Abstract
The growth hormone (GH)-insulin-like growth factor-I (IGF-I) axis regulates somatic growth during childhood and orchestrates tissue repair throughout the life span. Recently described inactivating mutations in Stat5b in humans with impaired growth have focused attention on this transcription factor as a key agent linking GH-stimulated signals to IGF-I gene expression, and several putative Stat5b sites have been identified in the IGF-I gene. Here, we define and characterize potential GH- and Stat5b-activated chromosomal enhancers that can regulate IGF-I gene transcription. Of 89 recognizable Stat5 sequences in 200 kb centering on the rat IGF-I gene, 22 resided within conserved regions and/or were identical among different species. Only 15 of these sites, organized into 7 distinct domains, were found to bind Stat5b by quantitative chromatin immunoprecipitation assays in liver chromatin of rats, but only after acute GH treatment. These sites could bind Stat5b in vitro, and individual domains could mediate GH- and Stat5b-stimulated IGF-I promoter activity in cultured cells. Further analyses revealed that four Stat5b domains possessed chromatin signatures of enhancers, including binding of co-activators p300 and Med1, and RNA polymerase II. These modifications preceded GH-stimulated recruitment of Stat5b, as did lysine 4 monomethylation of histone H3, which was enriched in 6/7 Stat5b-binding elements. In contrast, histone acetylation was induced by GH but was limited to Stat5b binding domains found within the IGF-I transcription unit. We conclude that GH stimulates recruitment of Stat5b to multiple dispersed regions within the igf1 locus, including several with properties consistent with long range transcriptional enhancers that collectively regulate GH-activated IGF-I gene transcription.
Keywords: Chromatin, Chromatin Immunoprecipitation (ChIP), Chromatin Histone Modification, Chromatin Regulation, Gene Expression, Gene Transcription, Genome Structure, Growth Factors, Signal Transduction, STAT Transcription Factor
Introduction
Signal transducers and activators of transcription include a family of seven related proteins (Stats 1–4, 5a, 5b, and 6) that function as key intermediates in signaling pathways of many cytokines, growth factors, and hormones (1–4). The first Stats were characterized nearly 20 years ago as effector molecules for interferons α/β and γ (5, 6), and subsequent studies have both broadened the biological significance of this protein family as critical agents in multiple physiological and pathophysiological processes and have further defined their mechanisms of action at biochemical, molecular, and atomic levels of resolution (1–4).
Although specific details differ for each Stat, these proteins are typically found in the cytoplasm of responsive cells prior to cytokine stimulation and are recruited to phosphorylated tyrosine residues found in intracellular segments of activated cytokine receptors, where they become phosphorylated on a tyrosine near the Stat COOH terminus by a receptor-associated tyrosine protein kinase, usually Jak1–3, or Tyk2 (1–3), depending on the receptor. The modified Stats then dissociate from the receptor-docking site, form dimers via reciprocal interactions of an Src homology 2 domain on one Stat molecule with the phosphorylated tyrosine on the other (1), and are translocated into the nucleus, where they bind as dimers to specific DNA sites in chromatin (1–4). Most Stats recognize the palindromic DNA sequence, 5′-TTCNxGAA-3′, where N is any deoxynucleotide, and x = 2–4, but with different preferences depending on the individual Stat (1, 7). Recent profiling of Stat1 interactions with chromatin of human HeLa cells has revealed a broad distribution of binding domains in DNA within the genome (8–10), with the majority being located in intragenic regions and in introns in some studies (9) but at or near promoters in others (8). More limited results focusing on Stat4 in mouse T lymphocytes have indicated that its binding sites in chromatin cluster near transcription start regions of target genes (11). Comparable experiments have not been reported for other Stats.
Pituitary-derived growth hormone (GH)2 plays a pivotal role in regulating somatic growth during childhood and adolescence (12–14), primarily by stimulating the biosynthesis of insulin-like growth factor-I (IGF-I), a 70-residue secreted growth-promoting protein (15, 16) that also is an important regulator of intermediary metabolism and tissue repair throughout the life span (14, 17, 18). The GH-IGF-I axis also has been linked in a negative way to aging and to the development of certain cancers in humans and other species (19, 20), implying that under normal physiological conditions its activity must be limited in scope and duration to preserve homeostasis. The single membrane-spanning GH receptor is a member of the cytokine receptor family, and hormone binding promotes activation of receptor-associated Jak2, leading to phosphorylation of a cohort of tyrosine residues on the intracellular part of the receptor (13, 21) and recruitment of several signaling molecules, including Stats 1, 3, 5a, and 5b, which collectively mediate the biological effects of GH (13, 21).
Recently identified inactivating molecular lesions in the STAT5B gene in humans with impaired growth (22, 23), targeted gene knockouts of Stat5b in mice (24, 25), and biochemical and molecular studies (26) have collectively implicated Stat5b as an essential intermediate in a signal transduction cascade leading from the hormone-activated GH receptor to induction of IGF-I biosynthesis by stimulating IGF-I gene transcription (16, 27). In further support of this hypothesis, two distinct GH-inducible Stat5b binding domains have been mapped to chromatin in human IGF1 and rat Igf1 loci (28–30), and both appear able to act as GH- and Stat5b-dependent regulators of IGF-I promoter function in cultured cells (28, 30). As results of more recent experiments have suggested the potential existence of multiple Stat5b-binding elements in the IGF-I gene (31, 32), we sought to identify and characterize putative GH-regulated and Stat5b-dependent chromosomal enhancers responsible for GH-activated IGF-I gene transcription. Our results show that GH acutely stimulates recruitment of Stat5b to a least seven distinct chromosomal domains found throughout the Igf1 locus coincident with induction of IGF-I gene transcription and that each can function in vitro to augment IGF-I promoter activity. Further mapping studies in rat liver chromatin demonstrate that these GH-activated Stat5b-binding elements include two distinguishable groups, one with the in vivo characteristics of chromosomal enhancers (33, 34), as evidenced by the presence of transcriptional co-factors p300 and Med1, and RNA polymerase II at these sites prior to hormonal stimulation, and the other lacking this signature. Taken together, our studies define a process whereby GH acutely induces binding of Stat5b to DNA at multiple dispersed sites in Igf1 chromatin that collectively regulate hormone-activated IGF-I gene expression.
EXPERIMENTAL PROCEDURES
Materials
The following reagents were purchased: recombinant rat GH, National Hormone and Pituitary Program, NIDDK, National Institutes of Health; fetal calf serum, Dulbecco's modified Eagle's medium, and phosphate-buffered saline, Mediatech-Cellgro (Herndon, VA); QuikChange site-directed mutagenesis kit, Stratagene (La Jolla, CA); SuperScript III reverse transcriptase system, protein A-Sepharose beads, SYBR Green platinum quantitative PCR mixture, and trypsin/EDTA solution, Invitrogen; whole genome amplification kit, Sigma; QIAquick PCR purification kit (Valencia, CA); Transit-LT1, Mirus (Madison, WI); protease inhibitor tablets, Roche Applied Sciences; okadaic acid, Alexis Biochemicals (San Diego); BCA protein assay kit, Pierce; and restriction enzymes, buffers, ligases, and polymerases, Roche Applied Sciences and Clontech. The following antibodies were purchased from commercial vendors: Stat5 (C17), RNA polymerase II (H-224), p300 (C-20), and Med1/TRAP 220 (C-19), Santa Cruz Biotechnology (Santa Cruz, CA); acetyl histone H3 (recognizes acetyl-Lys-9 and -Lys-14), acetyl histone H4 (recognizes acetyl-Lys-5, -Lys-8, -Lys-12, and -Lys-16), cAMP-response element-binding protein, and pStat5 (clone 8-5-2), Millipore (Billerica, MA); histone H3 trimethyl Lys-4 and histone H3 monomethyl Lys-4, Abcam (Cambridge, UK); and Stat5b, Invitrogen. Oligonucleotides were synthesized at the Oregon Health & Science University DNA Services Core. All other chemicals were reagent grade and were purchased from commercial suppliers.
Recombinant Plasmids
Expression plasmids in pcDNA3 for the mouse GH receptor and for FLAG epitope-tagged wild-type and constitutively active rat Stat5b (FLAG-Stat5bCA, N642H) in pcDNA3 have been described (28), as have firefly luciferase reporter gene plasmids derived from TK-Luc (thymidine kinase promoter in pGL2) containing HS7 (R34–35) and the 5′ distal (R8–9) regions of rat IGF-I (28). Other promoter-reporter plasmids were prepared as follows: DNA segments of 200–375 bp spanning putative Stat5b binding domains were amplified by PCR from rat genomic DNA and cloned 5′ to the TK promoter in TK-Luc by standard molecular methods. Cloning primers may be found in supplemental Table 1. A luciferase reporter gene plasmid in pGL2 containing 1711 bp of rat IGF-I promoter 1 plus 328 bp of exon 1 has been described (IGF-I P1-Luc), as have additional constructs with HS7 and 5′ distal domains located 5′ to P1 (30). An IGF-I promoter 2 luciferase fusion gene (IGF-I P2-Luc) was prepared by swapping a 124-bp genomic fragment cloned by PCR (80 bp of rat P2 plus 44 bp of exon 2) for P1 in IGF-I P1-Luc. The identical Stat5 binding domains were cloned immediately 5′ to each promoter in IGF-I P1-Luc and IGF-I P2-Luc (R2–3, R8–9, R13, R34–35, R53, R57–59, and R60–61), and are in the same location and orientation as in TK-Luc. Point mutations were introduced into selected TK-Luc plasmids using the QuikChange site-directed mutagenesis kit. All newly prepared DNA fragments in all reporter plasmids and all engineered mutations were confirmed by DNA sequencing.
Cell Culture, Transient Transfections, and Reporter Gene Assays
COS-7 cells (ATCC CRL-1651) were incubated in antibiotic-free Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in humidified air with 5% CO2 and were transfected with Stat5bCA or empty vector using Transit-LT1, as described previously (30). Cells were harvested following a 40-h incubation, and nuclear proteins were isolated (30). For promoter-reporter gene assays, cells on 6-well tissue culture dishes were co-transfected with mouse GH receptor (100 ng) and wild-type Stat5b (100 ng for TK promoter studies and 250 ng for IGF-I promoter studies), and the promoter-reporter plasmids are indicated in Figs. 5 and 6 and supplemental Fig. 1 (250 ng for TK-Luc and 500 ng for IGF-I P1-Luc and IGF-I P2-Luc). Cells were incubated for 24 h, followed by addition of serum-free medium containing 1% bovine serum albumin and either vehicle or rat GH (1 μg/ml = ∼ 40 nm) for 18 h. Cells were then harvested and lysates used for luciferase assays (28). All results were normalized to total cellular protein concentrations.
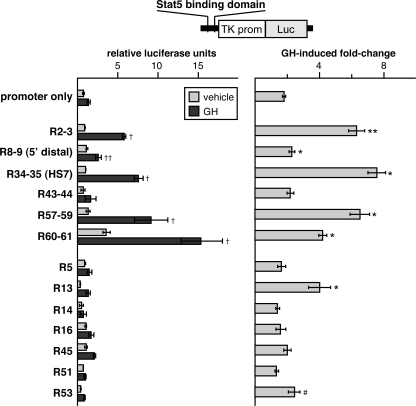
FIGURE 5.
Selected conserved Stat5b consensus sequences confer GH responsiveness to a neutral promoter. Left panel, results of luciferase (Luc) reporter gene assays in COS-7 cells transiently transfected with expression plasmids encoding the mouse GH receptor and rat Stat5b and incubated with vehicle (light bars) or rat GH (40 nm) (dark bars) for 18 h. Luciferase reporter plasmids contain the TK promoter (prom) plus an individual Stat5b binding domain as indicated. Details for each promoter plasmid are described under “Experimental Procedures.” The graph summarizes results of >3 independent experiments (mean ± S.E.), each performed in duplicate. Luciferase values for the TK promoter alone ranged from 5 to 20 × 103 relative light units/10 s (†, p < 0.02; ††, p < 0.05, versus TK promoter alone after GH). Right panel, graph depicts the mean (± S.E.) fold-change induced by GH for each promoter-reporter plasmid (*, p < 0.02; **, p < 0.05, # p < 0.20 versus TK promoter alone).
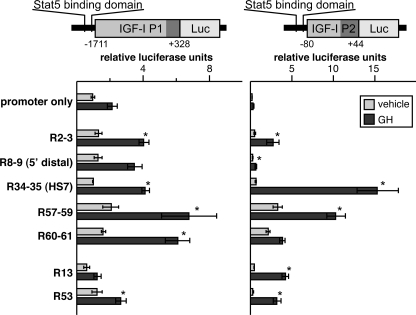
FIGURE 6.
Selected conserved Stat5b binding domains can confer GH responsiveness to one or both IGF-I promoters. Results of luciferase (Luc) reporter gene assays in COS-7 cells were transiently transfected with expression plasmids encoding the mouse GH receptor and rat Stat5b and incubated with vehicle (light bars) or rat GH (40 nm) (dark bars) for 18 h. The left panel shows results with IGF-I promoter 1 (P1) and the right panel with IGF-I promoter 2 (P2). Bars represent the mean ± S.E. of 3–6 independent experiments, each performed in duplicate (*, p < 0.02 versus fold-change P1 or P2). Details for each promoter plasmid are described under “Experimental Procedures.”
Animal Studies
Male Sprague-Dawley rats were purchased from Harlan Sprague-Dawley (Indianapolis, IN), after being hypophysectomized by a transauricular route at age 7 weeks. Rats were housed at the Oregon Health & Science University Animal Care Facility on a 12-h light/dark schedule with free access to food and water and received care according to National Institutes of Health guidelines. GH deficiency was confirmed by failure to grow during a >2-week observation period. Rats subsequently received a single intraperitoneal injection of vehicle (10 mm NaHCO3) or recombinant rat GH (1.5 mg/kg) and were sacrificed at 15 min to 6 h after hormone injection. The liver was removed, and chromatin, RNA, and nuclear proteins were isolated from individual animals as outlined below. For gene expression studies and ChIP experiments, up to four independent series of rats were used. The Oregon Health & Science University Committee on Animal Care and Use approved all studies using rats.
RNA Isolation and Analysis
Hepatic nuclear RNA was isolated as described previously (28). RNA integrity was assessed by agarose gel electrophoresis, and concentrations were determined spectrophotometrically at 260 nm. Nuclear RNA (5 μg) was reverse-transcribed with random hexamers in a final volume of 50 μl using a reverse transcription-PCR kit, and 1 μl of cDNA was used as template for PCR. Pilot studies were performed to establish a cycle number for each primer set that achieved amplification in the linear range (25–35 cycles depending on the primers). Products were visualized after agarose gel electrophoresis by ethidium bromide staining. Results presented in Fig. 1D are representative of three or more independent reverse transcription-PCR experiments and replicated with at least three different series of rats.
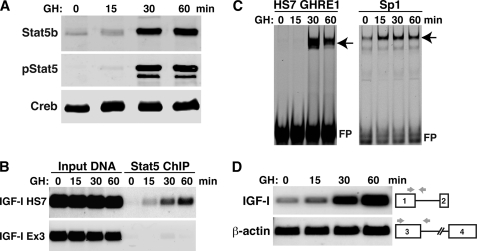
FIGURE 1.
GH acutely stimulates nuclear localization and activation of Stat5b, and promotes IGF-I gene transcription. Results are shown of time course studies using hepatic nuclei harvested from GH-deficient male rats treated with a single intraperitoneal injection of recombinant rat GH (1.5 mg/kg) for 15–60 min. A, immunoblots of nuclear proteins for Stat5b, phospho-Stat5 (pStat5), and cAMP-response element-binding protein (Creb). B, ChIP assays for Stat5b at HS7, a previously described Stat5b-binding element in IGF-I intron 2 (28), and at a site in IGF-I exon 3. C, results of gel mobility shift assays using IR-labeled double-stranded oligonucleotide probes for GHRE-1, one of two Stat5b-binding sites within HS7 (28), and for an Sp1 consensus binding sequence. Arrows indicate protein-DNA complexes, and FP indicates free probe. D, measurement by semi-quantitative reverse transcription-PCR of nascent nuclear RNA for IGF-I and β-actin gene transcription, using primers for an exon and adjacent intron, as diagrammed.
Protein Immunoblotting
Immunoblotting was performed as described previously (30), using hepatic nuclear protein extracts (5 μg) and the following dilutions of primary antibodies: anti-Stat5b-1:5000, anti-pStat5 −1:2000, and anti-cAMP-response element-binding protein −1:5000. Results were scanned and analyzed on a LiCoR Odyssey Infrared Imaging System, using software version 1.2 (LiCoR, Lincoln, NE).
Quantitative ChIP
Initial steps were modified from published protocols (28, 30). For each time point, an ∼600-mg fragment of rat liver was minced and incubated at 20 °C in 30 ml of Dulbecco's modified Eagle's medium plus 1% formaldehyde on a rotating platform for 15 min, followed by addition of 4.5 ml of 1 m glycine and incubation for an additional 5 min. After centrifugation at 200 × g for 5 min at 20 °C, the pellet was washed in phosphate-buffered saline, suspended in 1 ml of lysis buffer (50 mm Tris-HCl, 5 mm EDTA, 1% SDS, pH 8.1, plus protease inhibitors), and incubated for 15 min at 4 °C. Each sample was sonicated on ice with a Branson micro-tip sonicator (setting 10) using a total of 10 rounds of 10 pulses for 1 s each, interspersed with 30-s incubations on ice, followed by centrifugation at 14,000 × g for 10 min at 4 °C (these steps generated DNA fragments of average size of 500 ± 100 bp). The protein concentration of the supernatant (measured by BCA assay) was adjusted to 1 mg/ml in immunoprecipitation buffer (50 mm Tris-HCl, 5 mm EDTA, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, pH 8.1, plus protease inhibitors), and 1-ml aliquots were stored at −80 °C until use. Immunoprecipitations were performed as described previously (30), and DNA was extracted using the QIAquick PCR purification kit, suspended in 50 μl of 10 mm Tris-HCl, 1 mm EDTA, pH 8.1, and used as the template in quantitative PCRs. Primers were designed following guidelines from PrimerQuest (Integrated DNA Technologies, Coralville, IA) to be complementary to different regions of the rat IGF-I gene and to yield amplicons ranging in length from 70–110 bp (see supplemental Table 2). PCR experiments were performed using a Bio-Rad Chromo4 Real Time PCR detection system. Reactions (20 μl final volume) contained 1× SYBR Green mixture, 200 nm primers, and ChIP-enriched DNA and were performed in 8-well strips in a 96-well format. Standard curves contained 0.01–1.25 ng of genomic DNA and were included in each experiment for each primer set. Values are expressed as percent input necessary to achieve the identical cycle threshold. For Stat5-ChIP, the initial survey (Fig. 4A) represents a mean of independent immunoprecipitations using two rats, and the GH time course in Fig. 4B depicts the mean ± S.E. from independent immunoprecipitations from three different series of rats. Results presented in Figs. 8–10 are representative of three or more independent quantitative PCR experiments from independent immunoprecipitations.
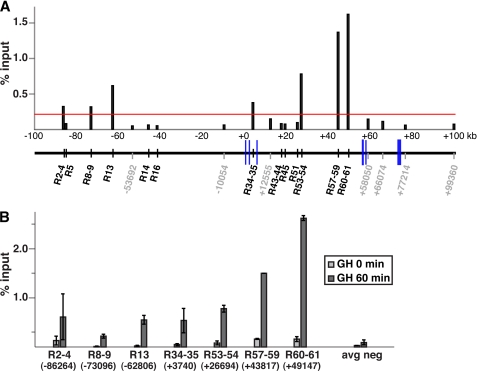
FIGURE 4.
GH induces recruitment of Stat5b to a subset of potential binding elements within the Igf1 locus. Results are shown of quantitative ChIP experiments for Stat5 using hepatic chromatin harvested from GH-deficient male rats treated with a single intraperitoneal injection of recombinant rat GH (1.5 mg/kg) for 0 or 60 min. Primer sets and their genomic locations are listed in supplemental Table 2. All studies were performed by quantitative PCR. A, analysis of binding of Stat5b after GH treatment to the 22 conserved putative Stat5-binding sites in 13 domains outlined in Fig. 3 plus 7 other control primer pairs dispersed throughout the rat Igf1 locus (the latter are outlined in gray and are designated by the chromatin position at the center of the amplicon relative to the 5′ end of the IGF-I gene). The red line depicts a value of 2.5 times the average for the seven controls. Results represent the mean of two independent experiments. B, binding of Stat5b to the 15 sites in seven segments termed positive in A prior to (light gray bars) and at 60 min after a single systemic GH injection (dark gray bars). Results reflect the mean ± S.E. of three independent experiments. The negative control (avg neg) represents the mean of the seven control regions in A.
FIGURE 8.
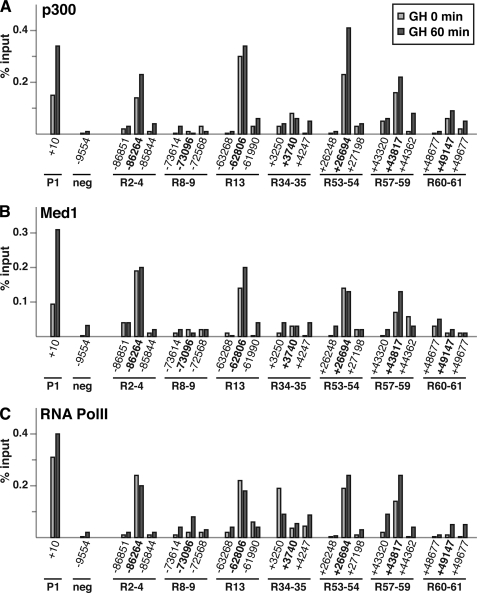
Binding of transcriptional co-factors to a subset of Stat5b binding domains within the Igf1 locus. Results are shown of quantitative ChIP experiments using hepatic chromatin harvested from GH-deficient male rats prior to (light gray bars) and at 60 min after a single systemic GH injection (dark gray bars). Primer sets and their genomic locations are listed in supplemental Table 2. A, ChIP for p300. B, ChIP for Med1. C, ChIP for RNA polymerase II (RNA pol II). For all experiments IGF-I promoter 1 (P1) serves as a positive control, and a region located ∼10 kb 5′ to the beginning of IGF-I exon 1 as a negative (neg) control. Results are representative of three independent experiments. The coordinates in boldface type correspond to the Stat5b-binding sites in Fig. 4B.
FIGURE 9.
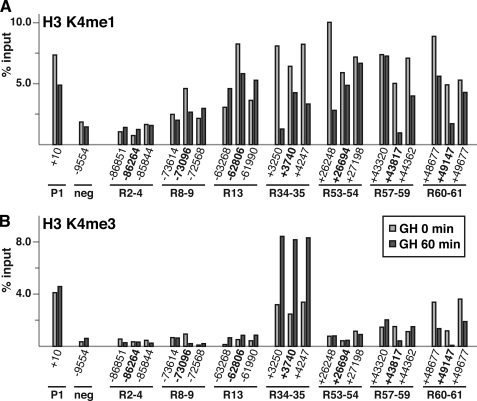
Variable patterns of histone lysine 4 methylation in chromatin at a subset of inducible Stat5b binding domains within the Igf1 locus. Results are shown of quantitative ChIP experiments using hepatic chromatin harvested from GH-deficient male rats prior to (light gray bars) and at 60 min after a single systemic GH injection (dark gray bars). A, ChIP for lysine 4 mono-methylation of histone H3 (H3K4me1). B, ChIP for lysine 4 tri-methylation of histone H3 (H3K4me3). For all experiments IGF-I promoter 1 (P1) serves as a positive control, and a region located ∼10 kb 5′ to the beginning of IGF-I exon 1 serves as a negative (neg) control. Results are representative of three independent experiments. The coordinates seen in boldface type correspond to the Stat5b-binding sites in Fig. 4B.
FIGURE 10.
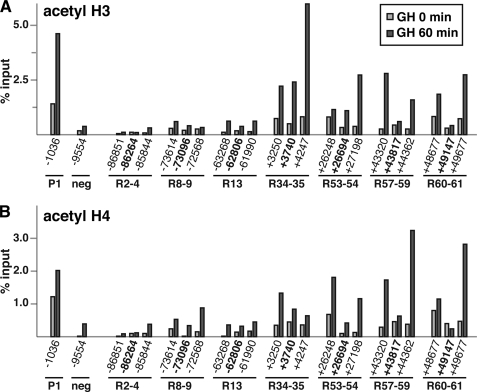
GH induces histone acetylation in chromatin at a subset of inducible Stat5b binding domains within the Igf1 locus. Results are shown of quantitative ChIP experiments using hepatic chromatin harvested from GH-deficient male rats prior to (light gray bars) and at 60 min after a single systemic GH injection (dark gray bars). A, ChIP for acetyl histone H3. B, ChIP for acetyl histone H4. For all experiments IGF-I promoter 1 (P1) serves as a positive control, and a region located ∼10 kb 5′ to the beginning of IGF-I exon 1 as a negative (neg) control. Results are representative of three independent experiments. The coordinates listed in boldface type correspond to the Stat5b-binding sites in Fig. 4B.
DNA-Protein Binding Studies
Electrophoretic gel mobility shift assays and antibody super-shift and DNA competition experiments were performed as described previously (30) with COS-7 (1–2 μg) or rat hepatic nuclear protein extracts (5 μg) and 5′-IR Dye700-labeled double-stranded oligonucleotides. Sequences for the top strand of the double-stranded DNA probes are shown in supplemental Table 3. After incubation of proteins and DNA for 60 min at 4 °C, products were separated by electrophoresis through nondenaturing 5% polyacrylamide gels in 1× Tris borate/EDTA (90 mm Tris, 90 mm boric acid, 2 mm EDTA, pH 8.3) at 200 V for 25–35 min at 20 °C. Results were scanned and analyzed on a LiCoR Odyssey Infrared Imaging System, using software version 1.2.
RESULTS
GH Acutely Stimulates Nuclear Localization and Activation of Stat5b and Induces IGF-I Gene Transcription
Stat5b plays a central role in GH-mediated somatic growth in large part via its ability to robustly stimulate IGF-I gene transcription (26, 28), yet its mechanisms of action have not been elucidated. In pituitary-deficient rats, a single systemic GH pulse rapidly activates Stat5b (35, 36), as seen by its tyrosine phosphorylation and accumulation in liver nuclei, as depicted in Fig. 1A, which shows the coordinate appearance of Stat5b and pStat5 in hepatic nuclear protein extracts within 30 min of hormone administration. GH-activated Stat5b is also found to associate with target gene DNA in liver chromatin with similarly rapid induction kinetics, as evidenced by binding by ChIP to a response element in IGF-I intron 2 termed HS7 (28), but not to a segment in nearby exon 3 (Fig. 1B). HS7 encodes two Stat5 sites separated by 60 bp of intervening DNA, and GH treatment promotes the ability of activated Stat5b to bind to these DNA elements (28), as shown by gel mobility shift assay for HS7 GHRE1 (Fig. 1C). GH also acutely stimulates IGF-I gene transcription (Fig. 1D), and the time course of IGF-I gene activation parallels the in vivo binding of Stat5b to HS7 (Fig. 1B). Thus, GH rapidly and potently activates Stat5b and promotes IGF-I gene transcription in pituitary-deficient male rats.
Defining Stat5b-binding Elements in the Igf1 Locus
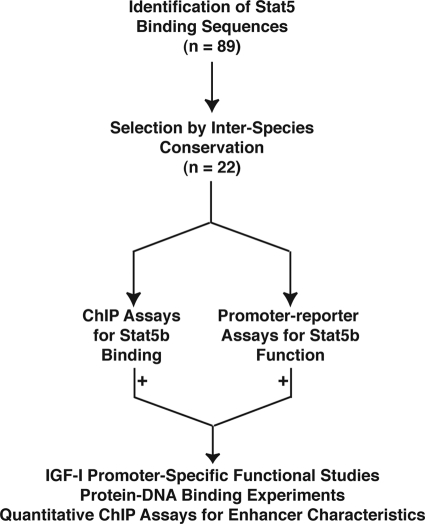
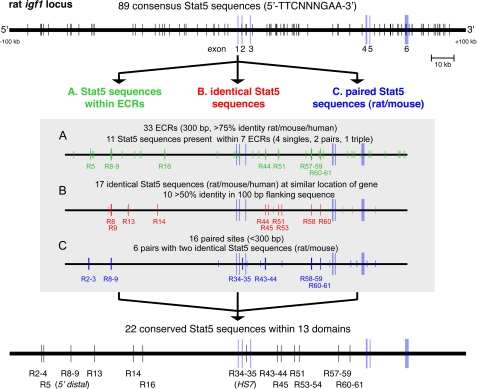
Since our discovery of HS7 as a GH-inducible Stat5b-binding element in IGF-I intron 2, and our demonstration that it could activate GH-mediated IGF-I gene transcription in transgenic mice and in tissue culture (28, 37), several other putative Stat5b-binding sites have been mapped to the Igf1 locus (29, 31, 32), although only one other region, termed 5′ distal, has been found to regulate IGF-I promoter activity in reconstitution experiments in cultured cells (29, 30). We thus devised a multistep approach combining genomics, in vivo protein- DNA binding assays in hepatic chromatin, and functional studies to identify and characterize putative Stat5b-binding sequences, including assessing their ability to regulate IGF-I gene expression. The overall strategy is outlined in Fig. 2. Using bio-informatics, we first identified and mapped a total of 89 Stat5 consensus sites, consisting of the DNA sequence, 5′-TTCNNNGAA-3′ (where N is G, A, T, or C) within an ∼200-kb genomic region encompassing the 75-kb 6-exon IGF-I gene and flanking DNA (Fig. 3, top panel). We evaluated the 89 sites using three independent criteria. First, based on studies showing that tissue-specific and developmentally acting enhancers were often found in conserved DNA (33, 34, 38, 39), we analyzed all conserved regions within the locus for the presence of Stat5 sequences (criteria for evolutionary conservation included nucleotide sequence identity >75% over 300 bp in human IGF1 and rat and mouse Igf1 loci) (40). Of 33 DNA segments meeting these criteria (Fig. 3A, green boxes), 7 contained a total of 11 conserved Stat5 sites (Fig. 3A, green vertical lines). Second, we assessed the 17 putative Stat5 sites in which all nine nucleotides were identical in human IGF1 and rat and mouse Igf1 loci, for flanking nucleotide sequence conservation, and we identified 10 in which adjacent DNA was >50% identical over at least 100 bp (Fig. 3B, red vertical lines). Finally, we looked for paired Stat5 sites, because these are common in Stat5-dependent gene promoters (41), and found 16. In six of these paired regions, the nine nucleotides of each site were identical in rat and mouse genomic DNA (Fig. 3C, blue vertical lines). When the three analytical approaches were combined, the results yielded 22 Stat5 elements grouped into 13 distinct domains in the rat Igf1 locus for further evaluation, including six single sequences (R5, R13, R14, R16, R45, and R51), five pairs (R8–9 (5′ distal (30)), R34–35 (HS7 (28)), R43–44, R53–54 (although R54 was not conserved), and R60–61), and two triple sites (R2-3-4 and R57-58-59 (although neither R4 nor R57 was conserved)) (Fig. 3, bottom panel).
FIGURE 2.
Plan for characterizing functional Stat5b elements in the rat Igf1 locus. Experimental scheme outlining steps to identify and characterize Stat5b-binding sites in chromatin within the rat Igf1 locus.
FIGURE 3.
Multiple putative Stat5b-binding sequences are dispersed throughout mammalian Igf1 loci. Top, map of the 200-kb genomic DNA segment on rat chromosome 7q13 containing the ∼75-kb IGF-I gene. The six numbered IGF-I exons are depicted by blue boxes. A scale bar is indicated. Vertical lines represent 89 putative Stat5-binding sites (top DNA strand, 5′-TTCNNNGAA-3′, where N is G, A, T, or C) that were identified by searching for consensus sequences. Outlined below the map and labeled A–C are the three approaches used to identify individual sites and Stat5 binding domains for further experimental study. The gray box outlines the details and results of each of the three analytical approaches. Bottom, 22 segments within 13 domains that were selected for further study are depicted on the rat Igf1 locus map and are numbered as indicated.
We used quantitative ChIP assays to measure Stat5b binding to each of the 13 domains in hepatic chromatin harvested from pituitary-deficient rats at 60 min after a single systemic GH injection, because at this time point strong binding was observed at HS7 (R34-R35) by qualitative ChIP (Fig. 1B). The results demonstrated that seven domains containing 15 of the 22 sites could bind Stat5b at levels >2.5 times higher than seven control DNA segments (lacking consensus Stat5 sites) that were dispersed throughout Igf1 chromatin (Fig. 4A). Additional quantitative ChIP experiments showed that GH treatment induced binding of Stat5b to each of the seven domains (Fig. 4B). The positive regions included one single site, R13, located at ∼63 kb 5′ to the start of IGF-I gene transcription, four paired elements, R8–9 (at ∼73 kb 5′ to IGF-I exon 1), R34–35 (in intron 2, at +3.7 kb), R53–54 (intron 3, +27 kb), and R60–61 (intron 3, +49 kb), and two triple segments, R2-3-4 (∼86 kb 5′ to exon 1) and R57-58-59 (intron 3, +44 kb). More extensive time course studies by qualitative ChIP revealed that GH treatment caused sustained recruitment of Stat5b to at least R34–35, R53–54, R57-58-59, and R60–61 (but not R14 or R43–44), with binding peaking between 1 and 4 h and lasting up to 6 h after systemic hormone administration (supplemental Fig. 1, other sites were not tested).
Functional Properties of Conserved Stat5b-binding Sequences
As outlined in our overall experimental strategy (Fig. 2), we assessed the functional properties of the conserved Stat5 sites in parallel with ChIP assays by examining their ability to stimulate the activity of a neutral promoter in response to GH-induced Stat5b in reconstitution experiments in cultured COS-7 cells. As depicted in Fig. 5, 6 of the 13 Stat5b domains tested significantly enhanced thymidine kinase promoter function in a GH- and Stat5b-dependent way (R2–3, R8–9, R34–35, R57–59, R60–61, and R13). Remarkably, the only positives in this gene regulation assay were those DNA segments to which GH recruited Stat5b in hepatic chromatin, although the domain containing R53 did not reach statistical significance, possibly because nonconserved R54 was not included in the reporter gene construct. Of particular note, the conserved paired site, R43–44, and the single sites, R5, R14, R16, R45, and R51, were negative, as they were for Stat5b quantitative ChIP assays.
As a further test that GH- and Stat5b-mediated transcription in this functional assay was dependent on the putative Stat5-binding sites, experiments were performed with promoter-reporter genes containing nucleotide substitutions that would abrogate binding of Stat5b (28). In the three domains tested, R2–3, R57–59, and R60–61, mutations all led to a significant reduction in GH-stimulated activity (supplemental Fig. 2), as was shown previously for R8–9 and R34–35 (28, 30). Thus, based on these results, there appears to be a strong correlation between GH-stimulated in vivo binding of Stat5b to sites within the Igf1 locus and the ability of each DNA segment to transmit GH- and Stat5b-regulated gene activity in functional promoter-based reconstitution assays.
In mammals the single copy IGF-I gene contains tandem promoters with distinct properties (42–44). Promoter 1 (P1) is active in all tissues in which IGF-I is expressed, whereas P2 is more restricted in its distribution, functioning primarily in liver, kidney, heart, uterus, and testes (42, 44, 45). We next looked at the effects of different Stat5b domains on each IGF-I promoter in reconstitution studies in COS-7 cells. Both promoters were responsive to addition of selected Stat5b domains (Fig. 6). For P1, all five double and triple domains could convey enhanced GH-stimulated and Stat5b-mediated reporter gene expression, as could single site R53 but not R13. For P2, R2–3, R34–35, R57–59, R13, and R53 each increased GH responsiveness, but R8–9 and R60–61 were ineffective. In addition, R57–59 was able to boost P2-driven reporter gene activity even in the absence of GH by up to 10-fold compared with P2 alone (Fig. 6). Taken together, the results in Figs. 4–6 demonstrate that all of the DNA domains shown by quantitative ChIP to bind Stat5b in a GH-dependent way in vivo were capable of mediating GH-stimulated transcriptional activity for at least two of the three promoters tested.
Assessing DNA-Stat5b Interactions
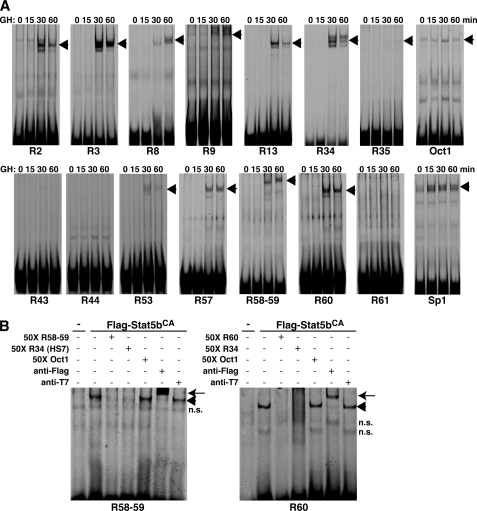
We next examined the Stat5b binding properties of individual sites in vitro by gel mobility shift assays using IR-labeled double-stranded oligonucleotide probes. Time course experiments with hepatic nuclear protein extracts from pituitary-deficient rats given a single injection of GH demonstrated inducible binding to all probes tested within 30 min after systemic hormone administration, except for R43 and R44, which also were negative for Stat5b binding by ChIP, and R61, which with adjacent R60 was positive (Fig. 4 and supplemental Fig. 1). As quality controls, probes containing Oct1 and Sp1 sites showed binding under all conditions (Fig. 7A). Relative binding strengths varied among the 12 positive Stat5b probes tested, with R9 and R35 appearing to be substantially weaker than the others, as noted previously (30). Quantitative binding experiments confirmed this impression (data not shown). Positive binding also was seen with nuclear protein extracts from COS-7 cells expressing constitutively active Stat5b (FLAG-Stat5bCA), but not with cells transfected with an empty vector (Fig. 7B), and DNA competition and antibody super-shift experiments confirmed specificity of the Stat5b-DNA interactions for R58–59 and R60 (Fig. 7B), as well as for the other probes (data not shown).
FIGURE 7.
Stat5b binds to selected consensus sequences found in the Igf1 locus with different affinities. Results of gel mobility shift assays using IR-labeled double-stranded oligonucleotide probes. A, time course studies using rat hepatic nuclear protein extracts harvested prior to and up to 60 min after systemic GH treatment. The probes are labeled below each individual assay, and their DNA sequences may be found in supplemental Table 3. Arrowheads indicate protein-DNA complexes. No binding was observed for R43, R44, or R61. B, competition and antibody super-shift experiments using nuclear protein extracts from COS-7 cells transfected with an expression plasmid for constitutively active NH2-terminal FLAG-tagged rat Stat5b (FLAG-Stat5bCA) and double-stranded IR-labeled probes for R58–59 and R60. Arrowheads indicate protein-DNA complexes, and arrows indicate antibody super-shifted complexes (n.s. = nonspecific band).
Defining Chromatin Modifications at Different Stat5b-binding Sites in the Igf1 Locus
Very little is known about the chromatin landscape of Stat5 binding domains in any vertebrate genome, although properties of Stat1 elements have been discerned in human HeLa cells (8, 9, 34) and Stat4 in mouse T lymphocytes (11). We thus performed a series of quantitative ChIP experiments to determine whether GH-induced binding of Stat5b led to selective histone alterations, recruitment of chromatin-modifying proteins, or binding of other transcriptional regulatory factors. For these studies, we broadened the scope of the region analyzed around each Stat5b domain tested by quantitative ChIP to ∼1 kb by using three adjacent primer pairs. Initial analyses focused on two proteins, the transcriptional co-activator p300 and Med1 or TRAP220, one component of the broadly functional transcriptional protein complex termed Mediator (48). Previous studies have shown that p300 interacts physically with Stat5a and Stat5b (46). Moreover, it has been implicated in the transcriptional control of several Stat5 regulated genes (46, 47) and has been found to be a common component of enhancers (33, 34, 38, 39). Med1 also has been recognized to be part of the chromatin signature of enhancers in some publications (33, 34). We also tested for the presence of RNA polymerase II (RNA pol II), as it has been found to associate with several transcription factors at sites distant from promoters (33, 38), and interacts with Mediator (48). We discovered that all three factors co-localized with each other at four Stat5b binding domains, R2–4, R13, R53–54, and R57–59 (Fig. 8). Binding of p300, Med1, and RNA pol II at these four locations was superimposed with Stat5b binding, was substantially greater than at any other chromosomal segment tested (up to 25 times higher than adjacent DNA or other Stat5 sites), except for proximal IGF-I P1, a positive control (49), and strikingly preceded GH-induced recruitment of Stat5b (compare Fig. 8, A–C with Fig. 4B). Moreover, GH treatment did not cause any major change in their concentration, but did broaden their localization slightly at some Stat5b domains (e.g. R57–59 for p300 and RNA pol II, R13 for Med1). By contrast, recruitment of these factors to R8–9, R34–35, and R60–61 was at best only slightly higher than the negative control, a DNA fragment located ∼10 kb 5′ to the start of IGF-I exon 1 (the exception being DNA flanking R34–35 for RNA pol II, as described previously (49)). Thus, there appears to be selective recruitment of transcription-associated proteins to a subset of GH-activated Stat5b binding domains in hepatic chromatin in the rat Igf1 locus, and unlike what is seen for Stat5b, this recruitment is not hormone-regulated.
We next examined chromatin around each Stat5b binding region for mono- and tri-methylation of lysine 4 in histone H3 (H3K4me1 and H3K4me3), as both modifications have been associated in some studies with transcriptional enhancers, including those that bind Stat1 (34, 50). H3K4me1 also is enriched in the body and 3′ end of genes, and H3K4me3 has been mapped to active promoters (51). Results were dramatically different, with H3K4me1 being enriched over six of seven Stat5b binding domains (the exception being R2–4) and H3K4me3 being found only at R34–35 and R60–61 (Fig. 9). Both histone modifications were more broadly distributed at each positive Stat5b region than was binding of p300, Med1, or RNA pol II (compare with Fig. 8). Moreover, GH treatment led to a selective decrease in abundance of H3K4me1 in Stat5b elements found within the body of the IGF-I gene (R34–35, R53–53, R57–59, and R60–61), but not those in 5′-flanking chromatin (R2–4, R8–9, and R13), and caused an increase in H3K4me3 at R34–35 and a decline in R60–61 (Fig. 9). Additional experiments interrogating other sites throughout the Igf1 locus confirmed a broader range of chromatin that was positive for the H3K4me1 modification than for H3K4me3 (supplemental Fig. 3). Few studies have examined the dynamics of change in H3K4 methylation status at promoters or enhancers during acute gene activation, and the mechanistic significance of these GH-induced alterations is unclear.
Recruitment of transcription factors to response elements is often accompanied by enhanced acetylation of core histone tails to “open” chromatin (51) that additionally provides evidence for co-recruitment and activation of histone acetyltransferases, and/or loss of deacetylases (51). We thus examined each Stat5b binding domain for changes in core histone acetylation by performing quantitative ChIP with antibodies to acetyl lysine histone H3 (recognizing acetyl-Lys-9 and -Lys-14) and H4 (recognizing acetyl-Lys-5, -Lys-8, -Lys-12, and -Lys-16) prior to and at 60 min after a single systemic GH pulse. In the absence of GH, core histone acetylation at each Stat5b domain was relatively low, being 2–12 times less than values measured for a positive control, IGF-I P1 (−1036 bp (49)) (Fig. 10, A and B, light gray bars), although even in hormone-deficient rats levels of H3 and H4 acetylation were generally higher in transcribed parts of the IGF-I gene than in 5′-flanking chromatin (Fig. 10, A and B, light gray bars, compare R34–35, R53–54, R57–59, and R60–61 with R2–4, R8–9, and R13). After GH administration, acetylation of histones H3 and H4 rose dramatically at segments within the IGF-I transcription unit (R34–35, R53–54, R57–59, and R60–61) but was much less at other sites (Fig. 10, A and B, dark gray bars). The correspondence between recruitment of p300 to a Stat5b binding domain and GH-stimulated histone acetylation was inconsistent, with a strong positive correlation at R53–54 and R57–59, but negative correlations at R2–4, and mixed at R13, where GH-induced increases were seen for acetyl histone H4 but not H3. At other sites, such as R34–35 and R60–61, there was little p300 on chromatin but strong enhancement of core histone acetylation by GH.
Taken together, the observations in Figs. 8–10 demonstrate dramatic variation in the chromatin signatures of GH-activated Stat5b binding domains in the Igf1 locus. These results also indicate that hormone treatment leads to rapid and extensive alterations in histone modifications throughout the locus, particularly within the transcribed portion of the IGF-I gene.
DISCUSSION
Genetic studies in mice and humans have established that the transcription factor Stat5b is a central mediator in the GH-IGF-I-growth axis (22, 25), and subsequent biochemical and molecular experiments have demonstrated that GH receptor-activated Stat5b can directly stimulate IGF-I gene transcription (26, 28). Here, we have employed a combination of approaches to identify and characterize seven distinct GH-regulated Stat5b binding domains in hepatic chromatin in the rat Igf1 locus. These include five sites identified previously (28–32), and two, R2–4 and R13, that are new. Four of these elements bear the potential epigenetic signatures of transcriptional enhancers, and all seven can stimulate IGF-I promoter function in a GH- and Stat5b-dependent way in cultured cells. Our results lead to the hypothesis that GH promotes IGF-I gene transcription via Stat5b through a combinatorial interplay of several long range enhancers.
Stat5a and Stat5b are structurally similar products of tandem and recently duplicated genes on human chromosome 17 and are ∼96% identical in amino acid sequence (2). The two Stat5s may serve distinct physiological functions largely because of different tissue-limited patterns of expression (2) but also may potentially interact with unique modifying factors, possibly through their more divergent COOH-terminal transcriptional activation domains (2). The majority of studies examining the roles of Stat5 in GH actions, particularly in vivo, have focused on effects in the liver, where Stat5b predominates over Stat5a (2), and loss of Stat5b alone leads to severe systemic growth defects in both humans and mice (22, 25), although a recent report has implicated Stat5a as a potential mediator of IGF-I gene transcription in the uterus (45). Although it has not been determined if Stat5a and Stat5b always recognize the same DNA sites in chromatin, it is likely that they usually do, because in vitro protein-DNA binding studies have yielded identical optimal sequence profiles (7). However, in one published report using interleukin-3-treated Ba/F3 cells, separate cohorts of genes were found to be associated with nearby transcription factor-binding sites in chromatin depending on whether an antibody to Stat5a or Stat5b was used for ChIP (52). As this experiment has not been replicated in other cells that express both Stat5 proteins, the degree of functional overlap between Stat5a and Stat5b remains an open question.
Recent molecular genetic analyses have demonstrated that a number of transcription factors can bind to a plethora of recognition sequences that are dispersed throughout the genome, being found in introns and in intergenic DNA, as well as near promoters of putative target genes (8, 9, 53, 54). Studies by ChIP-seq methods have shown that up to 50% of Stat1-binding sites in human HeLa cells treated with interferon γ map near the 5′ ends of genes (8), although other results have estimated this value to be ∼25%, with ∼50% of binding sites being intergenic and ∼25% being localized to introns (9). These data have not been analyzed for the occurrence of multiple Stat1 binding domains within or near single loci, similar to what we now find for Stat5b in Igf1, and it is not known how many other genes may resemble IGF-I in potentially being regulated by multiple enhancers that target the same transcription factor. Other results revealed that ∼75% of Stat1 binding domains in human HeLa cells were consensus sites (5′-TTC(T/C)N(G/A)GAA-3′, where N is G, A, T, or C), with half of these being single recognition sequences and the other half paired or triple elements (9), similar to what we now observe for the igf1 locus.
Only limited genome-wide screening results have been reported for Stat5a and Stat5b, but from the small number of binding elements analyzed to date, it appears that the majority are located near promoters or in introns (we find 4/7 for IGF-I) (55) and that most are paired sites (we find 6/7) (41, 55). Moreover, with the possible exception of Socs2 (32), there yet have been no examples similar to IGF-I in which multiple functional Stat5b binding domains have been associated with a single target gene. As analysis of the Igf1 locus on rat chromosome 7q13 revealed no other genes within ∼290 kb in the 5′ direction or within ∼120 kb in the 3′ direction of the 75-kb transcribed IGF-I gene (RGSC 3.4, updated September, 2009), it seems likely that the seven Stat5b domains mapped to the locus primarily interact with IGF-I.
ChIP-based whole genome assays have been employed recently to elucidate the chromatin characteristics of enhancers, although starting assumptions have varied among different investigative groups. One approach has been to use comparative genomics to define highly conserved intergenic regions among multiple species. In some cases these ultraconserved domains were prevalent in tissue-specific and developmentally specific enhancers, but so was DNA that was less conserved (38). Other studies looked for nuclease-hypersensitive sites as indices of open chromatin, as might be found in the landscape of an enhancer (56, 57), and others focused on selective histone modifications, particularly H3K4me1 and/or H3K4me3 (33, 34, 50), or binding of p300 and/or Med1 (33, 34). In the latter case, chromatin sites that were positive for p300, Med1, and H3K4me1 often were found to be embedded within evolutionarily conserved DNA (33), and when tested in promoter-reporter gene assays, the majority of these domains also functionally boosted transcriptional activity, a result consistent with their possessing enhancer-like properties (34). For example, in HeLa cells three interferon γ-inducible Stat1 elements that mapped in chromatin to regions with potential enhancer signatures mediated stimulatory transcriptional responses in promoter-reporter assays, although one that did not possess these characteristics was negative (33).
In contrast to these latter results, in our studies the main predictive determinant of functional enhancer properties appeared to be inducible Stat5b binding to DNA in hepatic chromatin after systemic GH treatment. All seven domains that were positive by Stat5b ChIP were able to stimulate activity of an IGF-I promoter and of a minimal TK promoter in a GH- and Stat5b-dependent way in cultured cells, but none that were negative by ChIP were able to do so, even though in each latter case a potential Stat5 site was present in the DNA (see Figs. 3–5). Moreover, only four of the seven Stat5b-positive domains were also positive for in vivo binding of p300 and Med1 (and RNA pol II) by ChIP, and there was no correlation between strength of transcriptional activity in reconstitution experiments and p300 or Med1 binding in vivo in rat liver chromatin. The discrepancy between our results with Stat5b and those outlined above for Stat1 may represent fundamental distinctions between the properties of the two transcription factors, may indicate differences between studies using terminally differentiated animal tissues as performed here versus experiments in cultured HeLa cells (33), or may reflect other as yet unrecognized features. It also is likely that reconstitution experiments that employ transiently transfected promoter-reporter plasmids do not fully replicate the chromatin environment of endogenous genes in mammalian cells. Clearly better characterization is needed to define the chromosomal properties of transcriptional enhancers, although it should be noted that p300 binding appears to be a strongly positive predictor of enhancers that are active during mouse development (39).
One additional feature of note in our results was the presence of p300, Med1, and RNA pol II on hepatic chromatin in four of seven domains in advance of GH signaling or recruitment of activated Stat5b. This observation contrasts with data from Nelson et al. (41), in which interleukin-2-mediated recruitment of Stat5a and Stat5b to a single enhancer in an intron of the NCAM2 gene in cultured cells also promoted binding of p300 and RNA pol II. It is not clear what mechanisms are responsible for pre-loading some chromatin domains in the Igf1 locus with transcriptional co-factors prior to GH-stimulated recruitment of Stat5b to the same sites. It is possible that this pre-loading reflects the selective binding of as yet unidentified transcription factors to some of the same regions that subsequently bind Stat5b in response to GH. Alternatively, perhaps the modifications that we now find at selective locations in the Igf1 locus, along with chromatin opening at the two IGF-I promoters that we have reported recently (49), may reflect the normal maturation of the liver, including constitutive binding of as yet unidentified transcription factors that are responsible for recruitment of p300, Med1, and RNA pol II to the putative enhancers identified here. Further investigations will be needed to define the processes that govern chromatin changes in the liver during postnatal development, and it will be of great interest to determine whether these alterations might be susceptible to environmental influences, including those mediated by systemic hormones, growth factors, or pharmacological agents.
Finally, the classification of GH-activated Stat5b binding domains in chromatin into two distinct groups based on the presence or absence of associated transcriptional co-factors raises a question about possible differential in vivo functions. If the four domains with bound p300, Med1, and RNA pol II can be shown to act as Stat5b-regulated long range transcriptional enhancers, then what functions might be ascribed to the three elements that lack these co-factors? Not only does GH play a pivotal role in promoting childhood growth by controlling production of IGF-I, but it also exerts similar effects in the adult, where IGF-I is an important mediator of tissue repair and regeneration, particularly in muscle and bone (17, 18). As evidenced by the linkage of GH and IGF-I with the development and propagation of several cancers (20), and with their postulated deleterious impact on aging (19), dysregulated expression of IGF-I may have potentially severe pathogenic consequences, implying that its production must be tightly controlled under normal physiological conditions. If true, then one potential role for the GH-induced Stat5b chromatin domains that lack the signature of enhancers may be to reduce availability of active Stat5b by acting as decoys to limit access to other more functional domains, such as those that promote GH-stimulated IGF-I gene transcription. One implication of this “sequestration hypothesis” would be that these alternative Stat5b-binding sites on DNA would function as allosteric modifiers of GH-activated and Stat5b-dependent target gene transcription through titration of available Stat5b from more productive interactions. Further evidence will be needed to support or refute this hypothesis and to elucidate the mechanisms of action of Stat5b to control IGF-I gene activity.
Supplementary Material
Acknowledgments
We thank Jennifer J. Young and April R. Mertens for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants K08 DK077897 and K12 HD057588 (to D. J. C.), T32 DK007674-16 (to B. V.-M.), and R01 DK069703 (to P. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1–3.
- GH
- growth hormone
- IGF-I
- insulin-like growth factor-I
- Stat
- signal transducers and activators of transcription
- ChIP
- chromatin immunoprecipitation
- TK
- thymidine kinase.
REFERENCES
- 1.Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 2.Hennighausen L., Robinson G. W. (2008) Genes Dev. 22, 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schindler C., Plumlee C. (2008) Semin. Cell Dev. Biol. 19, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H., Pardoll D., Jove R. (2009) Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindler C., Shuai K., Prezioso V. R., Darnell J. E., Jr. (1992) Science 257, 809–813 [DOI] [PubMed] [Google Scholar]
- 6.Shuai K., Schindler C., Prezioso V. R., Darnell J. E., Jr. (1992) Science 258, 1808–1812 [DOI] [PubMed] [Google Scholar]
- 7.Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. (2001) J. Biol. Chem. 276, 6675–6688 [DOI] [PubMed] [Google Scholar]
- 8.Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. (2007) Nat. Methods 4, 651–657 [DOI] [PubMed] [Google Scholar]
- 9.Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. (2008) Nucleic Acids Res. 36, 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman S. E., Bertone P., Nath A. K., Royce T. E., Gerstein M., Weissman S., Snyder M. (2005) Genes Dev. 19, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Good S. R., Thieu V. T., Mathur A. N., Yu Q., Stritesky G. L., Yeh N., O'Malley J. T., Perumal N. B., Kaplan M. H. (2009) J. Immunol. 183, 3839–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Roith D., Bondy C., Yakar S., Liu J. L., Butler A. (2001) Endocr. Rev. 22, 53–74 [DOI] [PubMed] [Google Scholar]
- 13.Lanning N. J., Carter-Su C. (2006) Rev. Endocr. Metab. Disord. 7, 225–235 [DOI] [PubMed] [Google Scholar]
- 14.Rosenfeld R. G., Hwa V. (2009) Horm. Res. 71, Suppl. 2, 36–40 [DOI] [PubMed] [Google Scholar]
- 15.LeRoith D. (2008) Pediatr. Endocrinol. Rev. 5, Suppl. 2, 739–743 [PubMed] [Google Scholar]
- 16.Rotwein P., Chia D. J. (2010) Pediatr. Nephrol. 25, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giustina A., Mazziotti G., Canalis E. (2008) Endocr. Rev. 29, 535–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velloso C. P. (2008) Br. J. Pharmacol. 154, 557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatar M., Bartke A., Antebi A. (2003) Science 299, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 20.Pollak M. (2008) Nat. Rev. Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 21.Waters M. J., Hoang H. N., Fairlie D. P., Pelekanos R. A., Brown R. J. (2006) J. Mol. Endocrinol. 36, 1–7 [DOI] [PubMed] [Google Scholar]
- 22.Kofoed E. M., Hwa V., Little B., Woods K. A., Buckway C. K., Tsubaki J., Pratt K. L., Bezrodnik L., Jasper H., Tepper A., Heinrich J. J., Rosenfeld R. G. (2003) N. Engl. J. Med. 349, 1139–1147 [DOI] [PubMed] [Google Scholar]
- 23.Hwa V., Little B., Adiyaman P., Kofoed E. M., Pratt K. L., Ocal G., Berberoglu M., Rosenfeld R. G. (2005) J. Clin. Endocrinol. Metab. 90, 4260–4266 [DOI] [PubMed] [Google Scholar]
- 24.Teglund S., McKay C., Schuetz E., van Deursen J. M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J. N. (1998) Cell 93, 841–850 [DOI] [PubMed] [Google Scholar]
- 25.Udy G. B., Towers R. P., Snell R. G., Wilkins R. J., Park S. H., Ram P. A., Waxman D. J., Davey H. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woelfle J., Billiard J., Rotwein P. (2003) J. Biol. Chem. 278, 22696–22702 [DOI] [PubMed] [Google Scholar]
- 27.Woelfle J., Chia D. J., Massart-Schlesinger M. B., Moyano P., Rotwein P. (2005) Pediatr. Nephrol. 20, 295–302 [DOI] [PubMed] [Google Scholar]
- 28.Woelfle J., Chia D. J., Rotwein P. (2003) J. Biol. Chem. 278, 51261–51266 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Jiang H. (2005) J. Biol. Chem. 280, 10955–10963 [DOI] [PubMed] [Google Scholar]
- 30.Chia D. J., Ono M., Woelfle J., Schlesinger-Massart M., Jiang H., Rotwein P. (2006) J. Biol. Chem. 281, 3190–3197 [DOI] [PubMed] [Google Scholar]
- 31.Eleswarapu S., Gu Z., Jiang H. (2008) Endocrinology 149, 2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laz E. V., Sugathan A., Waxman D. J. (2009) Mol. Endocrinol. 23, 1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 34.Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood T. J., Sliva D., Lobie P. E., Pircher T. J., Gouilleux F., Wakao H., Gustafsson J. A., Groner B., Norstedt G., Haldosén L. A. (1995) J. Biol. Chem. 270, 9448–9453 [DOI] [PubMed] [Google Scholar]
- 36.Gronowski A. M., Le Stunff C., Rotwein P. (1996) Endocrinology 137, 55–64 [DOI] [PubMed] [Google Scholar]
- 37.Ye P., Umayahara Y., Ritter D., Bunting T., Auman H., Rotwein P., D'Ercole A. J. (1997) Endocrinology 138, 5466–5475 [DOI] [PubMed] [Google Scholar]
- 38.Visel A., Rubin E. M., Pennacchio L. A. (2009) Nature 461, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., Pennacchio L. A. (2009) Nature 457, 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ovcharenko I., Nobrega M. A., Loots G. G., Stubbs L. (2004) Nucleic Acids Res. 32, W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson E. A., Walker S. R., Li W., Liu X. S., Frank D. A. (2006) J. Biol. Chem. 281, 26216–26224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamo M. L., Ben-Hur H., Roberts C. T., Jr., LeRoith D. (1991) Mol. Endocrinol. 5, 1677–1686 [DOI] [PubMed] [Google Scholar]
- 43.Hoyt E. C., Van Wyk J. J., Lund P. K. (1988) Mol. Endocrinol. 2, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 44.Hall L. J., Kajimoto Y., Bichell D., Kim S. W., James P. L., Counts D., Nixon L. J., Tobin G., Rotwein P. (1992) DNA Cell Biol. 11, 301–313 [DOI] [PubMed] [Google Scholar]
- 45.Hewitt S. C., Li Y., Li L., Korach K. S. (2010) J. Biol. Chem. 285, 2676–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfitzner E., Jähne R., Wissler M., Stoecklin E., Groner B. (1998) Mol. Endocrinol. 12, 1582–1593 [DOI] [PubMed] [Google Scholar]
- 47.Ye S. K., Agata Y., Lee H. C., Kurooka H., Kitamura T., Shimizu A., Honjo T., Ikuta K. (2001) Immunity 15, 813–823 [DOI] [PubMed] [Google Scholar]
- 48.Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W. (2005) Trends Biochem. Sci. 30, 250–255 [DOI] [PubMed] [Google Scholar]
- 49.Chia D. J., Young J. J., Mertens A. R., Rotwein P. (2010) Mol. Endocrinol. 24, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson A. G., Bilenky M., Tam A., Zhao Y., Zeng T., Thiessen N., Cezard T., Fejes A. P., Wederell E. D., Cullum R., Euskirchen G., Krzywinski M., Birol I., Snyder M., Hoodless P. A., Hirst M., Marra M. A., Jones S. J. (2008) Genome Res. 18, 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rando O. J., Chang H. Y. (2009) Annu. Rev. Biochem. 78, 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson E. A., Walker S. R., Alvarez J. V., Frank D. A. (2004) J. Biol. Chem. 279, 54724–54730 [DOI] [PubMed] [Google Scholar]
- 53.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 54.Kaneshiro K., Tsutsumi S., Tsuji S., Shirahige K., Aburatani H. (2007) Genomics 89, 178–188 [DOI] [PubMed] [Google Scholar]
- 55.Basham B., Sathe M., Grein J., McClanahan T., D'Andrea A., Lees E., Rascle A. (2008) Nucleic Acids Res. 36, 3802–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 57.Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T. Y., Peng W., Zhang M. Q., Zhao K. (2008) Nat. Genet. 40, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.