist S. J., Wolf D. H., Fröhlich K. U. (1999) J. Cell Biol. 145, 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abramovitch R. B., Kim Y. J., Chen S., Dickman M. B., Martin G. B. (2003) EMBO J. 22, 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam E., Kato N., Lawton M. (2001) Nature 411, 848–853 [DOI] [PubMed] [Google Scholar]
- 59.Ligterink W., Kroj T., zur Nieden U., Hirt H., Scheel D. (1997) Science 276, 2054–2057 [DOI] [PubMed] [Google Scholar]
- 60.Yang K. Y., Liu Y., Zhang S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S., Klessig D. F. (2001) Trends Plant Sci. 6, 520–527 [DOI] [PubMed] [Google Scholar]
- 62.Ren D., Yang H., Zhang S. (2002) J. Biol. Chem. 277, 559–565 [DOI] [PubMed] [Google Scholar]
- 63.Li S., Samaj J., Franklin-Tong V. E. (2007) Plant Physiol. 145, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaneda T., Taga Y., Takai R., Iwano M., Matsui H., Takayama S., Isogai A., Che F. S. (2009) EMBO J. 28, 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doukhanina E. V., Chen S., van der Zalm E., Godzik A., Reed J., Dickman M. B. (2006) J. Biol. Chem. 281, 18793–18801 [DOI] [PubMed] [Google Scholar]
- 66.Scheid M. P., Parsons M., Woodgett J. R. (2005) Mol. Cell Biol. 25, 2347–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyamoto S., Rubio M., Sussman M. A. (2009) Cardiovasc. Res. 82, 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiraishi I., Melendez J., Ahn Y., Skavdahl M., Murphy E., Welch S., Schaefer E., Walsh K., Rosenzweig A., Torella D., Nurzynska D., Kajstura J., Leri A., Anversa P., Sussman M. A. (2004) Circ. Res. 94, 884–891 [DOI] [PubMed] [Google Scholar]
- 69.Adini I., Rabinovitz I., Sun J. F., Prendergast G. C., Benjamin L. E. (2003) Genes Dev. 17, 2721–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S. B., Xuan Nguyen T. L., Choi J. W., Lee K. H., Cho S. W., Liu Z., Ye K., Bae S. S., Ahn J. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16584–16589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xuan Nguyen T. L., Choi J. W., Lee S. B., Ye K., Woo S. D., Lee K. H., Ahn J. Y. (2006) Biochem. Biophys. Res. Commun. 349, 789–798 [DOI] [PubMed] [Google Scholar]
- 72.Biondi R. M. (2004) Trends Biochem. Sci. 29, 136–142 [DOI] [PubMed] [Google Scholar]
- 73.Shan L., Thara V. K., Martin G. B., Zhou J. M., Tang X. (2000) Plant Cell 12, 2323–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.