Abstract
Microbacterium nematophilum causes a deleterious infection of the C. elegans hindgut initiated by adhesion to rectal and anal cuticle. C. elegans bus-2 mutants, which are resistant to M. nematophilum and also to the formation of surface biofilms by Yersinia sp., carry genetic lesions in a putative glycosyltransferase containing conserved domains of core-1 β1,3-galactosyltransferases. bus-2 is predicted to act in the synthesis of core-1 type O-glycans. This observation implies that the infection requires the presence of host core-1 O-glycoconjugates and is therefore carbohydrate-dependent. Chemical analysis reported here reveals that bus-2 is indeed deficient in core-1 O-glycans. These mutants also exhibit a new subclass of O-glycans whose structures were determined by high performance tandem mass spectrometry; these are highly fucosylated and have a novel core that contains internally linked GlcA. Lectin studies showed that core-1 glycans and this novel class of O-glycans are both expressed in the tissue that is infected in the wild type worms. In worms having the bus-2 genetic background, core-1 glycans are decreased, whereas the novel fucosyl O-glycans are increased in abundance in this region. Expression analysis using a red fluorescent protein marker showed that bus-2 is expressed in the posterior gut, cuticle seam cells, and spermatheca, the first two of which are likely to be involved in secreting the carbohydrate-rich surface coat of the cuticle. Therefore, in the bus-2 background of reduced core-1 O-glycans, the novel fucosyl glycans likely replace or mask remaining core-1 ligands, leading to the resistance phenotype. There are more than 35 Microbacterium species, some of which are pathogenic in man. This study is the first to analyze the biochemistry of adhesion to a host tissue by a Microbacterium species.
Keywords: Carbohydrate/Biosynthesis, Carbohydrate/Chemistry, Carbohydrate/Glycoconjugate, Glycoproteins/Carbohydrates, Glycoproteins/Secretion, Glycosylation, Receptors/Oligosaccharides, Carbohydrate Recognition, Mass Spectrometry, Surrogate Model
Introduction
Caenorhabditis elegans is a genetically and developmentally well characterized organism that has been used as a model to study host-pathogen interactions. The major habitat for C. elegans is soil, where it feeds on bacteria and may also come into contact with commensal or pathogenic bacteria. More than 40 pathogens are known to cause disease in C. elegans, and many of these (or their close relatives) are human pathogens. These pathogenic interactions have recently been reviewed (1).
Host-pathogen interactions require bidirectional recognition factors, and often at least one of these components is a glycoconjugate (2). Examples are recognition factors for bacterial toxins such as aerolysin from Aeromonas hydrophila, cholera from Vibrio cholerae, hemolysin from Escherichia coli, and the crystal proteins from Bacillus thuringiensis (3). The last bacterium can kill C. elegans by means of a crystal toxin that causes destruction of the intestine. The effect is directly analogous to the effect of this and other Bt toxins on Lepidoptera, Diptera, and Coleoptera species, many of which are agricultural pests (4). C. elegans mutants that are resistant to crystal toxin are defective in the bre gene family (5, 6), one of which is a homolog of Drosophila melanogaster egghead (egh) and encodes a GDP-Man:βGlc-Cer-β1,4-mannosyltransferase (bre-3), another encodes a UDP-GalNAc:β1,4N-acetylgalactosaminyltransferase (bre-4), and a third encodes is a homolog of Drosophila brainiac (brn) and encodes a UDP-GlcNAc:βMan N-acetylglucosaminyl transferase (bre-5). Identification of the BRE activities in C. elegans provides striking evidence of the utility of this nematode as a model organism to study the role of glycoconjugates in pathogenesis.
C. elegans possesses many attributes that make it an attractive model for the study of carbohydrate-dependent host-pathogen interactions. These include 1) a sequenced genome, 2) the complete mapping of all somatic cells through development, 3) a transparent body, which confers the possibility for visualization of microbial pathogens in situ and the capacity for direct monitoring of the distribution of glycoconjugates in tissues using lectins and antibodies, 4) the existence of many glycoconjugate-deficient strains, 5) sensitivity to more than 40 bacterial pathogens, many of which also infect humans or are their close relatives, and 6) an increasingly well characterized innate immune system. These properties, when combined with the increasing capability of modern mass spectrometry to determine glycan patterns and fine structural detail and the availability of highly sophisticated genetic and molecular techniques, now make it likely that the infection can be placed in the context of its glycoconjugate requirements at the nematode's “bedside.” From that point, the distribution and identity of the required carbohydrate entities can be revealed in a tissue-specific fashion and in fine structural detail. Genetic requirements needed to facilitate the infection and the innate immune components required to combat it can also be determined.
There are more than 35 members of the genus Microbacterium, some of which are human pathogens (7). The C. elegans pathogen M. nematophilum adheres to the rectal and anal region of the nematode, causing a distinct swelling in the region and surrounding tissues (8). The infection is usually non-lethal in wild type C. elegans but causes extensive larval mortality in related Caenorhabditis species or in immunocompromised C. elegans mutants (9, 10). Genetic screens for altered susceptibility to M. nematophilum infection yielded 19 complementation groups characterized by a loss of swelling in the tail region that led to a bacterially unswollen phenotype (bus) (11, 12). Among these were mutants of srf-2, srf-3, and srf-5, which have previously been isolated on the basis of their altered cuticle properties, as characterized by ectopic lectin binding (13). Although srf-2 and srf-5 have not been cloned, srf-3 encodes a UDP-Gal/UDP-GlcNAc transporter (14). The bus-2, bus-4, bus-8, bus-12, and bus-17 mutants each harbor a genetic lesion in a different component required for glycoconjugate biosynthesis (9, 10). The bus-2, bus-4, and bus-17 genes encode homologs of galactosyltransferases predicted to act in O-glycan biosynthesis. The bus-12 gene encodes a nucleotide sugar transporter homolog, and bus-8 encodes a mannosyltransferase homolog possibly active in N-glycan biosynthesis. In our previous study srf-3 strains were found to be deficient in both N- and -O-glycans (15). The most dramatic differences were seen in O-glycans, as they were reduced 65% in overall abundance compared with the parental strain. Five genes identified in the bus screen are likely to be involved in O-glycoconjugate synthesis, including three encoding core-1 galactosyltransferases homologs and two nucleotide encoding sugar transporters. Therefore, it is hypothesized that core-1 O-glycans are required for M. nematophilum infection. Here we further investigate this possibility and the molecular basis for the carbohydrate dependence of the C. elegans–M. nematophilum pathogenic relationship by the analysis of the bus-2 mutant in comparison to its parental strain N2 Bristol.
EXPERIMENTAL PROCEDURES
Culture Maintenance
The N2 Bristol wild type strain used in this work was provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. The bus-2 mutant strains used in this study, e2677, was generated by Tc1 transposon insertion into exon 5 of gene K08D12.5, which has been identified as bus-2.2 C. elegans strains were maintained at 16 °C on nematode growth medium agar seeded with E. coli OP50, as described previously (16).
Preparation of Mixed Bacterial Lawns
E. coli OP50 and M. nematophilum were grown in LB nutrient media at 37 °C for 24 and 48 h, respectively. Nematode growth medium agar plates were aseptically seeded with the E. coli strain OP50 containing 0.1% M. nematophilum.
Bacterial Adhesion Staining
C. elegans were stained with SYTO Green 13 (Molecular Probes, Carlsbad, CA), as previously reported (17).
Large Scale C. elegans Culture
Nematodes were cultured in large culture dishes in S-basal media (18). Approximately 10 ml of OP-50 E. coli were added as a nematode food source. The bacterial slurry contained 100 μl of 25 mg/ml cholesterol in ethanol, 50 μl of 35 mg/ml of streptomycin, and 37 ml of M9 media (18). Plates were gently rocked at 16 °C. Cultures were checked daily, and bacteria were added as needed at an average consumption rate of 2.5 ml of bacterial slurry per plate per day for 3–4 days.
Glycoprotein Preparation
Approximately 10–12 g of nematodes were used for glycan preparations. Flash frozen cultures were homogenized in a bead beater, lyophilized, and delipidated with CHCl3/MeOH/H2O (10:10:1). The glycan-containing pellet was dried and suspended in 50 mm citrate buffer. Glacial acetic acid was added to adjust the pH to ∼5.0. Free saccharide was removed from the mixture by extraction with 15–20 volumes of 50% MeOH/H2O at −20 °C for 4 h followed by centrifugation at 5000 × g for 15–20 min. The supernatant was removed, and this procedure was repeated 3–5 times until no neutral hexose was detectable by phenol sulfuric assay. The precipitate was suspended in 10 volumes 125 mm Tris-Cl (pH 6.8), 1% SDS, 5% β-mercaptoethanol and boiled for 10 min before the supernatant was collected. This procedure was repeated three times. The supernatants were combined and dialyzed against a solution of 10 mm sodium phosphate at pH 7 using a 10,000 molecular weight cutoff membrane. Dialyzed samples were lyophilized. Trypsin was added in a 1:50 trypsin-to-protein weight ratio, and the mixture was incubated for 4–8 h at 37 °C.
Glycan Release and Derivatization
C. elegans extracts were subjected to proteolytic digestion with trypsin, and then N-glycans were released by incubation of the peptide mixture with 4 milliunits/ml peptide N-glycosidase F (Prozyme, San Leandro, CA) overnight at pH 8.5 at 37 °C. The pH was adjusted to 5.5, and samples were precipitated in 50% MeOH, H2O at −20 °C. The supernatant containing the N-glycans was removed and evaporated to dryness, and the pellet was saved for subsequent O-glycan release. β-Elimination for the release of O-glycans was performed in 0.1 m NaOH, 1 m NaBH4 for 16 h at 45 °C. Purification of O-glycans was performed as previously described (15). Briefly, reactions were neutralized dropwise with acetic acid and applied to a column of Dowex 50W (hydrogen form), and the glycans were eluted with deionized water. The eluate was collected, and the borate was removed by repeated evaporation from acidified methanol. The resulting solution was concentrated by evaporation and applied to an AG-1X2 anion exchange column. Neutral glycans were eluted first with deionized water, and then the charged ones were eluted with a solution of 100 mm ammonium acetate. Released glycans were permethylated as previously described by Ciucanu et al. (19, 20).
Mass Spectrometric Analysis
Molecular weight profiles and relative abundances were calculated from MALDI-TOF mass spectra acquired on a Bruker Reflex IV fitted with a nitrogen laser (337 nm, 3-ns pulse width) and operated in the positive-ion reflectron mode. Samples were spotted on a ground steel target into 1-μl droplets of 2,5-dihydroxybenzoic acid solution (20 mg/ml in 20% acetonitrile). 200–300 shots were summed for each analysis, and each was performed in triplicate. The molecular ion intensities were averaged, and S.D. were calculated. Electrospray ionization collision-induced dissociation tandem mass spectrometry (ESI-CID-MS/MS)3 was utilized for structural determination of novel glycans. Sample solvent was a 50:50 MeOH/H2O solution containing 0.5 mm sodium acetate. The ESI-MS analyses were performed in the positive-ion mode on a Discovery model LTQ-Orbitrap mass spectrometer (Thermo-Fisher, San Jose, CA) equipped with a TriVersa NanoMate source (Advion Corp., Ithaca, NY). Measured mass accuracies were within 5 ppm of the calculated values. Fragment ion nomenclature is that of Domon and Costello (21).
Monosaccharide Analysis
Monosaccharide compositions of released glycans were determined by gas chromatography-MS analysis using a Finnigan TRACE GC 2000 series coupled to the GCQ plus electron ionization ion trap mass spectrometer. Monosaccharides generated by acid hydrolysis of glycans were trimethylsilylated before analysis (22).
Staining
Lectin staining was based on the method of Borgonie (23), except that C. elegans were stained intact and not frozen. Fluorescein isothiocyanate-conjugated lectins Ulex europaeus I, Agaricus bisporus, Helix pomatia, and Maackia amurensis (Sigma) were used. Nematodes were placed in 10 μg/ml lectin solutions in a phosphate-buffered salline-based buffer and incubated overnight at 4 °C before mounting.
Red Fluorescent Protein Rescue of bus-2
For the analysis of bus-2 tissue localization, a bus-2 RFP (mCherry) rescuing operon construct was made utilizing the previously described PCR fusion approach (24). The primers were designed so that the sequence between bus-2 and RFP was small enough that the RFP reporter was expressed as the second gene in an operon, under the control of the bus-2 promoter. Consequently, RFP was expressed in the same cells as BUS-2 but as a separate protein. Briefly, the initial PCR amplification used a forward primer corresponding to the bus-2 gene and a reverse primer corresponding to the poly(A) tail of bus-2 in addition to the polycistronic region of the pENTRY::mCherry plasmid, thus creating a 6.7-kb product. This was followed by the amplification of the 2.1-Kb mCherry region of the pENTRY plasmid and the amplification of both products to create a fusion PCR product of 8.8 Kb. Plasmids used were as follows: Bus-2 SpeI forward (GGA CTA GTC CCA CTT TGG TAG CTG ATA TCT CAG), Bus-2 pENTRY RFP reverse (GGT GAA AGT AGG ATG AGA CAG CGG CAA AAA AAT CCA TCA AGA TCG CCA CC), polycistRFP forward (CCG CTG TCT CAT CCT ACT TTC ACC; polycistronic construct as Cheung et al. (25)), polycistRFP reverse D (TCG ACG TTT CCC GTT GAA TAT G), Bus-2 A* forward (CAT CAA CTG ATA AGT TGT TGA TAT TGT TG), and polycistRFP reverse D* (GGC TCA TAA CAC CCC TTG TAT TAC TG. This construct was injected as the transgene eEx677 together with unc-119(+) into bus-2 and unc-119 mutants, rescuing both phenotypes.
RESULTS
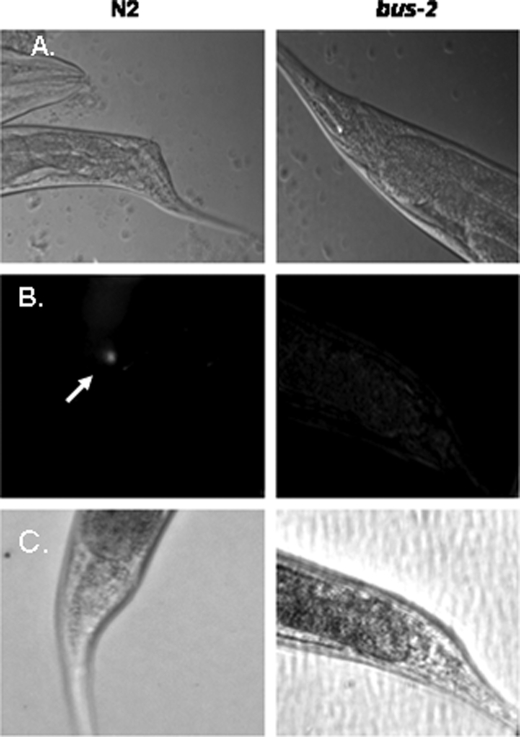
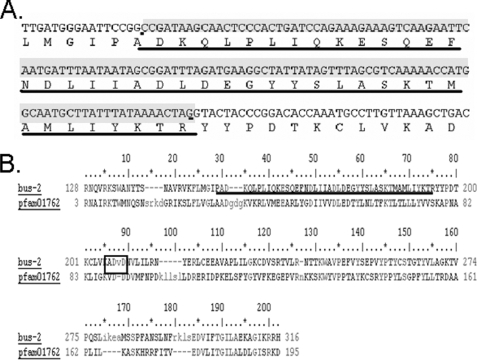
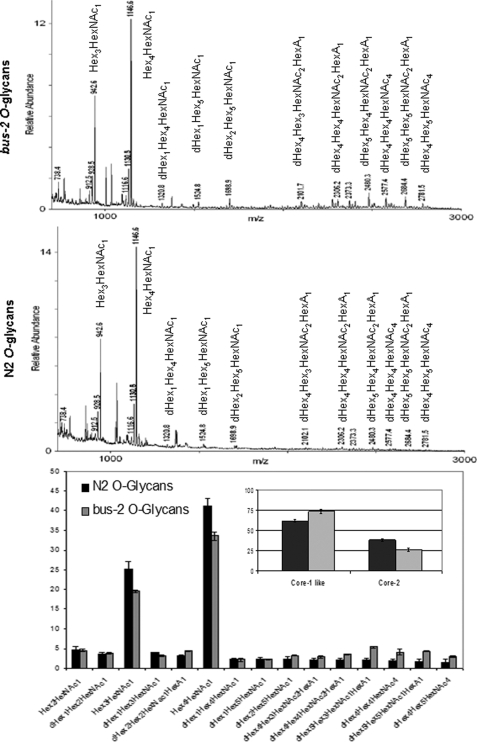
To summarize results the nucleic acid stain SYTO Green 13 was used to visually confirm that the bus-2 mutant had no detectable bacterial colonization in the rectal region compared with its parent strain N2 Bristol, as shown previously (11) and in Fig. 1, therefore, verifying the bus phenotype. The bus-2 coding region was cloned and sequenced, thereby revealing an error in the earlier sequence prediction. The NCBI Conserved Domain Data base (26) revealed that this correction led to an improvement in alignment from 63.8 to 98.0% of domain models used within the core-1 galactosyltransferase pfam01762 (Fig. 2). The improved alignment provided a score of 80.7 bits and an E value of 2e-16. Permethylation profiling by MALDI-TOF MS was used to provide robust and reproducible semiquantitative glycan data to compare abundances of these compounds in the bus-2 and parental strains (Fig. 3). Although the profiles of the N-glycans were nearly identical, those from O-glycans showed reduced abundances in Ce core-1-like O-glycans in the mutant. Additionally, a novel series of highly fucosylated O-glycans that contain an internal GlcA were discovered; in the following sections these will be referred to as Ce core-2 O-glycans. However, it must be noted that the linkages within these glycans have not been determined, and this designation is used to refer to C. elegans O-glycans containing a HexA that is proximal to the core reducing-end HexNAc. They are present at only low levels in the wild type nematode, but their abundances are increased substantially in the bus-2 strain. Tandem MS was used to determine the sequences and compositions of the new subclass of O-glycans (Table 1, Fig. 4 and supplemental Figs. S1–S3). Monosaccharide analyses of the bus-2 and parental N2 Bristol strains O-glycan pools yielded similar results; the oligosaccharide samples contained Gal, Glc, Man, GlcNAc, Fuc, and Rib, consistent with C. elegans glycoconjugates (data not shown). The GlcA and reducing end GalNAc-ol are less stable than other monosaccharide components under the conditions of analysis and were detected only in trace amounts. Rib was likely to be a contaminant from residual RNA. The amount of HexA could not be specifically determined due to the limited sample quantities, but it should be noted that C. elegans has been reported to use GlcA as its charged monosaccharide (15, 27). Localized glycoconjugate expression was analyzed using lectin staining experiments performed on whole mounted delipidated nematodes (Figs. 5 and 6 and supplemental Figs. S4–S6). These data indicated that, in comparison to N2 parent, the bus-2 strain was decreased in the abundance of core-1 glycans in the anus and tail region and increased in fucosylated glycans along the alimentary tract including the anal region. These alterations are significant as the tail, rectum, and anal region are normally infected. Expression analysis of bus-2 revealed that it is normally expressed in the posterior intestine as well as the hypodermal seam cells in the cuticle, both tissues that are close to and can affect the region normally infected (Fig. 7). A detailed discussion of these results appears in the following sections.
FIGURE 1.
Microscopy images of N2 Bristol and bus-2 strains cultured with M. nematophilum. The ventral view is shown. A, shown is Nomarski differential interference contrast of C. elegans grown on a mixed culture of M. nematophilum and E. coli. The dar phenotype is present in wild type but not bus-2 worms. B, SYTO green stained bacteria are seen in the anal region of N2 Bristol but not bus-2 nematodes; the arrow points to localization of florescence in the anal region. C, the light microscopy counterpart of fluorescence images seen in B is shown.
FIGURE 2.
The bus-2 sequence and alignment with Pfam 01762. A, underlined is a cDNA segment from the bus-2 gene that was incorrectly predicted as an intron by GeneFinder. B, the alignment with Pfam01762 using NCBI Conserved Domain Data base is shown. The corrected region is underlined.
FIGURE 3.
Permethylation profiles of bus-2 and N2 Bristol O-glycans. MALDI-TOF MS spectra of the permethylated O-glycans released from bus-2 and N2 Bristol and eluted under low salt conditions from anion exchange resin are shown in the top and center panels. Ion abundances for each composition are shown in the lower panel. The inset shows total Ce core-1 and Ce core-2 O-glycans. Error bars indicate S.D. over three analyses.
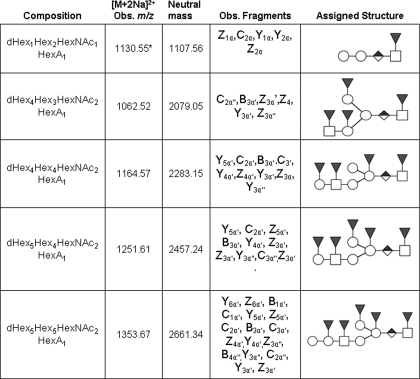
TABLE 1.
Composition and assigned structures of Ce core-2 O-glycans
See Fig. 4 for an explanation of symbols.
* Ion is [M + Na]+.
FIGURE 4.
Sequence assignment based on the ESI-CID-MS/MS fragmentation of bus-2 [M+Na]+m/z 1130.553. All fragments contain sodium. The Z2α product ion (circled) was isolated for ESI-CID-MS3 sequence analysis of bus-2 [M+Na]+ m/z 1130.553 → 690.328 as illustrated in the inset. Unlabeled peaks within the inset were observed in control spectra and correspond to electronic noise.
FIGURE 5.
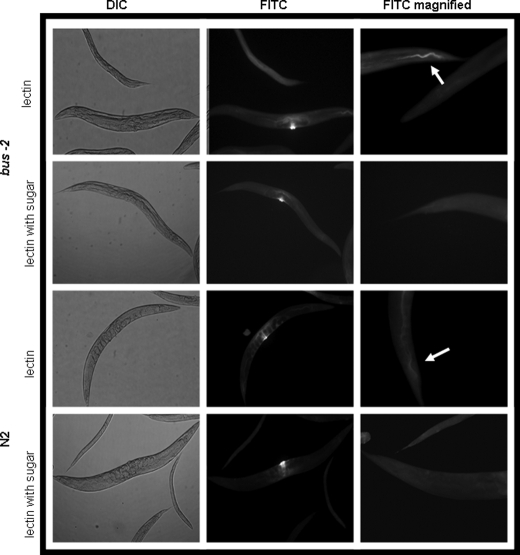
UEA I α-l- fucose specific lectin staining of whole mounted delipidated adult C. elegans. The left column shows control differential interference contrast (DIC) micrographs of lectin-treated nematodes with and without prior incubation with inhibitory concentrations (1 mm) of α-l-Fuc. The images in the center column show that vulva staining is background, as it is not affected by preincubation with α-l-Fuc. The right column shows fluorescence micrographs of UEA I. The bus-2 strains contain a higher abundance of fucosyl glycoproteins. FITC, fluorescein isothiocyanate.
FIGURE 6.
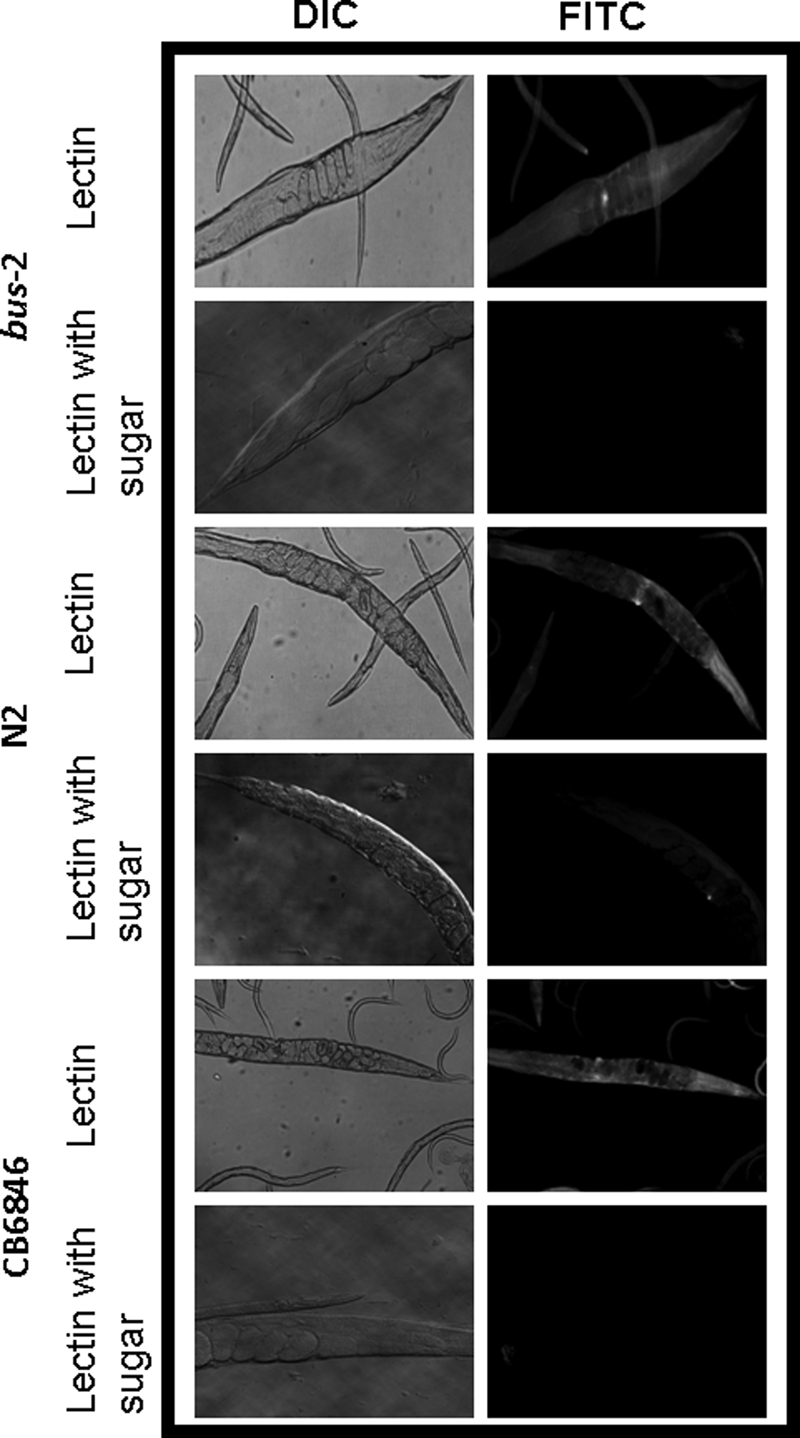
ABA Gal (β1,3) GalNAc - specific lectin staining of whole mounted delipidated adult C. elegans. The left column shows control differential interference contrast (DIC) micrographs of lectin-treated nematodes with and without prior incubation with an inhibitory sugar (β-d-galactose). The right column shows fluorescence micrographs of ABA-stained nematodes. FITC, fluorescein isothiocyanate.
FIGURE 7.
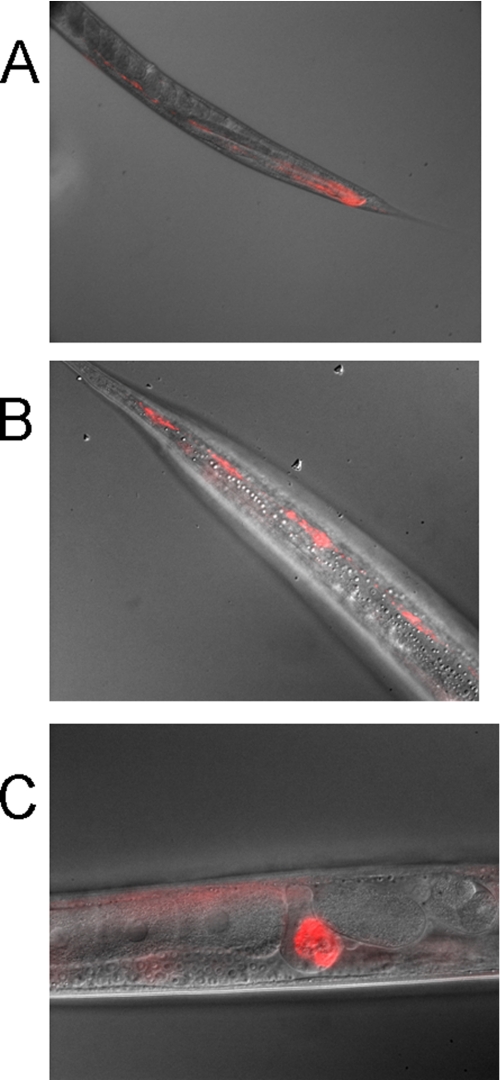
Tissue-specific expression of bus-2. A bus-2::RFP reporter construct is expressed in the posterior intestine (A), hypodermal seam cells (B), and the spermatheca (C).
Bus-2 Is Not Colonized by M. nematophilum
The fluorescent nucleic acid dye STYO Green 13 was used to visualize M. nematophilum colonization (see Fig. 1). This stain permeates cells in proportion to their surface areas and, therefore, under the conditions used here, stains the bacterial DNA preferentially. The bus-2 and wild type strains were incubated with a mixed bacterial lawn of M. nematophilum (0.1%) and OP50 for 72 h at room temperature. Nematodes were then extensively washed, incubated for an additional 60 min to assure complete digestion of microbes in the digestive tract, and extensively washed again before being stained. Fluorescent staining showed colonization of M. nematophilum in the anal region of the N2 Bristol strain but not in the bus-2 mutant, confirming that this bacterial infection cannot be detected in the bus-2 mutant.
Bus-2 Contains Conserved Domains of Core Type 1 Galactosyltransferases
The bus-2 gene, one of three bus genes to contain pfam01762 core type-1 galactosyltransferase conserved domains, is located on chromosome IV of the C. elegans genome (8). bus-2 is absent from the open reading frame library and was, therefore, cloned from a random-primed cDNA library (29)4; it was found to have an open reading frame different from that predicted by GeneFinder. After correction of the sequence the bus-2 alignment with the galactosyltransferase domain, pfam01762 was found to be 98.0% of domain models (rather than the earlier calculated value of 63.8%), as determined by the NCBI Conserved Domain Data base (see Fig. 2) (26). The score in the improved alignment was 80.7 Bits, and the expect value was 2e-16. The nucleotide and corrected protein sequence of bus-2 is deposited in GenBank™ (accession nos. Banklt 1331141; HM012823). In addition, BUS-2 contains a DXD motif (Fig. 2B), which is a hallmark of glycosyltransferases. It is conserved in 13 glycosyltransferase families (31) and is required for nucleotide sugar substrate binding through metal-coordinated binding of phosphate groups (32).
The bus-2 O-Glycosylation Profile Is Altered
Ce core-1 glycans have been previously described (15, 27, 33, 34). Permethylation renders the response of glycomers in a related series to be nearly identical with respect to detection by mass spectrometry and, thus, enables reproducible and semiquantitative comparison of biologically related samples (19). Permethylated glycan samples were analyzed in triplicate using MALDI-TOF MS, and the observed ion abundances were used to determine glycoform distribution. Analysis of peptide N-glycosidase F-released N-glycans did not reveal any significant differences between the two strains (data not shown). Through this analysis the presence of two O-glycan subfamilies was discovered. In addition to Ce core-1 compounds, the O-glycan profiles revealed the presence of a series of higher molecular weight O-glycans that were more abundant in the mutant than in the parent strain. In terms of total O-glycoform distribution, the bus-2 mutant had a significantly lower percentage of the Ce core-1 O-glycans as compared with the wild type and a greater content of the novel higher molecular weight glycans (see Fig. 3). As noted above, we refer to the new glycan series as Ce type-2 glycans. Total Ce core-1 glycans in N2 Bristol were found to constitute 73.5 ± 2.3% of the glycan pool and those from bus-2 represented 61.9 ± 1.4% of its pool, thus indicating that the latter had nearly a 12% lower content of these glycans. The level of Ce type-2 glycans increased in the bus-2 mutant from 26 ± 1.6 to 38 ± 1.5% of total ion abundance compared with the parent (see Fig. 3, inset), demonstrating a 12% higher content of this class. Individual compositions of Ce core-2 glycans were up to three times more abundant in the mutant. It should also be noted that acidic Ce core-1 glycans, which elute under higher salt conditions from the anion exchange resin used here, also had lower abundances in the bus-2 strain, indicating that all core-1 glycans were diminished in the mutant (data not shown).
Structural Detail of Novel Ce Type-2 O-Glycans
Permethylation renders a 14-atomic mass unit tag on all terminal residues, as all hydroxyls are replaced with O-methyl groups. At least one hydroxyl of internal residues (dependent on the number of its substituents) is involved in a glycosidic linkage and does not contain this mass tag. Because of this mass difference, it is possible to differentiate between terminal and internal substituents; this is an advantage and often key to structural assignment using permethylated oligosaccharide derivatives in contrast to derivatives that are modified only at the reducing end.
Tandem MS analysis revealed the presence of a series of related highly fucosylated glycomers whose presence has not been previously described (see Table 1). Although neutral fucosylated O-glycans have been reported in C. elegans (33), tandem MS analysis of the samples examined here revealed the presence of a series of related acidic highly fucosylated glycomers that have not been previously reported. Oligosaccharide sequence information was determined using MS2 and MS3 analyses when ion abundances were sufficient. The dHex1Hex2HexNAc1HexA1, [M+Na]+ m/z 1130.55, ion was analyzed, and the spectra are shown in Fig. 4. Assignment of the Hex2 branch was based on the C2α ion at m/z 463.213 and the Y3α and Y2α ions at m/z 912.437 and 708.339, respectively. These ions underwent sequential losses of two Hex residues from a linear sequence (see Table 1 and Fig. 4). The above assignments limit the position of the HexA and dHex to the core proximal HexNAc. To further define the position of these residues, MS3 experiments were performed on the Z2α fragment seen at m/z 690.328 (Fig. 4, inset). The HexA residue was thereby found to be internal and directly linked to the reducing end HexNAc on the basis of the C1β and Z1α fragments at m/z 229.175 and 472.211, respectively. The C1β fragment mass indicates that it is generated from a terminal dHex (since it contains the 14-atomic mass unit terminal tag). The Z1α fragment indicates loss of an internally linked HexA (loss of HexA not containing the 14-atomic mass unit tag) but retention of the terminal dHex linked to the core HexNAc. Together these assignments localize the HexA as internal.
The dHex4Hex3HexNAc2HexA1 [M+2Na]2+ m/z 1062.52 was analyzed, and the resulting spectrum is shown in supplemental Fig. S1. The presence of a dHex2Hex1HexNAc1 branch was confirmed by the loss of terminal dHex-HexNAc from the Z4α′ fragment at m/z 1272.615 and the Y3α′, B2α′/Z5α′, and Z3α′ fragments at m/z 1290.625, 628.292, and 1272.615, respectively. Assignment of the dHex-Hex branch is substantiated by the C2α″ ion at m/z 433.203. Fragmentation patterns of all the Ce type-2 saccharides studied (Table 1) were consistent with the presence of a conserved dHex1HexNAc1HexA1 core structure, as previously described herein as an internal component in the assignment of the dHex1Hex2HexNAc1HexA1 sequence above and in Fig. 4. The ion corresponding to dHex4Hex4HexNAc2HexA1, [M+2Na]2+ m/z 1164.870 (supplemental Fig. S2) was also subjected to CID, and the presence of a dHex1Hex1HexNAc1 branch was supported by observation of the product ions Z3α′, Y3α′, Z4α′, Y4α′, and Z5α′, Y5α′, which indicate the positions of dHex residues. The assignment of the terminal position of the dHexHex motif was further supported by the presence of C2α′ ion at m/z 433.203. The Y3α″ fragment at m/z 1055.508 (2+) indicated the presence of a terminal Hex. The dHex5Hex4HexNAc2HexA1, [M+2Na]2+ m/z 1251.61, had a mass shift corresponding to the addition of one fucose and no longer showed the loss of the non-fucosylated terminal Hex (Table 1). This supported the addition of a dHex residue to the Hex as reported here (compare the results obtained for compounds 3 and 4, Table 1). The ion at m/z 1353.67, corresponding to dHex5Hex5HexNAc2HexA1, [M+2Na]2+, was analyzed and found to have the same structure as the dHex5Hex4HexNAc2HexA1 with the addition of a terminal hexose on the HexNAc-containing branch, as supported by the reducing end fragments Y6α′ and Z6α′ and by the terminal fragments B1α′ and C1α′ (supplemental Fig. S3). A summary of the fragmentation data assignments for the above series of the fucosyl Ce type-2 O-glycans is presented in Table 1 and supplemental Figs. S1–S3. Data from all glycomers were consistent with the presence of an internal HexA and excluded the possibility of a terminal HexA indicating that these compounds are members of a biosynthetically related series and that HexA serves as a linkage point for further branch extension. C. elegans has been reported to use GlcA as its charged group (15, 27), so the HexA residue observed here is likely to be GlcA.
Tissue Expression Pattern of bus-2
Expression analysis using a bus-2::RFP operon reporter transgene revealed that bus-2 is expressed in the posterior intestine in addition to its expression in the hypodermal seam cells and spermatheca (see Fig. 7). These results are consistent with the notion that, in the absence of bus-2 expression, core-1 O-glycans are reduced in abundance in the seam cells and the posterior gut leading toward the rectum and exterior cuticle. This is important in the infective process for both Yersinia and Microbacterium pathogens if core-1 O-glycans are involved, as the head region and tail regions of the cuticle and posterior intestine would be deficient of core-1 O-glycans, and these are regions normally infected by these pathogens.
Lectin Studies Reveal Altered Glycosylation Patterns in the Region of Infection and Associated Tissues
Fluorescent lectin staining of whole mounted delipidated nematodes was performed with the fluorescein isothiocyanate conjugated α-l-fucose-specific lectin, U. europaeus I (UEA I) and Galβ1,3GalNAc-specific lectin, A. bisporus (ABA). UEA I lectin staining was observed along the length of the alimentary tract and concentrated in the posterior intestine in both bus-2 and N2 strains (see Figs. 5, supplemental Figs. S4 and S5) but was observed to be brighter in the bus-2 mutant, indicating that fucosyl glycoconjugates were more abundant in this region compared with the parental strain. Less intense staining was observed over the surface of the cuticle in both N2 and bus-2 strains but was more intense in the later strain. The buccal cavity flaps, sieve, and grinder stained brightly in the mutant. This staining was also seen in the wild type strain, but the staining was less intense. These tissues also contain a cuticle lining. The bus-2 rescued strain CB6846 demonstrated staining similar to wild type, as signal in the posterior intestine was diminished. However, the buccal and pharyngeal regions and overall cuticle staining remained more intense and were of an intermediate intensity between the N2 and bus-2 patterns (see supplemental Figs. S4 and S5). The bus-2-rescued transformant CB6846 (mCherry transformant, see Fig. 7) regained N2-like staining patterns of ABA (Fig. 6), thus confirming the role of bus-2 in the distributions of glycoconjugates.
Approximately 75% of nematodes were stained in this fashion with slightly fewer observed in wild type worms due to the overall lower level of staining. It should be noted that no changes in distribution were observed in fucosyl N-glycans in the bus-2 strain mass spectra. Because these nematodes had been delipidated, the staining could not have resulted (solely) from the presence of glycolipid. Therefore, the UEA I staining was consistent with the presence of a higher level of Ce core-2-O-glycoproteins in the posterior intestine and regions described above in the bus-2 strain.
In contrast, ABA lectin, with specificity for core-1 Galβ1,3GalNAc, showed the most intense staining in the anal region and surrounding tissues of N2 Bristol nematodes (see Figs. 6 and supplemental Fig. S6). Staining was also observed over the cuticle, although not as intensely as in the tail. The cuticle-lined structures of the buccal cavity and pharynx were also seen to stain, and this is consistent with seam cell expression of bus-2 as these structures are lined with cuticle and probably receive their carbohydrate-rich surface coat through seam cell secretion. ABA staining over the majority of the cuticle in the bus-2 strain was similar to wild type, but staining was greatly diminished in the tail region (see Fig. 6 and supplemental Fig. S6 for tail and head regions, respectively). Approximately 80% of nematodes were stained in the ABA lectin study. The specificity of each lectin was verified by incubating the lectin with an inhibitory concentration of the corresponding sugar before binding. Background staining was observed around the vulva with all lectins in both strains and was not inhibited by preincubation of free sugar (see Fig. 5). The same nonspecific vulval staining was observed in negative controls using a lectin for sialic acid, a residue absent in C. elegans (data not shown), and therefore, it was concluded that the vulva staining was background and not significant.
In the bus-2 strain, localization of fucosyl glycans to the posterior digestive tract leading up to the anus, approaching the site of infection, corresponds with the increased abundance of Ce fucosyl O-glycans detected by MALDI-TOF MS analysis. This result implies that an increase in the abundances of fucosyl glycans in conjunction with a decrease in core 1 type glycans, as revealed by ABA lectin observed in bus-2, may interfere with adherence of M. nematophilum in the mutant.
The bus-2 expression is seen in the hypodermal seam cells and posterior intestine. The seam cells probably secrete the carbohydrate-rich surface coat over the cuticle surface. This expression pattern predicts the presence of core 1 type glycoconjugates over the cuticle surface and in the posterior intestine. In such a case staining would appear to be most intense in the posterior region as both cuticle staining and intestinal staining would be coincident. Therefore, in the presence of bus-2 the region of highest ABA staining intensity should be coincident with the red fluorescent protein signal seen in the posterior intestine in CB6846 (Fig. 7), and this is the case (compare Figs. 6 and 7). Staining in the N2 Bristol and rescued strain CB6846 stain intensely in this region, whereas the bus-2 strain does not. If bus-2 and a glucuronosyltransferase, which gives rise to the Ce core-2 glycans, do compete for substrate in the seam cells and intestine, then in the absence of bus-2 fucosyl glycans should be increased and core 1 glycans decreased in those regions. This is the case as seen in Figs. 5, supplemental S4 and S5. Fucosyl glycan staining with UEA I is more intense over the cuticle surface and intestine in the bus-2 mutant compared with N2 and rescued strains. Therefore, bus-2 expression and lectin staining data are consistent with the idea that there is competition for a common substrate in bus-2 expressing tissues.
DISCUSSION
We have investigated the nature of M. nematophilum infection of C. elegans through comparative analysis of the resistant strain bus-2 and its parent strain N2 Bristol. In these studies we have used mass spectrometry, whole nematode mount staining, lectin studies, and bus-2::red fluorescent protein rescue. The gene has been cloned and the open reading frame corrected from that predicted by GeneFinder. BUS-2 aligns with galactosyltransferase domain pfam01762 to 98.0% of domain models used, as determined by the NCBI Conserved Domain Data base, predicting that bus-2 encodes a core-1 galactosyltransferase homolog. We have demonstrated a difference in O-glycan abundances and distribution between bus-2 and its parent. The bus-2 mutant was found to have a reduced level of Ce core-1 O-glycans, a result consistent with the absence of a predicted BUS-2 glycosyltransferase activity. Significant Ce core-1 O-glycan remains; this is also expected in whole nematode lysate chemical analysis as bus-4 and bus-17 also encode core-1 type galactosyltransferase homologs, and therefore, redundancy is likely to exist.
In this study the presence of a novel series of highly fucosylated O-glycans was discovered. These glycans contain internal core proximal GlcA, and this feature makes their overall structures novel. This new subclass of oligosaccharides has been termed Ce type-2 O-glycans. The relative abundances of peaks corresponding to this type of glycans were increased up to 3-fold in the mass spectra of the bus-2 mutant, and fluorescence labeling indicated that such glycans were more abundant near or at the site of infection, the posterior intestine leading to the anus, and the cuticle surface. An RFP reporter gene construct indicated that BUS-2 is normally expressed in the posterior intestine. This would predict the presence of a significant amount of Ce core-1 O-glycans in that region in wild type strains, and the results from the ABA lectin analysis were consistent with this prediction. It seems likely that in wild type C. elegans, the Ce core-1 glycans serve as ligands for M. nematophilum adhesion, as they have lower relative abundances in the resistant strains bus-2, srf-3 (15), and bus-4.5 In bus-2, Ce core-1 glycans have lower relative abundances, and the fucosyl Ce core-2 glycans exhibit higher relative abundances near the region of infection, suggesting that the altered distribution of glycoconjugates in this region may interfere with the capacity of M. nematophilum to adhere and infect.
Are there other possible explanations for the bus-2 resistant phenotype? Alterations of the cuticle surface other than changes in glycosylation in the area normally infected could also account for loss of M. nematophilum binding. In studies in C. elegans, lectin affinities are routinely used to monitor changes at the cuticle surface and underlying structures. Altered lectin binding patterns have been observed in strains that harbor genetic lesions in factors not directly active in glycosylation. For instance, mutations that lead to morphological defects in the sensory ray of the male tail have been shown to expose GalNAc epitopes (35). The affected genes, ram and mab, are critical components for male tail cuticle organization and are not involved in glycoconjugate biosynthesis. In the case of bus-2, there is no apparent disorganization of the tissues, and therefore, a ligand masking or suppressor un-masking phenotype affecting pathogen binding is not likely. However, it is still possible that subtle undetected structural changes or other molecular changes could contribute to the resistance phenotype.
How is the shift in O-glycan abundance explained? The bus-2 strain type-1 O-glycans were observed to be decreased in abundance, whereas core 2 O-glycans are increased in abundance. The Hex1HexNAc1 and Hex2HexNAc1 glycans are not significantly altered, although the Hex3HexNAc1 and Hex4HexNAc1 glycans are decreased, as are other downstream Core-1 glycans. It could be interpreted that the transferase between Hex2HexNAc1 and Hex3HexNAc1 is missing, and bus-2 encodes this enzyme. However, this explanation does not account for the increase seen in the core-2 glycans. Another explanation that would account for this shift is that bus-2 encodes a core-1 synthase giving rise to Galβ1,3GalNAc-Ser/Thr from the Tn antigen GalNAc-Ser/Thr. In the bus-2 background this activity is deficient in some cells. This results in shunting of Tn antigen for elongation by a GlcA transferase, giving rise to increased abundance of the Ce type-2 O-glycan seen here. In such a case it would be expected that the abundance of some Ce Core-1 glycans would be diminished, whereas those of Ce Core-2 glycans would increase. These changes would be observed in regions where the shift is most prominent. The second explanation would account for the shift in abundances and the localized change in fucosyl glycans as observed by lectin staining in and around the rectum, anus, and surrounding tissues. However, the presence of multiple cell types, the possibility of compensatory response, and an unknown enzymatic specificity make these arguments highly speculative.
How are the altered glycosylation pattern and distribution in the bus-2 strain related to the resistance phenotype? Although overall changes in O-glycan profiles may seem less than dramatic, Core-1 glycans are greatly diminished in the rectum, anus, and surrounding tissues in the bus-2 mutant. These glycans are likely to be a major target for M. nematophilum adhesion. These glycans were also observed to be decreased in the resistant strains srf-3 (15) and bus-4.5 Furthermore, it cannot be ignored that the Ce Core-2 glycans are increased in abundance in the posterior gut leading to the rectum and in the cuticle surface, which are the regions of infection. Their presence may further compromise bacterial adhesion. Some of these glycoconjugates may also be secreted, possibly further interfering with adhesion.
It should be noted that glycolipids were not analyzed in this study. However, lectin staining experiments performed after delipidation clearly show that glycoproteins are altered in the region of infection in the bus-2 strain. However, we cannot rule out the possibility that host glycolipids also may be involved in the infective process. Studies of the C. elegans glycolipids are presently under way.
Adhesion to the host cell surface is a common mechanism used by pathogens to initiate infection. ABO blood group antigens present in human intestinal mucous can serve as receptors for a wide range of pathogens such as Helicobacter pylori (36) and Campylobacter jejuni (37). Subtle differences in glycan structure, density, and presentation can be the deciding factors between resistance and susceptibility. The presence of secreted glycoconjugates in the case of secretor status can confer resistance, as these entities can serve as decoys for the invading pathogen. Rhesus monkeys with the secretor phenotype are resistant to gastric infection by H. pylori because their high mucous density prevents BabA adhesin from binding to cell surface ABO structures. Non-secretor infection rates have been reported to be higher, consistent with the notion that innate immunity conferred by the secretor phenotype has evolved to combat cell surface carbohydrate receptor targeting (38).
Norwalk virus infection is blood group-dependent in humans. Individuals with blood group O are more susceptible to infection than those with types A or B even when secretor type is taken into consideration. Terminal substitution of the H-antigen apparently has a modulatory effect on resistance, and the presence of α1,2Fuc appears to be required for infection as dependence on FUT-2 fucosyltransferase has been shown (30). The bus-2 study presented here may mimic blood group infection dependence as presentation of a different subclass of O-glycan may interfere with infection. In this way, C. elegans may serve as a model for innate genetic resistance phenotypes.
C. elegans is sensitive to more than 40 bacterial pathogens, many of which are human pathogens or their close relatives. Among these pathogens are Yersinia pestis and Yersinia peudotuberculosis (28). These bacteria secrete a biofilm that encases the head region, leading to starvation. This situation may mimic the bacteria infective modus operandi in the flea where biofilm blocks the proventriculus, thus compromising the ability of the insect to feed. The bus-2 mutants are resistant to this infection. Although part of the bus-2 expression is in the posterior intestine, it is also expressed in the hypodermal seam cells which are believed to contribute to the surface coat of the cuticle over much or all of the body, including both head and tail regions. Indeed, ABA lectin does stain over the entire nematode surface, albeit at a lower intensity than in the tail region. Interestingly, in genetic screens for resistance to Yersinia biofilm attachment, which yielded bah (biofilm absent on head) mutants, five of the eight bah complementation groups were also identified in the bus screen (10), suggesting that common host receptors are required for adhesion by both pathogens. It is likely that the altered O-glycan distribution seen in bus-2 is also responsible for resistance to these bacteria as well, although we cannot at this point speculate as to the exact changes in glycoform distribution that lead to Yersinia resistance.
M. nematophilum, although not a human pathogen, likely has mechanisms of virulence that are conserved within this genus. Little is known about adhesins in these bacteria. This work represents the first detailed analysis of the nature of Microbacterium adhesion to host tissue. Currently we are developing an affinity proteomic approach to aid in the identification of bacterial adhesion factors, which should allow further investigation of this pathogenic interaction. Efforts are also under way to extend the C. elegans model to the study of other bacterial pathogens.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P41RR010888 and S10RR015942 (to C. E. C.) and S10RR020946 (to J. Zaia). This work was also supported by Medical Research Council (United Kingdom) Grant G0600238 (to J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) HM012823.
D. Stroud and J. Hodgkin, manuscript in preparation.
D. Stroud, M. J. Gravato-Nobre, and J. Hodgkin, manuscript in preparation.
R. M. Mizanur, D. Stroud, J. Hodgkin, and J. F. Cipollo, unpublished data.
- ESI
- electrospray ionization
- CID
- collision-induced dissociation
- MS
- mass spectrometry
- UEA I
- U. europaeus I
- ABA
- A. bisporus
- RFP
- red fluorescent protein
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight.
REFERENCES
- 1.Sifri C. D., Begun J., Ausubel F. M. (2005) Trends Microbiol. 13, 119–127 [DOI] [PubMed] [Google Scholar]
- 2.O'Quinn A. L., Wiegand E. M., Jeddeloh J. A. (2001) Cell Microbiol. 3, 381–393 [DOI] [PubMed] [Google Scholar]
- 3.Rajamohan F., Lee M. K., Dean D. H. (1998) Prog. Nucleic Acid Res. Mol. Biol. 60, 1–27 [DOI] [PubMed] [Google Scholar]
- 4.Chungjatupornchai W., Höfte H., Seurinck J., Angsuthanasombat C., Vaeck M. (1988) Eur. J. Biochem. 173, 9–16 [DOI] [PubMed] [Google Scholar]
- 5.Griffitts J. S., Whitacre J. L., Stevens D. E., Aroian R. V. (2001) Science 293, 860–864 [DOI] [PubMed] [Google Scholar]
- 6.Griffitts J. S., Huffman D. L., Whitacre J. L., Barrows B. D., Marroquin L. D., Müller R., Brown J. R., Hennet T., Esko J. D., Aroian R. V. (2003) J. Biol. Chem. 278, 45594–45602 [DOI] [PubMed] [Google Scholar]
- 7.Gneiding K., Frodl R., Funke G. (2008) J. Clin. Microbiol. 46, 3646–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodgkin J., Kuwabara P. E., Corneliussen B. (2000) Curr. Biol. 10, 1615–1618 [DOI] [PubMed] [Google Scholar]
- 9.Partridge F. A., Tearle A. W., Gravato-Nobre M. J., Schafer W. R., Hodgkin J. (2008) Dev. Biol. 317, 549–559 [DOI] [PubMed] [Google Scholar]
- 10.Darby C., Chakraborti A., Politz S. M., Daniels C. C., Tan L., Drace K. (2007) Genetics 176, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravato-Nobre M. J., Nicholas H. R., Nijland R., O'Rourke D., Whittington D. E., Yook K. J., Hodgkin J. (2005) Genetics 171, 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yook K., Hodgkin J. (2007) Genetics 175, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Link C. D., Silverman M. A., Breen M., Watt K. E., Dames S. A. (1992) Genetics 131, 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeflich J., Berninsone P., Goebel C., Gravato-Nobre M. J., Libby B. J., Darby C., Politz S. M., Hodgkin J., Hirschberg C. B., Baumeister R. (2004) J. Biol. Chem. 279, 30440–30448 [DOI] [PubMed] [Google Scholar]
- 15.Cipollo J. F., Awad A. M., Costello C. E., Hirschberg C. B. (2004) J. Biol. Chem. 279, 52893–52903 [DOI] [PubMed] [Google Scholar]
- 16.Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas H. R., Hodgkin J. (2004) Curr. Biol. 14, 1256–1261 [DOI] [PubMed] [Google Scholar]
- 18.Lewis J. A., Fleming J. T. (1995) Methods Cell Biol. 48, 1–27 [Google Scholar]
- 19.Ciucanu I., Costello C. E. (2003) J. Am. Chem. Soc. 125, 16213–16219 [DOI] [PubMed] [Google Scholar]
- 20.Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 21.Domon B., Costello C. E. (1988) Glycoconj. J. 5, 397–409 [Google Scholar]
- 22.Kamerling J. P., Gerwig G. J., Vliegenthart J. F., Clamp J. R. (1975) Biochem. J. 151, 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgonie G., van Driessche E., Link C. D., de Waele D., Coomans A. (1994) Histochemistry 101, 379–384 [DOI] [PubMed] [Google Scholar]
- 24.Hobert O. (2002) Biotechniques 32, 728–730 [DOI] [PubMed] [Google Scholar]
- 25.Cheung B. H., Arellano-Carbajal F., Rybicki I., de Bono M. (2004) Curr. Biol. 14, 1105–1111 [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A., Bryant S. H. (2004) Nucleic Acids Res. 32, W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guérardel Y., Balanzino L., Maes E., Leroy Y., Coddeville B., Oriol R., Strecker G. (2001) Biochem. J. 357, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darby C., Hsu J. W., Ghori N., Falkow S. (2002) Nature 417, 243–244 [DOI] [PubMed] [Google Scholar]
- 29.Reboul J., Vaglio P., Tzellas N., Thierry-Mieg N., Moore T., Jackson C., Shin-i T., Kohara Y., Thierry-Mieg D., Thierry-Mieg J., Lee H., Hitti J., Doucette-Stamm L., Hartley J. L., Temple G. F., Brasch M. A., Vandenhaute J., Lamesch P. E., Hill D. E., Vidal M. (2001) Nat. Genet. 27, 332–336 [DOI] [PubMed] [Google Scholar]
- 30.Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. (2003) Nat. Med. 9, 548–553 [DOI] [PubMed] [Google Scholar]
- 31.Breton C., Mucha J., Jeanneau C. (2001) Biochimie 83, 713–718 [DOI] [PubMed] [Google Scholar]
- 32.Liu J., Mushegian A. (2003) Protein Sci. 12, 1418–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrows B. D., Haslam S. M., Bischof L. J., Morris H. R., Dell A., Aroian R. V. (2007) J. Biol. Chem. 282, 3302–3311 [DOI] [PubMed] [Google Scholar]
- 34.Costello C. E., Contado-Miller J. M., Cipollo J. F. (2007) J. Am. Soc. Mass Spectrom. 18, 1799–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko F. C., Chow K. L. (2000) Dev. Growth Differ. 42, 69–77 [DOI] [PubMed] [Google Scholar]
- 36.Borén T., Falk P., Roth K. A., Larson G., Normark S. (1993) Science 262, 1892–1895 [DOI] [PubMed] [Google Scholar]
- 37.Marionneau S., Cailleau-Thomas A., Rocher J., Le Moullac-Vaidye B., Ruvoën N., Clément M., Le Pendu J. (2001) Biochimie 83, 565–573 [DOI] [PubMed] [Google Scholar]
- 38.Lindén S., Mahdavi J., Semino-Mora C., Olsen C., Carlstedt I., Borén T., Dubois A. (2008) PLoS Pathog 4, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.